Abstract
Odorants are transmitted by small hydrophobic molecules that cross the aqueous sensillar lymph surrounding the dendrites of the olfactory neurons to stimulate the olfactory receptors. In insects, the transport of pheromones, which are a special class of odorants, is mediated by pheromone-binding proteins (PBPs), which occur at high concentrations in the sensillar lymph. The PBP from the silk moth Bombyx mori (BmPBP) undergoes a pH-dependent conformational transition between the forms BmPBPA present at pH 4.5 and BmPBPB present at pH 6.5. Here, we describe the NMR structure of BmPBPA, which consists of a tightly packed arrangement of seven α-helices linked by well defined peptide segments and knitted together by three disulfide bridges. A scaffold of four α-helices that forms the ligand binding site in the crystal structure of a BmPBP–pheromone complex is preserved in BmPBPA. The C-terminal dodecapeptide segment, which is in an extended conformation and located on the protein surface in the pheromone complex, forms a regular helix, α7, which is located in the pheromone-binding site in the core of the unliganded BmPBPA. Because investigations by others indicate that the pH value near the membrane surface is reduced with respect to the bulk sensillar lymph, the pH-dependent conformational transition of BmPBP suggests a novel physiological mechanism of intramolecular regulation of protein function, with the formation of α7 triggering the release of the pheromone from BmPBP to the membrane-standing receptor.
Male moths have an exquisitely sensitive olfactory system capable of detecting over great distances single molecules of pheromones emitted by the female, and they are capable of distinguishing highly selectively between closely similar compounds or isomers (1). These capabilities of male moths are because of olfactory sensory organs that consist of large branched antennae and can readily be isolated for biochemical characterization or electrophysiological recordings (2). The surface of the antennae is covered with hairlike protrusions composed of cuticle, which form a sheath (sensillum) surrounding one or several dendrite endings from olfactory neurons. The dendrites in each sensillum are bathed in a special fluid, the sensillum lymph. Odorant molecules gain access to the dendritic membrane through pores penetrating the sensillum wall and then crossing the sensillar lymph to interact with G protein-coupled olfactory receptors, which form a large eukaryotic protein family. Upon interaction with the olfactory receptors, a cascade of events leads to a change of the membrane potential (3), resulting in opening of ion channels and depolarization.
Common odorants are hydrophobic molecules that are poorly soluble in aqueous media, such as the sensillar lymph. Odorants overcome this barrier by binding to odorant-binding proteins (OBPs). OBPs are present at high concentrations in the lymph (up to 10 mM), bind the odorant, and transport it from the sensillum pore wall to the receptor at the dendritic membrane. It is not known whether the receptor recognizes either the odorant–OBP complex or the free odorant after its release by the OBP (4). A number of authors have proposed that OBPs might play an active role in odorant recognition, either by preferential binding of certain ligands or by forming odorant–OBP complexes with distinctive conformations that would support recognition of specified odorants by the receptor (5). Support for this hypothesis has been derived from the fact that a large number of different OBPs are usually isolated from an individual species, but that only one type of OBP is typically found in a given sensillum (6).
OBPs are small, highly water-soluble acidic proteins ranging from 14 to 16 kDa in size, which have been identified in many insect species and represent two different protein families. Pheromone-binding proteins (PBPs) are isolated from sensilla that are responsive specifically to pheromones produced by the females, whereas general OBPs have been found in sensilla that respond to a range of plant volatiles and odorants originating from food sources. Within a given species, several different general OBPs and one or several PBPs are typically identified. Sequence alignments reveal that six cysteines are strictly conserved in all OBPs. In moth species, the mature form of OBPs is 142 aa in length. Recently, it was demonstrated by NMR spectroscopy in solution that the PBP from the moth species Bombyx mori (BmPBP) undergoes a pH-dependent conformational transition (7). Above pH 6.0, BmPBP occurs in a basic form (BmPBPB), whereas below pH 4.9 an acidic form (BmPBPA) is observed. In the pH range of 4.9–6.0, a mixture of the A and B forms exists, which is in slow exchange on the NMR time scale.
Conformational heterogeneity observed in other OBPs indicates that pH-dependent conformational polymorphism may be a common feature of these proteins. This and the observation that a number of OBPs, including BmPBP, do not bind ligands below pH 5.0 (8, 9) suggests that pH variation of the folded conformation might play a physiological role in olfaction (10, 11). In this context, it is intriguing that near-UV circular dichroism measurements revealed that BmPBP undergoes a conformational transition when mixed with model membranes (12). Thus, as BmPBP approaches the dendritic membrane, it might undergo a conformational change to the BmPBPA form (7, 13) and thus trigger the release of the ligand to the receptor.
The structure of BmPBP in a complex with bombykol, which is a pheromone component emanated by female B. mori, was determined by x-ray crystallography in crystals grown at pH 8.2 (14). BmPBP in this complex with bombykol consists of six α-helices, which are knitted together by three disulfide bonds and completely surround the ligand, so that bombykol is protected from the aqueous solvent by a hydrophobic binding pocket. Sandler et al. (14) propose either that a loop covering the pheromone-binding pocket may move aside to allow entry of the ligand or that the entire protein may partially unfold to allow entry of the ligand, as there is no obvious entrance or exit for the ligand in the structure of the complex. To obtain more insight into the mechanisms of ligand binding and release, and the role played by the pH-dependent conformational transition in OBPs, we have started a project of NMR structure determination for all of the different conformations of BmPBP that seem to have a role in its physiological function. Here, we present the solution structure of the acidic form observed at pH 4.5, BmPBPA (7, 13), which reveals a striking conformational difference to the BmPBP conformation in the crystal structure of the bombykol complex. The C-terminal segment consisting of residues 131–141, which is in an extended conformation and located on the protein surface in the BmPBP–bombykol complex, forms an α-helix that is located in the protein core in BmPBPA. This suggests a novel intramolecular regulation mechanism of protein function by major conformational rearrangements involving residues that are widely separated in the amino acid sequence.
Materials and Methods
Sample Preparation.
The expression and purification of 13C,15N-labeled BmPBP in Escherichia coli has been described (7). The NMR sample used for the structure determination of BmPBPA was prepared by redissolving 9.75 mg of 13C,15N-labeled BmPBP lyophilized from H2O solution in 95% H2O/5% D2O containing 50 mM potassium phosphate buffer at pH 4.5 and 2 mM NaN3, resulting in ≈1 mM protein concentration.
NMR Data Collection and Structure Calculation.
All NMR data recorded to obtain structural constraints (Table 1) were performed at a 1H frequency of 750 MHz on a Bruker DRX 750 spectrometer at 20°C. Distance constraints were obtained from three nuclear Overhauser effect spectroscopy (NOESY) experiments with mixing times of 50 ms, i.e., a three-dimensional 15N-resolved [1H,1H]-NOESY spectrum and two three-dimensional 13C-resolved [1H,1H]-NOESY spectra with the 13C carrier frequency in the aliphatic and aromatic region, respectively. The three data sets were interactively peak-picked by using the program XEASY (15), and the peaks were automatically integrated by using SPCSAN (Ralf Glaser, personal communication), resulting in 4,653 NOE cross-peak intensities. On the basis of the BmPBPA sequence-specific resonance assignments (16), NOE cross-peak assignments were then obtained by using the module CANDID (T. Hermann, P.G., and K.W., unpublished data) within the program DYANA (17). CANDID/DYANA performs automated assignment and distance calibration of NOE intensities, removal of “meaningless” NOE distance constraints, and automatic NOE upper distance limit violation analysis. The CANDID/DYANA calculation consisted of seven cycles of iterative NOE assignment and structure calculation. During the first six CANDID cycles, ambiguous distance constraints (18) were used. For the final structure calculation, only distance constraints were retained that corresponded to unambiguously assigned NOE cross-peaks after the sixth CANDID cycle, resulting in 2,109 upper distance constraints; 478 torsion angle constraints obtained from short-range NOE data by using the module FOUND (19) were added in the last calculation. Energy refinement of the resulting DYANA conformers was done in a water shell with the program OPALP (20, 21), using the AMBER force field (22).
Table 1.
Input of conformational constraints and structure calculation for BmPBPA with the programs dyana and opal
| Quantity | Value |
|---|---|
| NOE upper distance restraints | 2,109 |
| Intraresidual | 857 |
| Sequential | 451 |
| Medium range | 384 |
| Long range | 417 |
| Dihedral angle constraints | 478 |
| dyana target function value (Å2)*† | 1.3 ± 0.2 (0.7–1.5) |
| Residual NOE distance constraint violations* | |
| Number > 0.1 Å | 31 ± 4 (24–38) |
| Sum (Å) | 25.3 ± 0.9 (23.3–27.3) |
| Maximum (Å) | 0.15 ± 0.01 (0.13–0.17) |
| Rms deviations (rmsd) (Å)*‡ | |
| Backbone (8–142) | 0.46 ± 0.07 (0.38–0.62) |
| Heavy atoms (8–142) | 0.78 ± 0.06 (0.7–0.92) |
| Ramachandran plot statistics§ | |
| Most favored regions (%) | 83.8 |
| Additional allowed regions (%) | 15.5 |
| Generously allowed regions (%) | 0.7 |
| Disallowed regions (%) | 0 |
The number given is the average ± the standard deviation for the set of 20 energy-refined conformers used to represent the NMR structure, with minimum and maximum values given in parentheses.
Before energy minimization.
rmsd values are relative to the mean coordinates.
As determined by procheck (29).
Results and Discussion
NMR Structure of BmPBPA.
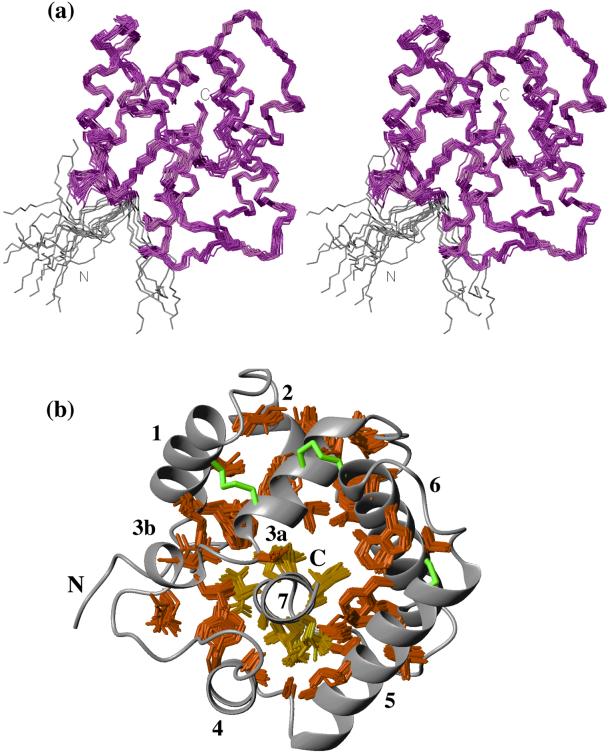
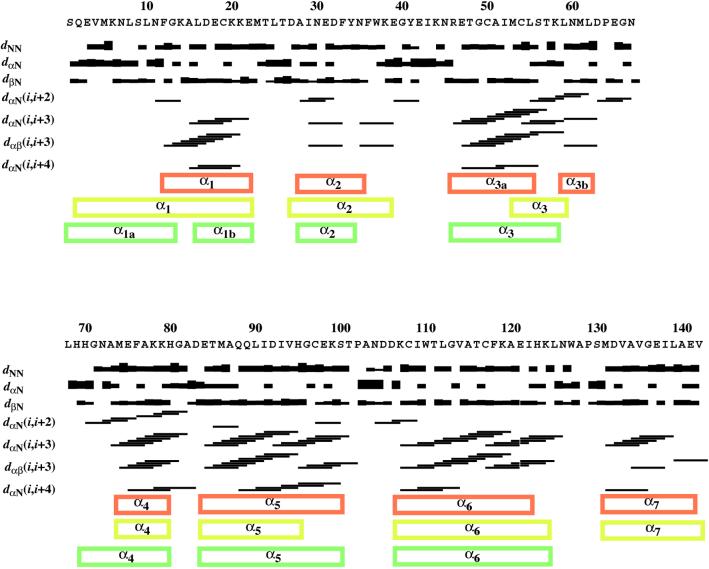
The A form of the 142-residue protein BmPBP consists of a tightly packed arrangement of seven α-helices consisting of the residues 12–22 (α1), 28–35 (α2), 46–55 (α3a),74–79 (α4), 84–100 (α5), 107–122 (α6), and 131–141(α7) (Fig. 1b). The N-terminal peptide segment of residues 1–7 is flexibly disordered (Fig. 1a). The peptide segments linking the helices are well defined (Fig. 1a) but have no regular secondary structure, except for the four-residue helix α3b in the 19-residue loop between α3a and α4. We use the notation α3a and α3b because of the overlap of these two helical structures with α3 in the crystal structure of a BmPBP–bombykol complex as well as with the third helix obtained from sequence-based secondary structure prediction with the program PHDsec (23) (Fig. 2). A prominent feature of the structure is the location of the C-terminal helix α7 in the hydrophobic core (Fig. 1b). The helix α7 is framed by the four helices α3a, α4, α5, and α6 and is covered by the loop of residues 56–73 that connects the helices α3a and α4 and includes the short helix α3b (Fig. 3b). The positioning of α7 in BmPBPA is determined by a dense network of more than 100 long-range NOEs between protons of all residues in the helix α7 and protons of 26 residues located in the helices α3, α4, α5, and α6 (Fig. 4a). Overall, BmPBPA thus has a well defined tertiary structure, which is in contrast to previous suggestions that BmPBP was unfolded at pH = 4.5 (12).
Figure 1.
Structure of BmPBPA. (a) A stereo view of the polypeptide backbone fold of the NMR structure of BmPBPA represented by a bundle of 20 energy-minimized dyana conformers. The superposition is for minimal rmsd of the backbone heavy atoms of residues 8–142 (magenta). The disordered residues 1–7 are indicated in gray. The chain ends are identified by the letters N and C. (b) Ribbon drawing of one of the 20 energy-minimized conformers, which was obtained after a rotation by about 90° relative to the orientation shown in a. The structure contains seven α-helices (see the text and Fig. 2 for the notation α3a and α3b) that are connected by linkers with nonregular secondary structure. The sequential order of the α-helices is indicated by the numbers 1 to 7, and the N and C termini are identified. The 53-aa side chains with solvent-accessible surface area below 15%, which form the hydrophobic core of the protein, are shown as bundles of 20 conformers, similar to the presentation in a. All but the first residue of helix α7 belong to this group and are indicated in yellow, whereas the other solvent-inaccessible side chains are drawn in red. The three disulfide bridges Cys-19–Cys-54, Cys-50–Cys-108, and Cys-97–Cys-117 are drawn in green for one of the 20 conformers. This and all other figures showing molecular models of BmPBP were generated with the program MOLMOL (26).
Figure 2.
Secondary structure information from sequential and medium-range NOEs for BmPBPA. The sequential NOE connectivities are indicated with thick or thin lines for strong or weak NOEs, respectively. The medium-range connectivities are shown by lines starting and ending at the positions of the residues related by the NOE. The locations of α-helices in BmPBPA are indicated by red boxes, the helical regions predicted by the secondary structure prediction program phdsec (23) from the BmPBP amino acid sequence by yellow boxes, and the α-helices in the crystal structure of a BmPBP–bombykol complex (14) by green boxes. We have chosen the notations α1a and α1b for the BmPBP–bombykol complex and α3a and α3b for BmPBPA because of the overlap with α1 and α3 in the other two data sets.
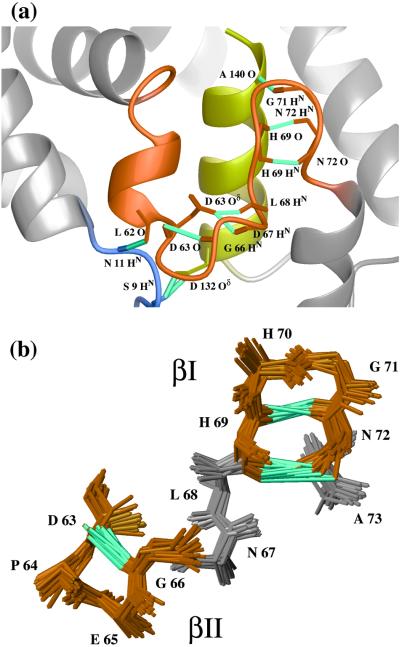
Figure 3.
Two close-up views of features in BmPBPA that differ from the conformation of the corresponding polypeptide segments in the crystal structure of a BmPBP–bombykol complex (14). (a) Surroundings of the C-terminal helix α7. The peptide segment linking α3a and α4 (residues 56–73), which includes the helix α3b, is indicated in orange, the residues 9–11 in blue, and the helix α7 with residues 131–142 in yellow. Hydrogen bonds identified in the loop region are indicated in cyan. (b) Close-up view of the loop region of residues 63–73, represented by a bundle of 20 energy-minimized dyana conformers. The backbone heavy atoms, HN and Cβ, have been locally superimposed for best fit, yielding a backbone rmsd for this region of 0.4 Å. A type II β turn of residues 63–66 and a type I β turn of residues 69–72 are indicated in orange, and the remaining residues are shown in gray. The hydrogen bonds are drawn in cyan.
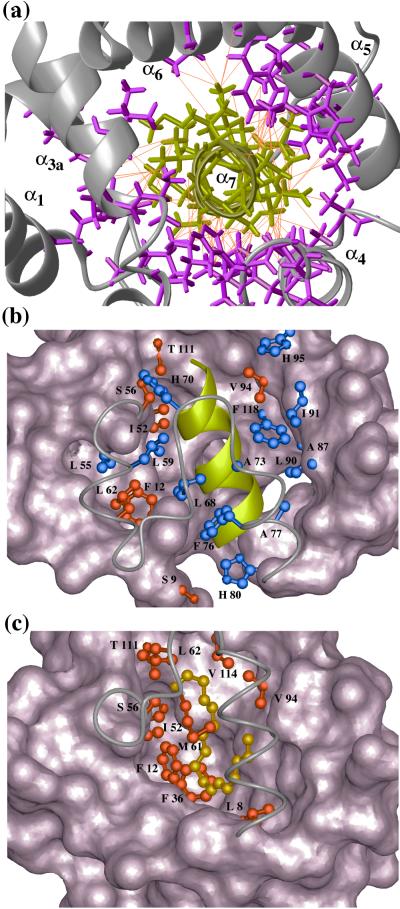
Figure 4.
Close-up views of the pheromone-binding site in a BmPBP–bombykol complex (14) and the corresponding molecular region of unliganded BmPBPA. (a) NOE network in the potential ligand-binding region in BmPBPA, viewed in the direction of the axis of the helix α7 (same viewing angle as in Fig. 1b). In total, 125 upper-distance limits between residues of α7 and the rest of the protein (shown in orange) define the position of α7 in the core of BmPBPA. α7 is shown in yellow, side chains from the remainder of the protein are magenta, and the backbone is gray. (b) View of the hydrophobic core of BmPBPA in a direction perpendicular to the axis of helix α7. The bottom of the cavity, with this viewing angle, is drawn as a space-filling surface. The cover of the cavity formed by residues 56–79 is drawn by a gray line representing the polypeptide backbone. The C-terminal helix α7 is represented by a yellow ribbon. Side chains that have heavy atoms within a distance of 4.0 Å to any atom in helix α7 are drawn as ball-and-stick models. Among these, the side chains that also show contacts to bombykol in the crystal structure of the BmPBP–bombykol complex are highlighted in red, whereas the other drawn side chains are blue. (c) Similar presentation as b of the crystal structure of the BmPBP–bombykol complex. The side chains that have heavy atoms closer than 4.0 Å to any pheromone atom are drawn in red (Ser-9 is not visible). The pheromone is drawn as a gold-colored ball-and-stick-model. Comparison of b and c shows particularly clearly that the position of the helix α7 in BmPBPA corresponds to the location of the pheromone in the structure of the BmPBP–bombykol complex described by Sandler et al. (14).
The Cβ chemical shifts of the six cysteinyl residues are equal to or larger than 35 ppm (16), showing that all of the cysteines are oxidized (24). The disulfide bonds determined chemically (25) are consistent with the network of NOE distance constraints. The three-dimensional arrangement of the helices is anchored by the three disulfide bonds as well as by a large number of noncovalent hydrophobic contacts. Two disulfide bonds, Cys-19–Cys-54 and Cys-50–Cys-108, anchor the relative positions of the helices α1, α3a and α5, and the third disulfide bond, Cys-97–Cys-117, connects α5 with α6 (Fig. 1b).
The conformation of the loop linking the helices α3a and α4 is defined by a dense network of long-range and medium-range NOE constraints, and hydrogen bonds were detected that stabilize its conformation (Fig. 3). The first half of the loop with residues 63–66 contains a classic type II β turn with a standard i,i+3 hydrogen bond, Gly-66 HN–Asp-63 O (Fig. 3b), as well as two backbone–side chain hydrogen bonds, Asn-67 HN–Asp-63 Oδ1 and Leu-68 HN–Asp-63 Oδ1 (Fig. 3a). A type I β turn is formed by the residues 69–72, which cover the C-terminal end of helix α7. This β turn is defined by the two hydrogen bonds His-69 HN–Asn-72 O and Asn-72 HN–His-69 O (Fig. 3b), and its position relative to the helix α7 is fixed by a third hydrogen bond, Gly-71 HN–Ala-140 O (Fig. 3a).
Two features highlighted in Fig. 3a show local differences between BmPBPA and the protein conformation in the crystal structure of the BmPBP–bombykol complex (14). First, the N-terminal part of the α1 helix seen in the crystal structure is replaced by an extended conformation in the NMR structure of BmPBPA, with long-range hydrogen bonds to the helices α3b, Asn-HN 11–Leu-62 O, and α7, Ser-9 HN–Asp-132 Oδ1 and Ser-9 HN–Asp-132 Oδ2. Second, in BmPBPA, the peptide segment corresponding to the C-terminal turn of the helix α3 in the crystal structure is replaced by a kinked turn connecting α3a and α3b, which has not been identified as part of a regular helical structure by the program MOLMOL (26).
The molecular surface of BmPBPA contains 14 glutamates, nine aspartates, 14 lysines, and one arginine, which are all charged at pH 4.5, and there are no hydrophobic patches on the surface. This surface structure provides a rationale for the observation that BmPBP is highly soluble and monomeric in aqueous media at physiological concentrations and low pH (12).
Helix α7 Fills the Binding Site of BmPBP.
The differences between the protein conformations in the unliganded solution structure of BmPBPA and the crystal structure of the BmPBP–bombykol complex (14) are most pronounced in the region of helix α1, which is N-terminally elongated in the structure of the complex (helix α1a), and in the C-terminal helix α7, which is present only in BmPBPA but has been indicated also by secondary structure prediction with the program PHDsec (23) (Fig. 2). The four helices α1, α3, α5, and α6 form the bombykol-binding cavity of the protein in the structure of the complex and occur in closely similar positions in BmPBPA. In the corresponding segments of α1b, α3a, α5, and α6 in the two structures, the backbone atoms can be superimposed with an rmsd of 1.1 Å, and superposition of only the backbone atoms of α3a, α5, and α6 results in an even better fit, with an rmsd of 0.6 Å. The most exciting observation is that the helix α7 in the hydrophobic core of BmPBPA (Fig. 1b) occupies a position that corresponds to the pheromone-binding site in the crystal structure of the BmPBP–bombykol complex (Fig. 4). Of the 20 residues in the A form that show contacts to helix α7 (Fig. 4b), the seven residues Ser-9, Phe-12, Ile-52, Ser-56, Leu-62, Val-94, and Thr-111 also show contacts to the pheromone in the crystal structure of the BmPBP–bombykol complex. Of these seven residues, Phe-12, Ile-52, Ser-56, and Val-94 are highly conserved among lepidopteran OBPs, which independently supports that they may be important for the mechanism of pheromone binding and release. The three residues Leu-8, Phe-36, and Met-61, which show direct contacts to bombykol in the crystal structure of the BmPBP–bombykol complex (Fig. 4c), do not contact helix α7 in the solution structure of the A form. Among these, Leu-8 and Phe-36 are highly conserved in lepidopteran OBPs, whereas position 61 shows little conservation across this class of proteins.
A Mechanism for Intramolecular Regulation of Protein Function.
The presently described differences between the solution structure of BmPBPA and the BmPBP conformation in the crystal structure of the BmPBP–bombykol complex imply that the previously described pH-dependent conformational change of BmPBP is related to the mechanism of pheromone binding and release. The BmPBP molecular architecture contains a rigid scaffold of the helices α3a, α4, α5, and α6, which is nearly invariant between the free and the ligand-bound protein, whereas the N-terminal decapeptide segment and the C-terminal dodecapeptide segment show extensive conformational changes between the solution structure of BmPBPA and the crystal structure of the BmPBP–bombykol complex (Fig. 3). An N-terminal helix of residues 1–13 forms one edge of the binding pocket for bombykol in the structure of the complex (14), whereas most of this segment is flexibly disordered in the solution structure of BmPBPA. A C-terminal helix α7 occupies the pheromone-binding site in the hydrophobic core of BmPBPA, whereas this segment is in an extended conformation on the surface of the BmPBP–bombykol complex. These observations explain the absence of pheromone binding at pH values below 5.0 (12) and indicate a likely passage way for the pheromone into its binding site.
Sandler et al. (14) proposed that the three residues His-69, His-70, and His-95 might contribute to the pH-dependent conformation change of BmPBP (7). Comparison of BmPBPA with the protein conformation in the crystal structure of the complex reveals that the histidine side chains are more widely separated in BmPBPA. This would reduce the charge repulsion resulting from histidine protonation at slightly acidic pH values and could thus destabilize the structure of the bombykol–BmPBP complex (14) in favor of BmPBPA. If one accepts the hypothesis that the pH near the membrane surface is lower than the pH value of 6.5 measured in the bulk sensillar lymph (27), these structural data would present a rationale for destabilization of the BmPBP–bombykol complex near the membrane-standing pheromone receptor, which would lead to ejection of the ligand and make it available to the receptor.
Overall, the present NMR structure determination of BmPBPA suggests an intramolecular regulation mechanism of protein function. It offers a plausible view of pheromone uptake, binding, release, and transfer to the membrane-standing receptor by the pheromone-binding protein of B. mori, which has long served as a paradigm for insect olfaction (28).
Acknowledgments
We thank Dr. Jon Clardy for kindly providing the coordinates of the crystal structure of the BmPBP–bombykol complex before release. This work was supported in part by the Schweizerischer Nationalfonds (Project 31.49047.96), the Komission für Technologie und Inovation KTI (Project Nr. 3392.1), and the Japanese Program for Promotion of Basic Research Activities for Innovative Biosciences.
Abbreviations
- OBP
odorant-binding protein
- PBP
pheromone-binding protein
- BmPBP
B. mori pheromone-binding protein
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- rmsd
rms deviation
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1GM0).
References
- 1.Hansson B S. Experientia. 1995;51:1003–1027. [Google Scholar]
- 2.Kaissling K E, Keil T A, Williams J L D. J Insect Physiol. 1991;37:71–78. [Google Scholar]
- 3.Steinbrecht R A. Ciba Found Symp. 1996;200:158–174. doi: 10.1002/9780470514948.ch13. [DOI] [PubMed] [Google Scholar]
- 4.Pelosi P, Maida R. Comp Biochem Physiol B. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- 5.Prestwich G D. Bioorg Med Chem. 1996;4:505–513. doi: 10.1016/0968-0896(96)00033-8. [DOI] [PubMed] [Google Scholar]
- 6.Maida R, Proebstl T, Laue M. Chem Senses. 1997;22:503–515. doi: 10.1093/chemse/22.5.503. [DOI] [PubMed] [Google Scholar]
- 7.Damberger F, Nikonova L, Horst R, Peng G, Leal W S, Wüthrich K. Protein Sci. 2000;9:1038–1041. doi: 10.1110/ps.9.5.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prestwich G D. Protein Sci. 1993;2:420–428. doi: 10.1002/pro.5560020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briand L, Nespoulous C, Huet J C, Takahashi M, Pernollet J C. Eur J Biochem. 2001;268:752–760. doi: 10.1046/j.1432-1327.2001.01927.x. [DOI] [PubMed] [Google Scholar]
- 10.Leal W S. Pure Appl Chem. 2001;73:613–616. [Google Scholar]
- 11.Nagnan-Le Meillour P, Huet J C, Maïbèche M, Pernollet J C, Descoins C. Insect Biochem Mol Biol. 1996;26:59–67. doi: 10.1016/0965-1748(95)00062-3. [DOI] [PubMed] [Google Scholar]
- 12.Wojtasek H, Leal W S. J Biol Chem. 1999;274:30950–30956. doi: 10.1074/jbc.274.43.30950. [DOI] [PubMed] [Google Scholar]
- 13.Leal W S. Biochem Biophys Res Commun. 2000;268:521–529. doi: 10.1006/bbrc.2000.2158. [DOI] [PubMed] [Google Scholar]
- 14.Sandler B H, Nikonova L, Leal W S, Clardy J. Chem Biol. 2000;7:143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- 15.Bartels C, Xia T H, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 16.Horst R, Damberger F, Peng G, Nikonova L, Leal W S, Wüthrich K. J Biomol NMR. 2001;19:79–80. doi: 10.1023/a:1008394615895. [DOI] [PubMed] [Google Scholar]
- 17.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 18.Nilges M, Macias M J, O'Donoghue S I, Oschkinat H. J Mol Biol. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- 19.Güntert P, Billeter M, Ohlenschläger O, Brown L R, Wüthrich K. J Biomol NMR. 1998;12:543–548. doi: 10.1023/A:1008391403193. [DOI] [PubMed] [Google Scholar]
- 20.Luginbühl P, Güntert P, Billeter M, Wüthrich K. J Biomol NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 21.Koradi R, Billeter M, Güntert P. Comput Phys Commun. 2000;124:139–147. [Google Scholar]
- 22.Cornell W D, Cieplak P, Bayly I, Gould I R, Merz K M, Ferguson D M, Spellmeyer T M, Caldwell J W, Kollman P A. J Am Chem Soc. 1996;118:2309–2309. [Google Scholar]
- 23.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 24.Wishart D S, Bigam C G, Yao J, Abildgaard F, Dyson J H, Oldfield E, Markley J L, Sykes D. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 25.Leal W S, Nikonova L, Peng G H. FEBS Lett. 1999;464:85–90. doi: 10.1016/s0014-5793(99)01683-x. [DOI] [PubMed] [Google Scholar]
- 26.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 27.Kaissling K E, Thorson J. In: Receptors for Neurotransmitters, Hormones and Pheromones in Insects. Sattelle D B, Hall L M, Hildebrand J G, editors. Amsterdam: Elsevier; 1980. pp. 261–282. [Google Scholar]
- 28.Schneider D. In: Mechanisms in Insect Olfaction. Payne T L, Birch M C, Kennedy C E J, editors. Oxford, U.K.: Clarendon; 1986. pp. 189–191. [Google Scholar]
- 29.Morris A L, MacArthur M W, Hutchinson E G, Thornton J M. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]