Abstract
Small GTPases are a family of low molecular weight GTP-hydrolyzing enzymes that cycle between an inactive state when bound to GDP and an active state when associated to GTP. Small GTPases regulate key cellular processes (e.g., cell differentiation, proliferation, and motility) as well as subcellular events (e.g., vesicle trafficking), making them key participants in a great array of pathophysiological processes. Indeed, the dysfunction and deregulation of certain small GTPases, such as the members of the Ras and Arf subfamilies, have been related with the promotion and progression of cancer. Therefore, the development of inhibitors that target dysfunctional small GTPases could represent a potential therapeutic strategy for cancer treatment. This review covers the basic biochemical mechanisms and the diverse functions of small GTPases in cancer. We also discuss the strategies and challenges of inhibiting the activity of these enzymes and delve into new approaches that offer opportunities to target them in cancer therapy.
Keywords: small GTPases, Arf1, inhibitors, cancer therapy, anticancer
1. Introduction
Small GTPases are a large family of hydrolases that bind and hydrolyze GTP to GDP in order to regulate many cellular activities (e.g., cell differentiation, proliferation, and motility) [1]. Inherently, small GTPases have a basal mild endogenous GTPase activity that is dependent on Mg2+ to weaken the bond between the last two phosphates in GTP in order to form GDP [2,3,4]. Small GTPases are normally maintained in an GDP-bound inactive state and are induced by guanine-nucleotide exchange factors (GEFs), which replace the GDP molecule that normally occupies the small GTPase binding pocket with GTP [4]. Equally important for the regulation of small GTPases are the GTPase-activating proteins (GAPs), which deactivate small GTPases through enhancing their endogenous hydrolytic activity, leading to the shifting of GTP to GDP and their subsequent inactivation [4].
Small GTPases have been grouped into five major classes according to their sequence homology and on their physiological functions [3]: Arf subfamily, Ras subfamily, Ras-homolog (Rho) subfamily, Ras-related in brain (Rab) subfamily, and Ras-related nuclear protein (Ran) subfamily [5,6]. However, the Ran and the Rab subfamilies have recently been fused due to the high homology that presents their components [7].
The Arf subfamily is involved in a broad spectrum of physiological roles, such as the organization of the cytoskeleton, the sorting of vesicle cargo, the recruitment of vesicle coat proteins, and the alteration of lipid membranes through the recruitment of key enzymes, including phosphatidylinositol-four-phosphate adapter protein 1 (FAPP1), FAPP2, and the ceramide transfer protein (CERT) (Figure 1) [8,9]. The Arf subfamily consists of 6 Arf isoforms, 22 different Arf-like proteins (ARL) and Sar1 [8,10]. Deregulation of some Arf isoforms has been shown to induce cancer formation and progression by enhancing cell proliferation through the activation of mitogen-activated protein kinases (MAPK) and ribosomal protein S6 kinase beta-1 (p70S6K) [11,12]. Furthermore, deregulation of certain Arf family members, such as Arf1 and Arf6, enhances cancer cell invasion and metastasis by stimulating Ras-related C3 botulinum toxin substrate 1 (Rac1), paxillin, talin or focal adhesion kinase (FAK) [12,13,14].
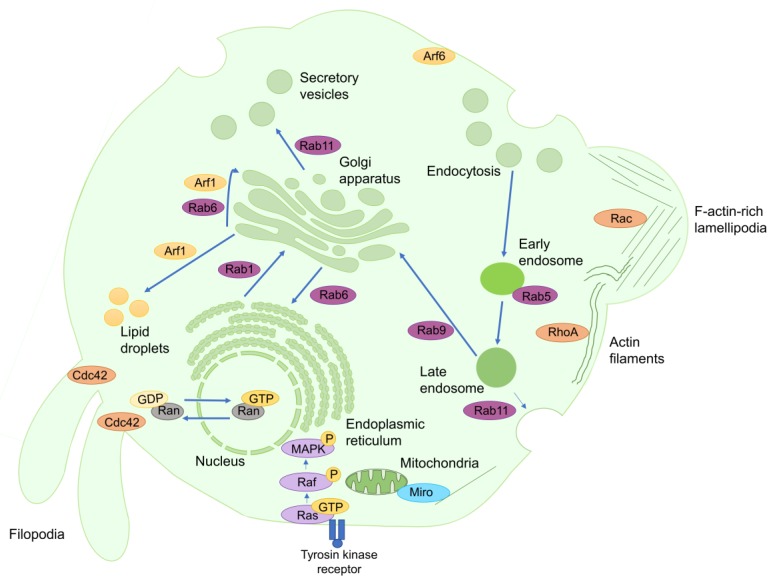
Figure 1.
Role of small GTPases in human cells. Most small GTPases are implied in the regulation of protein secretion, endocytosis and vesicle trafficking. For instance, Ran-activation gradient controls both the export and import of macromolecules between the nucleus and the cytoplasm. Additionally, Rab1 is responsible for regulation of vesicle trafficking between the endoplasmic reticulum and the Golgi apparatus, whereas Rab6 modulates the reverse transport, as well as through the different Golgi apparatus vesicles. Arf1 is implied in intra-Golgi transport, but also enables the accumulation of fatty acids inside the lipid droplets. Otherwise, Rab5 regulates endosome coating. The control of secretory vesicle formation is mainly mediated by Rab11. The products resulting from phagosome digestion can be carried to Golgi apparatus in a Rab9-dependent process, or return to the extracellular matrix in a Rab11-dependent mechanism. Arf6, which is associated with the plasma membrane when inactive, works as a master regulator of vesicle processes. On the other hand, other small GTPases are involved in the maintenance of cell shape and movement, such as Rac, which promotes the generation of lamellipodia, or Cdc42, which promotes the formation of filopodia. RhoA induces the formation of actin filaments in response to cellular stresses. Otherwise, Ras induces the phosphorylation and activation of MAPK, inducing prosurvival responses, such as cell proliferation and cell cycle progression, as well as limiting prodeath signals, such as apoptosis.
The Ras subfamily includes 36 different members divided into seven subgroups: Ras proteins, Ras-related proteins (RRAS), Ras-like proteins (Ral), Ras-proximal proteins (Rap), Ras-related associated with diabetes (Rad) and Gem-related proteins (GRE), Ras homolog enriched in brain (Rheb), and Ras-like in all tissues proteins (Rit) [7,15]. The members of the Ras subfamily are involved in the activation of intracellular signaling networks, resulting in enhancing cellular proliferation, adhesion, migration, and survival, as well as in limiting apoptosis (Figure 1) [15]. Ras overexpression has been found in more than 15% of human tumors [15], since its upregulation can promote cancer cell proliferation by the deregulation of the basal activation levels of the MAPK, phosphatidylinositol 3-kinase (PI3K), and phospholipase C epsilon (PLCε) pathways [15]. In addition, Ral overexpression stimulates tumorigenesis and tumor invasion in a Ras-dependent manner due to its ability to induce cell exocytosis by interacting with Sec5 and the exocyst complex component 84 (Exo84) [16]. The upregulation of certain Rap members can also promote cancer cell proliferation, migration and invasion due to their capability to alter integrin-mediated cell adhesion [17,18]. Rheb proteins promote carcinogenic cell proliferation and tumorigenesis by promoting the activation of the mechanistic target of rapamycin complex 1 (mTORC1) [19]. Finally, Rit proteins enhance neuronal survival and differentiation, thus their deregulation could be associated with neuronal tumor progression and with the onset of many nervous system pathologies, such as Parkinson’s disease, autism, or schizophrenia [20].
The Rho family consists of 22 proteins divided into seven subgroups: Ras-related C3 botulinum toxin substrate (Rac), Rho-related proteins (RhoA), cell division control protein 42 homolog (Cdc42), TC10 and T-cell leukemia/lymphoma protein 1A (TCL), Rho-related GTP-binding protein Rho6 precursor (Rnd), Rho-related BTB domain-containing protein (RhoBTB) and [21,22]. The members of the Rho family are involved in controlling actin turnover and in coordinating cell shape and movement through the regulation of the activity of the actomyosin complex (Figure 1) [23]. More in detail, Rho subfamily proteins can generate different actin structures to allow the displacement of the cell in response to several mechanical stimuli, such as Rac-dependent filamentous-actin-rich lamellipodia or Cdc42-dependent filopodia protrusions (Figure 1) [21,22]. On the other hand, the aberrant expression of several small GTPases of this family, such as Rac, Cdc42 and RhoA, in carcinogenic cells enhances cancer promotion and progression by facilitating cell cycle progression and mitosis, disrupting tight cellular junctions that prevent cell mobility and inducing epithelial to mesenchymal transition (EMT) to enhance the formation of secondary tumors [21,22]. Moreover, RhoA and RhoC can also promote the formation of new vessels towards the carcinogenic mass through inducing the release of several proangiogenic factors, such as vascular endothelial grown factor [21].
The Rab family consists of approximately 70 members whose main function is to manage vesicular trafficking between intracellular organelles (Figure 1) [24]. The Rab family members select and collect vesicle cargo by increasing the affinity of certain protein sorting receptors, such as the mannose-6-phosphate receptor (M6PR), to the nascent vesicle (Figure 1) [25]. These small GTPases modulate vesicle transport through actin filaments and microtubules by recruiting, respectively, myosin V and kinesins (Figure 1) [25]. Additionally, they induce vesicle fusion by interacting with certain members of the NSF-attachment protein receptors (SNAREs) family that tether the transport vacuole to the acceptor membrane (Figure 1) [25]. The upregulation of certain members of the Rab family (such as Rab25, Rab5 and Rab11) in carcinogenic cells induces tumorigenesis by increasing cell proliferation and migration, via the activation of the Akt/mTORC1, extracellular signal-regulated kinase 1 (ERK1), and Wnt/β-catenin pathways, as well as inhibits tumor cell apoptosis via the decrease of Bak and Bax expression [24,26].
Ran controls molecular export and import from the nucleus to the cytoplasm. Ran GEFs, which activate Ran, accumulate in the nucleus and their interaction with Ran allows the binding and transfer of the cargo from the nucleus to the cytoplasm (Figure 1) [27]. Once in the cytoplasm, the active Ran-GTPs are inhibited by their Ran GAPs, which are located in the cytoplasm, generating this distinct compartmentation of the Ran GEFs and Ran GAPs. The Ran-activation gradient between these two compartments is responsible for nuclear molecular exporting (Figure 1) [27,28]. Besides, more recent studies have demonstrated that Ran-activation gradient could be indispensable for nuclear import, since its disruption impedes the entrance of large proteins inside the nucleus [29]. Otherwise, Ran can also modulate the assembly of mitotic spindles that control chromosome spatial organization during cell division [30]. It has recently been claimed that Ran overexpression improves cancer aggressiveness by promoting tumor proliferation, progression and metastasis [31]. Therefore, the development of certain molecules that reduce its expression and activation in carcinogenic cells could prevent cell proliferation by disturbing mitotic spindle formation, leading to promote their death by apoptosis [31].
The outer mitochondrial membrane GTPase Miro, which contains four EF hands and two GTPase domains [32], enables the distribution of mitochondria within the cell due to its ability to associate with the kinesin heavy chain [33]. Besides, other studies have recently suggested that the Miro EF domains act as Ca2+ sensors, increasing the mitochondrial uptake of this ion [33].
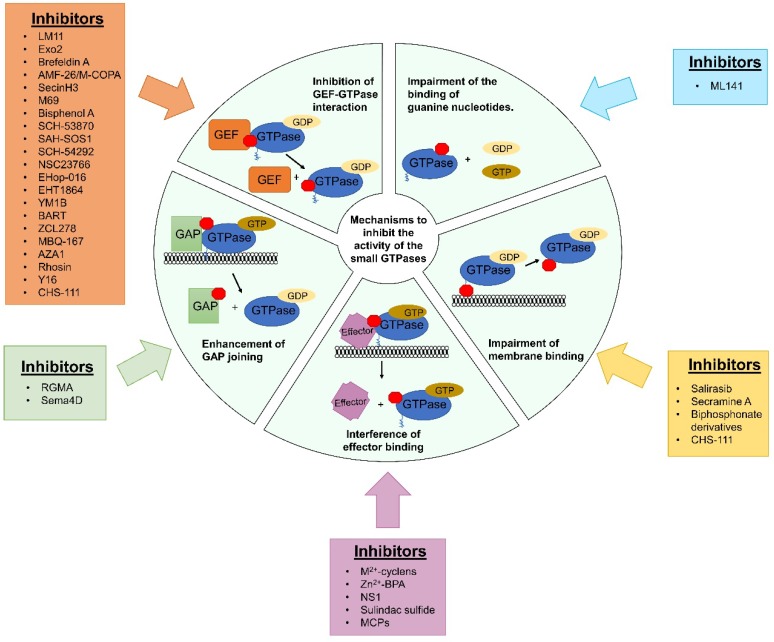
Therefore, the development of small GTPase inhibitors could be a useful new treatment strategy for both non-carcinogenic and carcinogenic diseases [34]. However, the generation of these inhibitors is a challenging issue owing to the fine regulatory roles assigned to each of the members of the small GTPases protein family (Figure 2) [34,35]. Possible mechanisms by which new inhibitors can be designed include the development of molecules that prevent the formation of the specific GEF-GTPases complex, the impairment of the binding of GTP to GTPases, the increase of GAP protein activity to reduce the pool of active small GTPases, the blocking of the transduction of the activation signal to their specific downstream effector and the inhibition of their membrane-binding domain (Figure 2) [34,36].
Figure 2.
New strategies to target small GTPases in human cancers. To improve the therapeutic efficacy of inhibitors of small GTPases, new approaches have been developed by different strategies. Red boxes represent the inhibitor of GTPases in each of the strategies. Those include generation of new molecules that can fill the specific GEF binding site in GTPases, disruption of GEF-mediated guanine nucleotide exchange, filling of nucleotide binding pocket of small GTPases, impairing nucleotide attachment, and the stimulation of GAP proteins. Given that most of small GTPases need to be attached to the organelle membrane to exert their actions, the development of novel molecules with the ability to abolish this binding has arisen recently as an innovative strategy to inhibit these molecules. Finally, the development of some drugs that interfere with these could also be great to inhibit small GTPases. A brief table situated next to each section of the graphic indicates the small GTPase inhibitors that work through that mechanism.
2. Arf1 and Its Inhibitors in Cancer Therapy
Arf1 overexpression stimulates tumor progression and invasion, thus the inhibition of this protein could be useful for restraining cancer progression [12]. The inhibitors that are currently available to block this protein are listed in Table 1
Table 1.
Action of Arf1 inhibitors in cancer treatment.
| Name of the Inhibitor | Mechanism of Action | Model | Global Outcomes | Reference |
|---|---|---|---|---|
| LM11 | Inhibition of ArfGEF binding to Arf1 | Breast cancer cell lines cultured in vitro and breast cancer xenografts in zebrafish | Inhibition of cell proliferation, invasion and metastasis | [38] |
| Breast adenocarcinoma cells cultured in vitro | Reduction of cell migration in a dose-dependent manner, cell adhesion to matrix and cell proliferation | [13] | ||
| Exo2 | Inhibition of ArfGEF activity | Prostate cancer cells cultured in vitro | Suppression of cell proliferation, invasion and migration and induction of programmed cell death through apoptosis | [44] |
| BFA | Hindering of Arf1 and GEF joining | Anaplastic large cell carcinoma in vitro | Reduction of cell proliferation | [49] |
| Lung, colon, melanoma, ovarian, renal, prostate, breast and central nervous system tumors in vitro | Increment of cell death and reduction of their proliferation | [50] | ||
| Acetylated BFA derivatives | Hindering of Arf1 and GEF joining | Esophagus squamous cell carcinoma in vitro | Increment of cell death in a sharper way than BFA | [51] |
| Ester derivatives of BFA | Hindering of Arf1 and GEF joining | Lung, colon, melanoma, ovarian, renal, prostate, breast and central nervous system tumors in vitro | Increment of cell death and reduction of their proliferation in a sharper way than BFA | [50] |
| C15 BFA derivatives | Hindering of Arf1 and GEF joining | Lung, colon, ovarian, renal, prostate, breast, leukemia, melanoma and central nervous system tumors in vitro | Increase of cell death, which is stronger than BFA | [52] |
| AMF-26/M-COPA | Impairment of ArfGEF activity | Breast cancer xenografts in vivo | Induction of complete reversion in the growth of these xenografts | [54] |
| Melanoma cells both in vitro and in vivo models | Inhibition of angiogenesis, proliferation and tumor growth through the suppression of VEGFR1/2. | [56] | ||
| Neoplastic mast cells cultured in vitro | Suppression of cell proliferation and resistance to imatinib through the abolishment of Kit signaling | [57] | ||
| SecinH3 | Inhibition of ArfGEF binding to Arf1 | Breast xenografts in vivo | Reduction of tumor growth, aggressiveness and metastasis | [63] |
| Non-small cell lung cancer cell lines in vitro | Inhibition of cell proliferation and reduction of cell resistance to gefitinib | [64] | ||
| Colorectal cancer models both in vivo and in vitro | Decrease cell proliferation, migration and proliferation through the abolishment of ARNO-dependent signaling | [65] | ||
| M69 | Block of ArfGEF activity | Acute T cell leukemia cells cultured in vitro | Disturbance of intracellular adhesion through restructuration of actin skeleton | [67] |
For instance, LM11 can abolish Arf1 activation by inhibiting its association in the Golgi apparatus with its specific GEF, Arf nucleotide-binding site opener (ARNO) [37,38]. The high specificity of LM11 to Arf1 ensures that it does not abrogate other analogs, such as Arf6, making it suitable to treat Arf1-overexpressing tumors [37]. For instance, the treatment of breast tumors that overexpress Arf1 with LM11 reduces their aggressiveness by decreasing cell proliferation and invasion, as well as by inducing apoptosis [13,38]. Furthermore, LM11 disrupts breast cancer adhesion to the extracellular matrix by inhibiting paxillin translocation to the cell membrane [13], which is essential to connect integrins with the actin cytoskeleton [39]. Curiously, LM11 seems to be ineffective when tumor cells carry a K38A substitution in Arf1 [37], thus it is essential that Arf1 overexpressing tumors be tested for variants before the use of this inhibitor.
The Arf1 inhibitor Exo2 prevents the activation of certain Arf1-specific GEF enzymes by binding to their Sec7 domain [40]. This inhibition impedes the release of secretory vesicles from the endoplasmic reticulum (ER) to the Golgi apparatus [41,42], as well as reduces cellular lipid storage by inhibiting perilipin-2 expression [43]. Besides, this molecule presents high specificity for vesicle cargo since it impedes the retrograde transport of the Shiga toxin from the early endosomes to the ER, but does not hinder cholera toxin transport between the same compartments [42]. On the other hand, our research group reveals that Exo2 has the potential to reduce prostate tumor growth and metastasis through inhibiting Arf1-mediated ERK1/2 activation [44]. Most recently, we showed for the first time that active GTP-bound Arf1 is much higher in metastatic head and neck squamous cell carcinoma (HNSCC) cells compared with their paired non-metastatic cells, supporting the critical role of Arf1 activation in HNSCC metastasis [45]. We further provided evidence that EGF induces HNSCC cell invasion through the EGFR-Arf1 signaling complex and interrupting it using Exo2 or histone deacetylase inhibitor TSA deters the progression of HNSCC, providing a rational basis for Arf1-targeted anti-HNSCC therapy [45].
Brefeldin A (BFA) is a lactone-derived compound isolated from Eupenicillium brefeldianum that impairs Arf1 activation by hindering its association with its GEF enzyme [46,47]. The 7-hydroxyl residue of BFA seems to be essential to this process because its loss disrupts its affinity for the Arf1-GEF complex, preventing its inhibitory action [48]. This molecule can reduce anaplastic large cell lymphoma proliferation through reducing Arf1-dependent signal transducer and activator of transcription 3 (STAT3) phosphorylation [49]. It also presents a slight cytotoxic activity in other types of cancers, such as in lung, colorectal, ovarian, breast, prostate, melanoma or central nervous system [50]. Nevertheless, BFA shows poor bioavailability and high toxicity while exhibiting a number of pleiotropic effects in non-target organs, preventing the development of phase 1 clinical trials [42,49,51]. Therefore, the generation of new chemical derivatives of BFA with higher anticarcinogenic activity and lower off-target effects is essential to improve its use in cancer therapy [50,51]. For instance, acetylated BFA derivatives can reduce the viability of esophagus squamous cell carcinoma cells in a 500-times greater manner than native BFA [51]. Furthermore, ester BFA derivatives present higher potency than native BFA against different cancer types, which ultimately can reduce their off-target effects by lowering administered doses [50]. Finally, the addition of vinyl or aromatic groups to the C15 of BFA increases its ability to reduce HeLa cell proliferation [52].
AMF-26, also known as M-COPA, which was isolated from some species of the Trichoderma genus, also impairs the formation of the Arf1-GEF complex by disrupting GEF activity [47,53,54]. This molecule has greater bioavailability than BFA, increasing its feasibility for being used in cancer treatment [54]. In fact, AMF-26 can induce complete tumor regression in breast cancer xenografts [54], reduce the proliferation of 39 different cancers in a variety of human organs (such as breast, colon, kidney, skin, central nervous system, lung, ovary, and stomach) [53,55], as well as diminish angiogenesis through suppressing the activation of the vascular endothelial growth factor receptor 1/2 (VEGFR1/2) and the nuclear factor-κB (NF-κB) pathways [56]. In addition, AMF-26 deactivates a mutant form of the endolysosomal Kit, leading to sensitizing carcinogenic mast cell to imatinib [57]. Finally, AMF-26 also prevents Shiga toxin-dependent apoptosis by decreasing its translocation into the Golgi apparatus [58].
Sec7 inhibitor H3 (SecinH3) is a non-specific Arf inhibitor, which abrogates both Arf6 and Arf1 signaling by binding and inhibiting the Sec7 catalytic domain of ARNO and deactivating cytohesins, which are small ARF-specific GEFs [59,60]. SecinH3 was firstly developed to analyze the harmful effects that insulin resistance generates in human cells, since Arf6 down-regulation hinders insulin response in hepatic cells [60] and impairs glucose-stimulated insulin secretion in pancreatic β cells [61]. Moreover, this inhibitor can also reduce Salmonella enterica invasion by decreasing the Arf-activated pool [62]. Otherwise, this inhibitor presents great therapeutic effects in some carcinogenic diseases. For example, it diminishes the growth of breast xenografts and reduces breast-related lung metastasis and tumor aggressiveness [63]. Furthermore, it can also reduce the proliferation of certain non-small-cell lung cancer cell types by decreasing epithelial growth factor receptor (EGFR) activation and inducing apoptosis in both in vivo and in vitro models [64]. These beneficial effects ultimately reduce non-small-cell lung cancer resistance to gefitinib [64]. Finally, SecinH3 abolishes the migration, invasion, and proliferation of colorectal cancer cells in both in vivo and in vitro models [65].
M69, which is a RNA aptamer (an oligonucleotide that recognizes and attaches to a specific target with high affinity) [66], can impede Arf effects by deactivating GEF enzymes through binding to their catalytic Sec7 domain [67]. Although few experiments have been currently done with this inhibitor, it seems to present anti-carcinogenic effects as its expression in T lymphocytes leads to the reorganization of their actin cytoskeleton and to decreasing their adhesion to the extracellular matrix [67].
3. Ras and Its Inhibitors in Cancer Therapy
The RAS oncogenes (HRAS, NRAS and KRAS) comprise the most frequently mutated class of oncogenes in human cancers, stimulating intensive effort in developing anti-Ras inhibitors in order to get them to the clinic (Table 2). However, there is no effective Ras inhibitor to be used for cancer treatment [35,68]. Therefore, other chemical and biological strategies should be developed to accomplish the inhibition of this small GTPase.
Table 2.
Action of Ras inhibitors in cancer treatment.
| Name of the Inhibitor | Mechanism of Action | Model | Global Outcomes | Reference |
|---|---|---|---|---|
| Bisphenol A | Disruption of the binding between Ras and SOS. | Cervical cancer cells cultured in vitro | Decrease in cell proliferation | [73] |
| SCH-53870 derivates | Disruption of the binding between Ras and SOS. | NIH3T3 mouse fibroblast in vitro | Decrease in cell proliferation both in normal and KRas-overexpressing cells | [74] |
| SAH-SOS1 | Disruption of the binding between Ras and SOS. | Pancreatic, lung and colon cancer cells cultured in vitro bearing different KRAS mutants | Decrease in cell proliferation in a dose-dependent manner, independently of the KRAS mutant which bears the cells. | [75] |
| SCH-54292 | Hindering of the binding between Ras and SOS | NIH3T3 mouse fibroblast in vitro | Inhibition of cell proliferation | [78] |
| MCP110 | Inhibition of Raf and Ras-binding | Colon cancer models both in vivo and in vitro | Impediment of cell proliferation both in vitro and in vivo and synergy with other chemotherapeutic drugs, such as paclitaxel or vincristine | [88] |
| Colon cancer cells cultured in vitro | Arrest of cell cycle in G1 phase through the abolishment of cyclin D1 levels | [87] | ||
| MCP1 | Inhibition of Raf and Ras binding | Multiple myeloma cells cultured in vitro | Reduction of cancer cell growth through the induction of intrinsic apoptosis | [87] |
| MCP1 and MCP110 | Inhibition of Raf and Ras binding | Multiple cancer cell lines defined by the National Cancer Institute (NCI) (Weinstein et al., 1997) | Reduction of cell proliferation | [87] |
| Enantiomeric iridium(III) metal-based compound | Inhibition of Ras and Raf interaction | Human kidney xenografts in vivo and kidney, breast, lung, prostatic, ovarian, melanoma and erythroleukemic cancer cell lines in vitro | Inhibition of cell cancer proliferation and reduction of tumor volume without affecting mice global weight | [83] |
| Sulindac sulfide | Hindering of Raf activation by Ras | NIH3T3 mouse fibroblast in vitro and Saos epithelial cells | Abolishment of Ras-dependent malignant transformation | [84] |
| Brest cancer cells in vitro | Inhibition of E2-derivated pro-proliferative outcomes | [85] | ||
| Sema4D | Stimulation of Ras-GAP activity | Adrenal gland phaeochromocytoma cells cultured in vitro | Reduction of cell migration through inhibition of β1 integrin activation | [91] |
| Salirasib | Inhibition of Ras anchorage to cytoplasmic membranes | Pancreatic cell xenografts in vivo | Inhibition of tumor growth dose-dependently and stimulation of gemcitabine antiproliferative effects | [94] |
| Lung cancer models both in vivo and in vitro | Inhibition of cell proliferation and tumor growth | [95] | ||
| Hepatocellular carcinoma models both in vivo and in vitro | Inhibition of cell proliferation through the arrest of cell cycle and the induction of apoptosis | [96] | ||
| Pancreatic cancer cells cultured in vitro | Reduction of cell proliferation through the arrest of cell cycle | [97] | ||
| Prostate cancer cells cultured in vitro | Enhancement of Exo2 effects on cell proliferation, migration and invasion. | [44] | ||
| Lung cancer patients | Common used doses and schedule failed in the inhibition of cell proliferation | [98] |
3.1. Chemical Strategies for Suppressing Ras Activity
Ras has a flat tertiary structure, which does not provide clear sites where small repressing ligands can bind, apart from its nucleotide binding site [69]. The picomolar affinity of Ras for GTP allows for this GTPase to be activated when the concentration of this nucleotide is low [70]. These two properties are responsible for the clinical failure of most potential inhibitors for Ras [34]. New strategies are now focusing on the creation of an imbalance between Ras-specific GAP and GEF activities, as well as on the prevention of Ras from transducing its downstream proteins (Figure 2) [69]. Despite these two setbacks, recent in silico techniques have identified new potential inhibitor binding sites in the Ras molecule, opening the possibility for the development of more effective Ras inhibitors [69].
Two of these regions in Ras, which are designated as sites 1 and 2 and are located between the switch 1 domain and the second α-helix (H2) structure, are the regions where the most important Ras GEF enzyme, son of sevenless homolog 1 (SOS1), binds and activates Ras [69,71]. Therefore, the occupancy of these sites with small inhibitory molecules could impede Ras guanine nucleotide exchange, preventing the activation of its downstream effectors [69,71]. Screening of molecular libraries has identified new inhibitory compounds with promising oncostatic effects on many carcinogenic cell lines [72]. For example, bisphenol A and its derivative 4,4’-biphenol can inhibit the proliferation of NIH3T3 cells [73] and some SCH-53870 derivatives can induce the death of NIH3T3 cell through the disruption of Ras and SOS binding [74]. The development of a synthetic α-helical structure which mimics SOS α helix and sequesters Ras-GDP molecules has been reported to be a possible new strategy for the treatment of Ras-overexpressing tumors [71]. Finally, some SAH-SOS1 inhibitors have been shown to be able to restrain the viability of Ras-overexpressing pancreatic, colon and lung cancer cells, demonstrating their potential use in cancer therapy [75].
Site 3, which is located between the switch 2 domain and the third α-helix of Ras, also corresponds to a GEF binding site [69]. SCH-54292, one of the first developed Ras inhibitors, binds this region in an inverse Mg2+ concentration-dependent manner [76,77] and can restrain the growth of NIH3T3 cells by inducing apoptosis [78].
Sites 4 and 5 are located near the GTP-binding site and loop 7 of Ras and are the preferential joining site for divalent metal-cyclens (M2+-cyclens) [69,79]. M2+-cyclens are constituted by a divalent metallic ion (such as Zn2+, Co2+ or Cu2+) attached to an organic cyclic structure [80,81]. These compounds can stabilize Ras-GTP in a pre-activated state, which is also known as state 1(T) [79]. The 1(T) intermediate displays lower affinity for its downstream effectors, as opposed to Ras-GTP in the state 2(T) conformation, which is able to effectively activate its effector molecules [79]. Therefore, the induction of Ras-GTP into state 1(T) by M2+-cyclens inhibits the Ras-related Raf activation pathway [79]. More potent organometallic compounds have been developed, such as Zn2+-bis (2-picolyl) amine (Zn2+-BPA), which blocks the activation of the downstream effectors of Ras through its specific binding to the loop L7 and switch 1 domains of this protein [79]. Zn2+-BPA can even inhibit the activation of some mutated Ras analogs without the previous requirement of having Ras bound to GTP [79]. All these mentioned properties allow the potential use of these compounds in the treatment of Ras-overexpressing tumors [79].
Another target with potential oncostatic effects in Ras-overexpressing tumors is the interaction of Ras and Raf, which is an essential step in the transduction of Ras signal [69]. For example, a derivative from enantiomeric iridium (III) that blocks the interaction of Ras with Raf, hinders the proliferation of different cancer cell lines and reduces tumor volume in mice kidney xenografts by inhibiting the Ras-Raf dependent activation of the MAPK pathway [82]. Similarly, R11.1.6, a poly-β sheet protein, can also block Ras and Raf interaction, as well as MAPK pathway activation in embryonic kidney cells, also making it a promising molecule in targeting Ras-overexpressing cancer [83]. Although NS1, a promising synthetic monobody has not been proven in preclinical models yet, it is a strong Ras inhibitory molecule that prevents Ras dimerization and Ras–Raf interaction due to its ability to specifically bind to the α4 and α5 helices of Ras [70]. Another method to inhibit the Ras–Raf interaction is the use of specific non-steroidal anti-inflammatory drugs (NSAIDs) [84,85]. Sulindac sulfide is an NSAID that can reduce tumorigenesis as well as colorectal and breast cancer proliferation in vitro [84,85]. Sulindac sulfide interacts with Ras in a non-covalent manner, which ultimately reduces its ability to transduce downstream cellular signaling [84,85]. Besides, other NSAIDs, such as aspirin or indomethacin, can also inhibit Ras and Raf protein interaction in vivo [86]. Finally, some members of MCP family, such as MCP1, MCP53 and MCP110, can also abolish Ras and Raf interaction by directly binding to the Ras effector domain and indirectly controlling Ras and Raf folding [87]. More in detail, MCP110 can inhibit the growth of colorectal tumor xenografts and sensitize colon cancer cells to different chemotherapeutical drugs such as paclitaxel, docetaxel, vincristine, and sorafenib, supporting its use in the treatment of Ras-overexpressing tumors [88].
Another approach to control the activity of Ras includes the activation of Ras-specific GAPs [89]. This process would increase the level of inactive Ras protein and consequently decrease downstream Ras signal transduction [89]. However, some Ras mutants have been reported to be insensitive to this inhibitory strategy because of the lack of certain amino acids inside their GAP-binding domain, impeding the action of these enzymes [69]. Screening of molecular libraries has identified several inducers that activate Ras GAPs [89]. For instance, repulsive guidance molecule A (RGMA), which is a synthetic protein that can extend neuronal axons, dissociates p120GAP from FAK, enhancing p120GAP activity and ultimately reducing cellular Ras-GTP content [90]. Semaphorin 4D (Sema4D), which is implied in the growth of neuronal system, can restrain integrin-mediated cancer cell invasion and migration through inducing Ras-specific GAP Plexin-B1 via direct protein–protein interaction [89,91].
Finally, another strategy for Ras inhibition is the disruption of its anchorage to the cytoplasmic membrane, which is an essential step during its activation [92]. For instance, salirasib (also known as trans-farnesylthiosalicylic acid) can act as the carboxyl-terminal farnesylcysteine carboxymethyl ester of Ras, exhibiting sufficient inhibitory properties to be used in Ras-overexpressing cancer treatment [92,93]. Indeed, this inhibitor reduces dose-dependently the growth of pancreatic and lung cancer xenografts [94,95], and restrains dose- and time-dependently the proliferation of pancreatic and liver cancer cells in vitro through arresting cell cycle progression and stimulating apoptotic cell death [96,97]. Besides, salisarib potentiates the restraining effects of Exo2 on prostate cancer proliferation, invasion and migration through inducing apoptotic cell death in both in vitro and in vivo models [44]. This inhibitor also stimulates gemcitabine-dependent reduction of tumor volume and weight, leading to increasing the survival rate of mice with pancreatic tumor xenografts [94]. Nevertheless, it has been claimed that the normally administered doses of salirasib are insufficient to inhibit the growth of lung carcinomas that carry mutations in Ras [98], failing in most in vivo assays due to the compensatory action of geranylgeranyl transferases [34,99]. In conclusion, more studies are required to determine its efficacy in cancer treatment.
3.2. Non-Chemical Mechanisms for Suppressing Ras Activity
Because of the difficulty finding effective Ras inhibitors, other more indirect anti-Ras strategies have recently arisen, such as the induction of certain Ras-related lethal genes or the restoration of the metabolism disturbances induced by this small GTPase [35].
One of these approaches consists in the detection of some specific genes that are essential for the growth of Ras-mutated cells, but not for Ras-wild type cells [35]. Therefore, the knockdown of these genes leads to reducing the effect of Ras mutations in tumor cell proliferation and ultimately emerging as an effective strategy in the prevention of tumorigenesis, as well as enhancing the efficacy of Ras inhibitors [35,100]. Currently, a great number of synthetic lethal interactors have been identified in Ras-mutated cells through siRNA screening [100,101,102]. Some of these interactors are implicated in Ras maturation, such as prenyl protein-specific endoprotease 2 (RCE1), and protein-S-isoprenylcysteine O-methyltransferase (ICMT); while others are Ras effectors, such as SHOC-2, phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 1 (PREX1) or RAF1; small GTPases, such as RAC1; or transcription factors, such as GATA-binding factor 1 (GATA1) [100,101,102]. However, the high differences existent among the different gene libraries makes this technique very inconsistent due to the high amount of false-negative results that occurs during the screening part [35]. Therefore, these protocols should be optimized to improve the sensitivity of this technique [35].
Another possible approach consists in the correction of the profound imbalances that the mutations in KRas generate in the metabolism of carcinogenic cells [35,103]. Indeed, KRas-mutated carcinogenic cells exhibit high levels of glucose uptake and glycolysis, fatty acid synthesis, glutaminolysis and nucleotide synthesis [103]. Due to the importance that these metabolic disturbances present in the maintenance of cancer promotion and progression, the methods that restore the metabolic rates to their basal levels could be an effective anti-cancer strategy [103]. In fact, tumorigenesis rate and pancreatic inflammation and fibrosis are clearly aggravated in mice carrying KRas tumors and are fed with high-fat diet [104]. The induction of autophagic response in KRas-mutated cells stimulates tumor aggressiveness and proliferation by accelerating glycolytic capability [105,106]. Indeed, autophagy inhibition in KRas-overexpressing tumors could decrease tumor promotion and progression [105,106].
4. Rac and Its Inhibitors in Cancer Therapy
Rac presents key functions in cancer promotion and progression since the aberrant expression of this small GTPase disrupts adherens cellular junctions, allowing cancer cells to undergo EMT and cell migration [21,107]. This alteration on Rac expression also enhances cellular proliferation by facilitating cell cycle progression from G1 to S phase and stimulating mitosis and cytokinesis [21,107]. Rac involvement in the reverse process of mesenchymal to epithelial transition (MET) facilitates the formation of secondary tumors [107]. These finding suggest that inhibition of Rac could be useful for cancer treatment [107]. The inhibitors that are currently available to block this protein, as well as their mechanism of action, are listed in Table 3
Table 3.
Action of Rho and Rab inhibitors in cancer treatment.
| Name of the Inhibitor | Mechanism of Action | Model | Global Outcomes | Reference |
|---|---|---|---|---|
| NSC23766 | Inhibition of RacGEF binding to Rac | Prostate cancer cells cultured in vitro | Reduction of cell proliferation and their invasive characteristics | [109] |
| Pancreatic cancer cells in vitro | Increase of sensibility to radiotherapy | [111] | ||
| NSCLC models both in vitro and in vivo | Inhibition of cell proliferation and migration. Increment of cell sensibility to gefitinib. | [110] | ||
| NSC23766 analogs | Inhibition of RacGEF binding to Rac | High-metastatic breast cancer cells cultured in vitro | Inhibition of cell proliferation in a sharper way than NSC23766 does | [112] |
| EHT 1864 | Inhibition of RacGEF activity | Breast cancer cells cultured in vitro | Inhibition of cell proliferation stimulated by estrogen signaling | [120] |
| Breast cancer cells cultured in vitro | Sensitization of cancer cells to tamoxifen | [120] | ||
| NIH3T3 mouse fibroblast in vitro | Inhibition of Rac1-derived malignant cell transformation | [119] | ||
| Breast cancer tumors biopsied from patients’ samples | Restraining of cell invasion and proliferation through programmed cell death induction | [121] | ||
| EHop-016 | Inhibition of Vav1 and -2 activity and its binding with Rac | Metastatic breast cancer cells cultured in vitro | Reduction of cell viability and migration through the inhibition of Rac-derived actin structures | [113] |
| Human and murine leukemic cell models both in vitro and in vivo and patient-derived cells | Increment of overall survival due to the inhibition of cell growth and survival | [115] | ||
| Myxofibrosarcoma cell lines cultured in vitro and xenografts tumors cultured in vivo | Inhibition of cell growth through the induction of apoptosis and suppression of the generation of lung metastasis | [116] | ||
| Breast cancer xenografts models with EHop-016 intraperitoneal administration | Repression of tumor growth, metastasis and angiogenesis | [114] | ||
| YM1B | Repression of RacGEF binding to Rac | Breast cancer cells cultured in vitro | Reduction of cell migration and invasion | [122] |
| BART | Repression of RacGEF activity | Pancreatic cancer cell lines cultured in vitro | Inhibition of cell motility and invasion through the regulation of actin cytoskeleton | [124] |
| Migrastatin analogs | Repression of Rac activity | High metastatic breast cancer cells in vivo xenograft models | Blockage of cell migration and metastasis through the inhibition of lamellipodia formation | [125] |
| Secramine A | Repression of Cdc42 shuttling between cytoplasm and cell membrane | ALCL cells cultured in vitro | Repression of cell proliferation through the induction of programmed cell death in ALK-positive cells | [133] |
| ZCL278 | Inhibition of ITSN and Cdc42 binding | Prostate cancer cell lines cultured in vitro | Inhibition of cell motility and migration mediated by actin filaments | [139] |
| ML141 or CID2950007 | Inhibition of GTP binding to Cdc42 | Ovarian cancer cells cultured in vitro | Inhibition of cell motility and invasion without affecting to its viability | [138] |
| MBQ-167 | Inhibition of GEF binding to Rac/Cdc42 | Breast cancer cells cultured in vitro and xenografts models in vivo | Repression of cell migration, metastasis and proliferation | [144] |
| AZA1 | Prevention of RacGEF binding to Cdc42/Rac | Prostatic cancer models both in vivo and in vitro | Decrease in cell proliferation through the induction of apoptosis in vitro. Reduction of tumor growth and improvement of mice survival in vivo | [145] |
| R-ketorolac | Inhibition of nucleotide docking | Ovarian cancer cell lines and primary patient-derived cells in vitro | Reduction in cell proliferation and growth | [146] |
| Rhosin | Inhibition of RhoAGEF binding to RhoA | Breast cancer cells cultured in vitro | Inhibition of cell proliferation, migration and invasion | [148] |
| Diffuse gastric cancer spheroids cultured in vitro | Inhibition of cell proliferation, migration and invasion. Sensitization of cells to cisplatin | [149] | ||
| Y16 | Hindering of RhoA and LARG joining | Breast cancer cells cultured in vitro | Reduction of cell proliferation and spheroid formation both alone and in combination with Rhosin | [150] |
| Biphosphonate derivatives | Inhibits Rab prenylation. | Melanoma cells cultured in vitro | Inhibition of cell proliferation through cell cycle arrest in S phase | [161] |
| Mesothelioma cells cultured in vitro | Induction of cell apoptosis due to the inhibition of topoisomerase II and Rab6 | [162] | ||
| Prostate and breast cancer cell lines cultured in vitro | Inhibition of cell adhesion to extracellular matrix | [163] | ||
| Prostate and breast cancer cell lines cultured in vitro | Inhibition of cell invasion and metastasis through the repression of MMPs activity | [164] |
NSC23766 was one of the first developed Rac1 inhibitors with capability to discern from other Rho family GTPases, such as Cdc42 or RhoA [108]. This synthetic compound binds to a zone located between the switch I, switch II and β1/β2/β3 regions of this protein [108], impeding its activation by occupying the location where two RacGEF enzymes, Trio and T-lymphoma invasion and metastasis-inducing protein 1 (Tiam1) join together with this GTPase [108,109]. Due to the restrictive effects that present in Rac activation, NSC23766 has been proven to impede cell proliferation in both carcinogenic and non-carcinogenic cells [109]. For instance, this inhibitor can restrain prostate cancer proliferation and mobility, sensitize prostate cancer cells to ionizing radiotherapy, and inhibit non-small cell lung cancer (NSCLC) proliferation and invasion both in vivo and in vitro [109,110,111]. Inversely, this inhibitor was unable to prevent high-metastatic breast cancer cells migration, thus some new NSC23766 derivatives should be developed to improve its efficiency to restrain these types of cancers, as well as to reduce the administered doses [112].
One derivative of NSC23766, EHop-016, inhibits Rac activation through disrupting its direct binding to Vav2 (a Rho-specific GEF) [113], giving it a 100-fold lower IC50 against this GTPase than NSC23766, which encourages its use instead its derivative in cancer therapy [113,114,115,116]. In fact, EHop-016 can inhibit the proliferation of breast, myxofibrosarcoma and leukemic cancer cells in vitro, as well as impair their invasive capabilities [113,115,116]. It can also reduce the proliferation, angiogenesis and invasion of breast xenografts, diminishing the generation of secondary tumors in lung, liver, spleen and kidneys [114]. Besides, it reduces the growth of myxofibrosarcoma xenografts by restraining Rac-derived Akt/mTORC1 and mTORC2 activation [116]. Alternatively, this inhibitor has also been proven useful in the treatment of some non-carcinogenic disorders since it prevents glucose-induced insulin secretion and actin remodeling in pancreatic β cells [117].
EHT 1864 impedes the formation of the Rac-Tiam1 complex, keeping this GTPase in an inactive state [118]. This inhibitor is unable to inhibit other Rac-related Rho-family GTPases, demonstrating high specificity [118,119]. EHT 1864 exhibits promising potential properties to be used during cancer treatment [119,120]. For example, it can repress estrogen-induced breast cancer cell proliferation through modulating the Rac-dependent estrogen receptor-alpha (ERα) expression, as well as hampers NIH3T3 Rac-dependent Ras-induced tumorigenesis [119,120]. Besides, it prevents breast cancer invasion and proliferation, as well as inducing apoptosis through inhibiting STAT3 in patient-derived samples [121]. Finally, it can also sensitize breast carcinogenic cells to tamoxifen, since Rac1 overexpression seems to be a hallmark in the acquisition of cell resistance to this drug [120].
YM1B, which is a monoclonal antibody against CCN1, can halt breast cancer migration and invasion through inhibiting Rac induction and actin reorganization [122]. Besides, the generation of aptamers that inhibit Tiam1 activity, by impeding the binding of Rac1 to this molecule and decreasing its activation, is a new strategy to inactivation of this GTPase in the treatment of the tumors that overexpress Rac1 [123]. Conversely, binder of Arl Two (BART) abrogated carcinogenic cell mobility due to its capability to restrain the activation of this small GTPase [124]. Finally, core macrolactam and core macroketone, which are two migrastatin analogs, can also abolish breast cancer migration through inhibiting Rac activation [125].
Nevertheless, the constant use of Rac inhibitors in cancer treatment could generate some off-target effects in platelets because of the indirect inhibition of p21-activated kinase 1/2 (PAK1/2) [126], thus the administration of those Rac inhibitors should be controlled and interrupted when these undesired effects arise.
5. Cdc42 and Its Inhibitors in Cancer Therapy
Cdc42, which belongs to the Rho family [127], takes part in the initiation of most human cancers [21,128] since it induces the proliferation of Ras-transformed carcinogenic cells by inducing cell cycle progression [129], as well as stimulates tumorigenesis through impeding the degradation of EGFR by ubiquitin-proteasome system [128,130]. In addition, Cdc42 also enhances metastasis, invasion and EMT [127,131]. These properties back up the development of Cdc42 inhibitors as a great strategy to be used in the treatment of several malignant diseases. The inhibitors that are currently available to block this protein, as well as their mechanism of action, are listed in Table 3.
Secramine A, a synthetic derivate of galantamine, can disturb the recruitment of prenylated Cdc42 to the Golgi apparatus membrane by blocking Rho guanine dissociation inhibitor 1 (RhoGDI1), which modulates its attachment to this membrane [132,133]. Indeed, the inhibition of Cdc42 activity by this molecule generates a significant reduction in the proliferation of anaplastic large cell lymphoma cells through inducing apoptotic cell death [134]. Curiously, the lack of anaplastic lymphoma kinase (ALK) in these cells hinders the capability of secramine A to induce this phenotype, thus its utilization should be restrained to the cells that express this protein [134]. Otherwise, this inhibitor has also been shown to restrain other non-carcinogenic pathophysiological processes [135,136,137]. For example, secramine A alters fertilization process by reducing progesterone-induced and spontaneous acrosome reaction in mammalian sperm [135], reduces the propagation of human immunodeficiency virus (HIV) in cluster of differentiation 4 (CD4)+ T lymphocytes [136] and disturbs the release of collagen I from vascular smooth muscle cells to the cell matrix [137]. Nevertheless, some harmful off-target effects have been reported during the administration of this compound, limiting its use in carcinogenic and non-carcinogenic diseases [138].
Another potential inhibitor of Cdc42 with therapeutic applications in cancer is ZCL278, a 4-bromine-2-chlorophenol derivative that disrupts the joining between Cdc42 and intersectin (ITSN), which is a Cdc42-specific GEF enzyme, leading to inhibiting the activation of this small GTPase [139,140]. ZCL278 presents promising properties for being used in cancer therapy, such as a high membrane permeability and low toxicity for non-carcinogenic cells [137]. For example, ZCL278 can restrain the migratory and invasive characteristics of the prostate cancer cell line PC3 in vitro [139]. In addition, it can also prevent the toxicity of sodium arsenite (NaAsO2) on astrocytes and cerebellar granule neurons, encouraging its use to alleviate arsenic poisoning on the nervous system [141,142].
ML141 (also known as CID2950007 or CID2995007) and its analog CID44216842 are selective Cdc42 inhibitors that can deactivate this small GTPase in a non-competitive and allosteric manner by locking this protein in an inactive conformation [34,143]. Although these molecules are unable to induce any anti-proliferative effects in ovarian cancer cells, they can restrain dose-dependently their mobility, which indicates their possible use as a therapeutic adjuvants to reduce the arising of secondary tumors [138].
Additionally, some double Rac/Cdc42 inhibitors have been developed due to the similar roles these two small GTPases perform in cancer promotion and progression [144]. For instance, MBQ-167, which seems to inhibit the activation of Rac and Cdc42 by occupying their effector domain, prevents breast cancer cell migration, reduces cell viability, and impedes EMT progression by disrupting cell polarity without affecting non-carcinogenic cells growth (Table 3) [144]. N*2*,N*4*-Bis-(2-methyl-1H-indol-5-yl)-pyrimidine-2,4-diamine (AZA1), which is a structural derivative of NSC23766, can repress tumor growth and abolish cell migration in prostate xenografts (Table 3) [145]. Otherwise, R-ketorolac, which is an enantiomer of the analgesic S-ketorolac, can diminish cell adhesion to extracellular matrix and invasion in ovarian cancer cells, validating its possible use in cancer therapy [146].
6. Targeting other Small GTPases in Cancer Therapy
RhoA is an oncogene that induces tumor progression through enhancing carcinogenic cell proliferation, angiogenesis, invasion and metastasis [21,147], suggesting the development of RhoA inhibitors could be a great strategy to restrain cancer promotion and progression [36]. However, the generation of these inhibitors present several challenges due to the absence of stable cavities in the surface of RhoA, apart from the nucleotide-binding pocket, as well as its ability to capture guanine nucleotides at sub-nanomolar range [36]. Therefore, new strategies should be carried out to inhibit RhoA in cancer cells (Figure 2) [36]. The inhibitors that are currently available to block this oncogene, as well as their mechanism of action, are listed in Table 3.
Despite these difficulties, some promising RhoA inhibitors have already been developed to be used in cancer treatment [36]. For instance, Rhosin, which disturbs RhoA activation by binding to its W58 and impeding the docking of GEF enzymes [148], can inhibit breast cancer cell proliferation, migration and invasion in vitro and restrain the formation of spheroid bodies in gastric cancer cells [148,149]. On the other hand, Y16, which also prevents the binding between RhoA and its GEF enzymes by binding between the DH and PH domains of Leukemia-associated RhoGEF (LARG), suppresses breast cancer proliferation, migration and invasion, as well as modifies the distribution of the actin cytoskeleton [150]. Both inhibitors synergize to more efficiently hinder the interaction between RhoA and LARG, intensifying the reduction of breast cancer proliferation and invasion and allowing their use in the treatment of RhoA-overexpressing tumors [150].
CHS-111 is a benzyl indazole derivative that prevents both the joining of RhoA to membranes and its activation by its GEF enzyme Vav by preventing its interaction with phospholipase D1 (PLD1) [151]. Although there is no CHS-111 preclinical study, it might be useful in the treatment of RhoA overexpressing cancers.
Rab family constituents are also involved in the pathogenesis of some cancer types, such as hepatocellular carcinoma, cholangiohepatoma, gastric cancer, lung cancer, renal cancer, breast cancer, tongue and oral squamous cell carcinoma, as well as ovarian cancer [26,152,153,154,155,156,157], because they can promote cancer progression and aggressiveness by inducing invasion, metastasis, proliferation and cell cycle progression [26,157,158]. Rab protein overexpression also increases the resistance of carcinogenic cells to chemotherapy [159]. Despite the role of this family of small GTPases in cancer, few effective Rab inhibitors are available for cancer treatment [160]. Among them, Rab geranylgeranyltransferase (RabGGTase) inhibitors are the most promising molecules to be used in Rab-overexpressing tumors [160]. These inhibitors prevent the addition of geranyl groups in the C-termini of Rabs, which are necessary for ensuring their attachment to cellular membranes [160]. RabGGTases present promising effects in cancer therapy because they can effectively restrain human myeloma and mesothelioma cell proliferation in vitro by inducing apoptosis and cell cycle arrest, as well as reduce prostate and breast cancer adhesion and metastasis by suppressing the proteolytic activity of matrix metalloproteinases (MMPs) [161,162,163,164]. Therefore, we can conclude that RabGGTases show promising in vivo effects, preventing the growth of different skeletal tumors [165].
7. Conclusions
Small GTPases are upregulated in a broad spectrum of human cancers since they have the potential to promote cell proliferation and mobility as well as to stimulate their invasive and metastatic characteristics. However, no inhibitors are currently available for its normal use in clinical therapy. Indeed, small GTPases are difficult to target because they present few stable cavities for inhibitors to bind on their surface and they can capture guanine nucleotides at sub-nanomolar range. To achieve the inhibition of these molecules, some new strategies have recently arisen, such as the inhibition of the binding between GTPase and GEF, the enhancement of GAP activity, the hindering of nucleotide attachment, the blocking of their binding to cellular membranes, or the inhibition of the activity of their downstream effectors. However, most of the new drugs that hamper these strategies have only been proven successful in in vitro studies, while demonstrating fewer promising abilities at the in vivo model and clinical trial stages. These discrepancies normally occur because of the enhancement of some compensatory mechanisms that mask the effect of the inhibitor. Therefore, more in vivo studies should be performed to further evaluate the capability of these inhibitors to impede cancer progression and reduce cancer chemoresistance.
Acknowledgments
We thank Austin Shull and Cooley Marion for their input. We apologize to numerous colleagues whose work could not be cited in this article due to space limitation.
Abbreviations
| ALK | Anaplastic lymphoma kinase |
| ARL | Arf-like proteins |
| ARNO | Arf nucleotide-binding site opener |
| AZA1 | N*2*,N*4*-Bis-(2-methyl-1H-indol-5-yl)-pyrimidine-2,4-diamine |
| BART | Binder of Arl Two |
| BFA | Brefeldin A |
| CD4 | Cluster of differentiation 4 |
| Cdc42 | Cell division control protein 42 homolog |
| CERT | Ceramide transfer protein |
| EGFR | Epithelial growth factor receptor |
| EMT | Epithelial to mesenchymal transition |
| ER | Endoplasmic reticulum |
| ERK | Extracellular signal-regulated kinase |
| ERα | Estrogen receptor-alpha |
| Exo84 | Exocyst complex component 84 |
| FAK | Focal adhesion kinase |
| FAPP1 | Phosphatidylinositol-four-phosphate adapter protein 1 |
| GAP | GTPase-activating proteins |
| GATA1 | GATA-binding factor 1 |
| GEF | Guanine-nucleotide exchange factors |
| GRE | Rad and Gem-related proteins |
| H2 | Second α-helix of Ras |
| HIV | Human immunodeficiency virus |
| HNSCC | Head and neck squamous cell carcinoma |
| IMCT | Protein-S-isoprenylcysteine O-methyltransferase |
| ITSN | Intersectin |
| LARG | Leukemia-associated RhoGEF |
| M2+-cyclens | Divalent metal-cyclens |
| M6PR | Mannose-6-phosphate receptor |
| MAPK | Mitogen-activated protein kinases |
| MET | Mesenchymal to epithelial transition |
| Miro | Mitochondrial Rho GTPase |
| MMP | Matrix metalloproteinase |
| mTORC1 | mechanistic target of rapamycin complex 1 |
| NF-κB | nuclear factor-κB |
| NSAID | Non-steroidal anti-inflammatory drug |
| NSCLC | Non-small cell lung cancer |
| p70S6K | Ribosomal protein S6 kinase beta-1 |
| PAK | p21-activated kinase |
| PI3K | Phosphatidylinositol 3-kinase |
| PLCε | Phospholipase C epsilon |
| PLD1 | Phospholipase D1 |
| PREX1 | Phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor |
| Rab | Ras-related in brain |
| RabGGTase | Rab geranylgeranyltransferase |
| Rac | Ras-related C3 botulinum toxin substrate |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 |
| Rad | Ras-related associated with diabetes |
| Ral | Ras-like proteins |
| Ran | Ras-related nuclear protein |
| Rap | Ras-proximal proteins |
| RCE1 | Prenyl protein-specific endoprotease 2 |
| RGMA | Repulsive guidance molecule A |
| Rheb | Ras homolog enriched in brain |
| Rho | Ras-homolog |
| RhoA | Rho-related proteins |
| RhoBTB3 | Rho-related BTB domain-containing protein 3 |
| RhoGDI1 | Rho guanine dissociation inhibitor 1 |
| Rit | Ras-like in all tissues proteins |
| Rnd | Rho-related GTP-binding protein Rho6 precursor |
| RRAS | Ras-related proteins |
| SecinH3 | Sec7 inhibitor H3 |
| Sema4D | Semaphorin 4D |
| SNARE | NSF-attachment protein receptor |
| SOS1 | Son of sevenless homolog 1 |
| STAT3 | Signal transducer and activator of transcription 3 |
| TCL | TC10 and T-cell leukemia/lymphoma protein 1A |
| Tiam1 | T-lymphoma invasion and metastasis-inducing protein 1 |
| VEGFR | VEGF receptor |
| VEGF | Vascular endothelial growth factor |
| Zn2+-BPA | Zn2+-bis (2-picolyl) amine |
Funding
This research was supported by NIH grant R03DE028387 (to YT). NPD conducted his study under supervision of YT at Augusta University supported by the Spanish Ministry of Education, Culture and Sports (“Becas FPU 2013”, reference FPU13/04173 and “Ayudas a la movilidad para estancias breves y traslados temporales 2016”, reference EST 16/00783).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Citi S., Spadaro D., Schneider Y., Stutz J., Pulimeno P. Regulation of small GTPases at epithelial cell-cell junctions. Mol. Membr. Biol. 2011;28:427–444. doi: 10.3109/09687688.2011.603101. [DOI] [PubMed] [Google Scholar]
- 2.Lu Q., Nassar N., Wang J. A mechanism of catalyzed GTP hydrolysis by Ras protein through magnesium ion. Chem. Phys. Lett. 2011;516:233–238. doi: 10.1016/j.cplett.2011.09.071. [DOI] [Google Scholar]
- 3.Shi G.X., Andres D.A., Cai W. Ras family small GTPase-mediated neuroprotective signaling in stroke. Cent. Nerv. Syst. Agents Med. Chem. 2011;11:114–137. doi: 10.2174/187152411796011349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson D.S., Chen Y.H. Ras family of small GTPases in immunity and inflammation. Curr. Opin. Pharmacol. 2012;12:458–463. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 6.Heider D., Hauke S., Pyka M., Kessler D. Insights into the classification of small GTPases. Adv. Appl. Bioinforma. Chem. 2010;3:15–24. doi: 10.2147/AABC.S8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donaldson J.G., Jackson C.L. Arf family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth M.G. Snapshots of ARF1: Implications for mechanisms of activation and inactivation. Cell. 1999;97:149–152. doi: 10.1016/S0092-8674(00)80723-0. [DOI] [PubMed] [Google Scholar]
- 10.Casalou C., Faustino A., Barral D.C. Arf proteins in cancer cell migration. Small GTPases. 2016;7:270–282. doi: 10.1080/21541248.2016.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannangai R., Vivekanandan P., Martinez-Murillo F., Choti M., Torbenson M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum. Pathol. 2007;38:639–644. doi: 10.1016/j.humpath.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Davis J.E., Xie X., Guo J., Huang W., Chu W.M., Huang S., Teng Y., Wu G. ARF1 promotes prostate tumorigenesis via targeting oncogenic MAPK signaling. Oncotarget. 2016;7:39834–39845. doi: 10.18632/oncotarget.9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlienger S., Ramirez R.A., Claing A. ARF1 regulates adhesion of MDA-MB-231 invasive breast cancer cells through formation of focal adhesions. Cell. Signal. 2015;27:403–415. doi: 10.1016/j.cellsig.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Li R., Peng C., Zhang X., Wu Y., Pan S., Xiao Y. Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci. 2017;182:80–84. doi: 10.1016/j.lfs.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Goitre L., Trapani E., Trabalzini L., Retta S.F. The Ras superfamily of small GTPases: The unlocked secrets. In: Trabalzini L., Retta S.F., editors. Ras Signaling: Methods and Protocols. Volume 1120. Springer; Berlin, Germany: 2014. pp. 1–18. [DOI] [PubMed] [Google Scholar]
- 16.Shirakawa R., Horiuchi H. Ral GTPases: Crucial mediators of exocytosis and tumourigenesis. J. Biochem. 2015;157:285–299. doi: 10.1093/jb/mvv029. [DOI] [PubMed] [Google Scholar]
- 17.Di J., Huang H., Qu D., Tang J., Cao W., Lu Z., Cheng Q., Yang J., Bai J., Zhang Y., et al. Rap2B promotes proliferation, migration, and invasion of human breast cancer through calcium-related ERK1/2 signaling pathway. Sci. Rep. 2015;5:12363. doi: 10.1038/srep12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gloerich M., Bos J.L. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011;21:615–623. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Armijo M.E., Campos T., Fuentes-Villalobos F., Palma M.E., Pincheira R., Castro A.F. Rheb signaling and tumorigenesis: mTORC1 and new horizons. Int. J. Cancer. 2016;138:1815–1823. doi: 10.1002/ijc.29707. [DOI] [PubMed] [Google Scholar]
- 20.Shi G.X., Cai W., Andres D.A. Rit subfamily small GTPases: Regulators in neuronal differentiation and survival. Cell Signal. 2013;25:2060–2068. doi: 10.1016/j.cellsig.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson R., Pedersen E.D., Wang Z., Brakebusch C. Rho GTPase function in tumorigenesis. Biochim. Biophys. Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Olson M.F. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases. 2018;9:203–215. doi: 10.1080/21541248.2016.1218407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadok A., Marshall C.J. Rho GTPases: Masters of cell migration. Small GTPases. 2014;5:e29710. doi: 10.4161/sgtp.29710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramani D., Alahari S.K. Integrin-mediated function of Rab GTPases in cancer progression. Mol. Cancer. 2010;9:312. doi: 10.1186/1476-4598-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 26.Wang S., Hu C., Wu F., He S. Rab25 GTPase: Functional roles in cancer. Oncotarget. 2017;8:64591–64599. doi: 10.18632/oncotarget.19571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 28.Cherfils J., Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 29.Dworak N., Makosa D., Chatterjee M., Jividen K., Yang C.S., Snow C., Simke W.C., Johnson I.G., Kelley J.B., Paschal B.M. A nuclear lamina-chromatin-Ran GTPase axis modulates nuclear import and DNA damage signaling. Aging Cell. 2019;18:e12851. doi: 10.1111/acel.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yudin D., Fainzilber M. Ran on tracks-cytoplasmic roles for a nuclear regulator. J. Cell Sci. 2009;122:587–593. doi: 10.1242/jcs.015289. [DOI] [PubMed] [Google Scholar]
- 31.Sheng C., Qiu J., Wang Y., He Z., Wang H., Wang Q., Huang Y., Zhu L., Shi F., Chen Y., et al. Knockdown of Ran GTPase expression inhibits the proliferation and migration of breast cancer cells. Mol. Med. Rep. 2018;18:157–168. doi: 10.3892/mmr.2018.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klosowiak J.L., Focia P.J., Chakravarthy S., Landahl E.C., Freymann D.M., Rice S.E. Structural coupling of the EF hand and C-terminal GTPase domains in the mitochondrial protein Miro. EMBO Rep. 2013;14:968–974. doi: 10.1038/embor.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K.S., Lu B. The myriad roles of Miro in the nervous system: Axonal transport of mitochondria and beyond. Front. Cell. Neurosci. 2014;8:330. doi: 10.3389/fncel.2014.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cromm P.M., Spiegel J., Grossmann T.N., Waldmann H. Direct modulation of small GTPase activity and function. Angew. Chem. Int. Ed. 2015;54:13516–13537. doi: 10.1002/anie.201504357. [DOI] [PubMed] [Google Scholar]
- 35.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y., Zheng Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin. Drug Discov. 2015;10:991–1010. doi: 10.1517/17460441.2015.1058775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flisiak S., Zeeh J.C., Guibert B., Cherfils J., Zeghouf M. An Arf1 GTPase mutant with different responses to GEF inhibitors. Biochem. Biophys. Res. Commun. 2008;377:156–160. doi: 10.1016/j.bbrc.2008.09.107. [DOI] [PubMed] [Google Scholar]
- 38.Xie X., Tang S.C., Cai Y., Pi W., Deng L., Wu G., Chavanieu A., Teng Y. Suppression of breast cancer metastasis through the inactivation of ADP-ribosylation factor 1. Oncotarget. 2016;7:58111–58120. doi: 10.18632/oncotarget.11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner C.E. Paxillin interactions. J. Cell Sci. 2000;113:4139–4140. doi: 10.1242/jcs.113.23.4139. [DOI] [PubMed] [Google Scholar]
- 40.Bourgoin S.G., El Azreq M.A. Small inhibitors of ADP-ribosylation factor activation and function in mammalian cells. World J. Pharmacol. 2012;1:55–64. doi: 10.5497/wjp.v1.i4.55. [DOI] [Google Scholar]
- 41.Spooner R.A., Watson P., Smith D.C., Boal F., Amessou M., Johannes L., Clarkson G.J., Lord J.M., Stephens D.J., Roberts L.M. The secretion inhibitor Exo2 perturbs trafficking of Shiga toxin between endosomes and the trans-Golgi network. Biochem. J. 2008;414:471–484. doi: 10.1042/BJ20080149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorieul M., Langhans M., Guetzoyan L., Hillmer S., Clarkson G., Lord J.M., Roberts L.M., Robinson D.G., Spooner R.A., Frigerio L. An Exo2 derivative affects ER and Golgi morphology and vacuolar sorting in a tissue-specific manner in Arabidopsis. Traffic. 2011;12:1552–1562. doi: 10.1111/j.1600-0854.2011.01258.x. [DOI] [PubMed] [Google Scholar]
- 43.Pauloin A., Adenot P., Hue-Beauvais C., Chanat E. The perilipin-2 (adipophilin) coat of cytosolic lipid droplets is regulated by an Arf1-dependent mechanism in HC11 mouse mammary epithelial cells. Cell Biol. Int. 2016;40:143–155. doi: 10.1002/cbin.10547. [DOI] [PubMed] [Google Scholar]
- 44.Lang L., Shay C., Zhao X., Teng Y. Combined targeting of Arf1 and Ras potentiates anticancer activity for prostate cancer therapeutics. J. Exp. Clin. Cancer Res. 2017;36:112. doi: 10.1186/s13046-017-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L., Gao L., Shay C., Lang L., Lv F., Teng Y. Histone deacetylase inhibitors suppress aggressiveness of head and neck squamous cell carcinoma via histone acetylation-independent blockade of the EGFR-Arf1 axis. J. Exp. Clin. Cancer Res. 2019;38:84. doi: 10.1186/s13046-019-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossessova E., Corpina R.A., Goldberg J. Crystal structure of ARF1•Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell. 2003;12:1403–1411. doi: 10.1016/S1097-2765(03)00475-1. [DOI] [PubMed] [Google Scholar]
- 47.Vigil D., Cherfils J., Rossman K.L., Der C.J. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeeh J.C., Zeghouf M., Grauffel C., Guibert B., Martin E., Dejaegere A., Cherfils J. Dual specificity of the interfacial inhibitor Brefeldin A for Arf proteins and Sec7 domains. J. Biol. Chem. 2006;281:11805–11814. doi: 10.1074/jbc.M600149200. [DOI] [PubMed] [Google Scholar]
- 49.Toda T., Watanabe M., Kawato J., Kadin M.E., Higashihara M., Kunisada T., Umezawa K., Horie R. Brefeldin A exerts differential effects on anaplastic lymphoma kinase positive anaplastic large cell lymphoma and classical Hodgkin lymphoma cell lines. Br. J. Haematol. 2015;170:837–846. doi: 10.1111/bjh.13508. [DOI] [PubMed] [Google Scholar]
- 50.Anadu N.O., Davisson V.J., Cushman M. Synthesis and anticancer activity of Brefeldin A ester derivatives. J. Med. Chem. 2006;49:3897–3905. doi: 10.1021/jm0602817. [DOI] [PubMed] [Google Scholar]
- 51.He B., Wang Y., Zheng Y., Chen W., Zhu Q. Synthesis and cytotoxic evaluation of acylated Brefeldin A derivatives as potential anticancer agents. Chem. Biol. Drug Des. 2013;82:307–316. doi: 10.1111/cbdd.12154. [DOI] [PubMed] [Google Scholar]
- 52.Seehafer K., Rominger F., Helmchen G., Langhans M., Robinson D.G., Özata B., Brügger B., Strating J.R.P.M., Van Kuppeveld F.J.M., Klein C.D. Synthesis and biological properties of novel Brefeldin A analogues. J. Med. Chem. 2013;56:5872–5884. doi: 10.1021/jm400615g. [DOI] [PubMed] [Google Scholar]
- 53.Shiina I., Umezaki Y., Ohashi Y., Yamazaki Y., Dan S., Yamori T. Total synthesis of AMF-26, an antitumor agent for inhibition of the Golgi system, targeting ADP-ribosylation factor 1. J. Med. Chem. 2013;56:150–159. doi: 10.1021/jm301695c. [DOI] [PubMed] [Google Scholar]
- 54.Ohashi Y., Iijima H., Yamaotsu N., Yamazaki K., Sato S., Okamura M., Sugimoto K., Dan S., Hirono S., Yamori T. AMF-26, a novel inhibitor of the Golgi system, targeting ADP-ribosylation factor 1 (Arf1) with potential for cancer therapy. J. Biol. Chem. 2012;287:3885–3897. doi: 10.1074/jbc.M111.316125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi Y., Okamura M., Hirosawa A., Tamaki N., Akatsuka A., Wu K.M., Choi H.W., Yoshimatsu K., Shiina I., Yamori T., et al. M-COPA, a Golgi disruptor, inhibits cell surface expression of MET protein and exhibits antitumor activity against MET-addicted gastric cancers. Cancer Res. 2016;76:3895–3903. doi: 10.1158/0008-5472.CAN-15-2220. [DOI] [PubMed] [Google Scholar]
- 56.Watari K., Nakamura M., Fukunaga Y., Furuno A., Shibata T., Kawahara A., Hosoi F., Kuwano T., Kuwano M., Ono M. The antitumor effect of a novel angiogenesis inhibitor (an octahydronaphthalene derivative) targeting both VEGF receptor and NF-κB pathway. Int. J. Cancer. 2012;131:310–321. doi: 10.1002/ijc.26356. [DOI] [PubMed] [Google Scholar]
- 57.Hara Y., Obata Y., Horikawa K., Tasaki Y., Suzuki K., Murata T., Shiina I., Abe R. M-COPA suppresses endolysosomal Kit-Akt oncogenic signalling through inhibiting the secretory pathway in neoplastic mast cells. PLoS ONE. 2017;12:e0175514. doi: 10.1371/journal.pone.0175514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hattori T., Watanabe-Takahashi M., Shiina I., Ohashi Y., Dan S., Nishikawa K., Yamori T., Naito M. M-COPA, a novel Golgi system disruptor, suppresses apoptosis induced by Shiga toxin. Genes Cells. 2016;21:901–906. doi: 10.1111/gtc.12386. [DOI] [PubMed] [Google Scholar]
- 59.Bi X., Schmitz A., Hayallah A.M., Song J.N., Famulok M. Affinity-based labeling of cytohesins with a bifunctional SecinH3 photoaffinity probe. Angew. Chem. Int. Ed. 2008;47:9565–9568. doi: 10.1002/anie.200803962. [DOI] [PubMed] [Google Scholar]
- 60.Hafner M., Schmitz A., Grüne I., Srivatsan S.G., Paul B., Kolanus W., Quast T., Kremmer E., Bauer I., Famulok M. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 61.Jayaram B., Syed I., Kyathanahalli C.N., Rhodes C.J., Kowluru A. Arf nucleotide binding site opener [ARNO] promotes sequential activation of Arf6, Cdc42 and Rac1 and insulin secretion in INS 832/13 β-cells and rat islets. Biochem. Pharmacol. 2011;81:1016–1027. doi: 10.1016/j.bcp.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davidson A.C., Humphreys D., Brooks A.B., Hume P.J., Koronakis V. The Arf GTPase-activating protein family is exploited by Salmonella enterica serovar Typhimurium to invade nonphagocytic host cells. MBio. 2015;6:e02253-14. doi: 10.1128/mBio.02253-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao H., Ahirwar D.K., Oghumu S., Wilkie T., Powell C.A., Nasser M.W., Satoskar A.R., Li D.Y., Ganju R.K. Endothelial Robo4 suppresses breast cancer growth and metastasis through regulation of tumor angiogenesis. Mol. Oncol. 2016;10:272–281. doi: 10.1016/j.molonc.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bill A., Schmitz A., König K., Heukamp L.C., Hannam J.S., Famulok M. Anti-proliferative effect of cytohesin inhibition in gefitinib-resistant lung cancer cells. PLoS ONE. 2012;7:e41179. doi: 10.1371/journal.pone.0041179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan T., Sun J., Hu J., Hu Y., Zhou J., Chen Z., Xu D., Xu W., Zheng S., Zhang S. Cytohesins/ARNO: The function in colorectal cancer cells. PLoS ONE. 2014;9:e90997. doi: 10.1371/journal.pone.0090997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Germer K., Leonard M., Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int. J. Biochem. Mol. Biol. 2013;4:27–40. [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer G., Blind M., Nagel W., Böhm T., Knorr T., Jackson C.L., Kolanus W., Famulok M. Controlling small guanine-nucleotide-exchange factor function through cytoplasmic RNA intramers. Proc. Natl. Acad. Sci. USA. 2001;98:4961–4965. doi: 10.1073/pnas.091100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Hattum H., Waldmann H. Chemical biology tools for regulating RAS signaling complexity in space and time. Chem. Biol. 2014;21:1185–1195. doi: 10.1016/j.chembiol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Wang W., Fang G., Rudolph J. Ras inhibition via direct Ras binding-is there a path forward? Bioorg. Med. Chem. Lett. 2012;22:5766–5776. doi: 10.1016/j.bmcl.2012.07.082. [DOI] [PubMed] [Google Scholar]
- 70.Spencer-Smith R., Li L., Prasad S., Koide A., Koide S., O’Bryan J.P. Targeting the α4-α5 interface of RAS results in multiple levels of inhibition. Small GTPases. 2018 doi: 10.1080/21541248.2017.1333188. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patgiri A., Yadav K.K., Arora P.S., Bar-Sagi D. An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 2011;7:585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu S., Jang H., Zhang J., Nussinov R. Inhibitors of Ras-SOS interactions. ChemMedChem. 2016;11:814–821. doi: 10.1002/cmdc.201500481. [DOI] [PubMed] [Google Scholar]
- 73.Schöpel M., Jockers K.F., Düppe P.M., Autzen J., Potheraveedu V.N., Ince S., Yip K.T., Heumann R., Herrmann C., Scherkenbeck J., et al. Bisphenol A binds to Ras proteins and competes with guanine nucleotide exchange: Implications for GTPase-selective antagonists. J. Med. Chem. 2013;56:9664–9672. doi: 10.1021/jm401291q. [DOI] [PubMed] [Google Scholar]
- 74.Peri F., Airoldi C., Colombo S., Martegani E., van Neuren A.S., Stein M., Marinzi C., Nicotra F. Design, synthesis and biological evaluation of sugar-derived Ras inhibitors. ChemBioChem. 2005;6:1839–1848. doi: 10.1002/cbic.200400420. [DOI] [PubMed] [Google Scholar]
- 75.Leshchiner E.S., Parkhitko A., Bird G.H., Luccarelli J., Bellairs J.A., Escudero S., Opoku-Nsiah K., Godes M., Perrimon N., Walensky L.D. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc. Natl. Acad. Sci. USA. 2015;112:1761–1766. doi: 10.1073/pnas.1413185112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taveras A.G., Remiszewski S.W., Doll R.J., Cesarz D., Huang E.C., Kirschmeier P., Pramanik B.N., Snow M.E., Wang Y.S., del Rosario J.D., et al. Ras oncoprotein inhibitors: The discovery of potent, Ras nucleotide exchange inhibitors and the structural determination of a drug-protein complex. Bioorg. Med. Chem. 1997;5:125–133. doi: 10.1016/S0968-0896(96)00202-7. [DOI] [PubMed] [Google Scholar]
- 77.Ganguly A.K., Wang Y.S., Pramanik B.N., Doll R.J., Snow M.E., Taveras A.G., Remiszewski S., Cesarz D., del Rosario J., Vibulbhan B., et al. Interaction of a novel GDP exchange inhibitor with the Ras protein. Biochemistry. 1998;37:15631–15637. doi: 10.1021/bi9805691. [DOI] [PubMed] [Google Scholar]
- 78.Palmioli A., Sacco E., Abraham S., Thomas C.J., Di Domizio A., De Gioia L., Gaponenko V., Vanoni M., Peri F. First experimental identification of Ras-inhibitor binding interface using a water-soluble Ras ligand. Bioorg. Med. Chem. Lett. 2009;19:4217–4222. doi: 10.1016/j.bmcl.2009.05.107. [DOI] [PubMed] [Google Scholar]
- 79.Kalbitzer H.R., Spoerner M. State 1(T) inhibitors of activated Ras. In: Tamanoi F., editor. Inhibitors of the Ras Superfamily G-Proteins, Part A. Volume 33. Elsevier Inc.; Amsterdam, The Netherlands: 2013. pp. 69–94. [DOI] [PubMed] [Google Scholar]
- 80.Spoerner M., Graf T., König B., Kalbitzer H.R. A novel mechanism for the modulation of the Ras-effector interaction by small molecules. Biochem. Biophys. Res. Commun. 2005;334:709–713. doi: 10.1016/j.bbrc.2005.06.144. [DOI] [PubMed] [Google Scholar]
- 81.Rosnizeck I.C., Graf T., Spoerner M., Tränkle J., Filchtinski D., Herrmann C., Gremer L., Vetter I.R., Wittinghofer A., König B., et al. Stabilizing a weak binding state for effectors in the human Ras protein by cyclen complexes. Angew. Chem. Int. Ed. 2010;49:3830–3833. doi: 10.1002/anie.200907002. [DOI] [PubMed] [Google Scholar]
- 82.Liu L.J., Wang W., Huang S.Y., Hong Y., Li G., Lin S., Tian J., Cai Z., Wang H.M.D., Ma D.L., et al. Inhibition of the Ras/Raf interaction and repression of renal cancer xenografts in vivo by an enantiomeric iridium(III) metal-based compound. Chem. Sci. 2017;8:4756–4763. doi: 10.1039/C7SC00311K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kauke M.J., Traxlmayr M.W., Parker J.A., Kiefer J.D., Knihtila R., McGee J., Verdine G., Mattos C., Wittrup K.D. An engineered protein antagonist of K-Ras/B-Raf interaction. Sci. Rep. 2017;7:5831. doi: 10.1038/s41598-017-05889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herrmann C., Block C., Geisen C., Haas K., Weber C., Winde G., Möröy T., Müller O. Sulindac sulfide inhibits Ras signaling. Oncogene. 1998;17:1769–1776. doi: 10.1038/sj.onc.1202085. [DOI] [PubMed] [Google Scholar]
- 85.Gilad L.A., Bresler T., Gnainsky J., Smirnoff P., Schwartz B. Regulation of vitamin D receptor expression via estrogen-induced activation of the ERK 1/2 signaling pathway in colon and breast cancer cells. J. Endocrinol. 2005;185:577–592. doi: 10.1677/joe.1.05770. [DOI] [PubMed] [Google Scholar]
- 86.Pan M.R., Chang H.C., Hung W.C. Non-steroidal anti-inflammatory drugs suppress the ERK signaling pathway via block of Ras/c-Raf interaction and activation of MAP kinase phosphatases. Cell. Signal. 2008;20:1134–1141. doi: 10.1016/j.cellsig.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 87.Khazak V., Eyrisch S., Kato J., Tamanoi F., Golemis E.A. A two-hybrid approach to identify inhibitors of the RAS-RAF interaction. In: Tamanoi F., editor. Inhibitors of the Ras Superfamily G-Proteins, Part A. Volume 33. Elsevier Inc.; Amsterdam, The Netherlands: 2013. pp. 213–248. [DOI] [PubMed] [Google Scholar]
- 88.Skobeleva N., Menon S., Weber L., Golemis E.A., Khazak V. In vitro and in vivo synergy of MCP compounds with mitogen-activated protein kinase pathway- and microtubule-targeting inhibitors. Mol. Cancer Ther. 2007;6:898–906. doi: 10.1158/1535-7163.MCT-06-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oinuma I., Katoh H., Negishi M. Semaphorin 4D/Plexin-B1–mediated R-Ras GAP activity inhibits cell migration by regulating β1 integrin activity. J. Cell Biol. 2006;173:601–613. doi: 10.1083/jcb.200508204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Endo M., Yamashita T. Inactivation of Ras by p120GAP via focal adhesion kinase dephosphorylation mediates RGMa-induced growth cone collapse. J. Neurosci. 2009;29:6649–6662. doi: 10.1523/JNEUROSCI.0927-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oinuma I., Ishikawa Y., Katoh H., Negishi M. The semaphorin 4D receptor plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- 92.Quah S.Y., Tan M.S., Teh Y.H., Stanslas J. Pharmacological modulation of oncogenic Ras by natural products and their derivatives: Renewed hope in the discovery of novel anti-Ras drugs. Pharmacol. Ther. 2016;162:35–57. doi: 10.1016/j.pharmthera.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 93.Laheru D., Shah P., Rajeshkumar N.V., McAllister F., Taylor G., Goldsweig H., Le D.T., Donehower R., Jimeno A., Linden S., et al. Integrated preclinical and clinical development of S-trans, trans-farnesylthiosalicylic acid (FTS, Salirasib) in pancreatic cancer. Investig. New Drugs. 2012;30:2391–2399. doi: 10.1007/s10637-012-9818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haklai R., Elad-Sfadia G., Egozi Y., Kloog Y. Orally administered FTS (salirasib) inhibits human pancreatic tumor growth in nude mice. Cancer Chemother. Pharmacol. 2008;61:89–96. doi: 10.1007/s00280-007-0451-6. [DOI] [PubMed] [Google Scholar]
- 95.Zundelevich A., Elad-Sfadia G., Haklai R., Kloog Y. Suppression of lung cancer tumor growth in a nude mouse model by the Ras inhibitor salirasib (farnesylthiosalicylic acid) Mol. Cancer Ther. 2007;6:1765–1773. doi: 10.1158/1535-7163.MCT-06-0706. [DOI] [PubMed] [Google Scholar]
- 96.Charette N., De Saeger C., Lannoy V., Horsmans Y., Leclercq I., Stärkel P. Salirasib inhibits the growth of hepatocarcinoma cell lines in vitro and tumor growth in vivo through Ras and mTOR inhibition. Mol. Cancer. 2010;9:256. doi: 10.1186/1476-4598-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McPherson R.A., Conaway M.C., Gregory C.W., Yue W., Santen R.J. The novel Ras antagonist, farnesylthiosalicylate, suppresses growth of prostate cancer in vitro. Prostate. 2004;58:325–334. doi: 10.1002/pros.10336. [DOI] [PubMed] [Google Scholar]
- 98.Riely G.J., Johnson M.L., Medina C., Rizvi N.A., Miller V.A., Kris M.G., Pietanza M.C., Azzoli C.G., Krug L.M., Pao W., et al. A phase II trial of Salirasib in patients with lung adenocarcinomas with KRAS mutations. J. Thorac. Oncol. 2011;6:1435–1437. doi: 10.1097/JTO.0b013e318223c099. [DOI] [PubMed] [Google Scholar]
- 99.Berndt N., Hamilton A.D., Sebti S.M. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., Chen W.W., Lander E.S., Sabatini D.M. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heid I., Lubeseder-Martellato C., Sipos B., Mazur P.K., Lesina M., Schmid R.M., Siveke J.T. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011;141:719–730. doi: 10.1053/j.gastro.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 102.Kumar M.S., Hancock D.C., Molina-Arcas M., Steckel M., East P., Diefenbacher M., Armenteros-Monterroso E., Lassailly F., Matthews N., Nye E., et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 103.Kawada K., Toda K., Sakai Y. Targeting metabolic reprogramming in KRAS-driven cancers. Int. J. Clin. Oncol. 2017;22:651–659. doi: 10.1007/s10147-017-1156-4. [DOI] [PubMed] [Google Scholar]
- 104.Chang H.H., Moro A., Takakura K., Su H.Y., Mo A., Nakanishi M., Waldron R.T., French S.W., Dawson D.W., Hines J., et al. Incidence of pancreatic cancer is dramatically increased by a high fat, high calorie diet in KrasG12D mice. PLoS ONE. 2017;12:e0184455. doi: 10.1371/journal.pone.0184455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lock R., Roy S., Kenific C.M., Su J.S., Salas E., Ronen S.M., Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell. 2011;22:165–178. doi: 10.1091/mbc.e10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M.S., Karantza V., et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mack N.A., Whalley H.J., Castillo-Lluva S., Malliri A. The diverse roles of Rac signaling in tumorigenesis. Cell Cycle. 2011;10:1571–1581. doi: 10.4161/cc.10.10.15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao Y., Dickerson J.B., Guo F., Zheng J., Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Akbar H., Cancelas J., Williams D.A., Zheng J., Zheng Y. Rational design and applications of a Rac GTPase-specific small molecule inhibitor. In: Balch W.E., Der C.J., Hall A., editors. Regulators and Effectors of Small GTPases: Rho Family. Volume 406. Elsevier Inc.; Amsterdam, The Netherlands: 2006. pp. 554–565. [DOI] [PubMed] [Google Scholar]
- 110.Kaneto N., Yokoyama S., Hayakawa Y., Kato S., Sakurai H., Saiki I. RAC1 inhibition as a therapeutic target for gefitinib-resistant non-small-cell lung cancer. Cancer Sci. 2014;105:788–794. doi: 10.1111/cas.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan Y., Hein A.L., Etekpo A., Burchett K.M., Lin C., Enke C.A., Batra S.K., Cowan K.H., Ouellette M.M. Inhibition of RAC1 GTPase sensitizes pancreatic cancer cells to γ-irradiation. Oncotarget. 2014;5:10251–10270. doi: 10.18632/oncotarget.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hernández E., de la Mota-Peynado A., Dharmawardhane S., Vlaar C.P. Novel inhibitors of Rac1 in metastatic breast cancer. P. R. Health Sci. J. 2010;29:348–356. [PubMed] [Google Scholar]
- 113.Montalvo-Ortiz B.L., Castillo-Pichardo L., Hernández E., Humphries-Bickley T., de la Mota-Peynado A., Cubano L.A., Vlaar C.P., Dharmawardhane S. Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J. Biol. Chem. 2012;287:13228–13238. doi: 10.1074/jbc.M111.334524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Castillo-Pichardo L., Humphries-Bickley T., de la Parra C., Forestier-Roman I., Martinez-Ferrer M., Hernandez E., Vlaar C., Ferrer-Acosta Y., Washington A.V., Cubano L.A., et al. The Rac inhibitor EHop-016 inhibits mammary tumor growth and metastasis in a nude mouse model. Transl. Oncol. 2014;7:546–555. doi: 10.1016/j.tranon.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin H., Mali R.S., Ma P., Chatterjee A., Ramdas B., Sims E., Munugalavadla V., Ghosh J., Mattingly R.R., Visconte V., et al. Pak and Rac GTPases promote oncogenic KIT-induced neoplasms. J. Clin. Investig. 2013;123:4449–4463. doi: 10.1172/JCI67509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Okada T., Lee A.Y., Qin L.X., Agaram N., Mimae T., Shen Y., O’Connor R., López-Lago M.A., Craig A., Miller M.L., et al. Integrin-α10 dependency identifies RAC and RICTOR as therapeutic targets in high-grade myxofibrosarcoma. Cancer Discov. 2016;6:1148–1165. doi: 10.1158/2159-8290.CD-15-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Veluthakal R., Tunduguru R., Arora D.K., Sidarala V., Syeda K., Vlaar C.P., Turmond D.C., Kowluru A. VAV2, a guanine nucleotide exchange factor for Rac1, regulates glucose-stimulated insulin secretion in pancreatic beta cells. Diabetologia. 2015;58:2573–2581. doi: 10.1007/s00125-015-3707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shutes A., Onesto C., Picard V., Leblond B., Schweighoffer F., Der C.J. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J. Biol. Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 119.Onesto C., Shutes A., Picard V., Schweighoffer F., Der C.J. Characterization of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. In: Balch W.E., Der C.J., Hall A., editors. Small GTPases in Disease, Part B. Volume 439. Elsevier Inc.; Amsterdam, The Netherlands: 2008. pp. 111–129. [DOI] [PubMed] [Google Scholar]
- 120.Rosenblatt A.E., Garcia M.I., Lyons L., Xie Y., Maiorino C., Désiré L., Slingerland J., Burnstein K.L. Inhibition of the Rho GTPase, Rac1, decreases estrogen receptor levels and is a novel therapeutic strategy in breast cancer. Endocr. Relat. Cancer. 2011;18:207–219. doi: 10.1677/ERC-10-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Katz E., Sims A.H., Sproul D., Caldwell H., Dixon J.M., Meehan R.R., Harrison D.J. Targeting of Rac GTPases blocks the spread of intact human breast cancer. Oncotarget. 2012;3:608–619. doi: 10.18632/oncotarget.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jim Leu S.J., Sung J.S., Huang M.L., Chen M.Y., Tsai T.W. A novel anti-CCN1 monoclonal antibody suppresses Rac-dependent cytoskeletal reorganization and migratory activities in breast cancer cells. Biochem. Biophys. Res. Commun. 2013;434:885–891. doi: 10.1016/j.bbrc.2013.04.045. [DOI] [PubMed] [Google Scholar]
- 123.Niebel B., Wosnitza C.I., Famulok M. RNA-aptamers that modulate the RhoGEF activity of Tiam1. Bioorg. Med. Chem. 2013;21:6239–6246. doi: 10.1016/j.bmc.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 124.Taniuchi K., Yokotani K., Saibara T. BART inhibits pancreatic cancer cell invasion by Rac1 inactivation through direct binding to active Rac1. Neoplasia. 2012;14:440–450. doi: 10.1593/neo.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shan D., Chen L., Njardarson J.T., Gaul C., Ma X., Danishefsky S.J., Huang X.Y. Synthetic analogues of migrastatin that inhibit mammary tumor metastasis in mice. Proc. Natl. Acad. Sci. USA. 2005;102:3772–3776. doi: 10.1073/pnas.0500658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dütting S., Heidenreich J., Cherpokova D., Amin E., Zhang S.C., Ahmadian M.R., Brakebusch C., Nieswandt B. Critical off-target effects of the widely used Rac1 inhibitors NSC23766 and EHT1864 in mouse platelets. J. Thromb. Haemost. 2015;13:827–838. doi: 10.1111/jth.12861. [DOI] [PubMed] [Google Scholar]
- 127.Gao L., Bai L., Nan Q.Z. Activation of Rho GTPase Cdc42 promotes adhesion and invasion in colorectal cancer cells. Med. Sci. Monit. Basic Res. 2013;19:201–207. doi: 10.12659/MSMBR.883983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arias-Romero L.E., Chernoff J. Targeting Cdc42 in cancer. Expert Opin. Ther. Targets. 2013;17:1263–1273. doi: 10.1517/14728222.2013.828037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stengel K.R., Zheng Y. Essential role of Cdc42 in Ras-induced transformation revealed by gene targeting. PLoS ONE. 2012;7:e37317. doi: 10.1371/journal.pone.0037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu W.J., Tu S., Cerione R.A. Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell. 2003;114:715–725. doi: 10.1016/S0092-8674(03)00688-3. [DOI] [PubMed] [Google Scholar]
- 131.Qadir M.I., Parveen A., Ali M. Cdc42: Role in cancer management. Chem. Biol. Drug Des. 2015;86:432–439. doi: 10.1111/cbdd.12556. [DOI] [PubMed] [Google Scholar]
- 132.Pelish H.E., Peterson J.R., Salvarezza S.B., Rodriguez-Boulan E., Chen J.L., Stamnes M., Macia E., Feng Y., Shair M.D., Kirchhausen T. Secramine inhibits Cdc42-dependent functions in cells and Cdc42 activation in vitro. Nat. Chem. Biol. 2006;2:39–46. doi: 10.1038/nchembio751. [DOI] [PubMed] [Google Scholar]
- 133.Xu B., Pelish H., Kirchhausen T., Hammond G.B. Large scale synthesis of the Cdc42 inhibitor Secramine A and its inhibition of cell spreading. Org. Biomol. Chem. 2006;4:4149–4157. doi: 10.1039/b609143a. [DOI] [PubMed] [Google Scholar]
- 134.Ambrogio C., Voena C., Manazza A.D., Martinengo C., Costa C., Kirchhausen T., Hirsch E., Inghirami G., Chiarle R. The anaplastic lymphoma kinase controls cell shape and growth of anaplastic large cell lymphoma through Cdc42 activation. Cancer Res. 2008;68:8899–8907. doi: 10.1158/0008-5472.CAN-08-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baltiérrez-Hoyos R., Roa-Espitia A.L., Hernández-González E.O. The association between CDC42 and caveolin-1 is involved in the regulation of capacitation and acrosome reaction of guinea pig and mouse sperm. Reproduction. 2012;144:123–134. doi: 10.1530/REP-11-0433. [DOI] [PubMed] [Google Scholar]
- 136.Nikolic D.S., Lehmann M., Felts R., Garcia E., Blanchet F.P., Subramaniam S., Piguet V. HIV-1 activates Cdc42 and induces membrane extensions in immature dendritic cells to facilitate cell-to-cell virus propagation. Blood. 2011;118:4841–4852. doi: 10.1182/blood-2010-09-305417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lengfeld J., Wang Q., Zohlman A., Salvarezza S., Morgan S., Ren J., Kato K., Rodriguez-Boulan E., Liu B. Protein kinase C δ regulates the release of collagen type I from vascular smooth muscle cells via regulation of Cdc42. Mol. Biol. Cell. 2012;23:1955–1963. doi: 10.1091/mbc.e11-06-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hong L., Kenney S.R., Phillips G.K., Simpson D., Schroeder C.E., Nöth J., Romero E., Swanson S., Waller A., Strouse J.J., et al. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem. 2013;288:8531–8543. doi: 10.1074/jbc.M112.435941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Friesland A., Zhao Y., Chen Y.H., Wang L., Zhou H., Lu Q. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. USA. 2013;110:1261–1266. doi: 10.1073/pnas.1116051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Muhoza D., Adams P.D. Two small molecules, ZCL278 and AZA197 show promise in influencing protein interactions involving the Ras-related protein cell division cycle 42 [Cdc42] to modulate its oncogenic potential. Open J. Biophys. 2017;7:71–81. doi: 10.4236/ojbiphy.2017.73006. [DOI] [Google Scholar]
- 141.An Y., Liu T., Liu X., Zhao L., Wang J. Rac1 and Cdc42 play important roles in arsenic neurotoxicity in primary cultured rat cerebellar astrocytes. Biol. Trace Elem. Res. 2016;170:173–182. doi: 10.1007/s12011-015-0456-7. [DOI] [PubMed] [Google Scholar]
- 142.Liu X., Gao Y., An Y., Fu X., Li Y., Sun D., Wang J. Neuroglobin plays a protective role in arsenite-induced cytotoxicity by inhibition of Cdc42 and Rac1GTPases in rat cerebellar granule neurons. Cell. Physiol. Biochem. 2015;36:1613–1627. doi: 10.1159/000430323. [DOI] [PubMed] [Google Scholar]
- 143.Biro M., Munoz M.A., Weninger W. Targeting Rho-GTPases in immune cell migration and inflammation. Br. J. Pharmacol. 2014;171:5491–5506. doi: 10.1111/bph.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Humphries-Bickley T., Castillo-Pichardo L., Hernandez-O’Farrill E., Borrero-Garcia L.D., Forestier-Roman I., Gerena Y., Blanco M., Rivera-Robles M.J., Rodriguez-Medina J.R., Cubano L.A., et al. Characterization of a dual Rac/Cdc42 inhibitor MBQ-167 in metastatic cancer. Mol. Cancer Ther. 2017;16:805–818. doi: 10.1158/1535-7163.MCT-16-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zins K., Lucas T., Reichl P., Abraham D., Aharinejad S. A Rac1/Cdc42 GTPase-specific small molecule inhibitor suppresses growth of primary human prostate cancer xenografts and prolongs survival in mice. PLoS ONE. 2013;8:e74924. doi: 10.1371/journal.pone.0074924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo Y., Kenney S.R., Muller C.Y., Adams S., Rutledge T., Romero E., Murray-Krezan C., Prekeris R., Sklar L.A., Hudson L.G., et al. R-Ketorolac targets Cdc42 and Rac1 and alters ovarian cancer cell behaviors critical for invasion and metastasis. Mol. Cancer Ther. 2015;14:2215–2227. doi: 10.1158/1535-7163.MCT-15-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen W., Mao K., Liu Z., Dinh-Xuan A.T. The role of the RhoA/Rho kinase pathway in angiogenesis and its potential value in prostate cancer (Review) Oncol. Lett. 2014;8:1907–1911. doi: 10.3892/ol.2014.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shang X., Marchioni F., Sipes N., Evelyn C.R., Jerabek-Willemsen M., Duhr S., Seibel W., Wortman M., Zheng Y. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol. 2012;19:699–710. doi: 10.1016/j.chembiol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoon C., Cho S.J., Aksoy B.A., Park D.J., Schultz N., Ryeom S.W., Yoon S.S. Chemotherapy resistance in diffuse type gastric adenocarcinoma is mediated by RhoA activation in cancer stem-like cells. Clin. Cancer Res. 2016;22:971–983. doi: 10.1158/1078-0432.CCR-15-1356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 150.Shang X., Marchioni F., Evelyn C.R., Sipes N., Zhou X., Seibel W., Wortman M., Zheng Y. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc. Natl. Acad. Sci. USA. 2013;110:3155–3160. doi: 10.1073/pnas.1212324110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chang L.C., Huang T.H., Chang C.S., Tsai Y.R., Lin R.H., Lee P.W., Hsu M.F., Huang L.J., Wang J.P. Signaling mechanisms of inhibition of phospholipase D activation by CHS-111 in formyl peptide-stimulated neutrophils. Biochem. Pharmacol. 2011;81:269–278. doi: 10.1016/j.bcp.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 152.He H., Dai F., Yu L., She X., Zhao Y., Jiang J., Chen X., Zhao S. Identification and characterization of nine novel human small GTPases showing variable expressions in liver cancer tissues. Gene Expr. 2002;10:231–242. doi: 10.3727/000000002783992406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shimada K., Uzawa K., Kato M., Endo Y., Shiiba M., Bukawa H., Yokoe H., Seki N., Tanzawa H. Aberrant expression of RAB1A in human tongue cancer. Br. J. Cancer. 2005;92:1915–1921. doi: 10.1038/sj.bjc.6602594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhao Z., Liu X.F., Wu H.C., Zou S.B., Wang J.Y., Ni P.H., Chen X.H., Fan Q.S. Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci. 2010;101:1454–1462. doi: 10.1111/j.1349-7006.2010.01558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li Y., Jia Q., Wang Y., Li F., Jia Z., Wan Y. Rab40b upregulation correlates with the prognosis of gastric cancer by promoting migration, invasion, and metastasis. Med. Oncol. 2015;32:126. doi: 10.1007/s12032-015-0562-6. [DOI] [PubMed] [Google Scholar]
- 156.Hou R., Jiang L., Yang Z., Wang S., Liu Q. Rab14 is overexpressed in ovarian cancers and promotes ovarian cancer proliferation through Wnt pathway. Tumor Biol. 2016;37:16005–16013. doi: 10.1007/s13277-016-5420-4. [DOI] [PubMed] [Google Scholar]
- 157.Da Silva S.D., Marchi F.A., Xu B., Bijian K., Alobaid F., Mlynarek A., Rogatto S.R., Hier M., Kowalski L.P., Alaoui-Jamali M.A. Predominant Rab-GTPase amplicons contributing to oral squamous cell carcinoma progression to metastasis. Oncotarget. 2015;6:21950–21963. doi: 10.18632/oncotarget.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hendrix A., Maynard D., Pauwels P., Braems G., Denys H., van den Broecke R., Lambert J., van Belle S., Cocquyt V., Gespach C., et al. Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. J. Natl. Cancer Inst. 2010;102:866–880. doi: 10.1093/jnci/djq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jin L., Huo Y., Zheng Z., Jiang X., Deng H., Chen Y., Lian Q., Ge R., Deng H. Down-regulation of Ras-related protein Rab 5C-dependent endocytosis and glycolysis in cisplatin-resistant ovarian cancer cell lines. Mol. Cell. Proteom. 2014;13:3138–3151. doi: 10.1074/mcp.M113.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Recchi C., Seabra M.C. Novel functions for Rab GTPases in multiple aspects of tumour progression. Biochem. Soc. Trans. 2012;40:1398–1403. doi: 10.1042/BST20120199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roelofs A.J., Hulley P.A., Meijer A., Ebetino F.H., Russell R.G., Shipman C.M. Selective inhibition of Rab prenylation by a phosphonocarboxylate analogue of risedronate induces apoptosis, but not S-phase arrest, in human myeloma cells. Int. J. Cancer. 2006;119:1254–1261. doi: 10.1002/ijc.21977. [DOI] [PubMed] [Google Scholar]
- 162.Okamoto S., Jiang Y., Kawamura K., Shingyoji M., Tada Y., Sekine I., Takiguchi Y., Tatsumi K., Kobayashi H., Shimada H., et al. Zoledronic acid induces apoptosis and S-phase arrest in mesothelioma through inhibiting Rab family proteins and topoisomerase II actions. Cell Death Dis. 2014;5:e1517. doi: 10.1038/cddis.2014.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boissier S., Magnetto S., Frappart L., Cuzin B., Ebetino F.H., Delmas P.D., Clezardin P. Bisphosphonates inhibit prostate and breast carcinoma cell adhesion to unmineralized and mineralized bone extracellular matrices. Cancer Res. 1997;57:3890–3894. [PubMed] [Google Scholar]
- 164.Boissier S., Ferreras M., Peyruchaud O., Magnetto S., Ebetino F.H., Colombel M., Delmas P., Delaissé J.M., Clézardin P. Bisphosphonates inhibit breast and prostate carcinoma cell invasion, an early event in the formation of bone metastases. Cancer Res. 2000;60:2949–2954. doi: 10.1080/09297040802385400. [DOI] [PubMed] [Google Scholar]
- 165.Fournier P.G.J., Daubiné F., Lundy M.W., Rogers M.J., Ebetino F.H., Clézardin P. Lowering bone mineral affinity of bisphosphonates as a therapeutic strategy to optimize skeletal tumor growth inhibition In vivo. Cancer Res. 2008;68:8945–8953. doi: 10.1158/0008-5472.CAN-08-2195. [DOI] [PubMed] [Google Scholar]