Abstract
Background: Lung cancer (LC) is the leading cause of cancer-related death in Eastern Asia. The prognosis of LC highly depends on tumor stages and early detection could substantially reduce LC mortality. Accumulating evidence suggested that circulating miRNAs in plasma or serum may have applications in early LC detection. We thus conducted a systematic literature review on the diagnostic value of miRNAs markers for LC in East Asian populations. Methods: PubMed and ISI Web of Knowledge were searched to retrieve relevant articles published up to 17 September 2018. Information on study design, population characteristics, investigated miRNAs and diagnostic accuracy (including sensitivity, specificity and area under the curve (AUC)) were independently extracted by two reviewers. Results: Overall, 46 studies that evaluated a total of 88 miRNA markers for LC diagnosis in East Asian populations were identified. Sixteen of the 46 studies have incorporated individual miRNA markers as panels (with 2–20 markers). Three promising miRNA panels with ≥90% sensitivity and ≥90% specificity were discovered, two of which were externally validated. Diagnostic performance of circulating miRNAs in East Asian populations was comparable to previously summarized performance in Western populations. Forty-four miRNAs were reported in both populations. No major differences in diagnostic performance by ethnicity of the same miRNA was observed. Conclusions: Circulating miRNAs or miRNA panels, possibly in combination with other promising molecular markers including epigenetic and genetic markers, may be promising candidates for noninvasive LC early detection. However, large studies with samples collected prospectively in true screening settings are required to validate the promising markers or marker panels.
Keywords: miRNA, lung cancer, early detection, East Asian populations
1. Introduction
Lung cancer (LC) is the leading cause of cancer mortality in Eastern Asia, with 950,015 cases and 815,635 deaths estimated in 2018 [1]. Although advances in therapy have led to improvements in survival of LC patients [2], the 5-year survival rate remains very low, mainly due to late diagnosis of disease [3]. Traditional screening methods such as chest radiography and sputum cytology have limited clinical applications as they display low sensitivity and specificity in detection of LC [4]. In recent years, Low-dose CT has been recommended for LC screening in high-risk smokers. However, potential hazards of CT screening, including radiation exposure, high false-positive rates, overdiagnosis and high cost, raise concerns [5,6]. Therefore, in order to reduce LC mortality, effective methods for early diagnosis of LC remain desirable.
MicroRNAs (miRNAs) are a class of single stranded RNAs composed of 18–22 nucleotides, which are involved in the regulation of gene transcription but have no protein coding function, and are widely present in eukaryotic cells [7]. At present, several studies have identified abnormally expressed miRNA patterns in blood specimens of LC patients, such as plasma, serum and exosome, suggesting that circulating miRNA may be useful for LC diagnosis [8,9,10].
Because of the genetic diversity of miRNA expression profiles of populations [11], we previously summarized the diagnostic performance of circulating miRNAs restricted to Western populations [12]. Herein, we systematically reviewed their diagnostic performance in East Asian populations, paying particular attention to the potential differences between the East Asian and Western populations.
2. Methods
This systematic review was conducted in accordance with PRISMA recommendations [13].
2.1. Literature Search
A systematic literature search was carried out to identify all studies that evaluated circulating miRNAs related to LC. We searched the PubMed and ISI Web of Science databases for relevant articles from inception to 17 September, 2018, using the following keyword combinations: ((lung OR pulmonary) AND (cancer OR carcinoma OR neoplasm OR tumor OR adenocarcinoma OR squamous carcinoma OR malignancy) AND (microRNA* OR miRNA* OR miR* OR let-7*) AND (detection OR diagnosis OR biomarker OR marker) AND (blood OR serum OR plasma)). Duplicate articles were removed.
2.2. Eligibility Criteria
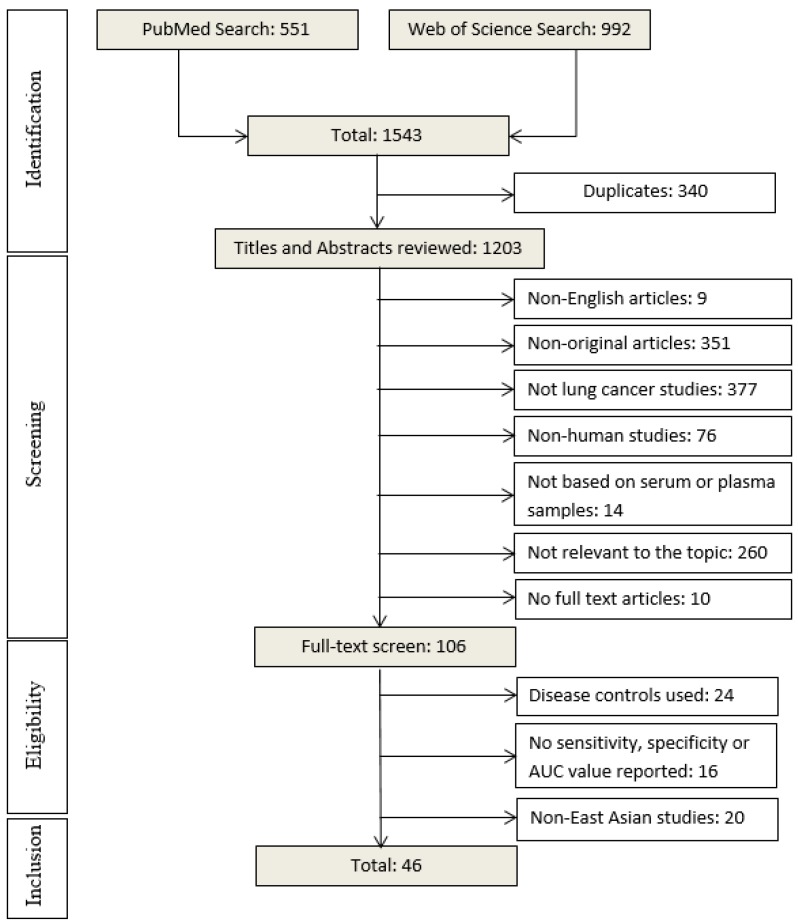
Based on the reading of titles and abstracts, we excluded: (1) non-English articles; (2) non-original articles; (3) not lung cancer studies; (4) non-human studies; (5) studies not based on plasma/serum samples; (6) studies not relevant to the topic; (7) no full text articles (Figure 1). The second round of screening was performed by reading full text of articles. In this step, the following studies were excluded: (1) studies using disease controls; (2) studies not reporting key data on diagnostic performance of miRNA markers (such as simple size, sensitivity, specificity, or area under the curve (AUC); (3) Non-East Asian studies.
Figure 1.
Overview of the literature search process (until 17 September 2018).
2.3. Data Extraction and Statistical Analysis
Two reviewers (H.Y. and Z.G.) independently read and extracted data from the studies that complied with the inclusion and exclusion criteria described above. From each study, we extracted available data on first author, publication year, country, study design, basic population characteristics (including size, age and sex distribution, and histological type and tumor stage for cases), type of bio-specimen, miRNA measurement method, targeted miRNA markers and diagnostic performance related indicators (including sensitivity, specificity, AUC and p-value). Individual miRNAs with p-value > 0.05 were dropped. Mean or median age and male proportion of included studies were calculated by statistical software R (version 3.3.3, R Foundation, Vienna, Austria) if this information was not reported but raw data were available. Different name versions of the same miRNAs were incorporated according to the accession number of miRBase provided (http://www.mirbase.org/). Any disagreements were discussed and resolved among the authors.
2.4. Quality Assessment
We employed QUADAS-2, an updated and widely recommended tool for quality assessment of diagnostic accuracy studies, to assess the quality of each included study. Four key domains of QUADAS-2 were assessed, i.e., patient selection, index test, reference standard and flow and timing. Each domain is evaluated for risk of bias and the first three domains are evaluated for applicability. QUADAS-2 assessment was conducted utilizing the software Review Manager (version 5.3.5, Cochrane Collaboration, Copenhagen, Denmark).
3. Results
3.1. Literature Search Result
Our literature search initially yielded 1543 articles using the search terms mentioned above, 551 from PubMed and 992 from Web of Science (Figure 1). After removal of 340 duplicate articles, titles and abstracts of 1203 articles were carefully reviewed. A total of 1097 articles were excluded according to the above described exclusion criteria. The remaining 106 articles were selected for full text reading, of which 60 articles were removed: 24 using disease controls, 16 not reporting sensitivity, specificity or AUC values, and 20 reporting studies in non-East Asian countries. In the end, 46 studies that evaluated diagnostic performance of circulating miRNAs for LC in East Asian populations and published between 2011 and 2018 (Table 1 and Table 2) were eligible for this systematic review [8,9,10,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56]. Of the included 46 studies, 45 were from China and one was from Japan [29].
Table 1.
Diagnostic performance of miRNA markers in lung cancer in East Asian populations.
| Study a | Cases vs. Controls | Specimen | Histology | Stage | miRNA | SEN | SPE | AUC | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age (y) | Male (%) | |||||||||
| Zhao, 2018 [14] b | 108/54 | / | 61/NA | serum | NSCLC | I–IV | miR-141 | / | / | 0.86 | <0.001 c |
| Sun, 2018 [15] b | 196/77 | / | 58/NA | serum | NSCLC | I–IV | miR-770 | 68 | 89 | 0.84 | <0.01 c |
| Shan, 2018 [16] b | 102/101 | / | 100/100 | plasma | SCC | I–III | miR-181a-5p | / | / | 0.73 | <0.001 c |
| miR-21-5p | / | / | 0.74 | <0.001 c | |||||||
| miR-106a-5p | / | / | 0.74 | <0.001 c | |||||||
| miR-93-5p | / | / | 0.69 | <0.001 c | |||||||
| Qin, 2018 [17] | 146/40 | 63/NA | 73/NA | serum | NSCLC | I–IV | miR-342-3p | 60 | 94 | 0.89 | <0.01 c |
| 78/40 | / | / | SCC | miR-342-3p | 83 | 76 | 0.87 | <0.01 c | |||
| 68/40 | / | / | ADC | miR-342-3p | 83 | 82 | 0.90 | <0.01 c | |||
| Bao, 2018 [18] b | 80/75 | / | 61/60 | serum | NSCLC | I–IV | miR-10a-5p | 66 | 73 | 0.71 | <0.0001 c |
| miR-196a-5p | 68 | 78 | 0.79 | 0.0018 c | |||||||
| Zhang, 2017 [20] b | 102/108 | / | 100/100 | Serum | SCC | I–III | miR-106a-5p | / | / | 0.83 | <0.001 c |
| miR-20a-5p | / | / | 0.80 | <0.001 c | |||||||
| miR-93-5p | / | / | 0.82 | <0.001 c | |||||||
| Wang, 2017 [24] | 127/60 | 58/59 | 62/58 | serum | NSCLC | I–IV | miR-98 | 80 | 82 | 0.86 | <0.01 c |
| Zhou, 2017 [19] b | 108/94 | / | 43/46 | plasma | ADC | I–IV | miR-19b-3p | / | / | 0.62 | <0.001 c |
| miR-21-5p | / | / | 0.69 | <0.001 c | |||||||
| miR-221-3p | / | / | 0.68 | <0.001 c | |||||||
| miR-409-3p | / | / | 0.61 | <0.001 c | |||||||
| miR-425-5p | / | / | 0.66 | <0.001 c | |||||||
| miR-584-5p | / | / | 0.69 | <0.001 c | |||||||
| Zhang, 2017 [21] | 129/83 | 60/60 | 63/58 | plasma | NSCLC | I–II | miR-145 | 81 | 89 | 0.89 | <0.0001 c |
| miR-20a | 80 | 88 | 0.89 | <0.0001 c | |||||||
| miR-21 | 78 | 86 | 0.84 | <0.001 c | |||||||
| miR-223 | 70 | 84 | 0.81 | <0.001 c | |||||||
| Yu, 2017 [22] b | 50/30 | / | 82/83 | plasma | SCLC | I–IV | miR-92a-2-5p | 56 | 100 | 0.76 | <0.001 c |
| Yang, 2017 [23] b | 113/30 | / | 52/NA | serum | ADC | I–IV | miR-31 | / | / | 0.84 | <0.01 c |
| Shang, 2017 [25] | 127/112 | 55/44 | 65/56 | serum | NSCLC | I–IV | miR-22 | 99 | 84 | 0.92 | <0.001 |
| miR-126 | 83 | 96 | 0.87 | <0.001 | |||||||
| Lv, 2017 [26] | 120/120 | 60/59 | 52/51 | serum | ADC | I–IV | miR-103 | / | / | 0.80 | <0.0001 c |
| miR-146a | / | / | 0.90 | <0.0001 c | |||||||
| miR-151 | / | / | 0.85 | <0.0001 c | |||||||
| miR-221 | / | / | 0.79 | <0.0001 c | |||||||
| miR-222 | / | / | 0.86 | <0.0001 c | |||||||
| miR-223 | / | / | 0.91 | <0.0001 c | |||||||
| Zhu, 2016 [9] | 112/40 | 59/58 | 54/55 | serum | NSCLC | 0–IIIB | miR-182 | 63 | 80 | 0.73 | <0.0001 |
| miR-183 | 41 | 83 | 0.63 | 0.0091 | |||||||
| miR-210 | 34 | 100 | 0.62 | 0.0121 | |||||||
| miR-126 | 61 | 93 | 0.79 | <0.0001 | |||||||
| Wang, 2016 [27] | 82/91 | / | / | plasma | ADC | I–II | miR-628-3p | 43 | 91 | 0.73 | <0.001 c |
| miR-339-3p | 65 | 71 | 0.72 | <0.001 c | |||||||
| miR-425-3p | 67 | 68 | 0.73 | <0.001 c | |||||||
| miR-532 | 54 | 80 | 0.66 | <0.001 c | |||||||
| Wang, 2016 [28] | 54/15 | 60/51 | / | serum | NSCLC | I–IV | miR-1244 | 82 | 80 | 0.83 | / |
| 26/15 | NA/51 | / | ADC | miR-1244 | 77 | 80 | 0.79 | <0.05 c | |||
| 18/15 | NA/51 | / | SCC | miR-1244 | 83 | 80 | 0.85 | <0.05 c | |||
| Sun, 2016 [30] | 50/60 | 67/62 | 76/78 | serum | NSCLC | I–IV | miR-21 | / | / | 0.87 | 0.000 |
| Su, 2016 [32] b | 100/100 | / | 65/70 | plasma | NSCLC | I–III | miR-195 | 78 | 86 | 0.89 | <0.01 c |
| Peng, 2016 [32] | 120/71 | 60/58 | 72/62 | serum | NSCLC | I–IV | miR-1254 | 97 | 39 | 0.68 | 0.000 |
| miR-485-5p | 95 | 63 | 0.79 | 0.000 | |||||||
| miR-574-5p | 99 | 28 | 0.64 | 0.002 | |||||||
| Gao, 2016 [10] | 90/90 | 62/62 | 80/78 | plasma | SCC | I | miR-324-3p | 72 | 76 | 0.79 | / |
| miR-1285 | 87 | 78 | 0.85 | / | |||||||
| Fan, 2016 [33] d | 94/58 | 60/58 | 54/53 | serum | NSCLC | I–IIIB | miR-15b-5p | 52 | 74 | / | 0.006 c |
| miR-16-5p | 88 | 86 | / | 0.0001 c | |||||||
| miR-17-5p | 82 | 68 | / | 0.0001 c | |||||||
| miR-19-3p | 83 | 67 | / | 0.0001 c | |||||||
| miR-20a-5p | 76 | 90 | / | 0.0001 c | |||||||
| miR-28-3p | 71 | 79 | / | 0.0001 c | |||||||
| miR-92a-3p | 87 | 66 | / | 0.0001 c | |||||||
| Zhou, 2015 [34] | 87/61 | 59/56 | 63/52 | serum | NSCLC | I–IV | miR-194 | / | / | 0.66 | / |
| miR-652 | / | / | 0.82 | / | |||||||
| miR-660 | / | / | 0.71 | / | |||||||
| 52/61 | NA/56 | NA/52 | ADC | miR-194 | / | / | 0.63 | 0.0192 c | |||
| miR-652 | / | / | 0.82 | <0.0001 c | |||||||
| miR-660 | / | / | 0.72 | <0.0001 c | |||||||
| 35/61 | NA/56 | NA/52 | SCC | miR-194 | / | / | 0.70 | 0.0007 c | |||
| miR-652 | / | / | 0.81 | <0.0001 c | |||||||
| miR-660 | / | / | 0.70 | 0.0003 c | |||||||
| Zhao, 2015 [35] | 80/60 | 58/55 | 61/52 | serum | NSCLC | / | miR-21 | 74 | 72 | 0.81 | 0.001 c |
| Yang, 2015 [36] b | 152/300 | / | 65/69 | serum | NSCLC | I–IV | miR-148a | 85 | 83 | 0.90 | <0.001 c |
| miR-148b | 83 | 83 | 0.90 | <0.001 c | |||||||
| miR-152 | 75 | 77 | 0.82 | <0.001 c | |||||||
| miR-21 | 69 | 71 | 0.81 | <0.001 c | |||||||
| Yan, 2015 [37] b | 300/300 | / | / | plasma | Any LC | I–IV | miR-31 | 77 | 75 | 0.79 | <0.001 c |
| Wang, 2015 [38] | 70/70 | 64/64 | 61/61 | serum | NSCLC | / | miR-125a-5p | 74 | 56 | 0.71 | <0.0001 c |
| miR-145 | 93 | 61 | 0.84 | <0.0001 c | |||||||
| miR-146a | 84 | 59 | 0.78 | <0.0001 c | |||||||
| Wang, 2015 [39] | 94/111 | NA/60 | NA/52 | serum | NSCLC | IA–IIB | miR-125a-5p | 88 | 75 | 0.83 | / |
| miR-25 | 83 | 75 | 0.82 | / | |||||||
| miR-126 | 83 | 63 | 0.79 | / | |||||||
| Wang, 2015 [40] e | 63/63 | 62/60 | 78/59 | serum | NSCLC | I–IV | miR-483-5p | / | / | 0.66 | 0.002 |
| miR-193a-3p | / | / | 0.81 | <0.0001 | |||||||
| miR-214 | / | / | 0.78 | <0.0001 | |||||||
| miR-25 | / | / | 0.84 | <0.0001 | |||||||
| miR-7 | / | / | 0.81 | <0.0001 | |||||||
| Li, 2015 [41] | 11/11 | 59/55 | 64/55 | plasma | NSCLC | I–IIIA | miR-486 | 91 | 82 | 0.93 | 0.0008 c |
| miR-150 | 82 | 82 | 0.75 | 0.0488 c | |||||||
| Guo, 2015 [42] b | 126/50 | / | 64/NA | plasma | NSCLC | I–IV | miR-204 | 76 | 82 | 0.81 | <0.001 c |
| Dou, 2015 [43] | 120/360 | 63/NA | 60/NA | plasma | NSCLC | I–IV | let-7c | 72 | 78 | 0.71 | 0.006 |
| miR-152 | 86 | 81 | 0.85 | 0.0002 | |||||||
| Zhu, 2014 [44] | 36/44 | / | / | serum | NSCLC | I | miR-125a-5p | 54 | 75 | 0.65 | 0.021 |
| let-7e | 50 | 83 | 0.64 | 0.0317 | |||||||
| Zhu, 2014 [45] | 70/48 | 59/NA | 80/NA | serum | NSCLC | I–IV | miR-29c | 66 | 74 | 0.68 | 0.0004 |
| miR-429 | 54 | 81 | 0.71 | <0.0001 | |||||||
| Li, 2014 [46] b | 514/54 | / | 53/NA | serum | NSCLC | I–IV | miR-499 | 74 | 93 | 0.91 | <0.001 c |
| Huang, 2014 [47] | 53/65 | 57/55 | / | serum | NSCLC | / | let-7i-3p | / | / | 0.89 | <0.001 c |
| miR-154-5p | / | / | 0.96 | <0.001 c | |||||||
| Geng, 2014 [48] b | 126/60 | / | 69/60 | plasma | NSCLC | I–II | miR-20a | 83 | 81 | 0.89 | <0.001 c |
| miR-223 | 87 | 86 | 0.94 | <0.001 c | |||||||
| miR-21 | 67 | 68 | 0.77 | <0.001 c | |||||||
| miR-155 | 86 | 84 | 0.92 | <0.001 c | |||||||
| miR-145 | 70 | 68 | 0.77 | <0.001 c | |||||||
| 45/60 | / | / | ADC | I–II | miR-20a | 87 | 83 | 0.90 | / | ||
| miR-223 | 80 | 82 | 0.91 | / | |||||||
| miR-21 | 60 | 60 | 0.63 | / | |||||||
| miR-155 | 87 | 87 | 0.93 | / | |||||||
| miR-145 | 71 | 70 | 0.77 | / | |||||||
| 64/60 | / | / | SCC | I–II | miR-20a | 97 | 97 | 0.98 | / | ||
| miR-223 | 95 | 93 | 0.98 | / | |||||||
| miR-21 | 91 | 90 | 0.97 | / | |||||||
| miR-155 | 92 | 90 | 0.96 | / | |||||||
| miR-145 | 95 | 95 | 0.97 | / | |||||||
| Gao, 2014 [49] | 36/32 | 55/53 | 31/31 | serum | ADC | I–IV | miR-155 | 72 | 69 | 0.76 | 0.000 |
| Tang, 2013 [50] | 34/32 | 65/66 | 65/81 | plasma | NSCLC | I–III | miR-21 | 53 | 72 | 0.71 | / |
| miR-145 | 56 | 56 | 0.66 | / | |||||||
| miR-155 | 68 | 66 | 0.74 | / | |||||||
| Li, 2013 [51] | 60/30 | 54/57 | 70/50 | serum | NSCLC | I–IV | miR-210 | 79 | 74 | 0.78 | <0.005 c |
| Wang, 2012 [52] | 31/39 | 61/46 | 71/23 | serum | Any LC | I–IV | miR-21 | 87 | 74 | 0.88 | <0.001 c |
| Le, 2012 [53] | 82/50 | 59/NA | 56/NA | serum | NSCLC | I–IV | miR-21 | 46 | 92 | 0.69 | 0.0058 c |
| miR-205 | 85 | 72 | 0.78 | 0.0298 c | |||||||
| miR-30d | 76 | 80 | 0.74 | 0.0147 c | |||||||
| miR-24 | 76 | 64 | 0.83 | <0.0001 c | |||||||
| Wei, 2011 [55] | 77/36 | 60/56 | 71/75 | plasma | NSCLC | I–IV | miR-21 | 61 | 83 | 0.73 | <0.0001 |
| Wei, 2011 [56] | 63/30 | 61/57 | 71/67 | plasma | NSCLC | I–IV | miR-21 | 76 | 70 | 0.78 | <0.0001 |
a all studies listed in the table were from China; b no mean age or median age but age distribution reported; c p-value represents the difference of miRNA levels between cases and controls (all other p-values represent the statistical significance of AUC values); d miRNAs detected with fluorescence quantum dots liquid bead array (all other studies detected with qRT-PCR); e USA validation set not included; SENs, SPEs and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation). Abbreviations: SEN: sensitivity; SPE: specificity; AUC: area under the curve; LC: lung cancer; NSCLC: non-small cell lung cancer; ADC: adenocarcinoma; SCC: squamous cell carcinoma; NA: not available.
Table 2.
Diagnostic performance of miRNA panels in lung cancer in East Asian populations.
| Study a | Cases vs. Controls | Specimen | Histology | Stage | miRNA | SEN | SPE | AUC | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age (y) | Male (%) | ||||||||
| Shan, 2018 [16] b | 15/15 | / | 100/100 | plasma | SCC | I–III | Panel A | / | / | 0.91 |
| Zhang, 2017 [20] b | 34/36 | / | 100/100 | serum | SCC | I–III | Panel B | / | / | 0.95 |
| Zhou, 2017 [19] b | 33/30 | / | 45/47 | plasma | ADC | I–IV | Panel C | 73 | 80 | 0.84 |
| Zhang, 2017 [21] | 129/83 | 60/60 | 63/58 | plasma | NSCLC | I–II | Panel D | 82 | 90 | 0.90 |
| LV, 2017 [26] | 120/120 | 60/59 | 52/51 | serum | ADC | I–IV | Panel E | 84 | 91 | 0.95 |
| 72/120 | NA/59 | NA/51 | I | / | / | 0.94 | ||||
| 31/120 | NA/59 | NA/51 | II | / | / | 0.97 | ||||
| 10/120 | NA/59 | NA/51 | III | / | / | 0.95 | ||||
| Jin, 2017 [8] | 47/13 | / | / | exosome | NSCLC | I | Panel F | 80 | 92 | 0.90 |
| Wang, 2016 [27] | 82/91 | / | / | plasma | ADC | I–II | Panel G | 90 | 99 | 0.98 |
| Panel H | 92 | 98 | 0.97 | |||||||
| Tai, 2016 [29] | 110/52 | 65/66 | 56/58 | serum | ADC | I–III | Panel I | 89 | 100 | 0.98 |
| Gao, 2016 [10] | 90/90 | 62/62 | 80/78 | plasma | SCC | I | -324-3p, -1285 | 85 | 82 | 0.89 |
| Fan, 2016 [33] c | 70/54 | 60/58 | 60/54 | serum | NSCLC | I–IIIB | Panel J | 94 | 94 | / |
| Zhou, 2015 [34] | 87/61 | 59/56 | 63/52 | serum | NSCLC | I–IV | -652, -660 | / | / | 0.86 |
| 52/61 | NA/56 | NA/52 | ADC | -652, -660 | / | / | 0.85 | |||
| 35/61 | NA/56 | NA/52 | SCC | -652, -660 | / | / | 0.87 | |||
| Yang, 2015 [36] b | 152/300 | / | 65/69 | serum | NSCLC | I–IV | Panel K | 96 | 91 | 0.98 |
| Wang, 2015 [39] | 142/111 | 61/60 | 61/52 | serum | NSCLC | IA–IV | Panel L | 88 | 83 | 0.93 |
| Wang, 2015 [40] d | 63/63 | 62/60 | 78/59 | serum | NSCLC | I–IV | Panel M | 89 | 68 | 0.82 |
| Tang, 2013 [50] | 34/32 | 65/66 | 65/81 | plasma | NSCLC | I–III | -21, -145 | 74 | 81 | 0.85 |
| -21, -145 | 56 | 66 | 0.73 | |||||||
| -145, -155 | 79 | 78 | 0.83 | |||||||
| Panel N | 77 | 81 | 0.87 | |||||||
| 40/60 | NA/66 | NA/77 | ADC | I–III | Panel N | 71 | 78 | / | ||
| 9/60 | NA/66 | NA/77 | SCC | I–III | Panel N | 67 | 78 | / | ||
| Chen, 2012 [54] | 200/110 | 60/59 | 79/72 | serum | NSCLC | I–IV | Panel O | 93 | 90 | 0.97 |
a All studies listed in the table were from China, except Tai [29] which was from Japan; b no mean age or median age but age distribution reported; c miRNAs detected with fluorescence quantum dots liquid bead array (all other studies detected with qRT-PCR); d USA validation set not included; SENs, SPEs and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation). Panel A: -106a-5p, -20a-5p, -93-5p; Panel B: -181a-5p, -21-5p, -106a-5p, -93-5p; Panel C: -19b-3p, -21-5p, -221-3p, -409-3p, -425-5p, -584-5p; Panel D: -145, -20a, -21, -223; Panel E: -146a, -222, -223; Panel F: let-7b-5p, let-7e-5p, -24-5p, -21-5p; Panel G: -628-3p, -339-3p, -425-3p, -532; Panel H: -628-3p, -425-3p, -532; Panel I (20 miRs): -451, -1290. -636, -30c, -22-3p, -19b, -486-5p, -20b, -93, -34b, -185, -126-5p, -93-3p, -1274a, -142-5p, -628-5p, -486-3p, -425, -645, -24; Panel J: -20a-5p, -16-5p, -15b-5p; Panel K: -148a, -148b, -152, -21; Panel L: -25, -125a-5p, -126; Panel M: -214, -483-5p, -193a-3p, -25, -7; Panel N: -21, -145, -155; Panel O (10 miRs): -20a, -24, -25, -145, -152, -199a-5p, -221, -222, -223, -320. Abbreviations: SEN: sensitivity; SPE: specificity; AUC: area under the curve; LC: lung cancer; NSCLC: non-small cell lung cancer; ADC: adenocarcinoma; SCC: squamous cell carcinoma; NA: not available.
3.2. Study Quality and Characteristics
QUADAS-2 assessment was completed by two reviewers (H.Y. and Z.G.) independently. Any inconsistencies were discussed and resolved between the investigators. High applicability concerns were found in the patient selection domain in 10 (22%) of the included studies. Unclear risk of bias were found in the patient selection domain and the index test domain in 24 (52%) and 6 (13%) of the included studies, respectively. No risk of bias or applicability concern was found in the reference standard domain, and the flow and timing domain. Details of QUADAS-2 results of the 46 studies are displayed in Figures S1 and S2.
All 46 studies were case-control studies in which blood samples were collected after disease diagnosis. Of the 46 studies, 43 evaluated individual miRNAs, eight of which conducted independent validation (Table 1). Sixteen studies assessed miRNA panels, 13 of which carried out independent validation (Table 2). Detailed information on each study, including the number of cases and controls, mean or median age, male proportion, specimen type, histological subtype, tumor stage, and diagnostic indicators, are summarized in Table 1 and Table 2. Table 1 additionally shows the p-value for testing the difference of each individual miRNA between cases and controls or the statistical significance of AUC values (indicated in the footnotes of Table 1).
The median numbers (range) of LC cases and controls were 94 (11–514) and 60 (11–360), respectively. Fifteen studies examined miRNAs in plasma [10,16,19,21,27,31,37,41,42,43,48,50,55,56,57], 30 in serum [9,14,15,17,18,20,23,24,25,26,28,29,30,32,33,34,35,36,38,39,40,44,45,46,47,49,51,52,53,54] and one in exosome [8]. Overall, 46 studies evaluated 69 miRNA markers and 19 miRNA panels (in total 88 miRNAs). All 46 studies quantified miRNA levels using quantitative real-time polymerase chain reaction (qRT-PCR), the most commonly used method for miRNA detection over the past 5 years. Only one study conducted by Fan et al. [33] also used fluorescence quantum dots liquid bead array to quantify miRNA levels.
3.3. Diagnostic Performance of miRNA Markers
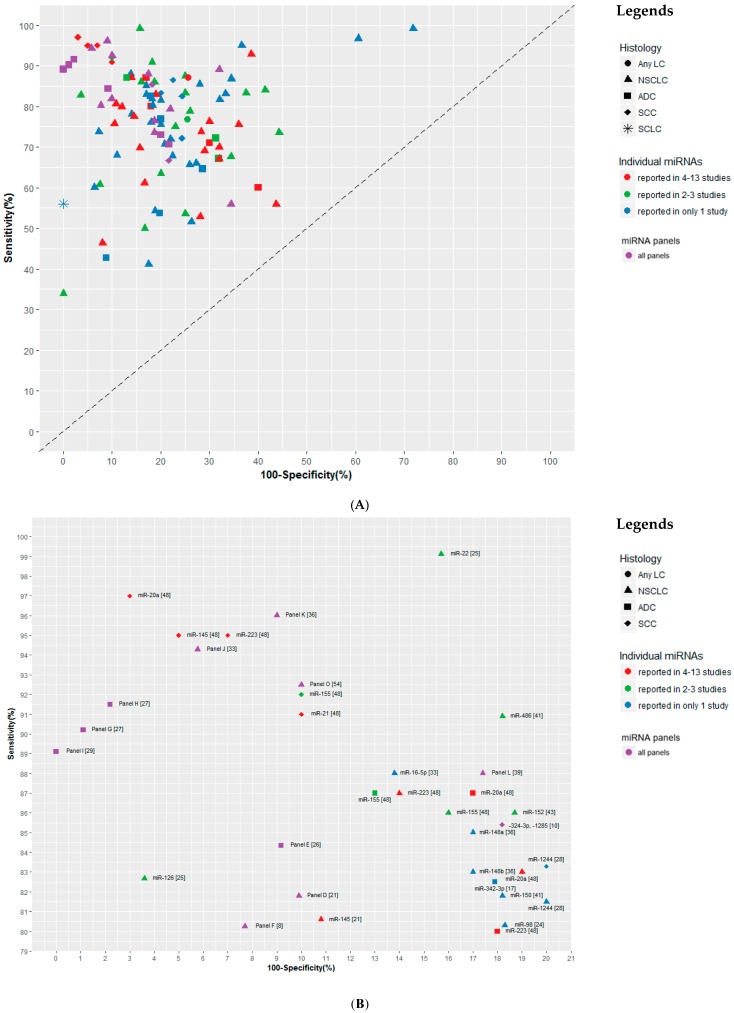
Of the 88 circulating miRNAs included in the 46 studies, 22 miRNAs were reported in ≥ 2 studies (Table 3). Most identified miRNAs were also included in panels, and only 31 miRNAs were not part of any panel (Table S1). The smallest panel included only two miRNAs [10,34,50], and the largest panel included 20 miRNAs [29]. An overview of the diagnostic performance of all reported miRNAs and miRNA panels is shown in Figure 2A. For individual miRNAs, the median (range) sensitivity and specificity were 76.9% (33.9–99.2%) and 80% (28.2–100%) respectively. The median (range) sensitivity and specificity of miRNA panels were 76.3% (55.9–96%) and 79.6% (65.6–100%) respectively. A more detailed representation of miRNAs and miRNA panels with ≥80% sensitivity and ≥80% specificity is given in Figure 2B (17 individual miRNAs and 11 miRNA panels). Even though several individual miRNAs, such as miR-16-5p [33] and miR-223 [48] showed comparable performance as some of the panels, such as a 3-miRNA panel in Wang’s study [39], miRNA panels generally outperformed individual miRNAs for non-small cell lung cancer (NSCLC), with three miRNA panels showing ≥90% sensitivity and ≥90% specificity (Figure 2B). Two of the three panels were also externally validated [33,54]. Two of 46 studies included LC cases of any histological subtypes [37,52], 34 studies included NSCLC cases [8,9,14,15,17,18,21,24,25,28,30,31,32,33,34,35,36,38,39,40,41,42,43,44,45,46,47,48,50,51,53,54,55,56], six studies included only ADC cases [19,23,26,27,29,49], three studies included only SCC cases [10,16,20], and one study included small cell lung cancer (SCLC) cases [57]. Regarding subgroup analyses, five studies performed histology-specific analyses (Table 1 and Table 2) [17,28,34,48,50], and eight studies performed analyses according to cancer stage (Table S2) [26,28,30,39,45,48,50,53]. In histology-specific analyses, several studies reported differential sensitivity, specificity or AUC values in different histological subtypes (ADC and SCC) for the same miRNA or miRNA panel [17,28,34,48,50]. This indicates that miRNAs could be differentially expressed in different histological subtypes of LC, but no histology-specific miRNA could be identified (Table 1 and Table 2). In analyses according to cancer stage, several studies showed that diagnostic efficacy of either miRNAs or miRNA panels in advanced stage of LC seems to be better compared to early stage of LC, however, the differences with respect to AUCs were rather limited (Table S2).
Table 3.
Summary of studies reporting associations of miRNAs with lung cancer in East Asian populations (only miRNAs that have been reported in ≥2 studies are shown).
| Study | miR-21 | miR-145 | miR-20a | miR-24 | miR-223 | miR-155 | miR-25 | miR-152 | miR-125a | miR-126 | miR-221 | miR-93 | miR-210 | miR-486 | miR-425 | miR-19b | miR-22 | let-7e | miR-146a | miR-222 | miR-31 | miR-106a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shan, 2018 [16] | ○↑ | ○↑ | ○↑ | |||||||||||||||||||
| Zhang, 2017 [20] | ○↑ | ○↑ | ○↑ | |||||||||||||||||||
| Zhou, 2017 [19] | ○↑ | ○↑ | ○↑ | |||||||||||||||||||
| Zhang, 2017 [21] | ○↑ | ○↑ | ○↑ | ○↑ | ||||||||||||||||||
| Yu, 2017 [22] | ||||||||||||||||||||||
| Yang, 2017 [23] | △↑ | |||||||||||||||||||||
| Shang, 2017 [25] | △↓ | △↑ | ||||||||||||||||||||
| Lv, 2017 [26] | ○↑ | △↑ | ○↑ | ○↑ | ||||||||||||||||||
| Jin, 2017 [8] | ○↓ | ○↑ | ○↓ | |||||||||||||||||||
| Zhu, 2016 [9] | △↓ | △↑ | ||||||||||||||||||||
| Wang, 2016 [27] | ○↑ | |||||||||||||||||||||
| Tai, 2016 [29] | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||||||
| Sun, 2016 [30] | △↑ | |||||||||||||||||||||
| Fan, 2016 [33] | ○↓ | |||||||||||||||||||||
| Zhao, 2015 [35] | △↑ | |||||||||||||||||||||
| Yang, 2015 [36] | ○↑ | ○↓ | ||||||||||||||||||||
| Yan, 2015 [37] | △↑ | |||||||||||||||||||||
| Wang, 2015 [38] | △↑ | △↑ | △↑ | |||||||||||||||||||
| Wang, 2015 [39] | ○↓ | ○↓ | ○↓ | |||||||||||||||||||
| Wang, 2015 [40] | ○↑ | |||||||||||||||||||||
| Li, 2015 [41] | △↑ | |||||||||||||||||||||
| Dou, 2015 [43] | △↓ | |||||||||||||||||||||
| Zhu, 2014 [44] | △↓ | △↓ | ||||||||||||||||||||
| Geng, 2014 [48] | △↑ | △↑ | △↑ | △↑ | △↑ | |||||||||||||||||
| Gao, 2014 [49] | △↑ | |||||||||||||||||||||
| Tang, 2013 [50] | ○↑ | ○↓ | ○↑ | |||||||||||||||||||
| Li, 2013 [51] | △↑ | |||||||||||||||||||||
| Wang, 2012 [52] | △↑ | |||||||||||||||||||||
| Le, 2012 [53] | △↑ | △↑ | ||||||||||||||||||||
| Chen, 2012 [54] | ○↑ | ○↑ | ○↑ | ○↑ | ○↑ | ○↑ | ○↑ | ○↑ | ||||||||||||||
| Wei, 2011 [55] | △↑ | |||||||||||||||||||||
| Wei, 2011 [56] | △↑ | |||||||||||||||||||||
| Total | 13 | 5 | 5 | 4 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
○ represents miRNAs which are part of a panel; △ represents miRNAs which have only been analyzed individually and not as a part of a miRNA panel; ↑ represents up-regulation; ↓ represents down-regulation; Table S1 provides all miRNAs for which significant associations with lung cancer have been reported.
Figure 2.
Graphical representation of sensitivity versus specificity of analyzed miRNAs. Sensitivity is plotted on the y-axis while on the x-axis the false positive rate is presented (100-Specificity). (A) Overview of all analyzed miRNAs and miRNA panels and (B) more detailed representation of miRNAs and miRNA panels with ≥80% sensitivity and ≥80% specificity. Panel D: -145, -20a, -21, -223; Panel E: -146a, -222, -223; Panel F: let-7b-5p, let-7e-5p, -24-5p, -21-5p; Panel G: -628-3p, -339-3p, -425-3p, -532; Panel H: -628-3p, -425-3p, -532; Panel I (20 miRs): -451, -1290. -636, -30c, -22-3p, -19b, -486-5p, -20b, -93, -34b, -185, -126-5p, -93-3p, -1274a, -142-5p, -628-5p, -486-3p, -425, -645, -24; Panel J: -20a-5p, -16-5p, -15b-5p; Panel K: -148a, -148b, -152, -21; Panel L: -25, -125a-5p, -126; Panel O (10 miRs): -20a, -24, -25, -145, -152, -199a-5p, -221, -222, -223, -320. Abbreviations: LC: lung cancer; NSCLC: non-small cell lung cancer; ADC: adenocarcinoma; SCC: squamous cell carcinoma; SCLC: small cell lung cancer.
Of the 22 miRNAs reported at least twice, miR-21 was most frequently reported (13 studies), followed by miR-145, miR-20a, miR-24, miR-223, miR-155, miR-25, miR-152, miR-125a-5p, miR-126, miR-221 and miR-93 (all three to five studies) (Table 3). Of note, higher frequency of reporting of the investigated markers does not go along with higher values of diagnostic performance parameters. For example, the median sensitivity and specificity of miR-21 were 69% (46.3–91%) and 71.9% (60–92%) respectively.
3.4. Direction of Dysregulation of Circulating miRNAs
Of the 46 studies, 45 studies described the direction of dysregulation of miRNAs in blood, and only one study did not report information on miRNA dysregulation (Table S1). Among the 22 miRNAs reported in ≥2 studies, inconsistent directions were reported for six miRNAs (Table S3), whereas consistent direction of dysregulation was observed for most of the markers. For example, up-regulation of miR-24, miR-223, miR-155 and miR-221, and down-regulation of miR-126 were consistently reported in all studies reporting on these miRNAs regardless of histological subtype, stage or sample type (Table S1). In addition, miR-21, the most frequently reported miRNA, was also up-regulated in both plasma and serum samples in all included studies, except one study reporting down-regulation of miR-21 in exosome [8].
3.5. Comparison of miRNAs Profiles for LC Detection between Eastern Asian and Western Populations
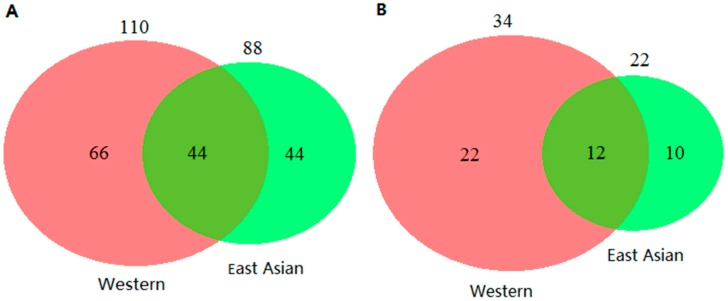
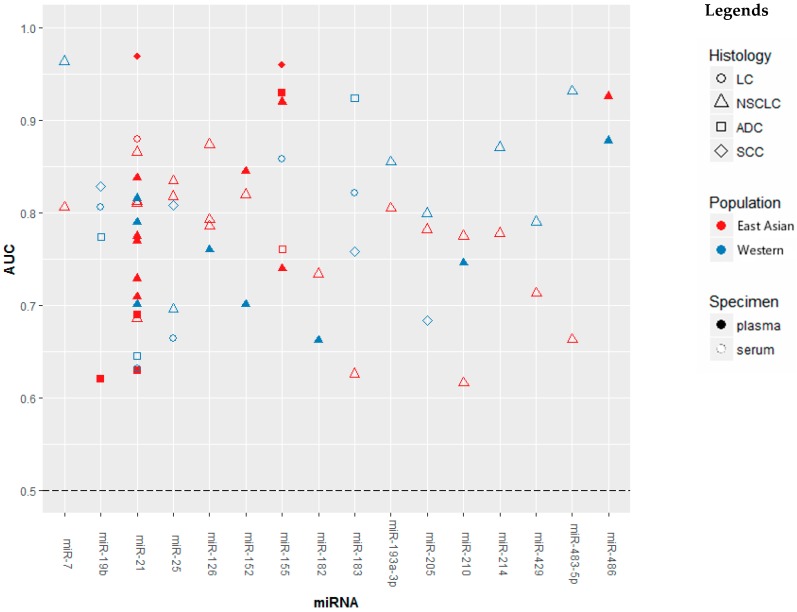
We previously performed a systematic review of literature on circulating miRNAs for lung cancer detection in Western populations [12]. A further update of this review until September 17th, 2018 identified a total number of 110 miRNAs, 34 of which were reported in ≥2 studies. We plotted Venn diagrams to illustrate the overlap of identified miRNAs between East Asian and Western populations. Forty-four miRNAs were reported in both populations (Figure 3A), which accounted for 50% of miRNAs reported in the East Asian populations, and 40% of miRNAs reported in the Western populations. Restricting miRNAs reported ≥2 times in each type of population, there were 12 overlapping miRNAs (Figure 3B), which account for 55% of miRNAs reported in East Asian and 35% of miRNAs reported in Western populations. Sixteen miRNAs were evaluated individually in both East Asian and Western populations, AUC values of which are shown in Figure 4. For some miRNAs showing differences in diagnostic performance were observed between East Asian and Western studies, but such differences were also seen within each type of study populations. Detailed information of the 16 miRNAs is provided in Table S4.
Figure 3.
Venn diagrams showing numbers of miRNAs with reported significant associations with lung cancer for Western and East Asian studies. (A) miRNAs reported in ≥1 study and (B) miRNAs reported in ≥2 studies.
Figure 4.
Graphical representation of Western studies versus East Asian studies of the diagnostic performance of miRNAs in lung cancer. Abbreviations: AUC: area under the curve; LC: lung cancer; NSCLC: non-small cell lung cancer; ADC: adenocarcinoma; SCC: squamous cell carcinoma.
4. Discussion
Our systematic literature review identified 46 studies that evaluated a total of 88 miRNA markers for LC diagnosis in East Asian populations. Sixteen of the 46 studies have incorporated individual miRNA markers as panels (with 2–20 markers). Three promising miRNA panels with ≥90% sensitivity and ≥90% specificity were discovered, two of which were verified externally [33,54]. Diagnostic performance of circulating miRNA in East Asian populations was comparable to diagnostic performance in Western populations. Forty-four miRNAs were reported in both populations. No major differences in diagnostic performance by ethnicity of the same miRNA was observed.
In general, the performance of the investigated miRNAs and miRNA panels for detecting LC in East Asian populations appears promising, and in most cases the sum of the sensitivity and specificity by far exceeded 100% (Figure 2A). There were 17 individual miRNAs and 11 miRNA panels for which both sensitivity and specificity above 80% were reported (Figure 2B). Several miRNA panels even showed rather good diagnostic efficiency. For instance, Fan et al. [33] used a panel composed of miR-20a-5p, miR-16-5p and miR-15b-5p in serum to discriminate NSCLC cases from healthy controls, and the sensitivity and specificity reached 94% and 94%, respectively, in the validation set. Chen et al. [54] used a 10-miRNA panel in serum, and reported 93% sensitivity and 90% specificity in the validation set. Several miRNA panels were reported to be useful for the detection of early stage LC, with verified AUC values over 0.85 [8,10,48]. However, most studies evaluated miRNAs or miRNA panels in samples of stage I–IV LC cases, the diagnostic efficiency of the reported markers or panels for early stage LC needs to be validated in true screening settings. Differences in the subgroup analyses for histology and stage of LC were rather small and relevant data were not sufficient to obtain robust results.
Aberrant expression of specific circulating miRNAs may provide important information for distinguishing LC histological subtypes or stages. However, subgroup analyses with respect to LC histological subtypes or stages have only been performed in a small proportion of the included studies [17,26,28,30,34,39,45,48,50,53]. Even though histology-specific analysis identified variations of diagnostic indicators of miRNA in different subtypes, no histology-specific miRNA was discovered due to the overlap in profiles. Consistent findings were also observed in studies conducted in the Western populations. Stage-specific analyses in both the Eastern Asian and Western populations showed that diagnostic efficacy was better in advanced stage compared to early stage; however, the differences with respect to AUC were quite small.
Over the past decade, studies focusing on miRNA and LC have been emerging rapidly and have identified a number of LC-specific miRNA expression profiles. However, these miRNA expression profiles are not consistent among different studies [58,59,60,61]. In this systematic review, the degree of overlap of LC-specific miRNAs reported by various studies was found to be low and inconsistent directions of dysregulation of miRNAs was observed. Similar findings were also noted among the Western studies [12]. To be used as biomarkers for LC screening, miRNAs should show consistent degrees and direction of dysregulation in different settings [62,63]. Possible sources of inconsistency of results across studies could include the use of different biospecimen or different analytical platforms. Ideally, future studies should involve different types of biospecimen and different types of analytical platforms in the same study population in order to further elucidate the role of these factors in miRNA profiling.
Further causes of the heterogeneity of reported miRNA biomarkers may be the differences in the characteristics of study populations. For the included studies in both East Asian and Western populations, we extracted demographic information such as age, sex and country, as these factors may affect the identification of miRNA markers [11,64,65]. Cigarette smoking could also induce the dysregulation of some circulating miRNAs, which might be associated with smoking-related LC [47,66,67]. However, information on smoking status was incomprehensive in most studied populations, especially in controls. Sample sizes of the majority of both East Asian and Western studies were rather small, which might have resulted in substantial random variation of LC-specific miRNAs identified among studies. In addition, miRNA profiles may change in different phases of cancer [68]. Most studies in both East Asian and Western populations however recruited LC cases with various stage proportions.
The use of different miRNA detection protocols could also affect the identification of LC-specific miRNAs. Pre-analytical factors such as speed and duration of centrifugation may affect the amount of cell debris remaining in the sample supernatant (especially in plasma samples) and cause miRNA contaminations [69,70,71,72]. Only a fraction of included studies in both East Asian and Western populations have applied a second high-speed centrifugation step to reduce the remaining cell debris (Table S5). Moreover, reports on most studies did not specifically address potential hemolysis, which could result in an increase of multiple miRNA levels [70,71,73,74]. Analytical factors such as differences in miRNA extraction and quantification methods could also contribute to the variability of identified LC-specific miRNAs [75]. The extraction methods in the included studies were diverse (Table S5), only few studies in both East Asian and Western populations have used miRNeasy kit which has been suggested to have better extraction efficiency compared to other kits [76,77]. In addition, a very important yet unresolved issue is the normalization of miRNA expression data. Circulating miR-16 is usually used as endogenous control for normalization (Table S5), however, it shows an unstable expression in the circulation of cancer patients as well as in hemolytic samples [71,78,79]. Several new bioinformatics tools such as miRNA ratios and differentially expressed miRNA pairs have been developed to build up miRNA panels to reduce the analytical bias [80,81,82]. These attempts so far have shown promising results but still require further validation.
The herein described circulating miRNAs have several advantages over some other markers of cancer detection: (i) miRNAs are extremely stable in cell-free fluids and in less than ideal sample handling conditions [79,83,84], (ii) they can be measured repeatedly over time in a non-invasive manner [85], (iii) they can be used to predict cancer in high-risk populations years in advance [80,82,86] and (iv) the cost of analysis is relative low. Still, other blood-based markers have been proposed for LC detection, such as cytokeratin 19 fragment 21-1 (CYFRA21-1), carcinoembryonic antigen (CEA), tissue polypeptide specific antigen (TPS) or neuron-specific enolase (NSE). However, these markers are primarily utilized for monitoring of disease progression and tend to show suboptimal diagnostic value for LC with sensitivity values usually under 50% [81,87]. Sanfiorenzo et al. [88] developed a signature of 11 plasma miRNAs to discriminate healthy individuals from NSCLC patients which yielded 85% sensitivity and 82.9% specificity. This estimate of sensitivity was compared favorably with an estimated sensitivity of blood-based biomarkers, such as CYFRA 21-1, TPS and CEA, which ranged from 34% to 53% in the same study population. Recently, DNA methylation markers have showed good diagnostic efficacy for LC. Zhang et al. [89] used a combination of F2RL3 methylation in whole blood and smoking exposure to predict LC incidence with an AUC value of 0.86 for participants ≥65 years. Since miRNA markers and DNA methylation markers both shown good diagnostic performance for LC, the combination of the two may offer an improvement of diagnostic efficiency in LC.
5. Conclusions
Our systematic review suggests a number of circulating miRNAs to be promising candidates for noninvasive LC detection in East Asian populations. The heterogeneity of reported LC-specific miRNA profiles in published studies needs to be addressed and protocols for the standardization of miRNA analysis procedures need to be put into place. Larger prospective studies, the improvement of miRNA detection technologies, the minimization of pre-analytical or analytical variability as well as the development of new analysis methods will be crucial to further reduce the bias in measurement and analysis and improve the diagnostic performance. Most importantly, however, will be the rigorous independent validation of identified promising miRNA algorithms or their combination with other biomarkers in prospective screening cohorts.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/3/415/s1, Figure S1: Risk of bias and applicability concerns graph: review authors’ judgements about each domain presented as percentages across included studies, Figure S2: Risk of bias and applicability concerns summary: review authors’ judgements about each domain for each included study, Table S1: Summary of studies reporting significant associations of miRNAs with lung cancer in East Asian populations, Table S2: Diagnostic performance of miRNAs and miRNA panels according to lung cancer stage in East Asian populations, Table S3: MiRNAs for which opposite directions of dysregulation were reported in lung cancer blood samples, Table S4: East Asian studies versus Western studies of miRNAs for lung cancer detection, Table S5: Protocols of blood miRNA detection.
Author Contributions
H.B., Y.Z. and H.Y. designed the study. H.Y. and Z.G. extracted the data. H.Y. wrote the manuscript. Y.Z., K.C. and H.B. revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported in part by the China Scholarship Council (CSC). The sponsor had no role in the study design and in the collection, analysis and interpretation of data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Jr., Wu Y.L., Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Siegel R.L., Jemal A. Lung cancer statistics. Adv. Exp. Med. Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Manser R., Lethaby A., Irving L.B., Stone C., Byrnes G., Abramson M.J., Campbell D. Screening for lung cancer. Cochrane Database Syst. Rev. 2013 doi: 10.1002/14651858.CD001991.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horeweg N., Scholten E.T., de Jong P.A., van der Aalst C.M., Weenink C., Lammers J.W.J., Nackaerts K., Vliegenthart R., ten Haaf K., Yousaf-Khan U.A., et al. Detection of lung cancer through low-dose ct screening (nelson): A prespecifi ed analysis of screening test performance and interval cancers. Lancet Oncol. 2014;15:1342–1350. doi: 10.1016/S1470-2045(14)70387-0. [DOI] [PubMed] [Google Scholar]
- 6.Shen J., Liao J., Guarnera M.A., Fang H., Cai L., Stass S.A., Jiang F. Analysis of micrornas in sputum to improve computed tomography for lung cancer diagnosis. J. Thorac. Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garzon R., Calin G.A., Croce C.M. Micrornas in cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 8.Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., Liu L., Lin B., Su H., Zhao L., et al. Evaluation of tumor-derived exosomal mirna as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 9.Zhu W.Y., Zhou K.Y., Zha Y., Chen D.D., He J.Y., Ma H.J., Liu X.G., Le H.B., Zhang Y.K. Diagnostic value of serum mir-182, mir-183, mir-210 and mir-126 levels in patients with early-stage non-small cell lung cancer. PLoS ONE. 2016;11:e0153046. doi: 10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X.J., Wang Y., Zhao H., Wei F., Zhang X.W., Su Y.J., Wang C.L., Li H., Ren X.B. Plasma mir-324-3p and mir-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma. Oncotarget. 2016;7:59664–59675. doi: 10.18632/oncotarget.11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang R.S., Gamazon E.R., Ziliak D., Wen Y., Im H.K., Zhang W., Wing C., Duan S., Bleibel W.K., Cox N.J., et al. Population differences in microrna expression and biological implications. RNA Biol. 2011;8:692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H., Guan Z., Cuk K., Brenner H., Zhang Y. Circulating microrna biomarkers for lung cancer detection in western populations. Cancer Med. 2018;7:4849–4862. doi: 10.1002/cam4.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moher D., Liberati A., Tetzlaff J., Altman D.G., Grp P. Preferred reporting items for systematic reviews and meta-analyses: The prisma statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y.Z. The diagnostic and prognostic role of circulating mir-141 expression in non-small-cell lung cancer patients. Int. J. Clin. Exp. Pathol. 2018;11:2597–2604. [PMC free article] [PubMed] [Google Scholar]
- 15.Sun B., Liu H.F., Ding Y., Li Z. Evaluating the diagnostic and prognostic value of serum mir-770 in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3061–3066. doi: 10.26355/eurrev_201805_15064. [DOI] [PubMed] [Google Scholar]
- 16.Shan X., Zhang H., Zhang L., Zhou X., Wang T.S., Zhang J.Y., Shu Y.Q., Zhu W., Wen W., Liu P. Identification of four plasma micrornas as potential biomarkers in the diagnosis of male lung squamous cell carcinoma patients in china. Cancer Med. 2018;7:2370–2381. doi: 10.1002/cam4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Y.Z., Zhou X.Y., Huang C., Li L., Liu H.S., Liang N.X., Chen Y.Y., Ma D.J., Han Z.J., Xu X.H., et al. Serum mir-342-3p is a novel diagnostic and prognostic biomarker for non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2018;11:2742–2748. [PMC free article] [PubMed] [Google Scholar]
- 18.Bao M., Pan S., Yang W.L., Chen S., Shan Y.B., Shi H.C. Serum mir-10a-5p and mir-196a-5p as non-invasive biomarkers in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2018;11:773–780. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X., Wen W., Shan X., Zhu W., Xu J., Guo R.H., Cheng W.F., Wang F., Qi L.W., Chen Y., et al. A six-microrna panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Shan X., Wang J., Zhu J., Huang Z., Zhang H., Zhou X., Cheng W.F., Shu Y.Q., Zhu W., et al. A three-microrna signature for lung squamous cell carcinoma diagnosis in chinese male patients. Oncotarget. 2017;8:86897–86907. doi: 10.18632/oncotarget.19666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H., Mao F., Shen T., Luo Q., Ding Z., Qian L., Huang J. Plasma mir-145, mir-20a, mir-21 and mir-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol. Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y.L., Zuo J.C., Tan Q., Thin K.Z., Li P., Zhu M., Yu M.X., Fu Z.M., Liang C.Z., Tu J.C. Plasma mir-92a-2 as a biomarker for small cell lung cancer. Cancer Biomark. 2017;18:319–327. doi: 10.3233/CBM-160254. [DOI] [PubMed] [Google Scholar]
- 23.Yang M., Xiao L.B., Zhang Y.H., Li G.J., Zhou J.W. Clinical significance of serum mir-31 as a predictive biomarker for lung adenocarcinoma. Int. J. Clin. Exp. Pathol. 2017;10:4668–4674. [Google Scholar]
- 24.Wang K.C., Dong L.Y., Fang Q.M., Xia H.W., Hou X.L. Low serum mir-98 as an unfavorable prognostic biomarker in patients with non-small cell lung cancer. Cancer Biomark. 2017;20:283–288. doi: 10.3233/CBM-170124. [DOI] [PubMed] [Google Scholar]
- 25.Shang A.Q., Xie Y.N., Wang J., Sun L., Wei J., Lu W.Y., Lan J.Y., Wang W.W., Wang L., Wang L.L. Predicative values of serum microrna-22 and microrna-126 levels for non-small cell lung cancer development and metastasis: A case-control study. Neoplasma. 2017;64:453–459. doi: 10.4149/neo_2017_317. [DOI] [PubMed] [Google Scholar]
- 26.Lv S.G., Xue J., Wu C.Y., Wang L., Wu J., Xu S.J., Liang X.H., Lou J.T. Identification of a panel of serum micrornas as biomarkers for early detection of lung adenocarcinoma. J. Cancer. 2017;8:48–56. doi: 10.7150/jca.16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Zhao H., Gao X.J., Wei F., Zhang X.W., Su Y.J., Wang C.L., Li H., Ren X.B. Identification of a three-mirna signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget. 2016;7:26070–26086. doi: 10.18632/oncotarget.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W.Z., Li W.L., Ding M.J., Yuan H.N., Yang J., Meng W., Jin E., Wang X.J., Ma S.L. Identification of mirnas as non-invasive biomarkers for early diagnosis of lung cancers. Tumor Biol. 2016;37:16287–16293. doi: 10.1007/s13277-016-5442-y. [DOI] [PubMed] [Google Scholar]
- 29.Tai M.C., Yanagisawa K., Nakatochi M., Hotta N., Hosono Y., Kawaguchi K., Naito M., Taniguchi H., Wakai K., Yokoi K., et al. Blood-borne mirna profile-based diagnostic classifier for lung adenocarcinoma. Sci. Rep. 2016;6:31389. doi: 10.1038/srep31389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun M.Z., Song J.X., Zhou Z.W., Zhu R., Jin H., Ji Y.Q., Lu Q., Ju H.X. Comparison of serum microrna21 and tumor markers in diagnosis of early non-small cell lung cancer. Dis. Markers. 2016;2016:3823121. doi: 10.1155/2016/3823121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su K.L., Zhang T.C., Wang Y.R., Hao G.J. Diagnostic and prognostic value of plasma microrna-195 in patients with non-small cell lung cancer. World J. Surg. Oncol. 2016;14:224. doi: 10.1186/s12957-016-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Peng H., Wang J., Li J., Zhao M., Huang S.K., Gu Y.Y., Li Y., Sun X.J., Yang L., Luo Q., et al. A circulating non-coding rna panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–242. doi: 10.1016/j.lfs.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Fan L.H., Qi H.W., Teng J.L., Su B., Chen H., Wang C.H., Xia Q. Identification of serum mirnas by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumor Biol. 2016;37:7777–7784. doi: 10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhou C.C., Chen Z.L., Dong J.S., Li J.G., Shi X.J., Sun N., Luo M., Zhou F., Tan F.W., He J. Combination of serum mirnas with cyfra21-1 for the diagnosis of non-small cell lung cancer. Cancer Lett. 2015;367:138–146. doi: 10.1016/j.canlet.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Zhao W., Zhao J.J., Zhang L., Xu Q.F., Zhao Y.M., Shi X.Y., Xu A.G. Serum mir-21 level: A potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int. J. Clin. Exp. Med. 2015;8:14759–14763. [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J.S., Li B.J., Lu H.W., Chen Y., Lu C., Zhu R.X., Liu S.H., Yi Q.T., Li J., Song C.H. Serum mir-152, mir-148a, mir-148b and mir-21 as novel biomarkers in non-small cell lung cancer screening. Tumor Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 37.Yan H.J., Ma J.Y., Wang L., Gu W. Expression and significance of circulating microrna-31 in lung cancer patients. Med. Sci. Monit. 2015;21:722–726. doi: 10.12659/MSM.893213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang R.J., Zheng Y.H., Wang P., Zhang J.Z. Serum mir-125a-5p, mir-145 and mir-146a as diagnostic biomarkers in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015;8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P., Yang D.W., Zhang H.L., Wei X.Y., Ma T.L., Cheng Z.L., Hong Q.Y., Hu J., Zhuo H.J., Song Y.L., et al. Early detection of lung cancer in serum by a panel of microrna biomarkers. Clin. Lung Cancer. 2015;16:313–319.e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Wang C., Ding M., Xia M.D., Chen S.D., Le A.V., Soto-Gil R., Shen Y., Wang N., Wang J.J., Gu W.J., et al. A five-mirna panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients. Ebiomedicine. 2015;2:1377–1385. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W.S., Wang Y., Zhang Q., Tang L.L., Liu X.P., Dai Y.H., Xiao L., Huang S.G., Chen L., Guo Z.M., et al. Microrna-486 as a biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS ONE. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo W.G., Zhang Y.X., Zhang Y., Shi Y., Xi J.J., Fan H., Xu S.T. Decreased expression of mir-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int. J. Mol. Med. 2015;36:1720–1726. doi: 10.3892/ijmm.2015.2388. [DOI] [PubMed] [Google Scholar]
- 43.Dou H.L., Wang Y., Su G., Zhao S. Decreased plasma let-7c and mir-152 as noninvasive biomarker for non-small-cell lung cancer. Int. J. Clin. Exp. Med. 2015;8:9291–9298. [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu W.Y., Luo B., An J.Y., He J.Y., Chen D.D., Xu L.Y., Huang Y.Y., Liu X.G., Le H.B., Zhang Y.K. Differential expression of mir-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Investig. 2014;32:394–401. doi: 10.3109/07357907.2014.922569. [DOI] [PubMed] [Google Scholar]
- 45.Zhu W.Y., He J.Y., Chen D.D., Zhang B.J., Xu L.Y., Ma H.J., Liu X.G., Zhang Y.K., Le H.B. Expression of mir-29c, mir-93 and mir-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS ONE. 2014;9:e87780. doi: 10.1371/journal.pone.0087780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M., Zhang Q., Wu L., Jia C.Y., Shi F.H., Li S.C., Peng A.M., Zhang G.L., Song X.L., Wang C.H. Serum mir-499 as a novel diagnostic and prognostic biomarker in non-small cell lung cancer. Oncol. Rep. 2014;31:1961–1967. doi: 10.3892/or.2014.3029. [DOI] [PubMed] [Google Scholar]
- 47.Huang J.K., Wu J.J., Li Y.Q., Li X., Yang T., Yang Q.Y., Jiang Y.G. Deregulation of serum microrna expression is associated with cigarette smoking and lung cancer. BioMed Res. Int. 2014;2014:364316. doi: 10.1155/2014/364316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geng Q., Fan T., Zhang B.Y., Wang W., Xu Y., Hu H. Five micrornas in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir. Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao F., Chang J.X., Wang H.Q., Zhang G.J. Potential diagnostic value of mir-155 in serum from lung adenocarcinoma patients. Oncol. Rep. 2014;31:351–357. doi: 10.3892/or.2013.2830. [DOI] [PubMed] [Google Scholar]
- 50.Tang D.F., Shen Y., Wang M.Z., Yang R.H., Wang Z.Z., Sui A.H., Jiao W.J., Wang Y.J. Identification of plasma micrornas as novel noninvasive biomarkers for early detection of lung cancer. Eur. J. Cancer Prev. 2013;22:540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 51.Li Z.H., Zhang H., Yang Z.G., Wen G.Q., Cui Y.B., Shao G.G. Prognostic significance of serum microrna-210 levels in nonsmall-cell lung cancer. J. Int. Med. Res. 2013;41:1437–1444. doi: 10.1177/0300060513497560. [DOI] [PubMed] [Google Scholar]
- 52.Wang B., Zhang Q.Y. The expression and clinical significance of circulating microrna-21 in serum of five solid tumors. J. Cancer Res. Clin. Oncol. 2012;138:1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PubMed] [Google Scholar]
- 53.Le H.B., Zhu W.Y., Chen D.D., He J.Y., Huang Y.Y., Liu X.G., Zhang Y.K. Evaluation of dynamic change of serum mir-21 and mir-24 in pre- and post-operative lung carcinoma patients. Med. Oncol. 2012;29:3190–3197. doi: 10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 54.Chen X., Hu Z.B., Wang W.J., Ba Y., Ma L.J., Zhang C.N., Wang C., Ren Z.J., Zhao Y., Wu S.J., et al. Identification of ten serum micrornas from a genome-wide serum microrna expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int. J. Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 55.Wei J., Liu L.K., Gao W., Zhu C.J., Liu Y.Q., Cheng T., Shu Y.Q. Reduction of plasma microrna-21 is associated with chemotherapeutic response in patients with non-small cell lung cancer. Chin. J. Cancer Res. 2011;23:123–128. doi: 10.1007/s11670-011-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei J., Gao W., Zhu C.J., Liu Y.Q., Mei Z., Cheng T., Shu Y.Q. Identification of plasma microrna-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin. J. Cancer. 2011;30:407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan P., Gao S.L. Management of breast cancer brain metastases: Focus on human epidermal growth factor receptor 2-positive breast cancer. Chronic Dis. Transl. Med. 2017;3:21–32. doi: 10.1016/j.cdtm.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H.Q., Wu S.N., Zhao L., Zhao J., Liu J.J., Wang Z.H. Clinical use of micrornas as potential non-invasive biomarkers for detecting non-small cell lung cancer: A meta-analysis. Respirology. 2015;20:56–65. doi: 10.1111/resp.12444. [DOI] [PubMed] [Google Scholar]
- 59.Huang Y.S., Hu Q.Y., Deng Z.H., Hang Y.L., Wang J., Wang K.Z. Micrornas in body fluids as biomarkers for non-small cell lung cancer: A systematic review. Technol. Cancer Res. Treat. 2014;13:277–287. doi: 10.7785/tcrt.2012.500377. [DOI] [PubMed] [Google Scholar]
- 60.He W.J., Li W.H., Jiang B., Wang Y.F., Xia Y.X., Wang L. Micrornas level as an initial screening method for early-stage lung cancer: A bivariate diagnostic random-effects meta-analysis. Int. J. Clin. Exp. Med. 2015;8:12317–12326. [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L., Jin H. Micrornas as novel biomarkers in the diagnosis of non-small cell lung cancer: A meta-analysis based on 20 studies. Tumor Biol. 2014;35:9119–9129. doi: 10.1007/s13277-014-2188-2. [DOI] [PubMed] [Google Scholar]
- 62.Griffith O.L., Melck A., Jones S.J.M., Wiseman S.M. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J. Clin. Oncol. 2006;24:5043–5051. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- 63.Chan S.K., Griffith O.L., Tai I.T., Jones S.J.M. Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol. Biomark. Prev. 2008;17:543–552. doi: 10.1158/1055-9965.EPI-07-2615. [DOI] [PubMed] [Google Scholar]
- 64.Sharma S., Eghbali M. Influence of sex differences on microrna gene regulation in disease. Biol. Sex Differ. 2014;5:3. doi: 10.1186/2042-6410-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hooten N.N., Fitzpatrick M., Wood W.H., De S., Ejiogu N., Zhang Y.Q., Mattison J.A., Becker K.G., Zonderman A.B., Evans M.K. Age-related changes in microrna levels in serum. Aging. 2013;5:725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oyetunji S.O., Xi S.C., Azoury S.C., Zhang M., Hong J., Inchauste S.M., Schrump D. Cigarette smoke induces epigenetic dysregulation of microrna expression during esophageal adenocarcinogenesis. J. Am. Coll. Surg. 2014;219:e68. doi: 10.1016/j.jamcollsurg.2014.07.562. [DOI] [Google Scholar]
- 67.Takahashi K., Yokota S., Tatsumi N., Fukami T., Yokoi T., Nakajima M. Cigarette smoking substantially alters plasma microrna profiles in healthy subjects. Toxicol. Appl. Pharmacol. 2013;272:154–160. doi: 10.1016/j.taap.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 68.Olson P., Lu J., Zhang H., Shai A., Chun M.G., Wang Y.C., Libutti S.K., Nakakura E.K., Golub T.R., Hanahan D. Microrna dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Gene Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duttagupta R., Jiang R., Gollub J., Getts R.C., Jones K.W. Impact of cellular mirnas on circulating mirna biomarker signatures. PLoS ONE. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDonald J.S., Milosevic D., Reddi H.V., Grebe S.K., Algeciras-Schimnich A. Analysis of circulating microrna: Preanalytical and analytical challenges. Clin. Chem. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 71.Pritchard C.C., Kroh E., Wood B., Arroyo J.D., Dougherty K.J., Miyaji M.M., Tait J.F., Tewari M. Blood cell origin of circulating micrornas: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng X.H., Cui C., Zhou X.X., Zeng Y.X., Jia W.H. Centrifugation: An important pre-analytic procedure that influences plasma microrna quantification during blood processing. Chin. J. Cancer. 2013;32:667–672. doi: 10.5732/cjc.012.10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becker N., Lockwood C.M. Pre-analytical variables in mirna analysis. Clin. Biochem. 2013;46:861–868. doi: 10.1016/j.clinbiochem.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 74.Kirschner M.B., Kao S.C., Edelman J.J., Armstrong N.J., Vallely M.P., van Zandwijk N., Reid G. Haemolysis during sample preparation alters microrna content of plasma. PLoS ONE. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mensah M., Borzi C., Verri C., Suatoni P., Conte D., Pastorino U., Orazio F., Sozzi G., Boeri M. Microrna based liquid biopsy: The experience of the plasma mirna signature classifier (msc) for lung cancer screening. J. Vis. Exp. 2017 doi: 10.3791/56326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.El-Khoury V., Pierson S., Kaoma T., Bernardin F., Berchem G. Assessing cellular and circulating mirna recovery: The impact of the rna isolation method and the quantity of input material. Sci. Rep. 2016;6:19529. doi: 10.1038/srep19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rice J., Roberts H., Burton J., Pan J., States V., Rai S.N., Galandiuk S. Assay reproducibility in clinical studies of plasma mirna. PLoS ONE. 2015;10:e0121948. doi: 10.1371/journal.pone.0121948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Appaiah H.N., Goswami C.P., Mina L.A., Badve S., Sledge G.W., Liu Y.L., Nakshatri H. Persistent upregulation of u6:Snord44 small rna ratio in the serum of breast cancer patients. Breast Cancer Res. 2011;13:R86. doi: 10.1186/bcr2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X., Ba Y., Ma L.J., Cai X., Yin Y., Wang K.H., Guo J.G., Zhang Y.J., Chen J.N., Guo X., et al. Characterization of micrornas in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 80.Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., Calabro E., Croce C.M., Pastorino U., Sozzi G. Microrna signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hennessey P.T., Sanford T., Choudhary A., Mydlarz W.W., Brown D., Adai A.T., Ochs M.F., Ahrendt S.A., Mambo E., Califano J.A. Serum microrna biomarkers for detection of non-small cell lung cancer. PLoS ONE. 2012;7:e32307. doi: 10.1371/annotation/bda2b1d8-1054-481a-bafd-2bcfa48514e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sozzi G., Boeri M., Rossi M., Verri C., Suatoni P., Bravi F., Roz L., Conte D., Grassi M., Sverzellati N., et al. Clinical utility of a plasma-based mirna signature classifier within computed tomography lung cancer screening: A correlative mild trial study. J. Clin. Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balzano F., Deiana M., Dei Giudici S., Oggiano A., Baralla A., Pasella S., Mannu A., Pescatori M., Porcu B., Fanciulli G., et al. Mirna stability in frozen plasma samples. Molecules. 2015;20:19030–19040. doi: 10.3390/molecules201019030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kroh E.M., Parkin R.K., Mitchell P.S., Tewari M. Analysis of circulating microrna biomarkers in plasma and serum using quantitative reverse transcription-pcr (qrt-pcr) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keller A., Leidinger P., Gislefoss R., Haugen A., Langseth H., Staehler P., Lenhof H.P., Meese E. Stable serum mirna profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011;8:506–516. doi: 10.4161/rna.8.3.14994. [DOI] [PubMed] [Google Scholar]
- 87.Hoseok I., Cho J.Y. Lung cancer biomarkers. Adv. Clin. Chem. 2015;72:107–170. doi: 10.1016/bs.acc.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 88.Sanfiorenzo C., Ilie M.I., Belaid A., Barlesi F., Mouroux J., Marquette C.H., Brest P., Hofman P. Two panels of plasma micrornas as non-invasive biomarkers for prediction of recurrence in resectable nsclc. PLoS ONE. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y., Schottker B., Ordonez-Mena J., Holleczek B., Yang R.X., Burwinkel B., Butterbach K., Brenner H. F2rl3 methylation, lung cancer incidence and mortality. Int. J. Cancer. 2015;137:1739–1748. doi: 10.1002/ijc.29537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.