Abstract
Interferons (IFNs) manifest their cellular functions by regulating expression of target genes known collectively as IFN-stimulated genes (ISGs). The repertoires of ISGs vary slightly between cell types, but routinely include a core of common ISGs robustly upregulated in most IFN-treated cells. Here, we review the regulation and cellular functions of 2 related ISGs, ISG12 (IFI27) and G1P3 (ISG 6–16), that are commonly induced by IFNs in most, if not all, IFN-responsive cells. On the basis of sequence similarity, they are grouped together within the newly defined FAM14 family. Emerging data on ISG12 and G1P3 suggest that both are mitochondrial proteins with opposing activities on apoptosis that may influence the innate immune responses of IFNs. The G1P3 gene encodes a low molecular weight mitochondrial protein that may stabilize mitochondrial function and oppose apoptosis. In contrast, ISG12 expression may sensitize cells to apoptotic stimuli via mitochondrial membrane destabilization. On the basis of these results and differences in induction kinetics between ISG12 and G1P3, we have proposed a model for the role of these genes in mediating cellular activity of IFNs.
Introduction
Interferon-stimulated genes (ISGs) are a diverse group of ∼300 genes that are induced by interferons (IFNs) (Staeheli 1990; Sen 1991; Stark and others 1998; Leaman and others 2003; Borden and others 2007; Cheriyath and others 2007). Studies of the mode of action of several ISGs have resulted in fundamental discoveries concerning translational control, regulation of RNA stability, editing, protein transport and turnover, and a host of other cellular processes (Staeheli 1990; Sen 1991; Stark and others 1998; Biron and Sen 2006). ISG15 and G1P3 were among the first ISGs identified (Friedman and others 1984; Kelly and others 1985; Kelly and others 1986), with ISG12 mRNA subsequently identified as an ISG related to 6–16/G1P3 (Rasmussen and others 1993). Although robust induction of ISG12 and G1P3 by IFNs suggests critical functions in innate immunity, they have been less thoroughly characterized than some other ISGs. The remainder of this article will focus on the regulation of ISG12 and G1P3, emerging roles of these proteins as apoptotic modulators and will propose a model about how these proteins may act in concert to mount a cellular response to IFNs and other stimuli.
FAM14 Family: Distribution and Evolution
Genes encoding proteins similar to G1P3 and ISG12 have been grouped together to form the newly defined FAM14 gene family (Parker and Porter 2004). Members of this family encode small hydrophobic proteins with at least 1 copy of an ∼80 amino acid motif, the ISG12 motif (Fig. 1A). So far, 46 members of the FAM14 family have been identified in 25 organisms ranging from higher mammals to single-celled amoeba (Parker and Porter 2004). Phylogenic distribution of higher eukaryotic proteins that contain an ISG12 motif illustrates that, while this common motif is highly conserved, sequences outside of the ISG12 motif are more divergent (Fig. 1B) but often include short, hydrophobic stretches enriched for amino acids with small side groups. FAM14 members in humans include FAM14D (ISG12a), FAM14A (ISG12b), FAM14B (ISG12c), and FAM14C (G1P3) (Parker and Porter 2004).
FIG. 1.
Putative ISG12 domain and phylogenic classification of FAM14 family members. (A) Alignment of putative ISG12 motifs of ISG12 and G1P3. The consensus sequence for the ISG12 motif (Pfam accession no. PF06140) of ISG12 and G1P3 are aligned. Black squares represent sequence identity and gray squares represent sequence similarity. (B) Phylogenetic alignment of the FAM14 family members with putative ISG12 motif. Phylogenetic alignment of FAM14 family members was curated from Tree families database (www.treefam.org). Boxes in the protein multialignments at right correspond to the putative ISG12 domains and thin lines to nondomain regions. Dark lines or boxes represent regions of identity; open or white areas indicate gaps. Vertical dark lines indicate locations of intron–exon splicing boundaries. ISG, interferon-stimulated genes.
The hydrophobic nature of the proteins of this family implies an association with membranes, which has been confirmed for both ISG12a and G1P3 (Tahara and others 2005; Cheriyath and others 2007; Rosebeck and Leaman 2008) and is consistent with ISGs IFITM1-3. On the basis of the presence of a single ISG12-like gene in Dictostelium discoideum and multiple ISG12 genes in mammals, it has been postulated that the ISG12 family arose from an ancestral gene that underwent an initial gene duplication event to form ISG12a and ISG12b between the emergence of amoeba and the divergence of fish (Parker and Porter 2004). In mice and in cattle the ISG12b gene exists as 2 genes, ISG12b1 and ISG12b2, which appear as nonorthologs that are not found in other species (Parker and Porter 2004). Hence, ISG12b2 and ISG12c genes probably arose recently. Unlike ISG12, G1P3 is identified only in higher mammals and appears to have arisen by interchromosomal duplication just before the divergence of ungulates and primates (Parker and Porter 2004). While both human G1P3 and ISG12a are strongly induced by IFNs, neither human ISG12b nor ISG12c was IFN responsive in the fibrosarcoma cell line HT1080 despite the presence of a putative IFN-stimulated response element (ISRE) in the promoter of each (Parker and Porter 2004). The presence of primordial ISG12-like sequences in D. discoideum, which lacks an IFN system and constitutive expression of FAM14 ISGs in various cancer histologies suggests that FAM14 family members are a part of the machinery that responds to cellular or environmental stress (Parker and Porter 2004; Tahara and others 2005; Cheriyath and others 2007; Weichselbaum and others 2008). They may have later evolved as a part of the IFN system to combat specific cellular insults such as viral infection and oxidative stress.
ISG12 (FAM14D, A & B) Genes and Proteins
ISG12a was first identified as an estrogen-induced transcript in a screen for differentially regulated genes (Rasmussen and others 1993), but has subsequently been implicated as a potent ISG that is regulated by IFNs and a variety of other innate immune stimuli (Gjermandsen and others 2000; Han and others 2002; Martensen and Justesen 2004). As mentioned above, multiple genes in humans, mice, cattle, and other higher eukaryotic species encode ISG12 orthologs. The 3 human ISG12 (hISG12) genes exist in tandem on band q32 of human chromosome 14, and are designated ISG12a-c (FAM14D, A, and B, respectively). Of the human ISG12 genes, only hISG12a has been characterized as IFN inducible (Parker and Porter 2004). Three ISG12 genes also exist in the mouse (mISG12a, b1, and b2) on the syntenic mouse chromosome (m12F1), all of which appear to be IFN inducible (Parker and Porter 2004).
The IFN-regulated hISG12a (FAM14D) gene encodes a putative 122 amino acid hydrophobic protein of 12 kDa. The protein has few recognizable features outside of the namesake “ISG12 motif” (hISG12a amino acid 39–100) but does include a putative N-terminal mitochondrial targeting sequence that is absent from hISG12b (FAM14A) and hISG12c (FAM14B) (Rosebeck and Leaman 2008) (Fig. 2). The other hISG12 proteins are predicted to be 130 amino acids (hISG12b) and 104 amino acids (hISG12c) in length. These proteins share 45% and 55% amino acid identity with hISG12a, respectively, and 50% with each other (Fig. 2). In addition to the transcription of these allelic variants, each ISG12 gene may also undergo alternative splicing to yield additional protein isoforms (Smidt and others 2003). The functional significance of these allelic or splice variants is currently unclear, but appears to be a common characteristic of FAM14 members (see later).
FIG. 2.
Alignment of ISG12 isoforms. Alignment of human ISG12a, b, and c to identify consensus amino acid sequence. Black squares represent sequence identity, gray squares represent sequence similarity, and numbers indicate amino acid position. Within the consensus row, amino acids conserved in all 3 proteins are highlighted with an asterisk, while positions in which amino acids are conserved in 2 of the 3 proteins are indicated with a dot.
ISG12a Expression and Protein Localization
Low basal ISG12a expression is observed in a wide variety of cells, but robust IFN-inducibility, sometimes approaching 1,000-fold over basal expression, occurs in nearly all cell types (Rosebeck and Leaman 2008). In published gene array studies, ISG12a is consistently among the genes most highly induced by type I IFNs (Certa and others 2001; Koike and others 2003; Leaman and others 2003; Sellebjerg and others 2009). Whereas most IFN-regulated genes are induced maximally by 8 or 18 h, hISG12a mRNA continues to accumulate out beyond 96 h post-treatment even if the IFN is removed after the first 2 h (Rosebeck and Leaman 2008). The underlying reasons behind the unusual kinetics of ISG12a induction are so far unexplained, but may reflect mRNA stabilization or secondary induction by IFN-regulated factors. hISG12a mRNA is also upregulated by IFN-γ in some cell types, but is not induced by other cytokines tested, including IL2, IL6, GM CSF, or TNFα (Gjermandsen and others 2000). Although induced strongly by double-stranded RNA (Gjermandsen and others 2000; Rosebeck and Leaman 2008), hISG12a induction requires IFN feedback, as demonstrated in IFN-unresponsive U4C cells that lack the JAK1 tyrosine kinase that is required for IFN responsiveness (Rosebeck and Leaman 2008). ISG12a mRNA expression is differentially regulated in response to various experimental manipulations such as paclitaxel treatment and gain-of-function JNK1 expression in Swiss 3T3 fibroblasts (Han and others 2002; Bani and others 2004). It was also upregulated in response to viral infections and immunological disorders such as psoriasis or inflammatory bowel disease (Dooley and others 2004; Saito and others 2004; Suomela and others 2004; Budhu and others 2007). As mentioned, hISG12b and hISG12c are both constitutively expressed, but are not upregulated by IFNs in cell types that have been examined thus far (Parker and Porter 2004). Whether they are induced by other stimuli has not been examined to date.
The precise subcellular distribution of the hISG12a protein is uncertain. One report found that ectopic ISG12 protein sedimented with the nuclear envelope fraction of insect cells, and immunocytochemical studies localized IFN-induced ISG12a protein within the nuclear membrane (Martensen and others 2001). However, 2 subsequent articles have instead implicated a mitochondrial localization. Transiently expressed hISG12a and mISG12b1 proteins exhibited clear mitochondrial distributions (Rosebeck and Leaman 2008; Li and others 2009). Whether the discrepancy between the earlier study as compared to more recent work reflects differences in detection procedures or altered distribution of the various gene products or isoforms remains to be determined.
ISG12 Function
Although much is known about ISG12 expression patterns in response to immune stimuli, only a few studies have assessed ISG12 protein biological activity. One report focused on a potential role in age-dependent resistance to Sindbis viral replication in mice (Labrada and others 2002). Forced expression of murine ISG12b1 was able to protect neonatal mice from lethal viral encephalitis infection (Labrada and others 2002). Although that study did not explore the biophysical or biochemical properties of the murine ISG12b1 protein, these data implicated ISG12 as a potentially important antiviral factor (Labrada and others 2002).
The mitochondrial distribution of hISG12a and mISG12b1 suggests a role in mitochondrial function. Upon localization to the mitochondria, the N-terminal signal peptide (SP) of hISG12a is cleaved, implicating insertion into the outer or inner membrane (Rosebeck and Leaman 2008). Cell culture-based studies demonstrated that transient expression of hISG12a led to decreased viable cell numbers and enhanced sensitivity to DNA-damage induced apoptosis (Rosebeck and Leaman 2008). ISG12a expression enhanced etoposide-induced cytochrome c release, Bax activation, and loss of mitochondrial membrane potential. The proapoptotic effects of hISG12a expression were blocked by Bcl-2 coexpression or treatment with a pan-caspase inhibitor. siRNA-mediated inhibition of hISG12a prevented sensitization of cells to etoposide-induced apoptosis by either ectopic ISG12a or IFN pretreatment, thereby implicating ISG12a in this process (Rosebeck and Leaman 2008). In adipose cells, forced mISG12b1 expression led to decreased mitochondrial function, including mitochondrial biogenesis and mitochondrial gene expression (Li and others 2009). In adipose cells, ISG12b1 also inhibited adipogenic differentiation of 3T3-L1 cells. Thus, ISG12b1 and, by extension, its impact on mitochondrial function may play a critical role in adipocyte development. It will be interesting to see if any of the hISG12 variants similarly contribute to adipocyte function.
Together, the above cell-based observations implicate ISG12 as a contributing regulator of IFN-induced apoptosis, which may in turn augment or support the antiviral activities of type I IFNs. ISG12 may function to decrease mitochondrial membrane integrity and/or enhance permeability. Alternatively, it may directly affect regulated ion transport in mitochondrial or other membranes. How these biological actions contribute to IFN-dependent antiviral effects is unclear, but may involve destruction of cells that are irreparably infected or that receive sustained IFN signals. In such cases, cell death can effectively short circuit viral replication as a means of preventing further dissemination throughout the host. Alternatively, mitochondrial perturbation may interfere with one or more steps in the viral replication pathway leading to decreased progeny production. The molecular mechanism by which ISG12 might regulate cell viability in coordination with G1P3 in the course of an IFN response is still unresolved, but will be discussed in subsequent sections of this review.
G1P3 (FAM14C) Gene and Proteins
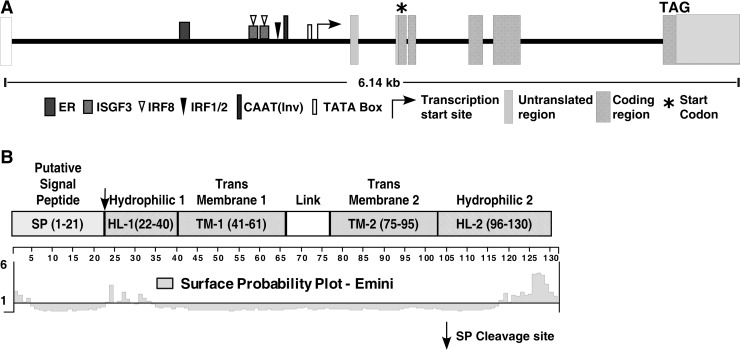
Unlike ISG12, G1P3 is encoded by a single gene and is so far identified only in higher eukaryotes (Parker and Porter 2004). The G1P3 gene is localized on chromosome 1p35 and spans ∼6 kb (Itzhaki and others 1992) (Fig. 3A). The genomic sequence of G1P3 has several distinctive features, including a minisatellite consisting of 26 tandemly repeated dodecanucleotides, a CpG island, and 5 Alu repeat sequences (Turri and others 1995). The repeat unit sequence, CAGGGTAAGGGTG, is positioned within a CpG island at the exon 2/intron 2 boundary and is similar to the consensus mammalian splice donor sequence. This repeat sequence causes 3 alternative splicing events extending exon 2 by 12 or 24 nucleotides, resulting in protein products that differ from each other by only 4–8 amino acids. The nonhypervariable nature of the minisatellite suggests a potential conserved function for this sequence, which is yet to be investigated. Although a CpG island is positioned in the center of the G1P3 gene, restriction enzyme analysis and studies with inhibitors of DNA methyl transferases in cancer cell lines suggested that G1P3 is not a methylation-silenced gene (Turri and others 1995; Cheriyath V, unpublished results). Nevertheless, the minisatellites and Alu sequences in the G1P3 gene might render this gene susceptible to homologous recombination.
FIG. 3.
Elements of G1P3 gene and protein. (A) Transcriptional and coding elements of the G1P3 gene. Graphic view of the genomic sequence of the G1P3 gene spanning ∼6.14 kb from the human genome browser. The promoter region of G1P3 is analyzed by ALLGEN promo (http://alggen.lsi.upc.es/) software. (B) Structural features of G1P3 protein. Putative mitochondrial targeting signal peptide, hydrophobic, and hydrophilic domains of G1P3. Surface probability analysis by Emini's method suggests the propensity of HL-1 and HL-2 amino acids to be exposed to solvent. Numbers in parentheses of each domain represent amino acid numbers of G1P3 protein.
G1P3 is a small protein with a molecular weight of ∼13 kDa. Primary structure analysis of G1P3 identified a 21 amino acid putative SP in its N-terminus that may target it to the mitochondria, followed by a 19 amino acid hydrophilic domain (HL-1), a 20 amino acid transmembrane region (TM-1), a linker region (L) that connects transmembrane 1 with a 20 amino acid transmembrane 2 domain (TM-2), and a 34 amino acid hydrophilic-2 (HL-2) region (Fig. 3B). Surface probability analysis with Emini's method suggested that parts of the SP and hydrophilic 1 and 2 regions have a higher propensity to be exposed to solvent (Fig. 3B). The polar HL-1 sequence is likely to form an extended coil, whereas the HL-2 domain is likely to form an amphipathic helix. Since HL-1 and HL-2 are separated by 2 transmembrane domains, they may not interact with each other to form a well-folded domain, but may instead protrude out as mostly unfolded tethers to which other proteins can bind. Using a yeast 2-hybrid system, calcium and integrin binding protein CIB, the γ-subunit of the eukaryotic cytosolic chaperonin-containing protein, TCP-1, and the calcium binding protein related S-100 (CAPL) were implicated as interacting partners of G1P3 (Tahara and others 2005). The interaction between G1P3 and CIB was further tested in vitro (Tahara and others 2005), but in vivo evidence for this interaction is lacking.
In accordance with the predicted molecular weight of G1P3, our studies have identified the molecular weight of both induced and constitutively expressed G1P3 as a 12–13 kDa protein in myeloma and breast cancer cell lines (Cheriyath and others 2007; Cheriyath and others 2010). In contrast, constitutively expressed G1P3 migrated as a 32 kDa polypeptide in TMK-1 gastric cancer cells and keratinocytes, due in part to the presence of O-linked glycosylation of serine and threonine residues (Tahara and others 2005; Szegedi and others 2010). However, the molecular weight of the IFN-induced form of G1P3 in these cell types has not yet been investigated.
G1P3 Expression
After identification of G1P3 as an ISG, the promoter element of G1P3 was used extensively in the identification of the critical components of the type I IFN signaling pathway, including the relevant STATs and JAKs (Friedman and others 1984). The promoter of G1P3 has 2 ISREs that are located tandemly within two 39-bp elements, an IRF-E and an inverted CAAT box (Porter and others 1988; Chernajovsky 1989; Dale and others 1989; Chernajovsky and Kirby-Sanders 1990) (Fig. 3). In in vitro binding assays, 2 ISGF3 factors bind cooperatively to the tandem ISREs (Li and others 1998). Additional in vitro studies of the G1P3 promoter suggested the formation of a triple-helical structure by 2 ISREs to interfere with IFN-induced transcription of G1P3 (Roy and Lebleu 1991; Roy 1993; 1994). However, in vivo evidence for the cooperative binding of ISGF3 factors and formation of triple-helical structures in IFNs treated cells is yet to be demonstrated. G1P3 induction by IFN-α2b or IFN-β is rapid (within 4 h) and is sustained in a manner similar to that observed with ISG12a, although for slightly less time (84 h versus >120 h) (Cheriyath V, unpublished results).
A number of other ligands, viruses, nuclear receptors, and disease conditions induce expression of G1P3. Among viruses, vesicular stomatitis virus (VSV), Hepatitis virus C, and human immunodeficiency virus (HIV) induce G1P3 (Martensen and Justesen 2004). Poly(I):poly(C) induced G1P3 (Kumar-Sinha and others 2002; Martensen and Justesen 2004), but like ISG12a direct induction was not observed in cells lacking IFN responses (Sun and Leaman 2005), suggesting indirect activation through IFN feedback. Other innate immune regulators, including lipopolysaccharide (LPS) and (TNF)-related apoptosis-inducing ligand (TRAIL), also induced G1P3 (Kumar-Sinha and others 2002; Martensen and Justesen 2004).
In the promyelocytic leukemia protein (PML)-positive cell line NB-4, all-trans-retinoic acid (RA), in combination with IFN-α, synergistically induced expression of G1P3 (Kumar and Korutla 1995). However, in PML-negative ME180 cells, RA failed to upregulate G1P3, despite the induction of other ISGs like IRF-1 and HLA-A2 mRNAs, suggesting the requirement of PML for RA-induced expression of G1P3 (Lancillotti and others 1995). Unlike ISG12, expression of G1P3 is not regulated by brahma-related gene (BRG1), a key component of ATP-dependent chromatin-remodeling SW12-SNF2 complex (Huang and others 2002). Basal and induced expression of G1P3 mRNA is also regulated by differentiation in some cell types. In the promyelocytic leukemia cell line HL-60, IFN-α strongly induced expression of G1P3 after granulocyte differentiation but not after macrophage differentiation or in the undifferentiated stage (Bandyopadhyay and others 1992). Additionally, expression of G1P3 in dendritic cells correlated with the stages of differentiation of these cells. Compared to immature dendritic cells expression of G1P3 and ISG12a was higher in mature dendritic cells (Hashimoto and others 2000).
A recent study implicated psoriasis susceptibility-related RNA gene induced by stress (PRINS), a noncoding RNA (ncRNA), in the regulation of G1P3 (Szegedi and others 2010). Comparative gene expression analysis of keratinocytes expressing PRINS ncRNA and PRINS knockdown in keratinocytes identified G1P3 as one of the genes induced by PRINS ncRNA (Szegedi and others 2010). These results suggest a potential role of ncRNA and miRNAs in expression of FAM14 family members or other ISGs during aberrant activation of immune system.
A search of GEO and Oncomine databases (www.oncomine.org) identified marked upregulation of G1P3 mRNA in a variety of neoplastic and autoimmune diseases (Sorbello and others 2003; Bani and others 2004; Tsai and others 2007), including malignancies with viral etiologies such as hepatocellular carcinomas, head and neck cancer, malignancies with chronic inflammation such as gastric cancers (Cheriyath V, unpublished observation), and in malignancies with endocrine deregulation such as breast and ovarian cancers. Semiquantitative RT-PCR studies of 18 primary breast tumors identified a marked correlation between expression of G1P3 with estrogen receptor positivity (Sorbello and others 2003). Additionally, in silico analysis of 2 pools of breast cancer and benign breast tissue libraries by SAGE digital gene expression displayer followed by virtual Northern and virtual microarray analysis of EST identified upregulation of G1P3 in breast cancers (Shen and others 2005). Results of these studies are in agreement with our studies in breast cancer where we identified marked upregulation of G1P3 at both RNA and protein levels in malignant epithelium relative to normal breast epithelium (Cheriyath and others 2010). We also observed the induction of G1P3 by estrogen signaling both in the ER-positive breast cancer cell line MCF-7 and in human mammary epithelial cells (Cheriyath and others 2010). In addition to the upregulation of G1P3 in malignant tissues, many cancer therapies induced its expression, including paclitaxel in ovarian cancers and radiation in breast, prostate, and gliomas (Bani and others 2004; Tsai and others 2007).
Deregulation of the immune system is a hallmark of autoimmune diseases such as systemic Lupus, psoriasis, and multiple sclerosis. In CD4+ T lymphocytes of systemic lupus patients, expression of G1P3 correlated with severity of lupus activity (Deng and others 2006). Similarly, in skin lesion biopsies from patients with psoriasis, G1P3 was markedly (4×) upregulated compared to adjacent skin (Quekenborn-Trinquet and others 2005). Another study in psoriasis identified ∼400-fold upregulation of G1P3 in hyperproliferative lesional and ∼9-fold upregulation in nonlesional psoriatic epidermis compared to healthy epidermis (Szegedi and others 2010). In multiple sclerosis, comparison of gene expression among 8 monotypic twins in which one had multiple sclerosis revealed marked G1P3 upregulation in affected siblings compared to their healthy twin (Sarkijarvi and others 2006).
G1P3 Function and Localization
Despite being one of the first identified ISGs, little is known about the cellular and biological functions of G1P3. Because of the high inducibility of G1P3 by IFNs, initial studies suggested that G1P3 was a positive mediator of the antiproliferative and antiviral effects of IFNs, but these studies were largely correlative (Tahara and others 1994, 1995; Kim and others 2002). The role of G1P3 in establishing the antiviral state elicited by IFNs was tested by knocking out G1P3 in HT1080 cells by homologous recombination (Yanez and Porter 2002). Although the induction of G1P3 was ablated in the G1P3−/− cells, there was no significant difference in the formation of plaques by encephalomyocarditis virus, semliki forest virus, or coca1 virus, suggesting that G1P3 was not required for establishing IFNs' antiviral state against these viruses in this cell type (Yanez and Porter 2002).
Contrary to the traditional view of IFNs as antiproliferative agents, several studies have highlighted the proliferative effects of IFNs on certain cell types (Ferlin-Bezombes and others 1998; Liu and others 1999). Data from recent functional studies, including ectopic expression and siRNA-mediated downregulation, characterized G1P3 as an antiapoptotic factor in IFNs survival pathways, and studies in gastric cancer and myeloma have identified antiapoptotic activities of G1P3 (Tahara and others 2005; Cheriyath and others 2007). TMK-1, a gastric cancer cell line without constitutive expression of G1P3, was sensitive to apoptosis induced by 5-flurauracil, cyclohexamide (CHX), and serum starvation. Ectopic expression of G1P3 in TMK-1 inhibited 5-flurauracil and CHX induced apoptosis. Further studies suggested that G1P3 inhibits apoptosis by suppressing mitochondrial membrane depolarization and by inhibiting the release of cytochrome c (Tahara and others 2005). These results are in agreement with the results of our study in myeloma where G1P3 inhibited the apoptotic activity of TRAIL to nullify the antiproliferative effects of IFN-α2b (Cheriyath and others 2007).
Depending on the experimental conditions, IFNs can either stimulate or inhibit cell survival (Ferlin-Bezombes and others 1998; Otsuki and others 1998; Chen and others 2001; Dimberg and others 2005; Gomez-Benito and others 2005; Cheriyath and others 2007). Despite the activation of STAT1 and the robust induction (>100-fold) of proapoptotic ISGs such as TRAIL and XIAP-associated factor 1 (XAF1), IFN-α2b had either an early antiapoptotic (24–32 h) or late proapoptotic effect (after 32 h) in myeloma (Cheriyath and others 2007). At early time points, cotreatment with IFN-α2b antagonized TRAIL induced activation of caspase 3 and suppressed the release of cytochrome c by inhibiting mitochondrial membrane depolarization (Cheriyath and others 2007). These observations led to the hypothesis that a mitochondrial-localized ISG with prosurvival activity might be mediating the inhibitory effects of IFNs on TRAIL-induced apoptosis. High-throughput gene expression analysis coupled with functional studies identified G1P3 as a mediator of the antiapoptotic activity of IFN-α2b (Cheriyath and others 2007). As in gastric cancer cell lines, G1P3 localized to the mitochondrial fraction of myeloma cells and stabilized mitochondrial potential under stress (Cheriyath and others 2007). The spontaneous apoptosis of keratinocytes upon downregulation of G1P3 demonstrates the antiapoptotic property of G1P3 in nonmalignant cells (Szegedi and others 2010). Together, these results suggest that either the upregulated or induced expression of G1P3 could lead to apoptosis resistance in malignant and nonmalignant cells. As discussed earlier, several cancer therapies also induced expression of G1P3, including paclitaxel in ovarian cancer and radiation in breast, prostate, and gliomas (Bani and others 2004; Tsai and others 2007), supporting the idea that G1P3 is a stress-induced protein and its antiapoptotic activity might play a role in the development of therapeutic resistance.
In normal cells, the antiapoptotic activity of G1P3 may positively affect IFN's antiviral and innate immune responses. During viral infection, delaying early apoptosis through the induction of a survival factor would protect surrounding healthy cells from infection, enhance IFN secretion, and overcome the proapoptotic activity of cytokines released into the surrounding milieu (Teodoro and Branton 1997; Hardwick 1998; Zhang and others 2003). Similarly, antiapoptotic pathways operating in immune cells may overcome activation induced cell death to maintain the balance of immune response (Zhang and others 2003). In agreement with this postulate, G1P3 enhanced the antiviral activity of IFN-α in a HCV-replicon cell culture system (Zhu and others 2003). In contrast to these results, large-scale semiquantitative RT-PCR analyses of treatment outcomes in patients with chronic hepatitis C virus associated the lack of induced expression of G1P3 with nonresponsiveness to combinations of pegylated IFNs and ribavirin (Cardoso and others 2010). On the basis of these results, G1P3 was described as a negative factor in mediating IFNs antiviral response (Cardoso and others 2010). Basal expression of G1P3 along with other ISGs was higher in nonresponding cohorts of patients, indicating that the antiviral pathways are already active in nonresponders and virus somehow overcame IFNs antiviral responses. Therefore, further treatment of these patients with an IFN-containing regimen is not beneficial. Viruses also may evade IFNs antiviral response by suppressing the activity of the mediators of IFNs antiviral response. The downregulation of G1P3 by respiratory syncytial virus (RSV) infection support the above scenario (Zhao and others 2008). Future work is critical to separate antiviral functions of G1P3 from its role simply as a highly sensitive biomarker of IFN activation in patients.
Summary
G1P3 and ISG12a, members of the FAM14 family, were among the first identified ISGs (Kelly and others 1986; Rasmussen and others 1993). The presence of ISG12 orthologs in D. discoideum suggests a primordial origin of the FAM14 family that predates the evolution of the IFN system and implicates functions outside of the innate immune response (Parker and Porter 2004). Although the G1P3 gene promoter played a crucial role in deciphering IFNs signaling pathways, studies are only beginning to uncover biological functions. The apparent evolutionary relationship, induction by IFNs, mitochondrial localization, and opposite effects on apoptosis by G1P3 and ISG12 suggest that these 2 proteins may functionally modulate IFNs pro- and antiproliferative effects by regulating mitochondrial permeability transition (Tahara and others 2005; Cheriyath and others 2007; Rosebeck and Leaman 2008).
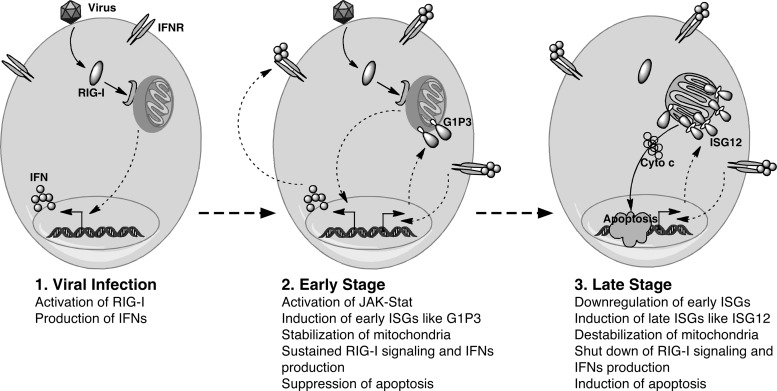
Recent studies have highlighted the critical role of mitochondria in the production of viral-induced IFNs. After viral infection the adaptor protein interferon-beta promoter stimulator 1 (IPS-1/MAVS) and/or RA-inducible gene I (RIG-I) relocalize into mitochondria to facilitate the activation of IRF-3 to produce IFNs (Seth and others 2005). Mitochondrial localized ISGs such as G1P3 might play a role in eliciting IFNs antiviral response by keeping mitochondria in a healthy state and allowing the formation of the MAVS-RIG-I complex (Baum and Garcia-Sastre 2010; Rehwinkel 2010; Takeuchi and Akira 2010; Wilkins and Gale 2010). In a virally infected cell, if the cell fails to clear the virus, it might be beneficial for it to undergo self-destruction to control the viral load and to prevent the overproduction of IFNs. This can be achieved through the depolarization of mitochondria through the induction of ISG12, which will uncouple RIG-I signaling and induce apoptosis. The immediate early induction of G1P3 and relatively late induction of ISG12 and their opposing activities on the mitochondrial membrane potential support this model (Fig. 4). Further studies on G1P3 and ISG12 will place these 2 proteins into an overall framework of ISGs that function coordinately to affect cell growth and pathogen resistance.
FIG. 4.
Model for the role of G1P3 and ISG12 in the modulation of IFNs antiviral response. In this model, G1P3 is induced at early time points after viral infection and serves to stabilize mitochondrial membrane potential, resist apoptosis, and sustain the production of IFNs through the RIG-I/MAVS pathway. ISG12 is induced at later time points of postinfection and destabilizes mitochondrial membrane potential, curtails RIG-I signaling, induces apoptosis, and downregulates IFNs production. IFN, interferon; RIG-I, RA inducible gene I.
Acknowledgments
The authors gratefully acknowledge Barbara Jacobs and Melissa Kuhns at the Cleveland Clinic for their critical reading of the article.
Author Disclosure Statement
Authors of this article have no competing financial interests in connection with this article.
References
- Bandyopadhyay SK. Kumar R. Rubin BY. Sen GC. Interferon-inducible gene expression in HL-60 cells: effects of the state of differentiation. Cell Growth Differ. 1992;3(6):369–375. [PubMed] [Google Scholar]
- Bani MR. Nicoletti MI. Alkharouf NW. Ghilardi C. Petersen D. Erba E. Sausville EA. Liu ET. Giavazzi R. Gene expression correlating with response to paclitaxel in ovarian carcinoma xenografts. Mol Cancer Ther. 2004;3(2):111–121. [PubMed] [Google Scholar]
- Baum A. Garcia-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids. 2010;38(5):1283–1299. doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron CA. Sen GC. Innate responses to viral infection. In: Knipe DM, editor; Howley PM, editor; Griffin DE, editor; Lamb RA, editor; Martin MA, editor; Roizman B, editor; Straus SE, editor. Fields Virology. Philadelphia: Lippincott, Williams & Wilkins; 2006. pp. 249–278. [Google Scholar]
- Borden EC. Sen GC. Uze G. Silverman RH. Ransohoff RM. Foster GR. Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A. Chen Y. Kim JW. Forgues M. Valerie K. Harris CC. Wang XW. Induction of a unique gene expression profile in primary human hepatocytes by hepatitis C virus core, NS3 and NS5A proteins. Carcinogenesis. 2007;28(7):1552–1560. doi: 10.1093/carcin/bgm075. [DOI] [PubMed] [Google Scholar]
- Cardoso AC. Moucari R. Figueiredo-Mendes C. Ripault MP. Giuily N. Castelnau C. Boyer N. Asselah T. Martinot-Peignoux M. Maylin S. Carvalho-Filho RJ. Valla D. Bedossa P. Marcellin P. Impact of peginterferon and ribavirin therapy on hepatocellular carcinoma: incidence and survival in hepatitis C patients with advanced fibrosis. J Hepatol. 2010;52(5):652–657. doi: 10.1016/j.jhep.2009.12.028. [DOI] [PubMed] [Google Scholar]
- Certa U. Seiler M. Padovan E. Spagnoli GC. High density oligonucleotide array analysis of interferon- alpha2a sensitivity and transcriptional response in melanoma cells. Br J Cancer. 2001;85(1):107–114. doi: 10.1054/bjoc.2001.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. Gong B. Mahmoud-Ahmed AS. Zhou A. Hsi ED. Hussein M. Almasan A. Apo2L/TRAIL and Bcl-2-related proteins regulate type I interferon-induced apoptosis in multiple myeloma. Blood. 2001;98(7):2183–2192. doi: 10.1182/blood.v98.7.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyath V. Glaser KB. Waring JF. Baz R. Hussein MA. Borden EC. G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J Clin Invest. 2007;117(10):3107–3117. doi: 10.1172/JCI31122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyath V. Kuhns MA. Jacobs BS. Evangelista P. Downs-Kelly E. Tubbs R. Huang TM. Borden EC. Aberrantly expressed G1P3, a survival factor of IFN-pathway, inhibits detachment induced mammary epithelial cell death and promote breast cancer. Under Review 2010 [Google Scholar]
- Chernajovsky Y. Constitutive in vitro binding of nuclear proteins to the 5'-flanking region of 6–16, a human gene inducible by alpha, beta-interferons. FEBS Lett. 1989;258(2):323–330. doi: 10.1016/0014-5793(89)81685-0. [DOI] [PubMed] [Google Scholar]
- Chernajovsky Y. Kirby-Sanders HM. A cis-acting sequence, located at −450 in the promoter of the human interferon-inducible gene 6–16, binds constitutively to a nuclear protein and decreases the expression of a reporter interferon-inducible promoter. Lymphokine Res. 1990;9(2):199–212. [PubMed] [Google Scholar]
- Dale TC. Rosen JM. Guille MJ. Lewin AR. Porter AG. Kerr IM. Stark GR. Overlapping sites for constitutive and induced DNA binding factors involved in interferon-stimulated transcription. EMBO J. 1989;8(3):831–839. doi: 10.1002/j.1460-2075.1989.tb03444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YJ. Huang ZX. Zhou CJ. Wang JW. You Y. Song ZQ. Xiang MM. Zhong BY. Hao F. Gene profiling involved in immature CD4 + T lymphocyte responsible for systemic lupus erythematosus. Mol Immunol. 2006;43(9):1497–1507. doi: 10.1016/j.molimm.2005.07.039. [DOI] [PubMed] [Google Scholar]
- Dimberg LY. Dimberg AI. Ivarsson K. Stromberg T. Osterborg A. Nilsson K. Oberg F. Jernberg Wiklund H. Ectopic and IFN-induced expression of Fas overcomes resistance to Fas-mediated apoptosis in multiple myeloma cells. Blood. 2005;106(4):1346–1354. doi: 10.1182/blood-2004-04-1322. [DOI] [PubMed] [Google Scholar]
- Dooley TP. Curto EV. Reddy SP. Davis RL. Lambert GW. Wilborn TW. Elson CO. Regulation of gene expression in inflammatory bowel disease and correlation with IBD drugs: screening by DNA microarrays. Inflamm Bowel Dis. 2004;10(1):1–14. doi: 10.1097/00054725-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Ferlin-Bezombes M. Jourdan M. Liautard J. Brochier J. Rossi JF. Klein B. IFN-alpha is a survival factor for human myeloma cells and reduces dexamethasone-induced apoptosis. J Immunol. 1998;161(6):2692–2699. [PubMed] [Google Scholar]
- Friedman RL. Manly SP. McMahon M. Kerr IM. Stark GR. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 1984;38(3):745–755. doi: 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- Gjermandsen IM. Justesen J. Martensen PM. The interferon-induced gene ISG12 is regulated by various cytokines as the gene 6–16 in human cell lines. Cytokine. 2000;12(3):233–238. doi: 10.1006/cyto.1999.0549. [DOI] [PubMed] [Google Scholar]
- Gomez-Benito M. Balsas P. Bosque A. Anel A. Marzo I. Naval J. Apo2L/TRAIL is an indirect mediator of apoptosis induced by interferon-alpha in human myeloma cells. FEBS Lett. 2005;579(27):6217–6222. doi: 10.1016/j.febslet.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Han SY. Kim SH. Heasley LE. Differential gene regulation by specific gain-of-function JNK1 proteins expressed in Swiss 3T3 fibroblasts. J Biol Chem. 2002;277(49):47167–47174. doi: 10.1074/jbc.M204270200. [DOI] [PubMed] [Google Scholar]
- Hardwick JM. Viral interference with apoptosis. Semin Cell Dev Biol. 1998;9(3):339–349. doi: 10.1006/scdb.1998.0243. [DOI] [PubMed] [Google Scholar]
- Hashimoto SI. Suzuki T. Nagai S. Yamashita T. Toyoda N. Matsushima K. Identification of genes specifically expressed in human activated and mature dendritic cells through serial analysis of gene expression. Blood. 2000;96(6):2206–2214. [PubMed] [Google Scholar]
- Huang M. Qian F. Hu Y. Ang C. Li Z. Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat Cell Biol. 2002;4(10):774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- Itzhaki JE. Barnett MA. MacCarthy AB. Buckle VJ. Brown WR. Porter AC. Targeted breakage of a human chromosome mediated by cloned human telomeric DNA. Nat Genet. 1992;2(4):283–287. doi: 10.1038/ng1292-283. [DOI] [PubMed] [Google Scholar]
- Kelly JM. Gilbert CS. Stark GR. Kerr IM. Differential regulation of interferon-induced mRNAs and c-myc mRNA by alpha- and gamma-interferons. Eur J Biochem. 1985;153(2):367–371. doi: 10.1111/j.1432-1033.1985.tb09312.x. [DOI] [PubMed] [Google Scholar]
- Kelly JM. Porter AC. Chernajovsky Y. Gilbert CS. Stark GR. Kerr IM. Characterization of a human gene inducible by alpha- and beta-interferons and its expression in mouse cells. EMBO J. 1986;5(7):1601–1606. doi: 10.1002/j.1460-2075.1986.tb04402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CD. Choe Y. Shim C. Kim K. Interferon beta secreted from human hair dermal papilla cells inhibits the growth of outer root sheath cells cultured in vitro. Biochem Biophys Res Commun. 2002;290(3):1133–1138. doi: 10.1006/bbrc.2001.6324. [DOI] [PubMed] [Google Scholar]
- Koike F. Satoh J. Miyake S. Yamamoto T. Kawai M. Kikuchi S. Nomura K. Yokoyama K. Ota K. Kanda T. Fukazawa T. Yamamura T. Microarray analysis identifies interferon beta-regulated genes in multiple sclerosis. J Neuroimmunol. 2003;139(1–2):109–118. doi: 10.1016/s0165-5728(03)00155-3. [DOI] [PubMed] [Google Scholar]
- Kumar R. Korutla L. Growth inhibition of human acute promyelocytic leukemia NB-4 cells by interferons and all-trans retinoic acid: trans-modulation of inducible gene expression pathways. Anticancer Res. 1995;15(2):353–360. [PubMed] [Google Scholar]
- Kumar-Sinha C. Varambally S. Sreekumar A. Chinnaiyan AM. Molecular cross-talk between the TRAIL and interferon signaling pathways. J Biol Chem. 2002;277(1):575–585. doi: 10.1074/jbc.M107795200. [DOI] [PubMed] [Google Scholar]
- Labrada L. Liang XH. Zheng W. Johnston C. Levine B. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J Virol. 2002;76(22):11688–11703. doi: 10.1128/JVI.76.22.11688-11703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancillotti F. Giandomenico V. Affabris E. Fiorucci G. Romeo G. Rossi GB. Interferon alpha-2b and retinoic acid combined treatment affects proliferation and gene expression of human cervical carcinoma cells. Cancer Res. 1995;55(14):3158–3164. [PubMed] [Google Scholar]
- Leaman DW. Chawla-Sarkar M. Jacobs B. Vyas K. Sun Y. Ozdemir A. Yi T. Williams BR. Borden EC. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J.Interferon Cytokine Res. 2003;23(12):745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- Li B. Shin J. Lee K. Interferon-stimulated gene ISG12b1 inhibits adipogenic differentiation and mitochondrial biogenesis in 3T3-L1 cells. Endocrinology. 2009;150(3):1217–1224. doi: 10.1210/en.2008-0727. [DOI] [PubMed] [Google Scholar]
- Li X. Leung S. Burns C. Stark GR. Cooperative binding of Stat1-2 heterodimers and ISGF3 to tandem DNA elements. Biochimie. 1998;80(8–9):703–710. doi: 10.1016/s0300-9084(99)80023-6. [DOI] [PubMed] [Google Scholar]
- Liu P. Oken M. Van NB. Interferon-alpha protects myeloma cell lines from dexamethasone-induced apoptosis. Leukemia. 1999;13(3):473–480. doi: 10.1038/sj.leu.2401334. [DOI] [PubMed] [Google Scholar]
- Martensen PM. Justesen J. Small ISGs coming forward. J Interferon Cytokine Res. 2004;24(1):1–19. doi: 10.1089/107999004772719864. [DOI] [PubMed] [Google Scholar]
- Martensen PM. Sogaard TM. Gjermandsen IM. Buttenschon HN. Rossing AB. Bonnevie-Nielsen V. Rosada C. Simonsen JL. Justesen J. The interferon alpha induced protein ISG12 is localized to the nuclear membrane. Eur J Biochem. 2001;268(22):5947–5954. doi: 10.1046/j.0014-2956.2001.02545.x. [DOI] [PubMed] [Google Scholar]
- Otsuki T. Yamada O. Sakaguchi H. Tomokuni A. Wada H. Yawata Y. Ueki A. Human myeloma cell apoptosis induced by interferon-alpha. Br.J.Haematol. 1998;103(2):518–529. doi: 10.1046/j.1365-2141.1998.01000.x. [DOI] [PubMed] [Google Scholar]
- Parker N. Porter AC. Identification of a novel gene family that includes the interferon-inducible human genes 6–16 and ISG12. BMC Genomics. 2004;5(1):8. doi: 10.1186/1471-2164-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter AC. Chernajovsky Y. Dale TC. Gilbert CS. Stark GR. Kerr IM. Interferon response element of the human gene 6–16. EMBO J. 1988;7(1):85–92. doi: 10.1002/j.1460-2075.1988.tb02786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quekenborn-Trinquet V. Fogel P. dana-Jammayrac O. Ancian P. Demarchez M. Rossio P. Richards HL. Kirby B. Nguyen C. Voegel JJ. Griffiths CE. Gene expression profiles in psoriasis: analysis of impact of body site location and clinical severity. Br J Dermatol. 2005;152(3):489–504. doi: 10.1111/j.1365-2133.2005.06384.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen UB. Wolf C. Mattei MG. Chenard MP. Bellocq JP. Chambon P. Rio MC. Basset P. Identification of a new interferon-alpha-inducible gene (p27) on human chromosome 14q32 and its expression in breast carcinoma. Cancer Res. 1993;53(17):4096–4101. [PubMed] [Google Scholar]
- Rehwinkel J. Exposing viruses: RNA patterns sensed by RIG-I-like receptors. J Clin Immunol. 2010;30(4):491–495. doi: 10.1007/s10875-010-9384-7. [DOI] [PubMed] [Google Scholar]
- Rosebeck S. Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13(4):562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- Roy C. Inhibition of gene transcription by purine rich triplex forming oligodeoxyribonucleotides. Nucleic Acids Res. 1993;21(12):2845–2852. doi: 10.1093/nar/21.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy C. Triple-helix formation interferes with the transcription and hinged DNA structure of the interferon-inducible 6–16 gene promoter. Eur J Biochem. 1994;220(2):493–503. doi: 10.1111/j.1432-1033.1994.tb18648.x. [DOI] [PubMed] [Google Scholar]
- Roy C. Lebleu B. DNA protein interactions at the interferon-responsive promoter elements: potential for an H-DNA conformation. Nucleic Acids Res. 1991;19(3):517–524. doi: 10.1093/nar/19.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. Ji G. Shinzawa H. Okumoto K. Hattori E. Adachi T. Takeda T. Sugahara K. Ito JI. Watanabe H. Saito K. Togashi H. Ishii K. Matsuura T. Inageda K. Muramatsu M. Kawata S. Genetic variations in humans associated with differences in the course of hepatitis C. Biochem Biophys Res Commun. 2004;317(2):335–341. doi: 10.1016/j.bbrc.2004.03.056. [DOI] [PubMed] [Google Scholar]
- Sarkijarvi S. Kuusisto H. Paalavuo R. Levula M. Airla N. Lehtimaki T. Kaprio J. Koskenvuo M. Elovaara I. Gene expression profiles in Finnish twins with multiple sclerosis. BMC Med Genet. 2006;7:11. doi: 10.1186/1471-2350-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellebjerg F. Krakauer M. Hesse D. Ryder LP. Alsing I. Jensen PE. Koch-Henriksen N. Svejgaard A. Soelberg Sorensen P. Identification of new sensitive biomarkers for the in vivo response to interferon-beta treatment in multiple sclerosis using DNA-array evaluation. Eur J Neurol. 2009;16(12):1291–1298. doi: 10.1111/j.1468-1331.2009.02716.x. [DOI] [PubMed] [Google Scholar]
- Sen GC. Transcriptional regulation of interferon-inducible genes. In: Cohen P, editor; Foulkes JC, editor. Hormonal Regulation of Transcription. Amsterdam: Elsevier; 1991. pp. 349–374. [Google Scholar]
- Seth RB. Sun L. Ea CK. Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shen D. He J. Chang HR. In silico identification of breast cancer genes by combined multiple high throughput analyses. Int J Mol Med. 2005;15(2):205–212. [PubMed] [Google Scholar]
- Smidt KC. Hansen LL. Sogaard TM. Petersen LK. Knudsen UB. Martensen PM. A nine-nucleotide deletion and splice variation in the coding region of the interferon induced ISG12 gene. Biochim Biophys Acta. 2003;1638(3):227–234. doi: 10.1016/s0925-4439(03)00087-5. [DOI] [PubMed] [Google Scholar]
- Sorbello V. Fuso L. Sfiligoi C. Scafoglio C. Ponzone R. Biglia N. Weisz A. Sismondi P. De BM. Quantitative real-time RT-PCR analysis of eight novel estrogen-regulated genes in breast cancer. Int J Biol Markers. 2003;18(2):123–129. doi: 10.1177/172460080301800205. [DOI] [PubMed] [Google Scholar]
- Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- Stark GR. Kerr IM. Williams BR. Silverman RH. Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Sun Y. Leaman DW. Involvement of Noxa in cellular apoptotic responses to interferon, double-stranded RNA, and virus infection. J Biol Chem. 2005;280(16):15561–15568. doi: 10.1074/jbc.M412630200. [DOI] [PubMed] [Google Scholar]
- Suomela S. Cao L. Bowcock A. Saarialho-Kere U. Interferon alpha-inducible protein 27 (IFI27) is upregulated in psoriatic skin and certain epithelial cancers. J Invest Dermatol. 2004;122(3):717–721. doi: 10.1111/j.0022-202X.2004.22322.x. [DOI] [PubMed] [Google Scholar]
- Szegedi K. Sonkoly E. Nagy N. Nemeth IB. Bata-Csorgo Z. Kemeny L. Dobozy A. Szell M. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19(3):269–278. doi: 10.1111/j.1600-0625.2010.01066.x. [DOI] [PubMed] [Google Scholar]
- Tahara E., Jr. Tahara H. Kanno M. Naka K. Takeda Y. Matsuzaki T. Yamazaki R. Ishihara H. Yasui W. Barrett JC. Ide T. Tahara E. G1P3, an interferon inducible gene 6–16, is expressed in gastric cancers and inhibits mitochondrial-mediated apoptosis in gastric cancer cell line TMK-1 cell. Cancer Immunol Immunother. 2005;54(8):729–740. doi: 10.1007/s00262-004-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara H. Hara E. Tsuyama N. Oda K. Ide T. Preparation of a subtractive cDNA library enriched in cDNAs which expressed at a high level in cultured senescent human fibroblasts. Biochem Biophys Res Commun. 1994;199(3):1108–1112. doi: 10.1006/bbrc.1994.1345. [DOI] [PubMed] [Google Scholar]
- Tahara H. Kamada K. Sato E. Tsuyama N. Kim JK. Hara E. Oda K. Ide T. Increase in expression levels of interferon-inducible genes in senescent human diploid fibroblasts and in SV40-transformed human fibroblasts with extended lifespan. Oncogene. 1995;11(6):1125–1132. [PubMed] [Google Scholar]
- Takeuchi O. Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Teodoro JG. Branton PE. Regulation of apoptosis by viral gene products. J Virol. 1997;71(3):1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MH. Cook JA. Chandramouli GV. DeGraff W. Yan H. Zhao S. Coleman CN. Mitchell JB. Chuang EY. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007;67(8):3845–3852. doi: 10.1158/0008-5472.CAN-06-4250. [DOI] [PubMed] [Google Scholar]
- Turri MG. Cuin KA. Porter AC. Characterisation of a novel minisatellite that provides multiple splice donor sites in an interferon-induced transcript. Nucleic Acids Res. 1995;23(11):1854–1861. doi: 10.1093/nar/23.11.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum RR. Ishwaran H. Yoon T. Nuyten DS. Baker SW. Khodarev N. Su AW. Shaikh AY. Roach P. Kreike B. Roizman B. Bergh J. Pawitan Y. van de Vijver MJ. Minn AJ. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C. Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22(1):41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez RJ. Porter AC. A chromosomal position effect on gene targeting in human cells. Nucleic Acids Res. 2002;30(22):4892–4901. doi: 10.1093/nar/gkf614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XR. Zhang LY. Devadas S. Li L. Keegan AD. Shi YF. Reciprocal expression of TRAIL and CD95L in Th1 and Th2 cells: role of apoptosis in T helper subset differentiation. Cell Death Differ. 2003;10(2):203–210. doi: 10.1038/sj.cdd.4401138. [DOI] [PubMed] [Google Scholar]
- Zhao D. Peng D. Li L. Zhang Q. Zhang C. Inhibition of G1P3 expression found in the differential display study on respiratory syncytial virus infection. Virol J. 2008;5:114. doi: 10.1186/1743-422X-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. Zhao H. Collins CD. Eckenrode SE. Run Q. McIndoe RA. Crawford JM. Nelson DR. She JX. Liu C. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology. 2003;37(5):1180–1188. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]