Abstract
In 1953, Watson and Crick proposed that rarely formed isomers of DNA bases cause spontaneous mutations to occur during the copying of DNA. Sixty-five years later, it looks as though they were right.
How do mutations arise when DNA is copied in cells? In a paper in Nature, Kimsey et al.1 combine observations of DNA structure with measurements of enzyme kinetics and computational modelling to provide a definitive explanation of a seminal mechanism.
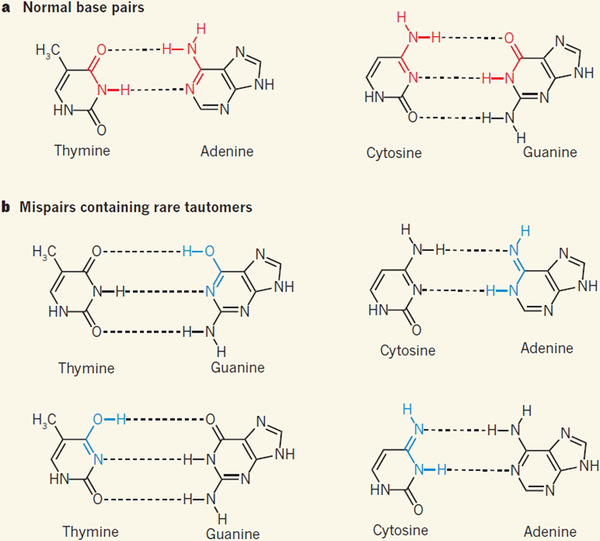
The elucidation of the structure of DNA reported in James Watson and Francis Crick’s classic 1953 paper2 is a monumental piece of work. The key finding was that DNA has a double-helix structure held together by specific interactions between pairs of bases, now known as Watson–Crick pairs: adenine (A) pairs up with thymine (T), whereas guanine (G) pairs with cytosine (C). The DNA bases exist as tautomers (readily interconvertible pairs of isomers), and the A·T and G·C base pairs contain each base in its predominant tautomeric form (Fig. 1a). The process by which DNA is assembled was not known at the time, but the discovery of base-pairing suggested that the sequence of nucleotides on one strand of a double helix could govern the sequence that was constructed on the complementary strand2.
Figure 1 |. Base-pair structures in DNA.
a, The double-helix structure of DNA is held together by specific interactions (dotted lines) between pairs of bases: adenine (A) pairs with thymine (T), and guanine (G) pairs with cytosine (C). b, The DNA bases form rare isomeric structures known as tautomers, which can allow the formation of mispairs; bonds shown in blue are the tautomeric forms of the bonds shown in red in a. Kimsey et al.1 have detected tautomeric G·T mispairs in DNA duplexes, and conclude from modelling studies that this explains the frequency with which G·T is misincorporated into DNA during DNA duplication by polymerase enzymes — as proposed3 by Watson and Crick in 1953.
The DNA structure had other far-reaching implications: it suggested a model for how mutations might arise spontaneously as DNA is made. Watson and Crick proposed3 that mutations could occur because of “a base occurring very occasionally in one of the less likely tautomeric forms, at the moment when the complementary chain is being formed”. In other words, G·T and A·C mispairs could occur if one of the bases is in a disfavoured tautomeric form (Fig. 1b). Such mutations would be easily accommodated because tautomeric mispairs do not distort the helical DNA structure. The disfavoured-tautomer model for spontaneous mutation formation (mutagenesis) was rapidly adopted by biologists and included in textbooks, despite the absence of supporting experimental evidence.
Mispaired structures other than those associated with tautomerization were discovered in the mid-1960s; these included the wobble pairs4 proposed by Crick, and Hoogsteen pairs5. In the mid-1980s, mispairs associated with charged forms of DNA bases were also identified6,7,8. But it wasn’t until 2011 that a C·A mismatch associated with a rare tautomer was finally observed in an X-ray crystal structure9. The mismatch was formed between bases of nucleotides bound in the active site of DNA polymerase (the enzyme that synthesizes DNA from nucleotides) when DNA synthesis was performed in the presence of manganese(ii) ions, which are known to cause mutations. A second X-ray structure10 reported that year identified an ionized G·T mismatch, also formed between substrates bound by DNA polymerase. In both cases, the mismatched pairs had the same geometry as Watson–Crick pairs.
In Watson and Crick’s model for mutagenesis, the rare occurrence of disfavoured tautomeric bases could account for the observed frequency with which DNA polymerases produce mismatches (about one per thousand to one per million base pairs formed11). But such tautomers and the associated base pairs were thought to be almost impossible to detect in duplexes. Then, in 2015, 62 years after the mutagenesis model was proposed, researchers from the same group as Kimsey et al. reported a tour de force of experimental work: they used nuclear magnetic resonance (NMR) spectroscopy to identify12 a long-lived wobble G·T structure that was in a dynamic equilibrium with transient, rarely formed G·T mispairs associated with disfavoured tautomers, and with ionized G·T− structures, both of which have Watson–Crick geometry.
The first step of the DNA-synthesis process that forms a G·T pair is the binding of dGTP (a G-containing nucleotide) in the polymerase’s active site. This is followed by the enzyme’s catalytic step, in which the DNA is elongated through incorporation of a new G·T base pair. Once dGTP is bound in the active site, the base pair formed between dGTP and T on the complementary strand assumes a distorted wobble conformation, but seemingly cannot make the conformational transition needed for the catalytic step10.
Kimsey and colleagues’ current study goes straight to the heart of the mutagenesis model by integrating structural analysis of G·T base pairs in duplexes with measurements of the kinetics of DNA polymerase reactions and computer modelling to show that tautomerism does indeed account for the misincorporation of base pairs. To ensure efficient catalysis, DNA polymerases require optimal geometrical alignment of nucleotide substrates with amino-acid residues in their active site13,14. Such alignment can occur when G·T adopts one of its Watson–Crick-like structures (one of the disfavoured tautomeric forms, or the ionized structure15). Kimsey et al. deduced from their studies that, at neutral pH, at least 99% of G·T misincorporation is attributable to the formation of G·T tautomers — rather than of the ionized structure — from an initially bound G·T wobble pair.
By successfully identifying a role for the disfavoured tautomeric forms of G·T in base-pair misincorporation, Kimsey and colleagues have solved half of the mystery of spontaneous mutagenesis. A solution for the other half now requires the disfavoured tautomeric forms of C·A to be characterized in duplexes and correlated with the rate of C·A misincorporation. So far, NMR and X-ray data have identified only charged C·A+ wobble structures in a DNA duplex7,8.
A related challenge would be to establish the mechanism by which 2-aminopurine, a base analogous to both adenine and guanine, induces mutagenesis. For example, 2-aminopurine is a potent mutagen of the virus bacteriophage T4, for which it increases the frequency of A·T to G·C mutations (and of the reverse G·C to A·T mutations) to 10–50 times the frequency of spontaneous mutation levels16. If 2-aminopurine was found to undergo a tautomeric shift much more frequently than A, it would implicate tautomerization in the mechanism, and thus povide the icing on the cake for the tautomerization model of mutagenesis.
References
- 1.Kimsey IJ et al. Nature 554, 195–201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson JD & Crick FHC Nature 171, 737–738 (1953). [DOI] [PubMed] [Google Scholar]
- 3.Watson JD & Crick FHC Cold Spring Harb. Symp. Quant. Biol 18, 123–131 (1953). [DOI] [PubMed] [Google Scholar]
- 4.Crick FHC J. Mol. Biol 19, 548–555 (1966). [DOI] [PubMed] [Google Scholar]
- 5.Hoogsteen K Acta Crystallogr. 16, 907–916 (1963). [Google Scholar]
- 6.Sowers LC, Eritja R, Kaplan B, Goodman MF & Fazakerly GV J. Biol. Chem 263, 14794–14801 (1988). [PubMed] [Google Scholar]
- 7.Sowers LC, Fazakerley GV, Kim H, Dalton L & Goodman MF Biochemistry 25, 3983–3988 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Hunter WN, Brown T, Anand NN & Kennard O Nature 320, 552–555 (1986). [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Hellinga HW & Beese LS Proc. Natl Acad. Sci. USA 108, 17644–17648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bebenek K, Pedersen LC & Kunkel TA Proc. Natl Acad. Sci. USA 108, 1862–1867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunkel TA & Bebenek K Annu. Rev. Biochem 69, 497–529 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW & Al-Hashimi HM Nature 519, 315–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echols H & Goodman MF Annu. Rev. Biochem 60, 477–511 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Tsai YC & Johnson KA Biochemistry 45, 9675–9687 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Eritja R, Bloom LB & Goodman MF J. Biol. Chem 268, 15935–15943 (1993). [PubMed] [Google Scholar]
- 16.Drake JW & Allen EF Cold Spring Harb. Symp. Quant. Biol 33, 339–344 (1968). [DOI] [PubMed] [Google Scholar]