Abstract
Antibiotic treatment can deplete the commensal bacteria of a patient’s gut microbiota and, paradoxically, increase their risk of subsequent infections. In allogeneic hematopoietic stem cell transplantation (allo-HSCT), antibiotic administration is essential for optimal clinical outcomes but significantly disrupts intestinal microbiota diversity, leading to loss of many beneficial microbes. Although gut microbiota diversity loss during allo-HSCT is associated with increased mortality, approaches to reestablish depleted commensal bacteria have yet to be developed. We have initiated a randomized, controlled clinical trial of autologous fecal microbiota transplantation (auto-FMT) versus no intervention and have analyzed the intestinal microbiota profiles of 25 allo-HSCT patients (14 who received auto-FMT treatment and 11 control patients who did not). Changes in gut microbiota diversity and composition revealed that the auto-FMT intervention boosted microbial diversity and reestablished the intestinal microbiota composition that the patient had before antibiotic treatment and allo-HSCT. These results demonstrate the potential for fecal sample banking and posttreatment remediation of a patient’s gut microbiota after microbiota-depleting antibiotic treatment during allo-HSCT.
INTRODUCTION
Antibiotic treatment damages the microbiota and increases risk of intestinal infection. Although this effect has been recognized for more than 60 years (1), remediation of the antibiotic-depleted gut microbiota has yet to become standard clinical practice. In patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT), antibiotics are routinely given to treat or reduce the risk of serious infection. Prospective studies of allo-HSCT patients demonstrated that the intestinal microbiota is markedly altered during treatment, with profound loss of obligate anaerobic bacteria including immunomodulatory species such as those belonging to the Clostridia class and Bacteroidetes phylum (2). The clinical consequences of these alterations are also apparent in allo-HSCT: Disruption of beneficial obligate anaerobes correlates with complications that include systemic infection with vancomycin-resistant Enterococcus (VRE), Clostridium difficile infection, and graft-versus-host disease (GVHD) (2–6). Overall, patients who lose gut microbiota diversity at the time of hematopoietic stem cell engraftment have higher rates of transplant-related death (7).
We explored whether the microbiota could be restored in allo-HSCT patients through the use of autologously derived fecal microbiota transplantation (auto-FMT). We chose to use each patient’s own feces, collected before allo-HSCT treatment, as opposed to feces from a heterologous donor, because of potential safety concerns. Human feces contain protozoa, bacteria, archaea, fungi, and viruses (8), and the overall composition can change from day to day depending on diet and other environmental exposures (9). Despite remarkable advances in recent years, current technologies are incapable of comprehensively determining fecal composition. Allo-HSCT patients remain immunocompromised for many months after engraftment, and although immunocompromised patients—including allo-HSCT recipients—have undergone heterologous FMT (10, 11), we reasoned that auto-FMT would be safer than heterologous FMT, thereby minimizing the risk of exposure to potentially pathogenic microorganisms not previously encountered by the patient.
We initiated a randomized controlled clinical trial to determine the feasibility of auto-FMT for restoring the pretransplant gut microbiota and for decreasing transplant-related complications. Here, we present an analysis of the gut microbiota compositional changes in 25 patients enrolled and randomized in this study from whom longitudinal fecal samples were collected. We demonstrate that auto-FMT is an effective intervention that restores gut microbiota diversity and can return microbial composition to the pre–allo-HSCT state.
RESULTS
Gut microbiota diversity is depleted after allo-HSCT
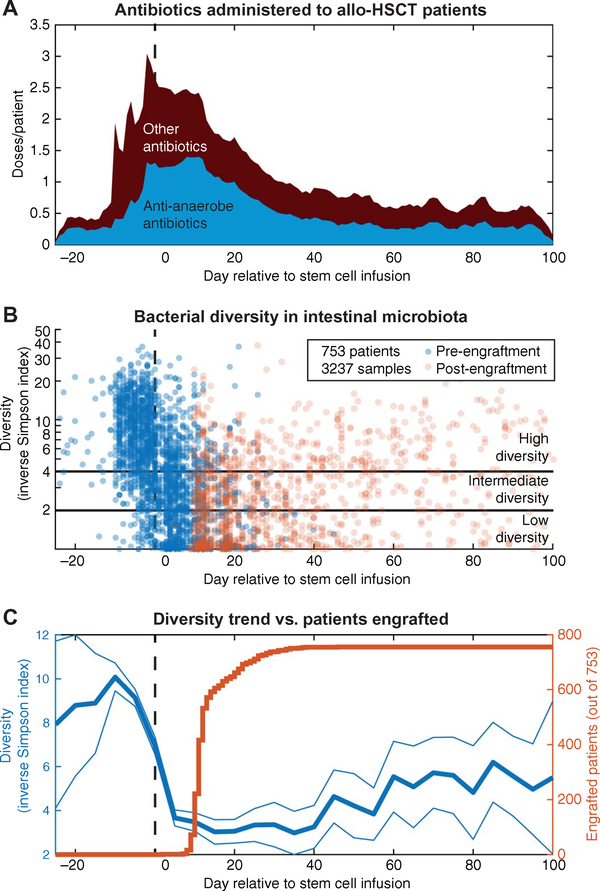
To establish the spectrum of gut microbiota changes that occur over the course of allo-HSCT, we analyzed fecal samples collected from our observational stool-banking cohort. Since 2009, we have collected fecal samples from patients undergoing allo-HSCT to characterize changes in the intestinal microbiota and their impact on clinical outcomes. We analyzed by 16S ribosomal RNA (rRNA) gene sequencing 3237 longitudinally collected fecal samples from 753 patients, obtained between day −25 (before hematopoietic cell infusion) and day +100 (after hematopoietic cell infusion). The average allo-HSCT patient’s microbiota diversity as measured by the inverse Simpson (IS) index was initially high but declined as they underwent treatment with antibiotics, either as routine infectious prophylaxis in the setting of immune suppression or as empiric treatment based on clinical setting and judgment (Fig. 1, A and B). Average diversity continued to decrease to a nadir at around day +5, and this persisted for about 6 weeks.
Fig. 1. The gut microbiota is disrupted during allo-HSCT.
A total of 3237 fecal samples from 753 patients who underwent allo-HSCT were obtained, and the microbiota composition was analyzed using high-throughput 16S rRNA amplicon sequencing. (A) Antibiotics with a varying spectrum of activity were given over the course of allo-HSCT, either as prophylaxis or for treatment of infections. Antibiotics with anti-anaerobic activity included β-lactams, carbapenems, metronidazole, clindamycin, and vancomycin (oral only); these antibiotics were defined as microbiota perturbing in the study protocol (see the Supplementary Materials). (B) The α-diversity of each microbiota sample was measured using the IS index. Intestinal microbiota diversity dropped sharply as patients progressed through allo-HSCT treatment, reaching very low levels (IS < 4 or lower). (C) The average diversity trend showed that the drop in average microbiota diversity began several days before hematopoietic stem cell infusion, during the conditioning regimen and antibiotic prophylaxis treatment.
For many patients, low microbiota diversity persisted beyond the time of hematopoietic cell engraftment (Fig. 1C), a critical period during which immune reconstitution begins. Previous studies demonstrated that mortality is higher for patients with low diversity (IS < 4) at the time of engraftment (7). Among the 753 patients, diversity was IS < 4 for 39% of the patients and IS < 2 for 25% of the patients at the time of engraftment. Average microbiota diversity recovered slowly starting approximately at day +50, but fecal samples collected up to day +100 rarely showed recovery of pre–allo-HSCT diversity (Fig. 1B).
A randomized controlled clinical trial of auto-FMT to restore the gut microbiota
We initiated a single-center, open-label, randomized controlled trial of gut microbiota reconstitution with auto-FMT versus no treatment, with the goal of determining feasibility, safety, and efficacy of the procedure for reconstitution of the gut microbiota. The study continues to accrue to ascertain its clinical benefits (ClinicalTrials.gov identifier: NCT02269150).
Participating subjects provided feces before initiation of allo-HSCT, which were frozen and stored. Fecal samples were screened for any intestinal pathogens, and adequate microbial diversity was determined by 16S rRNA sequencing. One to 5 weeks later (median, 13 days), upon hematopoietic cell engraftment, patients were reevaluated and another fecal sample was taken. If there was a paucity of healthy bacteria belonging to the Bacteroidetes phylum as determined by quantitative polymerase chain reaction (PCR), the patients proceeded to randomization. Eligible subjects were randomized to undergo auto-FMT versus no FMT. Subjects were followed clinically for 1 year after randomization, and fecal samples were collected longitudinally over this study period.
To assess the effect of auto-FMT on the intestinal microbiota, we analyzed fecal samples from the first 25 evaluatable randomized patients in the study: 14 from the treatment group and 11 from the control group. Patients were considered evaluatable if they provided a stool sample before randomization and a stool sample within 14 days of randomization. Of the first 31 randomized patients from the study (16 treatment and 15 control), 6 were not evaluatable due to postrandomization samples not being collected within the specified time period. The subjects and type of allo-HSCT performed varied in terms of underlying diagnosis, age, intensity of conditioning chemotherapy and radiation, and hematopoietic cell donor characteristics (table S1). Patients were randomized a median of 18 days after engraftment (range, 8 to 27) in the auto-FMT arm, and a median of 22 days after engraftment (range, 3 to 28) in the control arm.
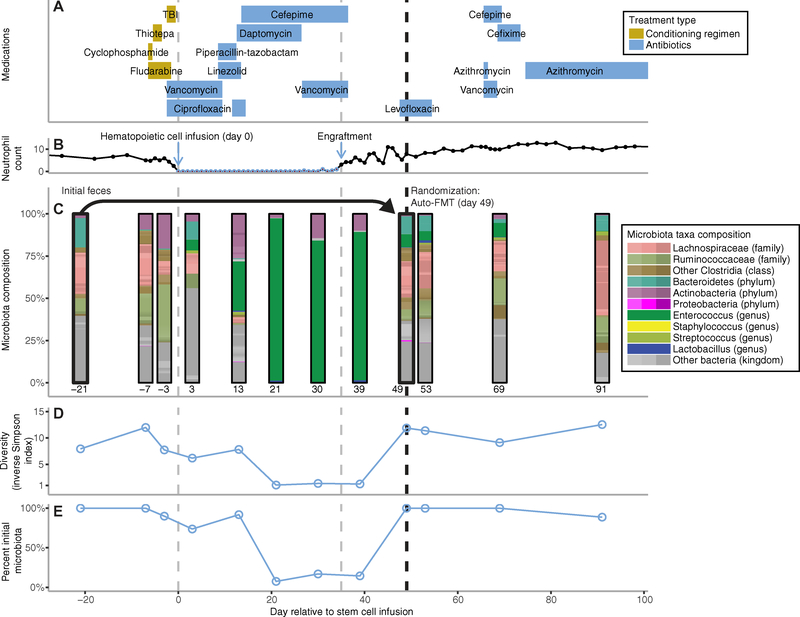
The gut microbiota profile of a representative patient demonstrates the complex compositional changes that occurred in patients during and after hematopoietic cell engraftment (Fig. 2). In this case, the patient’s microbiota composition was markedly disrupted by antibiotic treatment (Fig. 2A), with domination by Enterocccus clearly visible at day +21 (Fig. 2C). The patient was randomized to the treatment arm after engraftment and underwent auto-FMT (administered rectally by retention enema) 49 days after hematopoietic cell infusion (day +49) using the patient’s previously stored feces (Fig. 2B). Subsequent stool samples demonstrated restoration of the gut microbiota to pre–allo-HSCT levels in terms of diversity (Fig. 2D) and composition similarity (Fig. 2E).
Fig. 2. Timeline for a study patient undergoing allo-HSCT and randomized to receive auto-FMT.
(A) Allo-HSCT for study patient T5 was initiated with pretransplant conditioning [chemotherapy and total body irradiation (TBI)], followed by allogeneic hematopoietic stem cell infusion (day 0). Various antibiotics were given throughout this period for prophylactic and treatment purposes. (B) Neutrophil count reached a nadir but later recovered after hematopoietic stem cell engraftment. (C) The subject’s intestinal microbiota composition was altered as shown by sequencing of longitudinally collected fecal samples obtained before allo-HSCT and up to day 91 after transplant. After stem cell engraftment, randomization assigned patient T5 to the treatment arm and the patient received an auto-FMT on day 49 using the patient’s initial pretreatment feces, which had been collected and stored before allo-HSCT (initial feces collected at day −21). The intestinal microbiota was restored to that before transplant in terms of (D) its diversity (α-diversity quantified by the IS index) and (E) its percent similarity to the initial fecal microbiota.
Longitudinal assessment demonstrates successful reconstitution of the gut microbiota by auto-FMT
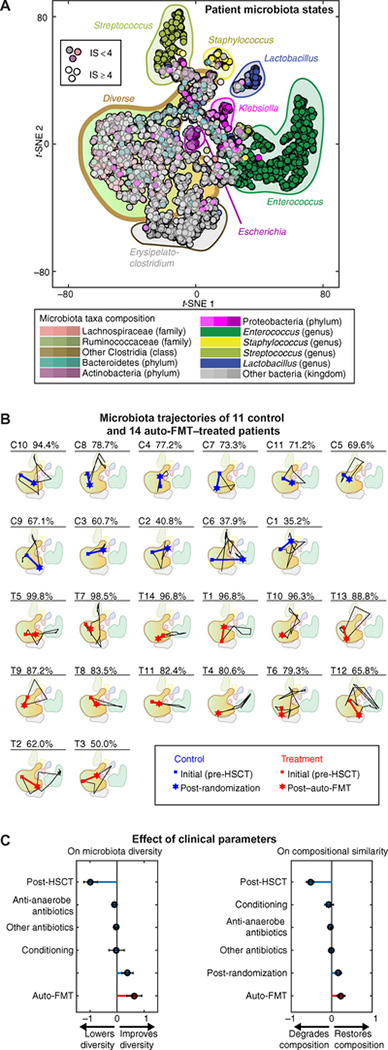
To track the gut microbiota dynamics of individual patients, we analyzed bacterial compositions from their collected stool samples by 16S rRNA gene sequencing and investigated how those compositions changed over time (fig. S1). The 16S rRNA sequencing identified ~5000 OTUs (operational taxonomic units) within our patient samples. To visualize the data, we reduced dimensionality by applying the t-distributed stochastic neighbor embedding (t-SNE) algorithm (12), which produced a two-dimensional (2D) scatter plot from the 3271 sequenced samples (756 allo-HSCT patients) from our observational cohort. In comparison to a more widely used PCoA (principal coordinate analysis) using Bray-Curtis dissimilarity [for example, (13)], the t-SNE plot could resolve multiple states of domination by specific bacterial taxa (Fig. 3A) better than 2D and even 3D PCoA (fig. S2). This comparison shows that t-SNE provided a clearer picture of gut microbiota dynamics in allo-HSCT patients, particularly with respect to progress to domination states that precede life-threatening bacteremias (2). Plotting the trajectories of the 25 randomized patients on the t-SNE space illustrated varying degrees of gut microbiota recovery after transplantation. Some patients reacquired nearly complete microbiota similarity (99.8% compositional similarity for patient T5), but others never regained pre–allo-HSCT diversity (35.2% compositional similarity for patient C1), and their gut microbiota similarity remained low even at day +100. The 14 patients receiving auto-FMT consistently regained pre–allo-HSCT diversity and composition, despite marked shifts in the microbiota after allo-HSCT (Fig. 3B). In contrast, the 11 control patients, who did not receive auto-FMT, did not recover gut microbial composition to pre–allo-HSCT levels, despite having started in a region of high diversity and undergone comparable reductions in gut microbiota composition in the post–allo-HSCT period.
Fig. 3. Auto-FMT improves gut microbiota diversity and composition in allo-HSCT patients.
(A) t-SNE plot provides visualization of gut microbiota composition during allo-HSCT. t-SNE plots were constructed from 3237 fecal samples collected from allo-HSCT patients since 2010; the patients were randomized in the auto-FMT trial. The plot shows a large region of high diversity (IS > 4) and clusters of low diversity corresponding to domination by specific bacterial species, for example, Enterococcus and Klebsiella. (B) Each patient followed a unique trajectory in the t-SNE space. The control patients (C) and those receiving auto-FMT treatment (T) were ranked by post-randomization gut microbiota recovery. The patient’s number and the percentage similarity of the gut microbiota to the initial fecal sample are shown. Patients started with a diverse gut microbiota composition, but then many progressed to domination states with loss of diversity due to the impact of antibiotic and chemotherapy administration. The microbiota composition in patients after auto-FMT (patients T1 to T14) moved closer to the initial composition than it did for control patients (patients C1 to C11). (C) Effect sizes of clinical parameters quantified by a mixed-effects model (along with a 95% CI) show that auto-FMT for allo-HSCT patients improved gut microbiota diversity (left-hand plot; P = 5 × 10−6) and recovery of the original gut microbiota composition (right-hand plot; P = 5 × 10−6).
To quantify the beneficial effect of auto-FMT on gut microbiota composition, we developed a mixed-effects model that took into account multiple clinical parameters, including antibiotics, and controlled for time and patient-specific microbiota. We fitted the model using data from 329 baseline patients for which we had complete metadata and the 25 randomized patients. The effect sizes computed (Fig. 3C) showed that auto-FMT provided a significant increase in microbiota diversity (P < 0.0001), boosting the IS index by an average of 63.8% [95% confidence interval (CI), 36.3 to 91.2%] on top of the baseline increase of 38.7% (95% CI, 17.9 to 59.6%) observed in control patients. The post–allo-HSCT compositional similarity in patients receiving auto-FMT also showed better recovery, indicating that the auto-FMT restored not only diversity but also the patient’s personal gut microbiota components (P < 0.0001). The mixed-effects model included antibiotics as covariates, which meant that the benefit of auto-FMT to microbiome recovery was significant despite variations in the antibiotic treatments given to each patient. Antibiotics with known impact on the commensal microbiota (listed in our clinical protocol, see the Supplementary Materials) had—as expected—a significant impact on both diversity (P < 0.0001) and composition (P = 0.007), whereas other antibiotics did not (P = 0.09 on diversity and P = 0.08 on composition).
The randomization process should obviate the need to control for other clinical parameters. Nevertheless, we sought to confirm that the beneficial effect detected for auto-FMT was not due to other factors known to affect gut microbiota composition. To control for the effect of dietary intake, which can rapidly affect gut microbiota composition (9), we obtained longitudinal data on dietary intake by randomized patients (that is, eating, not eating, or on a low dietary intake; fig. S3A). Then, to control for growth factors administered to allo-HSCT patients to enhance engraftment, which can also affect the gut microbiota (14), we obtained clinical data regarding granulocyte colony-stimulating factor (G-CSF) treatment (fig. S3B). We added the dietary intake and G-CSF administration as time-dependent covariates in a reworked mixed-effects model using only the 25 randomized patients. This model confirmed that the beneficial effect of auto-FMT remained significant both in boosting gut microbiota diversity (P < 0.001; fig. S3C) and in recovery of the original gut microbiota composition (P < 0.001; fig. S3D).
Auto-FMT remediates the antibiotic-disrupted gut microbiota
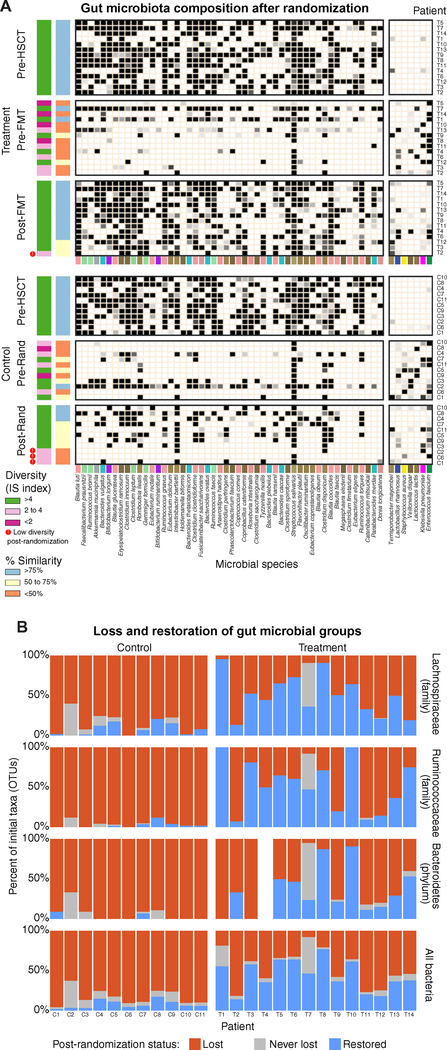
To determine how auto-FMT corrected the losses in specific gut microbiota commensals, we compared three time points from each auto-FMT–treated patient: (i) the initial stool sample collected before allo-HSCT and stored for possible later use in FMT (initial sample), (ii) the sample collected just before randomization (pre-randomization sample), and (iii) the sample collected after randomization (post-randomization sample). This comparison revealed many commensal members of the gut microbiota that were present initially but were lost after allo-HSCT treatment (pre-randomization sample) in both treatment and control groups. Auto-FMT, however, restored many lost commensals in the treatment group compared with the control group (Fig. 4A). We observed successful restoration of bacteria from beneficial groups, including Lachnospiraceae (family), Ruminococcaceae (family), and Bacteroidetes (phylum; Fig. 4B). These groups have each correlated with host benefit in previous studies and are important members of a healthy gut microbiota (4, 5, 15–17).
Fig. 4. Auto-FMT restores commensal members of the gut microbiota in allo-HSCT patients.
(A) Commensal species (ranked by their abundance) in the initial fecal sample from allo-HSCT patients disappeared after allo-HSCT before randomization to the auto-FMT or control arm (Pre-Rand); they reappeared after auto-FMT (Post-Rand) compared to the control group, who did not receive auto-FMT. Sixty percent of the patients enrolled in this trial (15 of 25) had a gut microbial diversity of IS < 4 before randomization; after randomization, 3 of 11 (27%) control patients but only 1 of 14 (7%) patients receiving auto-FMT retained this depleted diversity. The percent recovery also showed that auto-FMT helped to reconstitute the original gut microbial composition. Only 3 of 11 (27%) of the control patients had a composition 75% or more similar to their initial fecal sample, whereas 11 of 14 (79%) of the auto-FMT–treated patients showed 75% or more recovery. The bacteria in the left-hand grayscale heat map represent the top 50 bacterial strains present in the patient’s initial fecal sample; the right-hand grayscale heat map highlights seven bacterial species that increased after allo-HSCT (colors underlying the heat maps are defined in the key in Fig. 2). (B) Bacterial taxa from the patient’s initial fecal sample were classified on the basis of their presence in pre- and post-randomization fecal samples from the same patient. This is shown for all taxonomic groups (bottom panel) and for three important taxonomic groups (top three panels). A taxon was considered lost (red) if it was not detected in both pre- and post-randomization samples, restored (blue) if it was lost in the pre-randomization sample but was present in the post-randomization sample, and never lost (gray) if it remained detectable throughout the study (note that the initial sample of patient T4 did not have detectable Bacteroidetes).
To determine whether the auto-FMT also restored gut microbiota functions that were lost because of allo-HSCT, we used shotgun sequencing to analyze the metagenomes in the initial sample and in the post-randomization samples from four auto-FMT patients and four control patients. The analysis revealed a paucity of several microbial gene families, which suggested detrimental changes to the microbiome of control patients, but this loss was resolved in auto-FMT–treated patients (P < 0.05 after multiple hypothesis correction; fig. S4, A and B). Most notably, the post-randomization microbiome of control patients was enriched in antimicrobial-peptide resistance and pathways associated with microbial virulence, biofilm formation, and bacterial flagella assembly. Compared with auto-FMT–treated patients, the microbiota of control patients was also depleted in several microbial metabolic pathways (P < 0.05 by enrichment analysis; fig. S4C). Auto-FMT therefore appeared to have remediated alterations in the functional content of the gut microbiome in the setting of allo-HSCT treatment.
DISCUSSION
Antibiotics must remain as a vital and necessary component of allo-HSCT treatment, given that patients experience many infections over the course of transplantation. However, the unintended consequences of antibiotic treatment, which were long surmised, are now more apparent than ever before. Antibiotic-induced disruption of the gut microbiota leads to loss of colonization resistance, giving rise to infections such as VRE and C. difficile (2, 15). Disruption of microbes engaged in induction and modulation of host immunity may be linked to GVHD and transplant-related mortality (4–7, 18, 19).
In this randomized controlled interventional study, we examined auto-FMT as a strategy to remediate gut microbiota damage caused by antibiotic treatment during allo-HSCT. We found that the auto-FMT intervention reestablished the microbiota after disruption. We observed that important commensal groups such as Lachnospiraceae, Ruminococcaceae, and Bacteroidetes were successfully reestablished in subjects from the auto-FMT treatment arm. A formal comparison between auto-FMT, where patients provided their own sample for future transplant, and heterologous FMT, where a healthy donor provides the sample, is still lacking. However, when we compared the bacterial taxa recovered by auto-FMT in this study (Fig. 4) with those from another randomized trial that used heterologous FMT (20), we found no obvious difference in the taxa reinstated (fig. S5).
Our statistical analysis using mixed-effects models concluded that auto-FMT improved gut microbiota diversity and restored a patient’s gut microbiota composition. The model showed a minor role for other factors, including dietary intake, but this was expected. Whereas dietary intake can affect gut microbiota composition in healthy individuals, the effect is typically small (9) compared to the extreme compositional changes seen in allo-HSCT (2), which can cause losses of 100% of the diversity (Fig. 3C). Second, we randomized the patients to the treatment arm (auto-FMT, 14 patients) or control arm (no auto-FMT, 11 patients). This randomization should, in principle, eliminate the need to control for dietary and other variables. That said, a model that controlled for diet and other factors confirmed the beneficial effect (fig. S3, C and D), boosting our confidence that auto-FMT repaired the injured gut microbiota of antibiotic-treated allo-HSCT transplant patients.
Our study is limited by the fact that it was conducted at a single institution, and thus, our findings may not apply directly to patients undergoing allo-HSCT at other institutions or in clinical settings where the gut microbiota of patients may differ because of exposure to alternate antibiotics and chemotherapeutic agents. An additional limitation of the auto-FMT approach to reconstitute the gut microbiota of allo-HSCT patients is that the autologous “donor” FMT sample may be derived from patients who have been treated for cancer and exposed to antibiotics and who may be depleted of some commensal bacterial species. Despite these limitations, however, our study demonstrates that auto-FMT remediated acute microbiota losses associated with allo-HSCT and returned compositions toward the pre–allo-HSCT baseline while avoiding the potential introduction of viral or microbial agents that the patient had not previously encountered.
Existing and newly recruited patients to our randomized controlled trial remain under active monitoring to determine whether microbiota remediation improves clinical outcomes. For now, we can conclude that the auto-FMT procedure was well tolerated and effectively reestablished commensal bacterial populations at the critical early immune reconstitution stage after allo-HSCT. We have demonstrated the potential of auto-FMT as a clinical intervention to restore intestinal microbiota diversity to levels deemed safe in patients, thereby reversing the disruptive effects of broad-spectrum antibiotic treatment for patients undergoing allo-HSCT transplant.
MATERIALS AND METHODS
Study design
We conducted a phase 2, randomized, open-label, controlled study designed to assess the efficacy of auto-FMT for prevention of complications in patients who have undergone allo-HSCT. The primary efficacy end point of this trial was C. difficile infection, evaluated 1 year after randomization. Secondary end points included systemic and intestinal bacterial and viral infections and GVHD. The study is ongoing and continues to accrue patients (ClinicalTrials.gov identifier: NCT02269150). Adult patients (≥18 years) planning to undergo allo-HSCT at Memorial Sloan Kettering Cancer Center were enrolled in the study before their transplant hospitalization. The initial stool was collected before transplant conditioning and stored frozen (−80°C). This specimen served as the donor feces for auto-FMT if the subject was randomized to the treatment arm. The fecal sample was analyzed for microbial diversity by 16S rRNA gene sequencing and tested for the presence of potential intestinal pathogens including C. difficile. Subjects whose pre–allo-HSCT feces demonstrated low microbial diversity (IS index < 2.0) or tested positive for the presence of an intestinal pathogen, for example, C. difficile, were excluded from randomization. After successful hematopoietic cell engraftment (three consecutive blood neutrophil counts ≥500 per mm3), subjects underwent testing of a fecal specimen collected after engraftment to determine the presence of the Bacteroidetes phylum via quantitative PCR. Individuals with low abundance of Bacteroidetes (<0.1% total 16S) were eligible to proceed to randomization and treatment. Eligible subjects were 1:1 randomized to undergo auto-FMT with the subject’s stored pre–allo-HSCT feces versus no fecal transplantation. Randomization was stratified by cord blood source versus non–cord blood source. Subjects could be randomized within a 28-day window after engraftment. Subjects who were critically ill or required prolonged microbiota-perturbing antibiotics through the designated 28-day period were excluded from randomization. Auto-FMT consisted of a single procedure and was performed by rectal administration of the thawed fecal product through a retention enema. Subjects were followed for clinical end points for 1 year after randomization. Fecal specimens were then collected serially for correlative microbiome study analysis. Further details of the study design can be found in the clinical trial protocol in the Supplementary Materials.
Sample collection and DNA extraction
A frozen aliquot (≈100 mg) of each fecal sample was suspended, while frozen, in a solution containing 500 μl of extraction buffer [200 mM tris (pH 8.0), 200 mM NaCl, 20 mM EDTA], 200 μl of 20% SDS, 500 μl of phenol/chloroform/isoamyl alcohol (25:24:1), and 500 μl of 0.1-mm-diameter zirconia/silica beads (BioSpec Products). Microbial cells were lysed by mechanical disruption with a bead beater (BioSpec Products) for 2 min, after which two rounds of phenol/chloroform/isoamyl alcohol extraction were performed. DNA was precipitated with ethanol and resuspended in 50 μl of tris/EDTA buffer with ribonuclease (100 μg/ml). The isolated DNA was subjected to additional purification with QIAamp mini spin columns (Qiagen).
Human subjects
Sample collection from patients and analysis of the biospecimens were approved by the Memorial Sloan Kettering Cancer Center Institutional Review and Privacy Board. All participants provided signed informed consent for specimen collection and analysis, as well as auto-FMT treatment or control no auto-FMT.
Quantification 16S copy number density by real-time PCR
DNA extracted from feces was subjected to real-time PCR (rtPCR) of 16S rRNA, as described previously (21), using 0.2 μM concentrations of the broad-range bacterial 16S primers 517F (5′-GCCAGCAGCCGCGGTAA-3′) and 798R (5′-AGGGTATCTAATCCT-3′) and the DyNAmo SYBR Green rtPCR Kit (Finnzymes). Standard curves were generated by serial dilution of the PCR blunt vector (Invitrogen) containing one copy of the 16S rRNA gene derived from a member of the Porphyromonadaceae family. The cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min.
16S rRNA gene amplification and multiparallel sequencing
For each sample, duplicate 50-μl PCRs were performed, each containing 50 ng of purified DNA, 0.2 mM deoxynucleotide triphosphates, 1.5 mM MgCl2, 2.5 U Platinum Taq DNA polymerase, 2.5 μl of 10× PCR buffer, and 0.5 μM of each primer designed to amplify the V4-V5: 563F (5′-nnnnnnnn-NNNNNNNNNNNN-AYTGGGYDTAAAGNG-3′) and 926R (5′-nnnnnnnn-NNNNNNNNNNNN-CCGTCAATTYHTTTRAGT-3′). A unique 12-base Golay barcode (Ns) precedes the primers for sample identification (22), and one to eight additional nucleotides were placed in front of the barcode to offset the sequencing of the primers. Cycling conditions were 94°C for 3 min, followed by 27 cycles of 94°C for 50 s, 51°C for 30 s, and 72°C for 1 min. For the final elongation step, 72°C for 5 min was used. Replicate PCRs were pooled, and amplicons were purified using the QIAquick PCR Purification Kit (Qiagen). PCR products were quantified and pooled at equimolar amounts before Illumina barcodes and adaptors were ligated, using the Illumina TruSeq Sample Preparation protocol. The completed library was sequenced on an Illumina MiSeq platform following the Illumina recommended procedures with a paired-end 250 × 250 bp kit.
Sequence analysis
The 16S (V4-V5) paired-end reads were merged and demultiplexed. The UPARSE pipeline (23) was used for sequence processing and clustering. Sequences were error-filtered using maximum expected error (Emax = 1) (24). Singleton sequences were removed before clustering. Filtered sequences were grouped into OTUs of 97% distance-based similarity. Potentially chimeric sequences were identified and removed using both de novo and reference-based methods. Taxonomic assignment to species level was performed for representative sequences from each OTU; this was achieved using a custom Python script incorporating nucleotide BLAST (25), with National Center for Biotechnology Information (NCBI) RefSeq (26) as reference training set. We used a minimum E-value threshold of 1 × 10−10 for assignments. Sequence designations and identity scores were manually inspected for quality and consistency in terms of taxonomic structure and secondary matches. On the basis of our testing and comparisons using mock community data, we have found this approach to yield good robust species-level approximations for our candidate sequences. In particular, species-level classification of Clostridia species improved greatly compared with other routine classification methods.
Statistical analyses
We used MATLAB R2016a (MathWorks Inc.) and R 3.3 (R Development Core Team) for all data analysis and statistics. The t-SNE algorithm (12) was used on normalized OTU counts of samples collected at Memorial Sloan Kettering Cancer Center between August 2009 and November 2016 from hundreds of allo-HSCT patients. As in a previous application of t-SNE to biological data (27), we used the Barnes-Hut implementation through a MATLAB interface (function fast_tsne.m). We quantified the α-diversity of each sample using the logarithm of the IS index
where i is the sample and j is the OTU from a total of N. To quantify the similarity to initial microbiota, we calculated a matrix of Euclidean pairwise distances between every sample and then used the percentage of samples whose distance is higher or equal to the focal sample and the reference sample (the initial sample for that patient). For statistical analysis of the time series of α-diversity and similarity, we used a linear mixed-effects regression (function ftilme.m in MATLAB) with the following model defined in Wilkinson notation:
where Y is the dependent variable (the log of α-diversity or the value of fraction of similarity). The fixed-effects covariates are conditioning (true for days from −10 to 0, false otherwise), postBMT (true for days later than 0, false otherwise), anti_anaerobe_antibiotic (true if the patient received antibiotic with anti-anaerobic activity that day, false otherwise), other_antibiotic (true if the patient received another antibiotic on that day, false otherwise), postRandomization (true if this is a patient enrolled in the trial and the day of sample is post-randomization, false otherwise), and postFMT (true if this is a patient randomized to the treatment arm and the day of sample is post-randomization, false otherwise). To analyze the gene families determined by the Functional Mapping and Analysis Pipeline of the shotgun sequencing, we adopted an approach from gene set enrichment analyses commonly used on transcriptomics data (28, 29). We first determined the log2(fold change) of each of 7496 gene families for each patient (after adding pseudocounts to correct for missing values), and we compared those values in the control with those in the treatment group by computing the t statistics using a two-sided t test (function mattest.m in MATLAB). We corrected the P value obtained for each gene family using the Benjamini-Hochberg method to control for the false discovery rate (function mafdr.m in MATLAB). Finally, the gene set enrichment analysis was carried out using aggregate statistics, where the array of t statistics for the gene families in each gene family set were compared with those not in the set (function ttest2.m in MATLAB applied to each gene family and then corrected for multiple hypothesis using Benjamini-Hochberg procedure).
Supplementary Material
Fig. S1. Microbiota samples for the 25 patients (14 treatment and 11 control).
Fig. S2. PCoA of microbiota samples shown in Fig. 3.
Fig. S3. Mixed-effects model that controls for other clinical parameters confirms the beneficial effect of auto-FMT in remediating the microbiota of allo-HSCT patients.
Fig. S4. Shotgun sequencing shows that auto-FMT remediates the perturbed microbiome.
Fig. S5. Shown is an analysis of a heterologous FMT conducted in another study compared to the auto-FMT conducted in this study.
Table S1. Characteristics of patients included in this study (14 treated and 11 control). Clinical protocol
Data file S1. Source data for figures.
Acknowledgments
Funding: The auto-FMT clinical trial was supported by the Memorial Sloan Kettering Center for Microbes, Inflammation, and Cancer and the Leonard Tow Foundation. Sequencing of fecal samples for microbiota determination was supported by the National Institute of Allergy and Infectious Diseases of the NIH under award no. U01 AI12427, the Lymphoma Foundation, the Susan and Peter Solomon Divisional Genomics Program, NIH grant no. P30 CA008748 Memorial Sloan Kettering Cancer Center Support Grant/Core Grant, and the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center.
Competing interests: M.-A.P. is on the advisory boards of MolMed, NexImmune, Incyte, Novartis, Nektar, and Abbvie; consults for Merck; serves on the Data Safety Monitoring Boards of Servier and Medigene; and receives research funding from Incyte. J.B. consults for Angiocrine Bioscience, Gamida, Nohla Therapeutics, Guidepoint Consulting, and Magenta Therapeutics and receives research funding from Angiocrine Bioscience, Nohla Therapeutics, Gamida, and NYSTEM. E.P. has financial holdings in Aveo, BG Medicine, Biogen, Exelixis, Proteostasis, Regulus, Evelo, Apellis, and Leap Therapeutics. J.P. holds patents with or receives royalties from Seres Therapeutics Inc. R.R.J. is on an advisory committee for Seres Therapeutics Inc., has consulted for Ziopharm Oncology, and holds patents with or receives royalties from Seres Therapeutics Inc. M.R.M.v.d.B. is on the advisory board of and has financial holdings in Seres Therapeutics Inc., serves on the DKMS medical council, has received speaker honoraria from Merck and Acute Leukemia Forum, holds patents that receive royalties from Seres Therapeutics Inc., has received honorarium and research support (1 January 2017 to present) from Seres Therapeutics Inc., and IP licensing with Seres Therapeutics Inc. and Juno. S.A.G. has received research funding from Amgen, Actinium, Celgene, Johnson & Johnson, Miltenyi, and Takeda and serves on the advisory boards of Amgen, Actinium, Celgene, Johnson & Johnson, Jazz Pharmaceuticals, Takeda, Kite, and Spectrum Pharma. E.G.P. has received speaker honoraria from Bristol-Myers Squibb, Celgene, Seres Therapeutics, MedImmune, Novartis, and Ferring Pharmaceuticals; is an inventor on patent application no. WPO2015179437A1, entitled “Methods and compositions for reducing Clostridium difficile infection” and #WO2017091753A1, entitled “Methods and compositions for reducing vancomycin-resistant enterococci infection or colonization”; and holds patents that receive royalties from Seres Therapeutics Inc.
Footnotes
Data and materials availability: All data associated with this study can be found in the paper or the Supplementary Materials. 16S rRNA sequencing data have been deposited in the Sequence Read Archive, accession no. SUB4481714.
REFERENCES AND NOTES
- 1.Pamer EG, Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352, 535–538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M-A, Jenq RR, van den Brink MRM, Pamer EG, Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis 55, 905–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, Holler B, Wolff D, Edinger M, Andreesen R, Levine JE, Ferrara JL, Gessner A, Spang R, Oefner PJ, Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: Loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol. Blood Marrow Transplant 20, 640–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, Docampo MD, Peled JU, Arpaia N, Cross JR, Peets TK, Lumish MA, Shono Y, Dudakov JA, Poeck H, Hanash AM, Barker JN, Perales M-A, Giralt SA, Pamer EG, van den Brink MRM, Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol. Blood Marrow Transplant 21, 1373–1383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, Liu C, West ML, Singer NV, Equinda MJ, Gobourne A, Lipuma L, Young LF, Smith OM, Ghosh A, Hanash AM, Goldberg JD, Aoyama K, Blazar BR, Pamer EG, van den Brink MRM, Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med 209, 903–911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, Slingerland AE, Smith OM, Young LF, Gupta J, Lieberman SR, Jay HV, Ahr KF, Porosnicu Rodriguez KA, Xu K, Calarfiore M, Poeck H, Caballero S, Devlin SM, Rapaport F, Dudakov JA, Hanash AM, Gyurkocza B, Murphy GF, Gomes C, Liu C, Moss EL, Falconer SB, Bhatt AS, Taur Y, Pamer EG, van den Brink MRM, Jenq RR, Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med 8, 339ra71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pamer EG, Taur Y, Jenq R, van den Brink MRM, Impact of the intestinal microbiota on infections and survival following hematopoietic stem cell transplantation. Blood 124, SCI–48 (2014). [Google Scholar]
- 8.Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A, The gut microbiota and host health: A new clinical frontier. Gut 65, 330–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ, Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, Mimura I, Morita H, Sugiyama D, Nishikawa H, Hattori M, Hino Y, Ikegawa S, Yamamoto K, Toya T, Doki N, Koizumi K, Honda K, Ohashi K, Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood 128, 2083–2088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, Gordon S, Gluck M, Hohmann EL, Kao D, Kao JY, McQuillen DP, Mellow M, Rank KM, Rao K, Ray A, Schwartz MA, Singh N,Stollman N, Suskind DL, Vindigni SM, Youngster I, Brandt L, Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am. J. Gastroenterol 109, 1065–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Maaten L, Hinton G, Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605 (2008). [Google Scholar]
- 13.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL, Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, Kolls JK, Weiser JN, Worthen GS, The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med 20, 524–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YJ, Arguello ES, Jenq RR, Littmann E, Kim GJ, Miller LC, Ling L, Figueroa C, Robilotti E, Perales M-A, Barker JN, Giralt S, van den Brink MRM, Pamer EG, Taur Y, Protectivefactors in the intestinal microbiome against Clostridium difficile infection in recipients of allogeneic hematopoietic stem cell transplantation. J. Infect. Dis 215, 1117–1123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K,Mazmanian SK, Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB, Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect. Immun 80, 3786–3794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber D, Jenq RR, Peled JU, Taur Y, Hiergeist A, Koestler J, Dettmer K, Weber M, Wolff D, Hahn J, Pamer EG, Herr W, Gessner A, Oefner PJ, van den Brink MRM,Holler E, Microbiota disruption induced by early use of broad-spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol. Blood Marrow Transplant 23, 845–852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, Hahn J, Wolff D, Stammler F, Spang R, Herr W, Dettmer K, Holler E, Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 126, 1723–1728 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Kelly CR, Khoruts A, Staley C, Sadowsky MJ, Abd M, Alani M, Bakow B, Curran P, McKenney J, Tisch A, Reinert SE, Machan JT, Brandt LJ, Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: A randomized trial. Ann. Intern. Med 165, 609–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG, Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R, Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC, UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Edgar RC, Flyvbjerg H, Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I, RefSeq microbial genomes database: New representation and annotation strategy. Nucleic Acids Res. 42, D553–D559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amir E.-a. D., Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D, viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Q, Mukherjee S, Furey TS, GSAASeqSP: A toolset for gene set association analysis of RNA-Seq data. Sci. Rep 4, 6347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Wang C, Zhou Y, Speed TP, Gene set enrichment analysis made simple. Stat. Methods Med. Res 18, 565–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Microbiota samples for the 25 patients (14 treatment and 11 control).
Fig. S2. PCoA of microbiota samples shown in Fig. 3.
Fig. S3. Mixed-effects model that controls for other clinical parameters confirms the beneficial effect of auto-FMT in remediating the microbiota of allo-HSCT patients.
Fig. S4. Shotgun sequencing shows that auto-FMT remediates the perturbed microbiome.
Fig. S5. Shown is an analysis of a heterologous FMT conducted in another study compared to the auto-FMT conducted in this study.
Table S1. Characteristics of patients included in this study (14 treated and 11 control). Clinical protocol
Data file S1. Source data for figures.