Abstract
Resistance to chemotherapeutic drugs leads to a poor prognosis in gastric cancer (GC). The present study aimed to assess the association between pituitary homeobox paired homeodomain transcription 1 (PITX1) expression and the sensitivity of GC cells to the chemotherapeutic drugs 5-fluorouracil (5-FU) and cisplatin (CDDP). In the present study, the gastric cancer cell lines GES-1, AGS, BGC-823, MCG-803 and SGC-7901 were used. The expression of PITX1 was determined via reverse transcription-quantitative polymerase chain reaction in GC cell lines. AGS and BGC-823 cells, which exhibit a decreased PITX1 expression, were transfected with a PITX1 cDNA construct and its control vector. MCG-803 and SGC-7901 cells, which exhibit an increased PITX1 expression, were transfected with siRNA against PITX1 and its control scramble sequence. A Cell Counting kit-8 assay was performed to determine the impact of PITX1 expression on the sensitivity of GC cells to 5-FU and CDDP. The Cancer Genome Atlas database was used to analyze the expression of PITX1 with GC prognosis in the Asian population and to assess the potential mechanism of PITX1 in 5-FU and CDDP resistance. The results revealed that the overexpression of PIXT1 increased the sensitivity of GC cells to 5-FU/CDDP. The combination of 5-FU/CDDP and PITX1 overexpression also reduced the proliferation of GC cells. Additionally, PIXT1 knockdown decreased the sensitivity of GC cells to 5-FU/CDDP. TCGA data revealed that a lower expression of PITX1 is exhibited in Asian GC patients than in normal individuals. GC patients with a lower expression of PITX1 had a poor prognosis. The expression of PITX1 affected the sensitivity of GC cells to 5-FU/CDDP, indicating that PITX1 may increase the efficacy of treatment in GC patients.
Keywords: gastric cancer, pituitary homeobox paired homeodomain transcription 1, 5-fluorouracil, cisplatin, prognosis
Introduction
Although the incidence and mortality rates of gastric cancer (GC) are slowly declining due to the reduction of Helicobacter pylori infection rate and improvements in socioeconomic conditions (1), GC still poses a threat to public health, with >50% of cases occurring in Asia (2). Surgery remains the primary curative therapy for early gastric cancer; however, chemotherapy can improve the outcome of resectable GC (3). 5-fluorouracil (5-FU) is widely used for the treatment of GC as it exerts an anticancer effect by inducing apoptosis (4,5). Cisplatin (CDDP) is one of the most effective broad-spectrum anticancer drugs for GC, which functions by forming cross-links that activate apoptotic pathways causing cell death (6). However, frequent recurrences following surgical and medical therapies has resulted in the unsuccessful prognosis of patients with advanced GC (7). Furthermore, there are no effective biomarkers of GC that predict prognosis or treatment outcome, which may be a reason for the low survival rate or occurrence of unsuccessful treatment. Therefore, it is of great importance to assess novel molecular targeting agents for the improvement of patient prognosis, particularly in combination with cytotoxic agents.
Pituitary homeobox paired homeodomain transcription (PITX1) was originally considered to be a bicoid-associated transcription factor that was involved in the transcription of the proopiomelanocortin gene in the adult pituitary. However, PITX1 may serve a role in pituitary cell differentiation and pituitary formation (8). In addition, PITX1 is essential for limb development. The misexpression of PITX1 in the forelimb results in the transformation and translocation of specific muscles, tendons and bones such that they acquire a hindlimb-like morphology (9). Therefore, PITX1 is a key component for the identification and structure of the hindlimb (10,11).
PITX1 has been revealed to be a tumor suppressor that functions by downregulating the Ras pathway (12). Various studies have also indicated that PITX1 expression is downregulated in certain types of malignant cancer, including oral squamous cell carcinoma (13), esophageal (14), gastric (15,16), lung (17), breast (18), hepatic (19), colorectal (20), pancreatic (21) and prostatic cancer (22), as well as malignant melanoma (16,23). The strong expression of PITX1 is associated with a favorable outcome in human osteosarcoma (24). PITX1 expression also serves as a novel biomarker for the prediction of patient prognosis in oral epithelial dysplasia (25) and may be a predictive biomarker of human head and neck squamous cell carcinoma chemosensitivity (26). However, the association between PITX1 expression and GC patient survival remains unclear. Additionally, whether the expression of PITX1 is associated with the sensitivity of GC cells to chemotherapy is poorly understood.
The present study assessed the effect of PITX1 on the sensitivity of GC cells to 5-FU and CDDP. The results revealed that increased PITX1 enhanced the sensitivity of GC cells (AGS and BGC-823) to 5-FU and CDDP. The knockdown of PITX1 weakened the sensitivity of GC cells (MCG-803 and SGC-7901) to 5-FU and CDDP. These results may provide a potential therapeutic target for GC in future studies.
Materials and methods
GC cell lines
GC cell lines (AGS, BGC-823, MCG-803 and SGC-7901) and a gastric epithelial cell line (GES-1) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in RPMI-1640 medium (Wisent, Inc., St Bruno, QC, Canada) supplemented with 10% fetal bovine serum (Wisent, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified incubator with 5% CO2 at 37°C.
Plasmid construction and PITX1 small interfering (si)RNA
PITX1 cDNA was isolated via reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and then cloned into the HindIII/EcoRI sites of pcDNA3.1. The primer sequences were as follows: PITX1-pcDNA3.1-HindIII, 5′-CCCCAAGCTTATGGACGCCTTCAAGGGGG-3′ and PITX1-pcDNA3.1-EcoRI, 5′-GGTGGAATTCGGCGGTCAGCTGTTGTACTGG-3′.
PITX1 siRNA and a scrambled/non-targeting siRNA were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: PITX1 siRNA1 forward, 5′-AUCGCCGUGUGGACCAACCUCACCGAGCC-3′ and reverse, 5′-GGCUCGGUGAGGUUGGUCCACACGGCGAU-3′; PITX1 siRNA2 forward, 5′-CACUUCACAAGCCAGCAGUUGCAAGAGCU-3′ and reverse, 5′-AGCUCUUGCAACUGCUGGCUUGUGAAGUG-3′; Scrambled/nontargeting siRNA forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′.
Cell transfection
AGS and BGC-823 cells were transiently transfected with 2 µg of the control pcDNA3.1 (Guangzhou RiboBio Co., Ltd., Guangzhou, China) or PITX1-pcDNA3.1 using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. PITX1 siRNA and negative control siRNA were synthesized and purified by Shanghai GenePharma Co., Ltd. (Shanghai, China) as aforementioned and transfected into cells at a final concentration of 50 nM using Lipofectamine 2000 according to the manufacturer's protocol. After the gastric cancer cells were transfected in 6-well plates for 48 h, transfection efficiency was evaluated and subsequent experiments were performed.
RT-qPCR
Total RNA from the gastric cancer cell lines GES-1, AGS, BGC-823, MCG-803 and SGC-7901 was isolated using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). First strand cDNA was synthesized from 1 µg of total RNA using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) with the StepOne Plus system (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Each PCR was performed in triplicate and β-actin was used as the internal control. The thermocycling conditions were as follows: Pre-denaturation at 95°C for 5 min, denaturation at 98°C for 10 sec, annealing at 59°C for 5 sec, extension at 72°C for 2 min and 35 cycles. The relative expression of target RNA was evaluated via the comparative 2−ΔΔCq method (27). The primer sequences of each gene were as follows: PITX1 forward, 5′-TCCACCAAGAGCTTCACCTT-3′ and reverse, 5′-CGGTGAGGTTGTTGATGTTG-3′; β-actin forward, 5′-AAAGACCTGTACGCCAACAC-3 and reverse, 5′-GTCATACTCCTGCTTGCTGAT-3.
Cell viability and proliferation assays
The 30% inhibitory concentration (IC30) is a measure of the potency of a substance in inhibiting a specific biological or biochemical function and it represents the concentration of a drug that is required for 30% inhibition in vitro. To calculate this value for 5-FU and CDDP, AGS, BGC-823, MCG-803 and SGC-7901 cells were seeded into 96-well plates at a density of 1,000 cells/well, treated with 5-FU (0, 25, 50, 100, 200, 400 or 800 µM) and CDDP (0, 1, 2.5, 5, 7.5, 10, 15 or 20 µM and cultured for 48 h to determine the appropriate concentration for subsequent experiments. Subsequently, 10 µl CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) solution was added to each well of the plate and incubated at 37°C for 3 h. AGS or BGC-823 cells (1,000 cells per well) were plated and transiently transfected with the pPITX1 construct. MCG-803 and SGC-7901 cells (1,000 cells/well) were transiently transfected with PITX1 siRNA in 96-well plates. Cell proliferation was determined using a Cell Counting Kit-8 (CCK-8) assay for 4 days (Dojindo Molecular technologies, Inc.). Optical density was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and each experiment was performed in triplicate. Cells (3.5×103 per well) were plated in 96-well plates in triplicate at 37°C for 24 h. The cells were then incubated with 5-FU (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany; AGS in 25 µM, BGC-823 in 25 µM, MCG-803 in 12.5 µM and SGC7901 in 6.25 µM) or CDDP (Sigma-Aldrich; Merck KGaA; AGS in 20 µM, BGC-823 in 5 µM, MCG-803 in 2.5 µM and SGC-7901 in 2.5 µM) at 37°C for 24 h. Following treatment at the indicated times, CCK-8 was added to the culture for 3 h prior to the measurement of absorbance at 450 nm. Each experiment was performed in triplicate.
Bioinformatics analysis of RNA sequencing data from The Cancer Genome Atlas
The Cancer Genome Atlas (TCGA) dataset (http://ualcan.path.uab.edu/analysis.html), a large cancer dataset with high-throughput sequencing data for protein coding genes, was used to assess the expression of PITX1 mRNA in stomach adenocarcinoma based on race. The Bioconductor package edgeR (https://www.bioconductor.org/packages/release/bioc/html/edgeR.html) was used to compute the P-values and fold changes of the RNA-sequence read count data in the R platform (version 3.2.3; http://www.r-project.org/). PITX1 expression was extracted from HTSeq-kilobase per million mapped reads (FPKM) data (https://portal.gdc.cancer.gov/), excluding those patients lacking complete clinical information or with low-quality data. A box plot was used to present the differential expression of PITX1 (PITX1 mRNA levels with an FPKM value ≤17.1 were considered as low-expression) between 87 Asian GC tissues and 34 normal gastric tissues. The 60-month survival time was assessed from the TCGA data.
Bioinformatics analysis obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG)
The correlation between the expression level of PITX1 and each protein coding gene (PCG) was calculated using two-sided Pearson correlation coefficients and the z-test. PCGs positively or negatively correlated with PITX1 were considered as PITX1-associated PCGs (r≥0.4 and P<0.01). KEGG pathway analysis (28) of PITX1-related PCGs was analyzed using the clusterProfiler R package (https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html), with P-value <0.05 and adjusted P-value <0.05.
Statistical analysis
The proliferation of the single reagent and combined therapy group was compared using a paired Student's t-test. An independent Student's t-test was used to compare the results (expressed as the mean ± standard deviation) between any two preselected groups. One-way analysis of variance, followed by a Dunnett's test, were performed to compare data between three or more groups. P<0.05 was considered to indicate a statistically significant difference. Patients were divided into low or high PITX1 expression groups according to the PITX1 median value. P-values for survival curves were determined from the Kaplan-Meier survival curves by use of the log-rank test. The correlation between the expression of PITX1 and each protein-coding gene was assessed by performing a two-sided Pearson correlation coefficient test and a z-test. If the protein-coding gene positively or negatively correlated with PITX1, it was considered to be a PITX1-associated protein-coding gene.
Results
Expression analysis of PITX1 mRNA in GC cell lines
To select the appropriate cell model for each experiment, levels of PITX1 were determined in several GC cell lines. RT-qPCR was performed to analyze PITX1 expression (differentiated fold) between four GC cell lines and one normal gastric mucosa cell line (GES-1). The expression of PITX1 in AGS and BGC-823 was significantly lower compared with GES-1. Additionally, PITX1 was significantly increased in the MCG-803 cell line and markedly higher in the SGC-7901 cell line compared with the GES-1 cell line (Fig. 1).
Figure 1.

The expression of PITX1 in gastric cancer cell lines determined via reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. GES-1. PITX1, pituitary homeobox paired homeodomain transcription 1.
Establishment of the IC30 of each cell line treated with 5-FU and CDDP
To determine the cytotoxic activity of 5-FU and CDDP as single agents, the cell viability of four GC cell lines treated with 5-FU and CDDP was determined (Fig. 2A and B, respectively). The cells were treated with single 5-FU and CDDP treatments at different concentrations. The results revealed that the cell viability of each experimental group was markedly inhibited by 5-FU and CDDP in a dose-dependant manner. In addition, the IC30 of 5-FU in AGS, BGC823, MCG-803 and SGC-7901 was 21.6, 27.7, 15.7 and 5.9 µM, respectively. Therefore, the concentrations of 5-FU at 25 µM in AGS, 25 µM in BGC-823, 12.5 µM in MCG-803 and 6.25 µM in SGC-7901 were selected to perform the following experiments. Similarly, the IC30 of CDDP in AGS, BGC823, MCG-803 and SGC-7901 was 17.8, 4.4, 2.7 and 3.0 µM, respectively. The selected concentrations of CDDP were 20 µM in AGS, 5 µM in BGC-823, 2.5 µM in MCG-803 and 2.5 µM in SGC-7901.
Figure 2.
The viability of four gastric cancer cell lines was determined following treatment with different concentrations of 5-FU and CDDP. The viability of four gastric cancer cells treated with different concentration of (A) 5-FU and (B) CDDP was measured using a Cell Counting Kit-8 assay. 5-FU, 5-fluorouracil; CDDP, cisplatin; IC30, 30% inhibitory concentration.
Overexpression of PITX1 enhances the sensitivity of AGS and BGC-823 to 5-FU
To assess the effect of PITX1 on the chemosensitivity of GC cells, PITX1 was overexpressed in AGS and BGC-823. The expression of PITX1 was significantly increased in AGS and BGC-823 cells transfected with pcDNA-PITX1 compared with that of pcDNA3.1-transfected cells (Fig. 3A). Cells were then treated with 5-FU, which is clinically used in the treatment of gastrointestinal cancer. In AGS and BGC-823 cells, proliferation was measured via a CCK-8 assay in 5 groups: The control group (pcDNA 3.1), the 5-FU treatment group (5-FU), the overexpression of PITX1 group (pPITX1), the group treated with empty pcDNA3.1 vector+5-FU (pcDNA 3.1+5-FU) and 5-FU treatment combined with the overexpression pPITX1 group (pPITX1+5-FU). In AGS and BGC-823 cells, it was demonstrated that cell proliferation was inhibited in the 5-FU, pPITX1, pcDNA3.1+5-FU and pPITX1+5-FU groups compared with the pcDNA3.1 group (Fig. 3B). Furthermore, compared with 5-FU treatment alone, pPITX1+5-FU treatment inhibited proliferation significantly. These results indicate that the overexpression of PITX1 enhances GC cell line sensitivity to 5-FU treatment.
Figure 3.
PITX1 overexpression affects the sensitivity of AGS and BGC-823 cell lines to 5-FU. (A) The expression of PITX1 was detected in AGS-pPITX1 cells via western blotting and RT-qPCR, and in BGC-823-pPITX1 cells via RT-qPCR. PITX1 expression was normalized to that of endogenous β-actin. (B) A Cell Counting kit-8 assay was utilized to assess the proliferation of AGS and BGC-823 cells treated with 5-FU, the pPITX1 construct, the 5-FU+pcDNA3.1 vector, the 5-FU+pPITX1 construct and the pcDNA3.1 vector. *P<0.05. All experiments were performed in triplicate and repeated at least three times. PITX1, pituitary homeobox paired homeodomain transcription 1; 5-FU, 5-fluorouracil; pPITX1, overexpression of PITX1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; OD, optical density.
Silenced PITX1 impairs the sensitivity of MCG-803 and SGC-7901 cells to 5-FU
PITX1 expression was determined via the transient transfection of PITX1 siRNA in MCG-803 and SGC-7901 cells (Fig. 4A). In the absence of 5-FU, PITX1 knockdown (siPITX1 group) significantly promoted the proliferation of MGC-803 cells compared with the control. Similarly, PITX1 knockdown significantly increased the proliferation of SGC-7901 cells. The other 3 groups (5-FU, siCtrl+5-FU and siPITX1+5-FU) exhibited a lower proliferation in MCG-803 and SGC-7901 cell lines compared with the control (Fig. 4B). Furthermore, the proliferation of MCG-803 and SGC-7901 cells treated with siPITX1+5-FU were not significantly inhibited compared with the 5-FU group. The results revealed that the silencing of PITX1 in GC cells attenuated the sensitivity of GC cells to 5-FU treatment.
Figure 4.
Silenced PITX1 reduces the sensitivity of MCG-803 and SGC-7901 cells to 5-FU. (A) The expression of PITX1 was detected via western blotting and RT-qPCR in MCG-803 cells. SGC-7901 PITX1 expression was also determined via RT-qPCR The PITX1 mRNA level was normalized to that of endogenous β-actin. (B) A Cell Counting kit-8 assay was performed to analyze the proliferation of MCG-803 and SGC-7901 cells treated with 5-FU, siPITX1, siCtrl+5-FU, siPITX1+5-FU and siCtrl. *P<0.05. All experiments were performed in triplicate and repeated at least three times. PITX1, pituitary homeobox paired homeodomain transcription 1; 5-FU, 5-fluorouracil; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si, small interfering RNA; Ctrl, control; OD, optical density.
Ectopic PITX1 expression increases the sensitivity of AGS and BGC-823 cells to CDDP
CDDP is also commonly used for the clinical treatment of GC. Cell proliferation was assessed via a CCK-8 assay following transfection with a PITX1 construct. The impact of PITX1 on CDDP treatment was assessed by dividing the cells into five groups. The proliferation of AGS and BGC-823 cells was markedly reduced in the pPITX1, CDDP, pcDNA3.1+CDDP and the pPITX1+CDDP group (Fig. 5A) compared with the pcDNA3.1 group. Furthermore, pPITX1+CDDP treated cells induced a lower reduction of AGS and BGC-823 cell proliferation compared with the CDDP group.
Figure 5.
Differentially expressed PITX1 affects the sensitivity of gastric cancer cell lines to CDDP. (A) A Cell Counting kit-8 assay was performed to analyze the proliferation of (A) AGS and BGC-823, and (B) MCG-803 and SGC-7901 cells treated with CDDP, a pPITX1 construct, a CDDP+pcDNA3.1 vector, a CDDP+pPITX1 construct and a pcDNA3.1 vector. *P<0.05. All experiments were performed in triplicate and repeated at least three times. PITX1, pituitary homeobox paired homeodomain transcription 1; CDDP, cisplatin; pPITX1, overexpression of PITX1; OD, optical density.
The knockdown of PITX1 weakens the sensitivity of MCG-803 and SGC-7901 cells to CDDP
Following the successful transfection of siPITX1 into MCG-803 and SGC-7901 cells the effect of downregulated PITX1 on CDDP sensitivity in these cell lines were assessed (Fig. 5B). The cells were divided into 5 groups: The control group (siCtrl), the CDDP-treated group (CDDP), the PITX1 knockdown group (siPITX1), the CDDP with scrambled/non-targeting siRNA group (siCtrl+CDDP) and the CDDP with PITX1 knockdown group (siPITX1+CDDP). Compared with the siCtrl group, it was revealed that the CDDP, siCtrl+CDDP and siPITX1+CDDP groups inhibited the proliferation of MCG-803 cells. However, the siPITX1 group significantly promoted the proliferation of MCG-803 (Fig. 5B). Furthermore, compared with the CDDP group, the inhibition of cell proliferation was reduced in the siPITX1+CDDP group. Additionally, a similar result was demonstrated in the SGC-7901 cell line (Fig. 5B). These data indicate that the knockdown of PITX1 in GC cells weakens the sensitivity of GC cells to CDDP treatment.
The decreased expression of PITX1 exhibits a negative outcome in human GC based on TCGA data
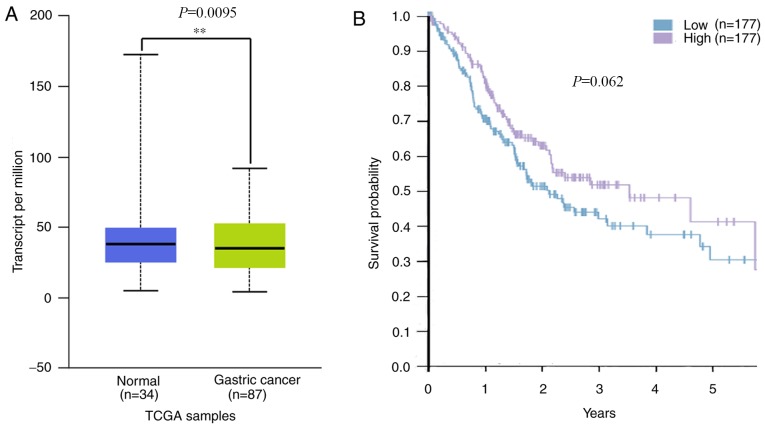
A previous study revealed that patients with GC that exhibited lower PITX1 protein levels tended to have a poorer prognosis than those with higher levels (29). To assess whether PITX1 serves a prognostic role in GC of Asian populations, mRNA levels of PITX1 in the TCGA database were analyzed. The results revealed that PITX1 was markedly decreased at the transcriptional level in the 87 Asian GC tissues compared with 34 normal gastric tissues (Fig. 6A). Although there was no significant difference in the survival rates of patients with GC between the low and the high PITX1 mRNA expression groups (Fig. 6B), the data indicates that the patient's survival was affected by the low expression of PITX1.
Figure 6.
Evaluation of PITX1 expression and the survival of patients with GC in Asian populations based on TCGA data. (A) PITX1 mRNA expression was assessed in Asian patients with GC from the TCGA database. (B) Kaplan-Meier survival curve analysis for the overall survival of Asian patients with GC separated by PITX1 expression. PITX1 mRNA levels were measured as FPKM. PITX1, pituitary homeobox paired homeodomain transcription 1; GC, gastric cancer; TCGA, The Cancer Genome Atlas; FPKM, kilobase per million mapped reads. **P<0.01.
To assess the functional similarity of genes co-expressed with PITX1, a Pearson correlation coefficient was performed. Co-expressed genes were selected at a certain cutoff threshold and then filtered to 1,620 genes by considering correlation coefficients at r≥0.4 and significances at P<0.01. A KEGG analysis was utilized to assess the potential function of PITX1 in the chemotherapeutic resistance to 5-FU and CDDP in GC cells. The majority of these co-expressed genes were revealed to participate in the regulation of necroptosis and certain genes were also associated with the p53 signaling pathway (Fig. 7A). Certain genes, which are widely thought to be involved in the necroptosis pathway, including caspase-8, Fas-associated protein with death domain (FADD), receptor-interacting serine/threonine-protein kinase 3 (RIP3) and mixed lineage kinase domain like pseudokinase (MLKL), were positively correlated with PITX1 (Fig. 7B).
Figure 7.
Kyoto Encyclopedia of Genes and Genomes analysis of PITX1 co-expressed genes based on the TCGA database. (A) Functional analysis of the genes related to PITX1 from the TCGA database according to differential gene screening. (B) The represented genes that participated in necroptosis were positively correlated with PITX1. PITX1, pituitary homeobox paired homeodomain transcription 1; TCGA, The Cancer Genome Atlas; CASP8, caspase-8; FADD, Fas-associated protein with death domain; MLKL, mixed lineage kinase domain like pseudokinase; RIP3, receptor-interacting serine/threonine-protein kinase 3; Cor, correlation.
Discussion
Although PITX1 was first reported in a study on the development of hindlimb morphology (30,31), an increasing number of reports have determined that decreased PITX1 levels in various types of human cancer serve important roles in carcinogenesis. The downregulation of PITX1 expression may contribute to the progression of cutaneous malignant melanoma by promoting cell proliferative activity (23). Furthermore, a decreased PITX1 expression has been revealed in bronchial (17,18), hepatic (19), colorectal (20), pancreatic (21), prostatic (22) and gastric malignancies (15,16). It has also been demonstrated that the level of PITX1 is correlated with patient survival; patients with higher PITX1 levels exhibit a longer median overall survival in human osteosarcoma (24). The reduced expression of PITX1 is also independently correlated with shorter patient survival and may serve as a prognostic marker in colorectal carcinoma (32). In a previous study (29), it was revealed that the expression of PITX1 is decreased in GC tissues and that the increased expression of PITX1 significantly suppresses the proliferation of GC cells and tumorigenesis in vitro and in vivo. The present study assessed the role of PITX1 expression in GC cell sensitivity to the chemotherapeutic drugs 5-FU and CDDP, which are clinically used in the treatment of gastrointestinal cancer.
The current study first assessed the connection between PITX1 expression and the sensitivity of GC cells to chemotherapy and the prognosis of patients with GC. To determine whether PITX1 expression was correlated with GC cell sensitivity to chemotherapeutic drugs, 5-FU and CDDP were utilized. 5-FU inhibits DNA synthesis and is used to treat colorectal, breast and head and neck cancer (33). CDDP is an inorganic compound that exerts cytotoxicity by inducing apoptosis (34) and is commonly administered in ovarian (35), testicular (36) and esophageal cancer (37). The present study transiently transfected a PITX1 construct into the GC cell lines, AGS and BGC-823. The results revealed that 5-FU/CDDP and PITX1 significantly suppressed GC cell proliferation. Compared with the 5-FU/CDDP treatment, pPITX1+5-FU/CDDP treatment significantly inhibited cell proliferation. These results indicated that the overexpression of PITX1 in the GC cell lines, AGS and BGC-823, enhanced the efficacy of 5-FU/CDDP treatment. Furthermore, compared with the 5-FU/CDDP group, the inhibition of cell proliferation in the siPITX1+5-FU/CDDP group was reduced, indicating that the knockdown of PITX1 in the GC cell lines, MCG-803 and SGC-7901, weakens the sensitivity of GC cells to CDDP and 5-FU treatment.
To determine the correlation of PITX1 with GC prognosis, the TCGA dataset, which consists of high-throughput sequencing data for protein-coding gene expression, was considered in the further analysis. The current study demonstrated that PITX1 mRNA expression was significantly lower in 87 Asian GC tissues than in 34 normal gastric mucous tissues. A Kaplan-Meier survival curve of patients with GC classified into 2 groups depending on the high and low expression of PITX1 from the TCGA database. The results revealed that a high and low expression of PITX1 influenced patient survival. Those with a high PITX1 mRNA expression had a lower survival than those with a low PITX1 mRNA expression. Combining the results of a previous study (29), the results indicate that patients with higher PITX1 levels have a longer survival time than those with a lower PITX1 level. The expression of PITX1 may therefore be a reliable biomarker for the prediction of GC patient prognosis. To further assess the mechanism by which PITX1 contributes to chemotherapy insensitivity, all known co-expressed genes were categorized using a KEGG analysis. A total of ~1620 target genes were screened, the biological processes of which were primarily implicated in necroptosis. The kinase RIP3, the adaptor protein FADD and the proximal initiator caspase-8, have been identified as fundamental regulators of the necroptotic cell death pathway (38–40). In addition, MLKL, a key component downstream of RIP3, is suggested to be a terminal executor of necroptosis (41). Previous studies also revealed that the four aforementioned genes were positively correlated with necroptosis (42,43). The present study hypothesized that PITX1 enhances the cytotoxicity of 5-FU and CDDP in GC cells partially by inducing necroptosis; however, the mechanism requires further exploration.
In summary, a high PITX1 expression increases the sensitivity of GC cell lines to 5-FU and CDDP. However, the precise molecular mechanism by which this occurs requires further study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81472548 and 81672414).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XS conceived the study and was a major contributor in writing the manuscript. GY, SY, YM and YZ contributed to cell culture and CCK-8 assay. PG was responsible for bioinformatics analysis. YL, FQ and ZZ were involved in drafting the manuscript and revising it critically for important intellectual content. HF designed the research protocols and gave final approval of the version to be published. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20:4483–4490. doi: 10.3748/wjg.v20.i16.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinou JC, Desagher S, Antonsson B. Cytochrome c release from mitochondria: All or nothing. Nat Cell Biol. 2000;2:E41–E43. doi: 10.1038/35004069. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y, Tang XM, Half E, Kuo MT, Sinicrope FA. Cyclooxygenase-2 overexpression reduces apoptotic susceptibility by inhibiting the cytochrome c-dependent apoptotic pathway in human colon cancer cells. Cancer Res. 2002;62:6323–6328. [PubMed] [Google Scholar]
- 6.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 7.Wagner AD, Syn NL, Moehler M, Grothe W, Yong WP, Tai BC, Ho J, Unverzagt S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2017;8:CD004064. doi: 10.1002/14651858.CD004064.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamonerie T, Tremblay JJ, Lanctôt C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 9.DeLaurier A, Schweitzer R, Logan M. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev Biol. 2006;299:22–34. doi: 10.1016/j.ydbio.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 10.Shang J, Li X, Ring HZ, Clayton DA, Francke U. Backfoot, a novel homeobox gene, maps to human chromosome 5 (BFT) and mouse chromosome 13 (Bft) Genomics. 1997;40:108–113. doi: 10.1006/geno.1996.4558. [DOI] [PubMed] [Google Scholar]
- 11.Shang J, Luo Y, Clayton DA. Backfoot is a novel homeobox gene expressed in the mesenchyme of developing hind limb. Dev Dyn. 1997;209:242–253. doi: 10.1002/(SICI)1097-0177(199706)209:2<242::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 13.Libório TN, Acquafreda T, Matizonkas-Antonio LF, Silva-Valenzuela MG, Ferraz AR, Nunes FD. In situ hybridization detection of homeobox genes reveals distinct expression patterns in oral squamous cell carcinomas. Histopathology. 2011;58:225–233. doi: 10.1111/j.1365-2559.2011.03751.x. [DOI] [PubMed] [Google Scholar]
- 14.Lord RV, Brabender J, Wickramasinghe K, DeMeester SR, Holscher A, Schneider PM, Danenberg PV, DeMeester TR. Increased CDX2 and decreased PITX1 homeobox gene expression in Barrett's esophagus and Barrett's-associated adenocarcinoma. Surgery. 2005;138:924–931. doi: 10.1016/j.surg.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Chen YN, Chen H, Xu Y, Zhang X, Luo Y. Expression of pituitary homeobox 1 gene in human gastric carcinogenesis and its clinicopathological significance. World J Gastroenterol. 2008;14:292–297. doi: 10.3748/wjg.14.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi DL, Ohhira T, Fujisaki C, Inoue T, Ohta T, Osaki M, Ohshiro E, Seko T, Aoki S, Oshimura M, Kugoh H. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol Cell Biol. 2011;31:1624–1636. doi: 10.1128/MCB.00470-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Knösel T, Ye F, Pacyna-Gengelbach M, Deutschmann N, Petersen I. Decreased PITX1 homeobox gene expression in human lung cancer. Lung Cancer. 2007;55:287–294. doi: 10.1016/j.lungcan.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Stender JD, Stossi F, Funk CC, Charn TH, Barnett DH, Katzenellenbogen BS. The estrogen-regulated transcription factor PITX1 coordinates gene-specific regulation by estrogen receptor-alpha in breast cancer cells. Mol Endocrinol. 2011;25:1699–1709. doi: 10.1210/me.2011-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knösel T, Chen Y, Hotovy S, Settmacher U, Altendorf-Hofmann A, Petersen I. Loss of desmocollin 1–3 and homeobox genes PITX1 and CDX2 are associated with tumor progression and survival in colorectal carcinoma. Int J Colorectal Dis. 2012;27:1391–1399. doi: 10.1007/s00384-012-1460-4. [DOI] [PubMed] [Google Scholar]
- 21.Hamidov Z, Altendorf-Hofmann A, Chen Y, Settmacher U, Petersen I, Knösel T. Reduced expression of desmocollin 2 is an independent prognostic biomarker for shorter patients survival in pancreatic ductal adenocarcinoma. J Clin Pathol. 2011;64:990–994. doi: 10.1136/jclinpath-2011-200099. [DOI] [PubMed] [Google Scholar]
- 22.Kwok SC, Liu X, Mangel P, Daskal I. PTX1(ERGIC2)-VP22 fusion protein upregulates interferon-beta in prostate cancer cell line PC-3. DNA Cell Biol. 2006;25:523–529. doi: 10.1089/dna.2006.25.523. [DOI] [PubMed] [Google Scholar]
- 23.Osaki M, Chinen H, Yoshida Y, Ohhira T, Sunamura N, Yamamoto O, Ito H, Oshimura M, Kugoh H. Decreased PITX1 gene expression in human cutaneous malignant melanoma and its clinicopathological significance. Eur J Dermatol. 2013;23:344–349. doi: 10.1684/ejd.2013.2021. [DOI] [PubMed] [Google Scholar]
- 24.Kong G, Liu Z, Wu K, Zhang Y, Deng Z, Feng W, Chen S, Wang H. Strong expression of paired-like homeodomain transcription factor 1 (PITX1) is associated with a favorable outcome in human osteosarcoma. Tumour Biol. 2015;36:7735–7741. doi: 10.1007/s13277-015-3512-1. [DOI] [PubMed] [Google Scholar]
- 25.Nakabayashi M, Osaki M, Kodani I, Okada F, Ryoke K, Oshimura M, Ito H, Kugoh H. PITX1 is a reliable biomarker for predicting prognosis in patients with oral epithelial dysplasia. Oncol Lett. 2014;7:750–754. doi: 10.3892/ol.2013.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takenobu M, Osaki M, Fujiwara K, Fukuhara T, Kitano H, Kugoh H, Okada F. PITX1 is a novel predictor of the response to chemotherapy in head and neck squamous cell carcinoma. Mol Clin Oncol. 2016;5:89–94. doi: 10.3892/mco.2016.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao F, Gong P, Song Y, Shen X, Su X, Li Y, Wu H, Zhao Z, Fan H. Downregulated PITX1 modulated by MiR-19a-3p promotes cell malignancy and predicts a poor prognosis of gastric cancer by affecting transcriptionally activated PDCD5. Cell Physiol Biochem. 2018;46:2215–2231. doi: 10.1159/000489590. [DOI] [PubMed] [Google Scholar]
- 30.Klopocki E, Kähler C, Foulds N, Shah H, Joseph B, Vogel H, Lüttgen S, Bald R, Besoke R, Held K, et al. Deletions in PITX1 cause a spectrum of lower-limb malformations including mirror-image polydactyly. Eur J Hum Genet. 2012;20:705–708. doi: 10.1038/ejhg.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Infante CR, Park S, Mihala AG, Kingsley DM, Menke DB. Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev Biol. 2013;374:234–244. doi: 10.1016/j.ydbio.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gajewska A, Herman AP, Wolinska-Witort E, Kochman K, Zwierzchowski L. In vivo oestrogenic modulation of Egr1 and Pitx1 gene expression in female rat pituitary gland. J Mol Endocrinol. 2014;53:355–366. doi: 10.1530/JME-14-0092. [DOI] [PubMed] [Google Scholar]
- 33.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 34.Marullo R, Werner E, Degtyareva N, Moore B, Altavilla G, Ramalingam SS, Doetsch PW. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PLoS One. 2013;8:e81162. doi: 10.1371/journal.pone.0081162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozols RF, Bookman MA, du Bois A, Pfisterer J, Reuss A, Young RC. Intraperitoneal cisplatin therapy in ovarian cancer: Comparison with standard intravenous carboplatin and paclitaxel. Gynecol Oncol. 2006;103:1–6. doi: 10.1016/j.ygyno.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 36.Burger AM, Double JA, Newell DR. Inhibition of telomerase activity by cisplatin in human testicular cancer cells. Eur J Cancer. 1997;33:638–644. doi: 10.1016/S0959-8049(96)00521-7. [DOI] [PubMed] [Google Scholar]
- 37.Kies MS, Rosen ST, Tsang TK, Shetty R, Schneider PA, Wallemark CB, Shields TW. Cisplatin and 5-fluorouracil in the primary management of squamous esophageal cancer. Cancer. 1987;60:2156–2160. doi: 10.1002/1097-0142(19871101)60:9<2156::AID-CNCR2820600906>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 38.Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–232. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kantari C, Walczak H. Caspase-8 and bid: Caught in the act between death receptors and mitochondria. Biochim Biophys Acta. 2011;1813:558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Fan C, Zhang H, Zhao Q, Liu Y, Xu C, Xie Q, Wu X, Yu X, Zhang J, Zhang H. MLKL and FADD are critical for suppressing progressive lymphoproliferative disease and activating the NLRP3 inflammasome. Cell Rep. 2016;16:3247–3259. doi: 10.1016/j.celrep.2016.06.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver Metzig M, Fuchs D, Tagscherer KE, Gröne HJ, Schirmacher P, Roth W. Inhibition of caspases primes colon cancer cells for 5-fluorouracil-induced TNF-α-dependent necroptosis driven by RIP1 kinase and NF-κB. Oncogene. 2016;35:3399–3409. doi: 10.1038/onc.2015.398. [DOI] [PubMed] [Google Scholar]
- 43.Ratovitski EA. Phospho-ΔNp63α-responsive microRNAs contribute to the regulation of necroptosis in squamous cell carcinoma upon cisplatin exposure. FEBS Lett. 2015;589:1352–1358. doi: 10.1016/j.febslet.2015.04.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.