Abstract
Background
Schistosome parasites lay up to a thousand eggs per day inside the veins of their mammalian hosts. The immature eggs deposited by females against endothelia of venules will embryonate within days. Approximately 30% of the eggs will migrate to the lumen of the intestine to continue the parasite life-cycle. Many eggs, however, are trapped in the liver and intestine causing the main pathology associated with schistosomiasis mansoni and japonica, the liver granulomatous response. Excretory-secretory egg proteins drive much of egg-induced pathogenesis of schistosomiasis mansoni, and Schistosoma japonicum induce a markedly distinct granulomatous response to that of S. mansoni.
Methods
To explore the basis of variations in this responsiveness, we investigated the proteome of eggs of S. japonicum. Using mass spectrometry qualitative and quantitative (SWATH) analyses, we describe the protein composition of S. japonicum eggs secretory proteins (ESP), and the differential expression of proteins by fully mature and immature eggs, isolated from faeces and ex vivo adults.
Results
Of 957 egg-related proteins identified, 95 were exclusively found in S. japonicum ESP which imply that they are accessible to host immune system effector elements. An in-silico analysis implies that ESP are able of stimulating the innate and adaptive immune system through several different pathways. While quantitative SWATH analysis revealed 124 proteins that are differentially expressed by mature and immature S. japonicum eggs, illuminating some important aspects of eggs biology and infection, we also show that mature eggs are more likely than immature eggs to stimulate host immune responses.
Conclusions
Here we present a list of potential targets that can be used to develop better strategies to avoid severe morbidity during S. japonicum infection, as well as improving diagnosis, treatment and control of schistosomiasis japonica.
Electronic supplementary material
The online version of this article (10.1186/s13071-019-3403-1) contains supplementary material, which is available to authorized users.
Keywords: Eggs, ESP, Mass spectrometry Schistosoma japonicum, Schistosomiasis, SWATH analysis
Background
Schistosomiasis is a chronic parasitic disease affecting more than 250 million people in tropical and subtropical countries [1, 2]. Eggs of the three main Schistosoma species (Schistosoma mansoni, S. haematobium and S. japonicum) have been demonstrated to play a major role in human disease attributable to schistosomiasis, being involved in its morbidity, pathology, host immune modulation, diagnosis and transmission [3, 4]. A single female S. japonicum lays approximately 1000 eggs per day inside the veins of its mammal host [5]. Some 30% of those eggs will reach the faeces, whereas the remainder will be trapped in the liver and intestinal tissues, where they induce the main pathology of schistosomiasis, the host granulomatous response [6]. The presence of eggs in the tissues leads to a continuous antigenic stimulation that induces chronic inflammation, subsequent liver fibrosis and portal hypertension, which often evolve into conditions such as hepatosplenomegaly and ascites [2, 6, 7].
When first released into the host circulation, the eggs of S. mansoni and S. japonicum are immature and, therefore, smaller and less complex than mature eggs [8, 9]. Subsequent egg maturation takes seven days. During this process, the eggs develop an extra-embryonic envelope beneath the shell, the inner envelope, that is highly active metabolically and is thought to be the primary source of the egg’s secretions [8]. The shell has numerous pores through which these secretions escape, releasing in S. mansoni at least, highly immunogenic molecules such as Kappa-5, IL-4-inducing principle (IPSE/ alpha-1), and the T2 ribonuclease Omega-1 [10–12]. Over recent years, total and specific proteins from whole eggs, egg secretory proteins (ESP), and soluble egg antigens (SEA) have been implicated in processes such as modulation of the host immune response towards a T helper (Th2) pattern [6, 13, 14]. Moreover, specific egg-derived proteins, including Sm-P40 and thioredoxin peroxidase-1 (Sm-TPx-1), were shown to induce liver lesions and peri-ovular reactions [15, 16]. Other proteins, such as Sjp40 have shown potential to be used for early diagnosis of schistosomiasis [17].
Egg ESP provoke and maintain the granulomatous response of the host and, therefore, play an important role in pathogenesis. For the parasite, however, this response is adaptive, in that it pushes eggs across endothelial boundaries into the intestinal lumen for excretion from the host, an extremely important step for the parasite to continue its life-cycle, but one still poorly understood [18, 19]. Indeed, the stimulation of the host immune response generates a cascade of reactions that leads to the activation of molecules, such as Interleukins, surface-adhesion molecules of vascular endothelia (ICAM-1, E-selectin and VCAM-1) and plasma factors that appear to help the eggs cross the intestinal barrier by causing modifications of the tight junctions [19–22].
Of significant interest in relation to the host-schistosome parasite interaction is the nature of the proteins that are released from the eggs. Different proteomic analyses of the schistosome eggs and derived secretions have revealed their complexity and variability [23, 24]. Whereas Liu et al. [23] evaluated the whole S. japonicum egg and identified a set of 258 proteins in this type of sample, two distinct investigations of the ESP derived from S. mansoni showed that the eggs release as few as six proteins [25], or as many as 188 [26]. This wide discrepancy might be explained by the different proteomic methodologies applied in each investigation and indicate the necessity of a careful comparison between studies.
Nonetheless, a more specific proteomic approach of the S. japonicum eggs could result in a better understanding of its biology, as well as the severe pathology of schistosomiasis associated with the granulomatous response and the host-parasite interactions. Moreover, these data could lead to improvement of the diagnosis of schistosomiasis, which is currently primarily based on direct or indirect detection of eggs and their quantification [27–29]. To address these questions, we investigated the protein composition of ESP derived from S. japonicum eggs, as well as the differential expression of proteins in fully mature and immature eggs through mass spectrometry qualitative and quantitative (SWATH) analyses. Through these approaches we explored the potential role of different proteins in the host-parasite relationship, immune modulation and pathogenesis of schistosomiasis japonica and generated a list of suitable molecular targets with potential for diagnosis and treatment of this disease.
Methods
Purification of S. japonicum eggs
ARC Swiss mice were infected percutaneously with 40 cercariae of S. japonicum (Anhui strain, China) shed from field isolates of Oncomelania hupensis snails. After 6 weeks, feces were collected and the mice were perfused to collect the livers and worms.
Embryonated S. japonicum eggs from mice faeces (herein designated mSj eggs for mature S. japonicum eggs) were purified from samples of at least 15 infected animals by serial passage through meshes of 500, 150 and 45 µm, respectively. The eggs retained in the last sieve were submitted to a 60% iodixanol (w/v) (OptiPrep, Sigma, Gillingham, UK) density gradient and centrifuged at 2000× rpm for 2 min. The layer containing the purified eggs in feces was verified by optical microscopy in order to ensure that debris were removed (Additional file 1: Figure S1). Liver eggs were obtained by Collagenase B (Sigma) digestion, according to the method of Dalton et al. [30], and were further purified using 60% iodixanol density gradient and verified by optical microscopy, as described above.
After perfusion, the S. japonicum worms were cultured in Sterile RPMI 1640 Cell Culture Media (Gibco, Dublin, Ie), supplemented with 10 µl/ml of penicillin-streptomycin (10,000 U/ml, Gibco), for 24 h at 37 °C, 5% CO2. Immature eggs (iSj eggs) were then collected within the whole culture media, washed three times with sterile phosphate buffer solution (PBS 1×), verified by optical microscopy (Additional file 2: Figure S2) and frozen at -20 °C. All the procedures for isolation of liver eggs and iSj eggs were repeated at least three times and each time involved a group of three animals. For further analyses, all samples from mSj eggs, liver eggs or iSj eggs were pooled together.
Egg secretory proteins (ESP)
To obtain ESP, approximately 500 eggs purified from liver were added per well in a 96-well plate with sterile RPMI 1640 media (Gibco), supplemented with 10 µl/ml of penicillin-streptomycin (10,000 U/ml, Gibco), and incubated at 37 °C, 5% CO2. The supernatant was collected each hour in the first 3 h incubation. To determine whether eggs hatched and if eggs were viable post-incubation, the percentage of hatched eggs were observed under microscopy before and after incubation. The viability of the eggs after incubation was verified by making the eggs hatch (Additional file 3: Figure S3). Supernatants collected in the first 3 h were then centrifuged at 4 °C, 14,000× g for 30 min and the supernatant was frozen at -20 °C. These procedures were repeated at least three times (eggs obtained from different group of animals).
Protein extraction and fractionation
In order to extract the total proteins from mSj, iSj, and liver eggs, eggs were ruptured by freezing in liquid nitrogen, after which lysis buffer was added (1% SDS, 10 mM CHAPS, 0.5 M MgCl2, and protease inhibitor cocktail 1× in 100 mM TEAB). The samples were agitated at 4 °C for 40 min, centrifuged at 14,000× g for 10 min, and the supernatant was collected and frozen at -20 °C. The pellets were verified under an optical microscope to verify the rupture of the eggs. The proteins were concentrated and the buffer was exchanged using Millipore Amicon filters 10K. The protein concentration of each sample was verified using the Bicinchoninic acid assay (Pierce, Thermo Fisher Scientific, Paisley, UK) following the manufacturer’s protocol.
Proteomic analysis: in-gel fractionation
Triplicate samples (20 µg) of proteins extracted from S. japonicum liver eggs and ESP, were resolved in 12% one-dimension (1DE) SDS-PAGE gels. After gel fixation, the proteins were stained with EZBlue G-250 colloidal Coommassie Stain (Sigma), and each line was fractionated into 12 gel bands. In-gel trypsin digestion was performed [31], and the mixture of peptides was then collected, dried in speed vacuum and frozen at -20 °C.
Proteomic analysis: filter-aided sample preparation method (FASP)
The total proteins from mSj and iSj were processed by the FASP method [32]. Briefly, triplicate samples of 30 µg of protein were reduced (final concentration of 20 mM DTT, at 95 °C, 10 min) and alkylated (final concentration of 40 mM iodoacetamide, at room temperature for 30 min in the dark). The proteins were then transferred into a 10K Millipore Amicon filter (Merck Millipore, Hertfordshire, UK) and washed first with 8 M urea in 100 mM TEAB and then 50 nM TEAB. Trypsin was added into the samples (ratio 1:50) and tryptic digestion was performed at 37 °C for 18 h. The peptides were recovered in 50 mM TEAB, dried in speed vacuum, and frozen at -20 °C.
Mass spectrometry (MS) analyses: information-dependent acquisition (IDA)
The peptide fractions from the S. japonicum liver eggs and ESP were suspended in 0.1% TFA and desalted on ZipTip C18 pipette tips, 5 µg (Millipore). The C18 tips were activated with 70% ACN/ 0.1% TFA and then equilibrated with 0.1% TFA before loading the peptides. Loaded peptides were washed with 0.1% TFA and eluted with 80% ACN/ 0.1% TFA before drying in speed vacuum, resuspended in 20 µl of 0.2% ACN/ 0.1% TFA complemented with iRT calibrant (Biognosys, Zurich, Switzerland) [33], and analyzed by reverse phase chromatography with an Eksigent nanoflex cHiPLC coupled to a TripleTOF 5600 mass spectrometer (ABSCIEX, Canada) equipped with a nano-electrospray ion source. The peptides were separated on the eksigent analytical cHiPLC column (3 μm, ChromXP C18CL, 120 Å, 15 cm × 200 µm) with a method using three consecutive linear gradients: 5–10% solvent B (acetonitrile/0.1% formic acid) over 2 min; 10–40% solvent B over 58 min; 40–50% solvent B over 5 min, at a 500 nl/min flow rate. Eluted peptides were acquired in positive mode by electrospray (voltage 2300 V) using IDA method, and those ions exceeding a threshold of 50 counts and possessing a charge state of +2 to +4, were set to trigger the acquisition of product ion spectra for the 10 the most intense ions with 10 second exclusion after one occurrence.
MS: Sequential window acquisition of all theoretical fragment ion spectra (SWATH)
Peptides from mSj and iSj samples (triplicates), obtained from FASP were resuspended in 0.1% TFA, normalized and desalted on ZipTip C18 pipette tips, 5 µg (Millipore) as described above, dried in speed vacuum and resuspended in 30 µl of 2% ACN/0.1% FA complemented by iRT calibrant (Biognosys, Zurich, Switzerland) [33]. The peptides were separated on an Eksigent analytical cHiPLC column (3 m, ChromXP C18CL, 120 Å, 15 cm × 200 µm) using three linear gradients: 5–10% solvent B (ACN/0.1% formic acid) over 2 min, 10–40% solvent B over 58 min and 40–50% solvent B over 5 min at a 300 nl/min flow rate, and then directly introduced into a TripleTOF 5600 mass spectrometer (ABSCIEX, Ontario, Canada), as previously described. A rolling collision energy method was used to fragment all ions in a set of 26 sequential overlapping windows of 25 AMU over a mass range coverage of 350–1000 (m/z). An accumulation time of 100 ms was used for each fragment ion scan resulting in a total cycle time of 2.9 s. Data were acquired and processed using Analyst TF 1.7 software (AB SCIEX).
Data analysis
The detected protein threshold was set as 0.1 and the false-discovery rate (FDR) was calculated using searches against a decoy database comprised of reversed sequences. All searches were conducted against Schistosoma sp. specific protein databases downloaded from WormBase Parasite (http://www.ebi.ac.uk), UniProt (http://www.uniprot.org), and Scientific Data Sharing Information Bioinformation (http://lifecenter.sgst.cn/protein). The set comprised a total of 39,768 non-redundant protein sequences.
X! Tandem Jackhammer TPP (2013.06.15.1) was used to search all IDA files (in triplicate for each sample) against a target/decoy version of the Schistosoma databases reference proteome set additionally containing a reversed version of each sequence and the iRT sequences. The spectral library was generated accordingly following Schubert et al. [62]; results were assessed for statistical validity in the Trans Proteomic Pipeline (TPP) using PeptideProphet and iProphet, and the false discovery rate was estimated using Mayu. SpectraST (version 5.0) in the TPP was used to normalize the retention time and generate a consensus library, while the OpenSWATH was used to build a target/decoy spectral library [33]. The final lists of proteins were generated based on the results of the triplicates, and included all proteins identified with at least two unique peptides.
For SWATH analysis, ProteoWizard msconvert was used to convert the files to mzML format, and then searches against the spectral library were performed using OpenSWATH with a FDR < 0.1%. Feature alignment was performed using the TRansition of Identification Confidence (TRIC) algorithm (https://pypi.python.org/pypi/msproteomicstools). Next, the R package SWATH2stats was used to filter low scoring peak groups and remove proteins with less than two associated peptides [34]. Then, the data were normalized (‘equalizeMedians’ method), summarized (Tukey’s median polish’ parameter estimation method), and finally differential expression analysis was performed with MSstats [35]. Differential expression values with an adjusted P-value ≤ 0.05 were considered significant.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012835, available at http://www.proteomexchange.org/submission/index.html.
Bioinformatics analysis
The gene ontology and enrichment analyses were made using FunRich (Version 3.1.3) (http://funrich.org/download) and the gene ontology (GO) database (http://www.geneontology.org/). Pathways analysis was performed using the Reactome database (Version 3.5) (https://reactome.org/PathwayBrowser/) and SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/), with mammalian parameters, was used to determine whether the proteins identified were predicted to be secreted through classical or non-classical pathways.
Results
Processing effects on egg viability
Matheson & Wilson [25] suggested that purification of schistosome eggs using the Dalton et al. method [30] could lead to their rupture and death. However, at the end of incubation, the eggs were verified as alive by hatching experiments, conducted in de-ionized water, which revealed hatching rates of over 90%, indicating that our methods of purification of liver eggs did not adversely affect their integrity or viability.
Library: total proteins from S. japonicum eggs
To improve the sensitivity of the LC-MS/MS analysis, we fractionated the total proteins extracted from whole S. japonicum eggs purified from livers by resolving them in 1-DE SDS-PAGE gels. Of the total proteins identified using IDA analysis, excluding repeats and redundancies, we identified 862 proteins, filtered to 95% sequence identity, with a unique peptide count ≥ 2 (Additional file 4: Table S1). This list of proteins was used as our reference library for SWATH analysis. SecretomeP analysis (SecP) revealed that 524 (61%) of the proteins found in S. japonicum eggs contained a predicted signal or a non-classical sequence (Additional file 4: Table S1).
Proteins found exclusively in the ESP
A total of 461 proteins were identified in the analysis of S. japonicum ESP. Of these, some 62% were predicted to contain classical or non-classical secretory sequences (Additional file 4: Table S1). In order to distinguish those proteins primarily secreted, the set of proteins identified in ESP was compared with the set of proteins identified in the total extract of whole S. japonicum eggs (Additional file 4: Table S1); those proteins found in both extracts were separated from proteins only present in ESP, resulting in a list of 95 proteins in ESP (Table 1).
Table 1.
List of proteins identified exclusively in ESP from Schistosoma japonicum eggs
| Accession | Protein name | Unused | % Cov | Peptides (95%) | SecP |
|---|---|---|---|---|---|
| tr|Q1HDV2 | Fructose-bisphosphate aldolase | 66.55 | 88.98 | 116 | – |
| tr|B2LXU1 | Enolase | 0.20 | 65.89 | 47 | – |
| tr|C1LRC5 | Carbonyl reductase 1 | 0.00 | 81.95 | 45 | – |
| tr|Q5DFP8 | Putative uncharacterized protein | 2.00 | 75.83 | 43 | – |
| tr|C1LKA0 | Purine nucleoside phosphorylase | 2.17 | 72.13 | 40 | NC |
| tr|G4VHK8 | Tubulin beta chain | 40.95 | 6.24 | 33 | NC |
| tr|C1LIX7 | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog | 31.78 | 45.39 | 23 | NC |
| tr|C1LIA3 | Dihydrolipoyl dehydrogenase | 36.56 | 57.74 | 22 | NC |
| tr|C1LAW4 | Major egg antigen (P40) | 19.2 | 48.01 | 22 | – |
| tr|A0A183N4E4 | Uncharacterized protein | 0.00 | 43.66 | 19 | SP |
| tr|Q5DBS1 | SJCHGC07012 protein | 29.51 | 51.88 | 18 | NC |
| tr|C1LNQ7 | Protein disulfide-isomerase | 27.32 | 43.20 | 17 | SP |
| tr|Q5DEE6 | Peptidyl-prolyl cis-trans isomerase | 10.54 | 55.48 | 15 | – |
| tr|G4V9B9 | Putative heat-shock protein | 0.03 | 26.78 | 15 | – |
| tr|C1LS07 | Cell wall integrity and stress response component 1 | 0.00 | 51.16 | 15 | – |
| tr|C1L675 | Arginase OS=Schis | 25.96 | 52.20 | 14 | – |
| tr|C1LWQ9 | Elongation factor 1-alpha | 17.02 | 64.45 | 14 | – |
| tr|A0A094ZS46 | Spectrin alpha chain | 25.99 | 33.16 | 12 | – |
| tr|C7TYX3 | Putative uncharacterized protein | 0.00 | 4.30 | 12 | SP |
| tr|A0A183QGT0 | Tubulin alpha chain | 0.00 | 29.92 | 11 | – |
| tr|G4VSJ5 | Putative glycogen phosphorylase | 0.00 | 37.74 | 11 | – |
| tr|Q7Z0T1 | Phosphoglycerate kinase | 15.49 | 49.75 | 9 | – |
| tr|C1LLK8 | Peptidase M8, leishmanolysin,domain-containing protein | 9.56 | 33.91 | 9 | SP |
| tr|C7TQR6 | Ribonuclease X25 | 8.96 | 53.29 | 9 | SP |
| tr|Q5DHF8 | Histone H3 | 5.64 | 41.17 | 9 | NC |
| tr|C1L4U6 | 3’(2’), 5’-bisphosphate nucleotidase | 16.83 | 36.62 | 8 | SP |
| tr|C1LYD5 | Ubiquitin C | 7.32 | 7.80 | 8 | – |
| tr|C1LQN5 | Uncharacterized protein | 0.08 | 44.90 | 8 | SP |
| tr|A0A094ZSL4 | Spectrin beta chain | 13.2 | 21.92 | 7 | – |
| tr|C1L6J2 | Uncharacterized protein | 12.2 | 47.83 | 7 | SP |
| tr|Q86DZ2 | Clone ZZZ395 mRNA sequence | 10.59 | 39.91 | 7 | – |
| tr|C1LEJ4 | Histone H2A | 5.28 | 56.80 | 7 | NC |
| tr|C1LUP9 | Peptidase inhibitor 16 | 0.00 | 46.02 | 7 | SP |
| tr|C1L9E0 | Receptor expression-enhancing protein | 8.30 | 35.44 | 6 | NC |
| tr|G4VS58 | Putative prohibitin | 7.97 | 34.02 | 6 | NC |
| tr|A0A183L8B7 | Uncharacterized protein | 1.70 | 35.49 | 6 | NC |
| tr|G4VG96 | Cell polarity protein | 10.72 | 18.52 | 5 | – |
| tr|Q5DAE9 | SJCHGC09095 protein | 8.60 | 43.95 | 5 | – |
| tr|C1LRF9 | Fatty acid binding protein 7, brain | 8.27 | 80.29 | 5 | – |
| tr|Q7JNB4 | Aspartic protease (Fragment) | 6.14 | 27.34 | 5 | SP |
| tr|A0A183MKN4 | Uncharacterized protein | 5.08 | 3.04 | 5 | – |
| tr|Q5DF47 | Putative uncharacterized protein | 8.77 | 41.81 | 4 | – |
| tr|A0A183NI42 | Uncharacterized protein | 7.00 | 31.92 | 4 | – |
| tr|G4VFV4 | Putative rap1 | 4.97 | 30.97 | 4 | NC |
| tr|A0A183PY58 | Uncharacterized protein | 4.02 | 15.36 | 4 | NC |
| tr|A0A183NZF1 | Peptidyl-prolyl cis-trans isomerase | 0.07 | 30.52 | 4 | SP |
| tr|C1LID9 | LAMA-like protein 2 | 6.72 | 1.90 | 3 | SP |
| tr|C1LA47 | Cathepsin B-like cysteine proteinase | 6.18 | 25.15 | 3 | SP |
| tr|Q5C774 | SJCHGC02069 protein (Fragment) | 5.38 | 3.84 | 3 | SP |
| tr|Q5D9E0 | SJCHGC01621 protein | 4.86 | 2.87 | 3 | NC |
| tr|C1LCS9 | Phosphatase 2A inhibitor I2PP2A | 4.86 | 52.49 | 3 | – |
| tr|C1LKV5 | Rho GDP-dissociation inhibitor 2 | 4.85 | 45.73 | 3 | – |
| tr|Q5DBC3 | SJCHGC06819 protein | 4.57 | 17.52 | 3 | NC |
| tr|Q5DFY2 | SJCHGC06929 protein | 4.40 | 25.61 | 3 | SP |
| tr|Q5DFR4 | ATP:ADP antiporter | 4.01 | 40.83 | 3 | – |
| tr|A0A183QZZ1 | Uncharacterized protein | 4.00 | 19.41 | 3 | NC |
| tr|Q8I8A1 | Rho2 GTPase (Fragment) | 3.82 | 32.80 | 3 | NC |
| tr|Q03528 | 22.6kd tegumental associated antigen | 3.75 | 35.60 | 3 | NC |
| tr|G4VDJ6 | Putative adp-ribosylation factor, arf | 0.06 | 45.86 | 3 | NC |
| tr|A0A183NLB0 | T-complex protein 1 subunit gamma | 0.03 | 24.59 | 3 | – |
| tr|C1LNJ0 | Putative Retinol dehydrogenase 11 | 4.92 | 39.01 | 2 | NC |
| tr|A0A183K1U2 | Uncharacterized protein | 4.81 | 19.40 | 2 | SP |
| tr|A0A183JQD4 | Uncharacterized protein | 4.48 | 31.16 | 2 | – |
| tr|C7TUC3 | Putative uncharacterized protein | 4.05 | 71.74 | 2 | SP |
| tr|G4V9G5 | Putative 60s ribosomal protein L12 | 4.03 | 45.44 | 2 | NC |
| tr|C1L7G5 | Ferritin | 4.00 | 31.97 | 2 | – |
| tr|Q5BT38 | SJCHGC02867 protein (Fragment) | 3.73 | 21.87 | 2 | – |
| tr|C1LTL1 | Universal stress protein | 3.65 | 35.22 | 2 | – |
| tr|C1LKU6 | Erythrocyte band 7 integral membrane protein | 3.24 | 15.64 | 2 | NC |
| tr|C1LNJ7 | Egg protein CP1531 | 3.18 | 17.52 | 2 | SP |
| tr|G4V7S8 | Serine hydroxymethyltransferase | 3.09 | 69.43 | 2 | – |
| tr|C1L857 | Chaperonin containing TCP1, subunit 5 (Epsilon) | 2.98 | 35.98 | 2 | – |
| tr|A0A094ZXB9 | Aminoacylase-1 | 2.89 | 17.02 | 2 | – |
| tr|Q9B8Z2 | Cytochrome c oxidase subunit 2 | 2.82 | 84.16 | 2 | SP |
| tr|G4VJ08 | Acyl-CoA thioesterase-related | 2.47 | 10.39 | 2 | NC |
| tr|C1LEX9 | Transmembrane protease, serine 6 | 2.21 | 13.34 | 2 | SP |
| tr|G4M1G4 | REVERSED Dynein heavy chain, putative | 2.14 | 57.13 | 2 | – |
| tr|A0A183P199 | Uncharacterized protein | 2.06 | 13.37 | 2 | NC |
| tr|A0A0R4I956 | Myosin 2 heavy chain | 2.05 | 15.50 | 2 | – |
| tr|G4VB27 | Putative uncharacterized protein | 2.02 | 95.39 | 2 | – |
| tr|G4V6J0 | Putative copine | 2.02 | 56.00 | 2 | NC |
| tr|A0A094ZYC9 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2 | 2.01 | 9.40 | 2 | SP |
| tr|C1LP66 | Aquaporin-3 (AQP-3) | 2.00 | 15.24 | 2 | – |
| tr|A0A183LXE3 | Uncharacterized protein | 2.00 | 32.67 | 2 | NC |
| tr|G4LXF1 | Rab6, putative | 1.87 | 33.98 | 2 | NC |
| tr|A0A183KN75 | Uncharacterized protein | 1.68 | 17.46 | 2 | NC |
| tr|Q7Z1I7 | GTP-binding protein-like protein | 1.37 | 14.06 | 2 | – |
| tr|A0A183NZ41 | Uncharacterized protein | 1.37 | 79.14 | 2 | – |
| tr|Q5DI05 | Elongation factor Tu | 1.24 | 16.21 | 2 | SP |
| tr|A0A183MXQ1 | Uncharacterized protein | 1.24 | 27.05 | 2 | NC |
| tr|A0A183QCJ7 | Uncharacterized protein | 1.16 | 95.11 | 2 | NI |
| tr|A0A183LI65 | Uncharacterized protein | 0.77 | 19.61 | 2 | – |
| tr|Q7Z1I5 | 40S ribosomal protein S8 | 0.64 | 23.57 | 2 | – |
| tr|G4VQJ4 | Putative adenine phosphoribosyltransferase | 0.09 | 96.77 | 2 | – |
| tr|A0A095ACH1 | REVERSED Uncharacterized protein (Fragment) | 0.00 | 24.43 | 2 | NC |
Abbreviations: ESP, egg secretory proteins; Unused, quantification for proteins in the ProteinPilot software; %Cov, ratio of the protein sequence covered by the matched peptides; Peptides (95%) total number of detected peptides with 95% of confidence; SecP, (Secretome P 2.0) results described as: SP, indicate presence of predicted signal sequence; NC, indicate non-classical secreted proteins; (–), non-detected
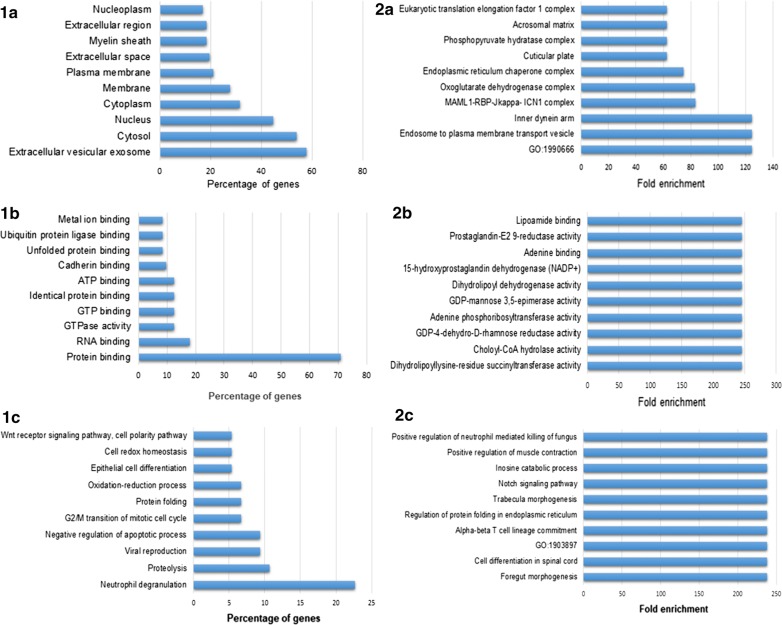
The complete gene ontology and enrichment analyses of the 95 proteins primary present in ESP is shown in Fig. 1 that presents the ten predominant cellular components, molecular functions and biological processes in which the proteins were classified. The analysis, based on homology of the S. japonicum proteins with human proteins, indicated that proteins present in the ESP were those commonly associated with extracellular vesicles, which is ultimately in line with the verification of the presence of classical or non-classical secretory sequences in 53 (54%) of these proteins (Table 1). Most of these proteins contained non-classical signal sequence that could be associated with protein secreted inside vesicles. In relation to the molecular function, most proteins were verified to be involved in “binding” and “catalytic activity”, while in terms of biological processes, the proteins in ESP were primary associated with neutrophil degranulation and proteolysis (Fig. 1).
Fig. 1.
Gene ontology and enrichment analyses of the S. japonicum ESP. The panels show the top 10 classes of: Cellular component and related enrichment analysis (1a and 2a); Molecular function and related enrichment analysis (1b and 2b); Biological process and related enrichment analysis (1c and 2c). Analyses performed using FunRich (V 3.1.3)
Proteomic comparison between immature (iSj) and mature (mSj) eggs
SWATH/MS is a type of data-independent acquisition method of analysis used to evaluate quantitatively complex samples with high reproducibility [36]. Using this method, we compared the expression level of each protein identified in immature (iSj) and mature (mSj) eggs. Our spectra libraries were constructed through analyses of the whole S. japonicum eggs from liver and comprised 862 identified proteins (Additional file 4: Table S1).
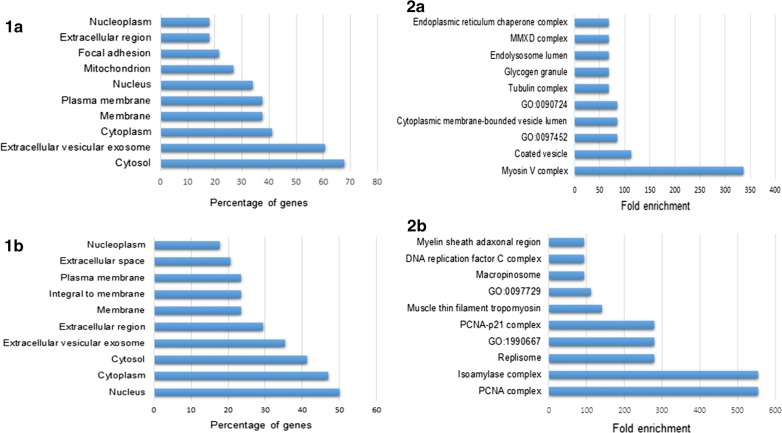
The relative expression levels of proteins were calculated using OpenSWATH and MStats and revealed 124 proteins expressed differentially between the two samples. Of these, 74 were shown to be upregulated, while 50 were downregulated proteins in iSj eggs when compared with mSj ones (Additional file 5: Table S2). According to gene ontology analysis (Figs. 2, 3 and 4), both up- and downregulated proteins in iSj are associated with a range of intracellular organelles and the cytosol. The enrichment analysis demonstrated that, in general, the proteins of iSj are components of organelles and structures participating in cell organization. Analysis of proteins predominantly expressed by mSj eggs showed they are associated with structures responsible for motility and the uptake of molecules (Fig. 2).
Fig. 2.
Cellular components and enrichment analyses of proteins that are up- and downregulated in S. japonicum immature eggs when compared to mature ones. The top 10 classes of cellular components and related enrichment analysis of proteins upregulated (1a and 2a) and proteins down-regulated (1b and 2b) in iSj. Analyses performed using FunRich (V 3.1.3)
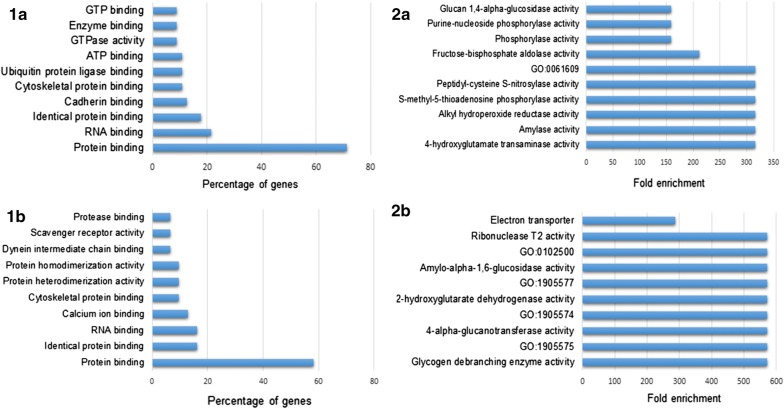
Fig. 3.
Molecular functions and enrichment analyses of proteins up- and downregulated in S. japonicum immature eggs when compared to mature ones. The top 10 classes of molecular functions and related enrichment analysis of proteins upregulated (1a and 2a) and proteins downregulated (1b and 2b) in iSj. Analyses performed using FunRich (V 3.1.3)
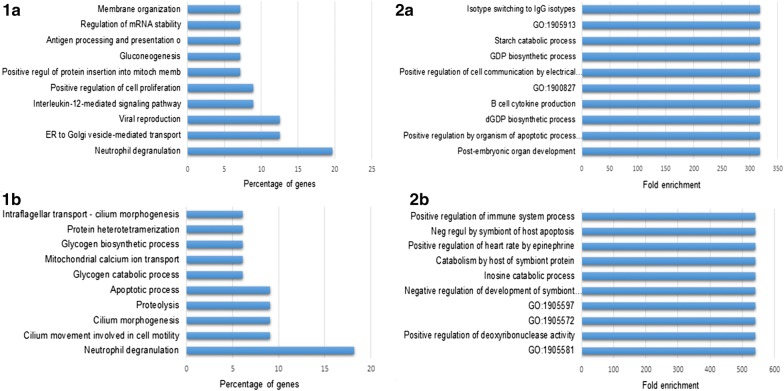
Fig. 4.
Biological processes and enrichment analyses of proteins up- and downregulated in S. japonicum immature eggs when compared to mature ones. The top 10 classes of biological processes and related enrichment analysis of proteins upregulated (1a and 2a) and proteins downregulated (1b and 2b) in iSj. Analyses performed using FunRich (V 3.1.3)
In relation to molecular functions, proteins up- and downregulated in eggs were classified with similar functions mainly related to binding and catalytic activity (Figs. 1a, b, 3). The enrichment analysis of iSj eggs revealed that the upregulated proteins are mostly involved in protein and energy production, while proteins predominantly in mSj eggs are associated with cell signaling, movement, transport and regulatory functions (Figs. 2a, b, 3).
Gene ontology analysis also demonstrated that both iSj and mSj eggs seem to be capable of stimulating the host immune system, since their proteins were predicted to be mainly involved in the process of neutrophil degranulation. On the other hand, the other main processes associated with each egg stage indicated that iSj and mSj eggs are performing quite different processes (Fig. 4). While proteins of immature eggs were primary involved in cell proliferation, those of mature eggs were mostly associated with energetic metabolism, especially glycogen degradation, movement and proteolytic activities.
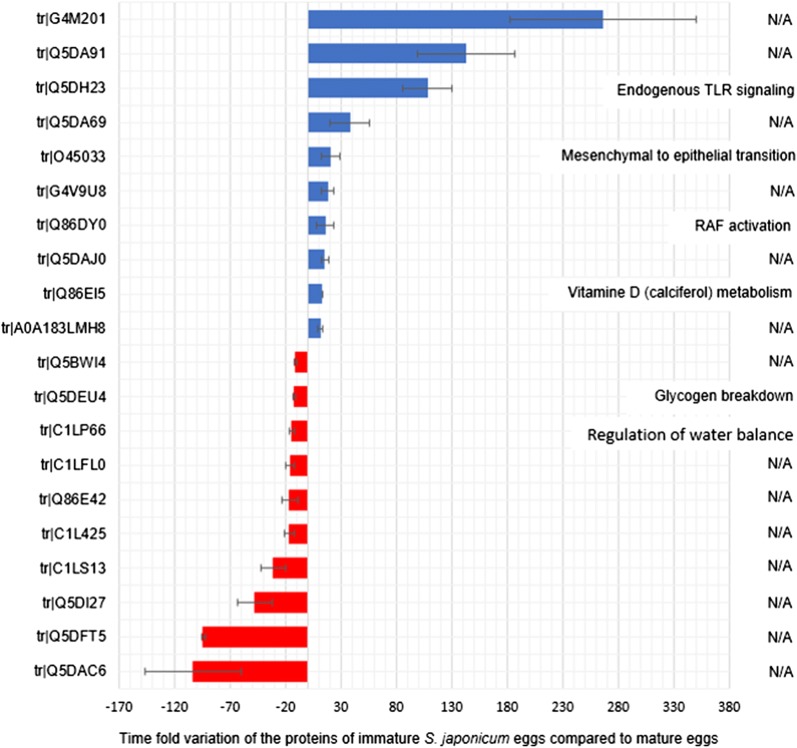
The 10 most significantly up- and downregulated proteins and the respective biological pathways in which they were predicted to be involved are shown in Fig. 5. Considering proteins previously characterized, the stromal cell-derived factor 2-like (G4M201) was the most significantly abundant protein in iSj eggs, while the most abundant protein in mSj eggs was the cell wall integrity and stress response component 1 protein. Many proteins could not be classified according to their biological pathway, but those that were indicated that iSj eggs prioritize pathways related to development, organization and immunomodulation. On the other hand, mSj eggs seemed to be expressing proteins with roles in energy production and regulation of water (Fig. 5), which could be related to the presence of a highly active miracidium and the environmental conditions that mature eggs have to face.
Fig. 5.
Top 10 proteins that are differentially expressed in immature and mature S. japonicum eggs. Proteins analyzed by SWATH method. Results presented as average ± standard deviation of the negative (red) or positive (blue) variation of the proteins of S. japonicum immature eggs compared with the mature eggs. The bars on the left, accession number of the proteins. The text of the right, the respective predicted biological pathway in which the protein is involved. Biological pathway analyses performed using FunRich (V 3.1.3)
Proteins from ESP, immature and mature eggs stimulate the host immune system in different ways
Reactome analysis, concentrating only on those pathways related to the immune system, was performed in order to investigate how the ESP and proteins differentially expressed by eggs at different stages of development are potentially able to interact with and stimulate the host immune system (Table 2). The analysis showed that all the samples contain proteins able to stimulate the host immune system, although it is evident that ESP is capable of doing it via many different ways. ESP is likely to play a central role in driving the immune response towards a Th2 pattern, since it is predicted to stimulate the production of several interleukins, including IL1, IL3, IL5, IL4 and IL13.
Table 2.
Immune system pathways predicted by Reactome 3.5 analysis to be stimulated by proteins present in ESP, immature and mature eggs
| Type of response | ESP | Immature eggs | Mature eggs |
|---|---|---|---|
| Cytokine signalling in immune response | Interferon signalling: Antiviral mechanism | – | – |
| Signalling by Interleukins: IL1, IL3, IL5, GM-CFS, IL4, IL13 | – | – | |
| TNF alpha non-canonical NF-Kappaβ pathway | – | – | |
| Innate immune system | – | Toll-like receptor cascade: Trafficking and processing of endosomal TLR | – |
| Neutrophil degranulation | Neutrophil degranulation | Neutrophil degranulation | |
| Fc gamma receptor (FCGR) dependent phagocytosis | – | – | |
| Fc epsilon receptor (FCERI) signalling; Toll-like receptor cascade | – | – | |
| C-type lectin receptors (CLR’s): CLEC7A (Dectin-1) | – | – | |
| Nucleotide-Binding domain, leucine rich repeat containing receptor (NLR) signalling pathways | – | – | |
| DDX58/IFIHI mediated induction of IFN alpha/beta pathways | – | – | |
| Cytosolic sensors of pathogens associated DNA | – | – | |
| Adaptive immune system | MHC class II antigen presentation | MHC class II antigen presentation | MHC class II antigen presentation |
| RAP-1 signalling | RAP-1 signalling | – | |
| MHC class I mediated antigen processing and presentation | – | – | |
| Signalling by the B cell receptor | – | – | |
| TRL signalling | – | – |
Note: Analysis performed using the software Reactome 3.5
Discussion
Proteomic analysis of a whole eggs extract used eggs purified from mice livers at six weeks post-infection and, therefore, consisted of eggs at different stages of development. It was important to include eggs ranging in development from freshly laid, early zygotic stages, through to fully mature eggs, since this set of data was used as a reference library for subsequent differential analysis of immature and mature eggs. A total of 862 proteins was identified from this hepatic eggs proteome, a composition consistent with other reports of the S. mansoni and S. japonicum egg proteomes (Additional file 4: Table S1) [23–25].
Although excretory-secretory extracts from various life-cycle stages of S. mansoni and S. japonicum have been published [8, 25, 26, 37–39], to our knowledge, the work described here is the first to investigate and identify proteins in the ESP of S. japonicum eggs. In general, the set of 461 proteins we found in S. japonicum ESP is comparable to those components previously characterized in ES products from different developmental stages of S. japonicum [39, 40]. Likewise, Liu et al. [23], used high-throughput proteomics to characterize the protein profiles of different developmental stages of S. japonicum and identified 1441 egg-associated proteins, with 473 of these being exclusively present in eggs when compared with other life-cycle stages. Although Liu et al. [23] verified an overlapping of proteins across the life-cycle, their analysis showed that certain proteins are stage-enriched or probably expressed in response to environmental stimuli.
When we refined the ESP data through comparison with proteins present in a total egg extract, the complexity of S. japonicum ESP, consisting of 95 proteins, still differed considerably from the published data obtained for S. mansoni ESP. For example, Cass et al. [26] identified 188 proteins using shotgun proteomic analysis. By contrast, Ashton and colleagues [8] and Mathieson & Wilson [25], allied 1- and 2-DE gels with mass spectrometry, and identified very few proteins in S. mansoni ESP, notably Omega-1 and IPSE/alpha-1 proteins, and a few proteases. In another study comparing soluble proteins across four life-cycle stages of S. mansoni, including eggs, only 32 distinct proteins were identified [37].
It is important to explain such discrepancies in the number of reported proteins. Mathieson & Wilson [25] attributed the paucity of proteins in ESP products they obtained to the rigorous but gentle processing schedule they used to prepare their egg samples for incubation, notably through the use of saline solutions and trypsin to digest liver samples for egg extraction. The authors further argued that the high egg hatching rates they obtained in S. mansoni egg samples after incubation and collection of ESP was evidence of a superior processing schedule that did not kill or rupture eggs. In our study of S. japonicum eggs, we also performed hatching assays after incubations to obtain ESP and found high hatching rates similar to those of Mathieson & Wilson [25] (Additional file 3: Figure S3). We therefore consider that the variation of the MS sensitivity, the use of different databases, and the application of different methods to prepare and analyze the samples, e.g. straight shotgun MS analysis [26] or MS analysis of 2-DE gel spots [25, 37], could be reasonable explanations for the differences observed between the studies. We argue that even the most carefully undertaken protocol cannot guarantee that a small number of eggs will not hatch, resulting in contamination, albeit minimal, with contents from inside the eggs. It is also possible that a small number of eggs will hatch or rupture within a host.
As expected, most of the proteins identified in S. japonicum ESP were also detected in the proteome of the whole hepatic egg extracts (Additional file 4: Table S1). The fact that 95 proteins were identified exclusively in the ESP does not mean that they are absent in the whole egg, but indicates that these proteins are primarily secreted and therefore transitory in the eggs. Consequently, their presence in a very low concentration in eggs could prevent the identification by the methods used. Proteins such as actin, HSPs, enolase, Glyceraldehyde-3-phosphate dehydrogenase, thioredoxin, proteasome subunits enolase, GLPR1-like protein 1 and cathepsins were among the proteins found in both the whole egg and ESP proteomes. Several proteins identified in the extracts of S. japonicum eggs were previously shown to be present in Schistosoma spp. eggs, e.g. thioredoxin, tetraspanin, cathepsins, serpins, Sjp40 and SM22.6 [23, 26], although the main antigenic proteins found in S. mansoni egg-derived extracts, namely Omega-1, IPSE/ alpha-1 and Kappa-5, were not found in any of the S. japonicum extracts. Additionally, various motor proteins (tubulin, actin and myosin), chaperones, structural and binding proteins, as well as proteins involved in metabolism and in signal transduction were identified. Remarkably, most of the proteins present in ESP were associated with extracellular vesicles in our analysis (Fig. 1) which, allied to the prior evidence of cytoskeleton proteins in ES products, suggest that these proteins possibly follow unconventional routes of excretion in parasites [26, 39, 41].
Egg-related proteins were previously shown to have central roles in polarizing the host immune responses to prevent adaptive responses during schistosomiasis. ESPs, such as Sjp40 and tetraspanin, modulate dendritic cells (DC) through specific interaction with Toll-Like receptor (TLR), stimulating cytokines such as IL-4 and IL-10, thereby driving the immune response towards a Th2 pattern that favors parasite survival [12, 42–44]. Reinforcing these data, our pathway analysis demonstrated the ability of S. japonicum ESP to stimulate the human immune system through several ways, including elicit secretion of interleukins related to the Th2 response (Table 2).
Of note, S. japonicum ESP has been predicted to interact with C-type lectin receptors (CLR’s) in a specific way involving Dectin-1. Although this mechanism is still poorly understood, it was recently demonstrated that S. mansoni SEA can induce a Th2 response through specific interactions involving Dectin 1/2 [45]. Notably, Kaiser and colleagues also showed that SEA depleted of Omega-1 glycoprotein was able to trigger a Th2 response [45]. By implication, this observation suggests that S. japonicum ESP could stimulate the immune system in a similar Omega-1-independent way.
Gene ontology analyses revealed that S. japonicum ESP proteins are mainly associated with neutrophil degranulation (Fig. 1). It is well established that neutrophils play a central role during granuloma formation in response to the presence of schistosome eggs. Indeed, the relative abundance of neutrophils in granulomas is variable in infections with different schistosome species. For example, with schistosomiasis japonica neutrophils accumulate around the eggs eight days after egg deposition starts and the granulomas formed contain higher number of neutrophils than those observed in S. mansoni infections [46–48]. Since IPSE/alpha-1 glycoproteins can inhibit the infiltration of neutrophils, the absence of such a protein in S. japonicum eggs might explain the elevated number of neutrophils in granulomas around these eggs [6, 49].
Recently Zheng et al. [50] reported that during S. japonicum infection, Vγ2 T cells can recruit neutrophils and aggravate liver fibrosis by secreting IL-17A, thereby causing the eggs to be trapped. In addition, it has been shown that neutrophils associated with S. japonicum hepatic granuloma, but not S. mansoni, release neutrophil extracellular traps (NETs) [6, 51], strongly suggesting that there are major differences in ESP constituency between the two species causing hepato-intestinal schistosomiasis and that further studies may lead to a better understanding of the differential pathogenesis of the two infections.
Major immunomodulatory ESP components of S. mansoni eggs are Omega-1 (Ribonuclease-RNase T2) and ISPE [11, 12]. Orthologues of Omega-1 protein have been identified in the genome of S. japonicum [52], but it does not appear to be part of the ESP of its eggs. While we did not identify either protein for S. japonicum ESP, we did find a Ribonuclease T2 (C1LS35) in both S. japonicum eggs and ESP. After BLAST analysis against protein sequences from trematode parasites, we determined that C1LS35 has 32% (score 166, E-value 1.2E-14) identity with the putative hepatotoxic ribonuclease Omega-1 protein from S. mansoni (UniProt G4V5C6).
Of interest, recently Ke et al. [4] identified a S. japonicum RNase T2 family member, CP1412 protein, as a component of ESP of eggs. Alignment and BLAST analysis have shown that C1LS35 and CP1412 are the same protein, since they have 99% identity. Additional characterization of the CP1412 revealed that, as observed for Omega-1, this Ribonuclease T2 also stimulates polarization of the host Th2 immune response. Further information on the importance of this molecule in the processes of infection and pathogenesis in schistosomiasis japonica might be demonstrated using RNA interference methods.
ESP of schistosome eggs provokes an immune attack to drive the eggs from tissues into the intestinal lumen for excretion from the host. Proteins such as serine, cysteine, aspartyl and metallo-proteases that we identified in S. japonicum ESP participate in these fundamental processes, including invasion, escape, and modulation of the host immune system [53, 54], making them suitable targets for the development of vaccines and new treatments [55]. Although most of the proteases identified here remain uncharacterized in terms of egg function, it was previously demonstrated that serine proteases can interfere with host coagulation and the immune response. A 27 kDa serine protease with fibrinolytic activity was characterized from S. mansoni eggs and hypothesized to block intravascular fibrin deposition by platelets activated by the eggs [56].
Our analyses demonstrated that protease inhibitors, including serpin, cystatin, serine and cysteine proteases inhibitors respectively, are differentially expressed in mature and immature eggs (Fig. 5). These molecules have central roles in host-parasite interactions [57, 58] and could also represent important targets for future development of drugs and vaccines. Serpins can activate aggregation of platelets and inhibition of fibrin deposition in the vascular endothelium [56], both fundamental processes to guarantee successful infection. The Serpin B6 here demonstrated upregulated in mSj eggs has predicted anti-thrombin activity that might help in parasite propagation, since inhibition of thrombin would culminate in less fibrin available to for clot formation, ultimately helping eggs escape through the intestinal wall. In addition, cystatins contribute to immune evasion and modulation of the Th2 response by activating macrophages and inducing IL-10 production [58–60], and also suppressing exogenous-antigen presentation by DCs, affirming their immunosuppressive role [61].
The comparison of the levels of expression of proteins between immature and mature S. japonicum eggs demonstrated important differences in terms of metabolic processes occurring in eggs at different stages of development. Immature eggs primarily produce proteins related to structure and cell organization (e.g. tubulin beta chain, putative dynein and actin depolymerizing factor), mature eggs mostly express proteins involved with energy metabolism and homeostasis regulation. While both immature and mature S. japonicum eggs expressed proteins highly involved in neutrophil degranulation process, mSj eggs expressed proteins associated with movement and proteolytic processes, which is likely linked to the presence of an active miracidium and the need for eggs to escape the host and ultimately to hatch.
Mathieson & Wilson [25] compared S. mansoni eggs at two different stages of development and showed some 80% similarity in expressed proteins expressed in the two stages. These authors demonstrated that Sm_p40 was an abundant protein in mature S. mansoni eggs, while the heat-shock protein-70 (HSP70) was the most abundant protein identified in immature eggs. We also found that proteins related to response to stress were enriched in mSj eggs (e.g. cell wall integrity and stress response component 1) compared with immature, which probably has to do with changes in the environment that mature schistosome eggs have to confront.
Another remarkable difference that we found between immature and mature eggs reflects their predicted ability to stimulate the host immune system (Table 2). Previously, Ashton et al. [8] highlighted that immature S. mansoni eggs induce a weaker immune response than the mature forms. By in vitro analyzing those proteins more highly expressed by iSj or mSj eggs in terms of their ability to interact and trigger the immune response we found that mature eggs were more likely to activate an innate response by activating T cells (Table 2). The mSj eggs also may elicit adaptive responses through at least two different pathways, including that involving Rap-1 signaling. Rap-1 is a member of the mitogen-activated protein kinase (MAPK) signaling pathways that are pivotal transmitters of extracellular signals, including cytokines. Therefore, once activated, MAPKs can regulate key cellular processes including T cell responses [62, 63]. Taking together the strong potential of ESP for stimulating host immune responses, our data highlight the complexity and significance of immunomodulatory events that S. japonicum eggs trigger in order to maintain the life-cycle of the parasite.
Conclusions
In the present study, we present a list of 957 egg and egg-derived excretory-secretory proteins from S. japonicum of which 95 were exclusively found in S. japonicum ESP; such proteins are completely accessible to the host immune system effector elements, and therefore in general represent interesting targets for diagnosis and as vaccines. Often, proteins from ESP are involved in key functions that permit schistosome eggs withstand stressful environmental features, including host immune responses that lead to the granulomatous inflammatory reaction observed around the eggs trapped in tissues, but also assist their passage across the gut wall. Further investigation of some of the key proteins differentially expressed in S. japonicum eggs and ESP, and even in immature and mature eggs, can lead to better strategies to avoid severe morbidity during schistosomiasis japonica, as well as improving diagnosis, treatment and control of this persistent parasitic infection.
Additional files
Additional file 1: Figure S1. S. japonicum mature eggs isolation. Eggs isolated from mice feces after six weeks of infection.
Additional file 2: Figure S2. S. japonicum immature eggs isolation. a S. japonicum female worm in the media laying eggs. b S. japonicum immature eggs isolated after 24 h incubation.
Additional file 3: Figure S3. S. japonicum egg secretory proteins production. a S. japonicum liver eggs incubated in a well with RPMI media (time 0 h). White arrow indicates immature eggs, black arrow indicates mature eggs. b S. japonicum liver eggs incubated in a well with RPMI media, at 37 °C, 5% CO2 (time 3 h). c S. japonicum miracidia. After incubation in RPMI media the eggs were induced to hatch by being placed in water and exposed to direct light for 1 h.
Additional file 4: Table S1. List of proteins identified in the whole eggs and those shared with ESP from S. japonicum eggs.
Additional file 5: Table S2. Proteins identified with significant variation in S. japonicum immature eggs compared with mature eggs, listed in descending order of variation.
Acknowledgements
We would like to thank the Mary Duke for helping with the animal work.
Funding
CDMV received an Endeavour Scholarships and Fellowships from Australian Government.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012835, available at http://www.proteomexchange.org/submission/index.html.
Authorsʼ contributions
CDMV and JP isolated the samples, analyzed and interpreted the data regarding the MS analyses. JM helped with the mass spectrometry analysis. CDMV and MKJ were a major contributor in writing the manuscript. HY and DPM helped in the contextualization of the ideas and writing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Animal Ethics Committee of the QIMR Berghofer Medical Research Institute approved all work involving the use of animals under project P1289.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CLR’s
C-type lectin receptors
- DC
Dendritic cells
- ESP
Egg secretory proteins
- FASP
Filter Aided Sample Preparation Method
- ICAM-1
Intercellular Adhesion Molecule 1
- IDA
Information Dependent Acquisition
- IL
Interleukins
- IPSE/ alpha-1
IL-4-inducing principle
- iSj
Immature S. japonicum eggs
- LC-MS/MS
Liquid chromatography with mass spectrometry
- mSj
Mature S. japonicum eggs
- NETs
Neutrophil extracellular traps
- SEA
Soluble egg antigens
- SecP
SecretomeP analysis
- SWATH
Sequential window acquisition of all theoretical fragment ion spectra
- Th2
T helper 2
- TLR
Toll-Like receptor
- VCAM-1
Vascular cell adhesion molecule 1
Contributor Information
Carolina De Marco Verissimo, Email: c.demarcoverissimo@qub.ac.uk.
Jeremy Potriquet, Email: jeremy.potriquet@gmail.com.
Hong You, Email: Hong.You@qimrberghofer.edu.au.
Donald P. McManus, Email: don.mcmanus@qimrberghofer.edu.au
Jason Mulvenna, Email: Jason.Mulvenna@qimrberghofer.edu.au.
Malcolm K. Jones, Email: m.jones@uq.edu.au
References
- 1.Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014;383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou X-N. Schistosomiasis. Nature Rev Dis Primers. 2018;4:13. doi: 10.1038/s41572-018-0013-8. [DOI] [PubMed] [Google Scholar]
- 3.Xu T, Chen J, Zhu D, Chen L, Wang J, Sun X, Duan Y. Egg antigen p40 of Schistosoma japonicum promotes senescence in activated hepatic stellate cells via SKP2/P27 signaling pathway. Sci Rep. 2017;7:275. doi: 10.1038/s41598-017-00326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke XD, Shen S, Song LJ, Yu CX, Kikuchi M, Hirayama K, et al. Characterization of Schistosoma japonicum CP1412 protein as a novel member of the ribonuclease T2 molecule family with immune regulatory function. Parasit Vectors. 2017;10:89. doi: 10.1186/s13071-016-1962-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheever AW, Macedonia JG, Mosimann JE, Cheever EA. Kinetics of egg production and egg excretion by Schistosoma mansoni and S. japonicum in mice infected with a single pair of worms. Am J Trop Med Hyg. 1994;50:281–295. doi: 10.4269/ajtmh.1994.50.281. [DOI] [PubMed] [Google Scholar]
- 6.Chuah C, Jones MK, Burke ML, McManus DP, Gobert GN. Cellular and chemokine-mediated regulation in schistosome-induced hepatic pathology. Trends Parasitol. 2014;30:141–150. doi: 10.1016/j.pt.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Wilson MS, Mentink-Kane MM, Pesce JT, Ramalingam TR, Thompson R, Wynn TA. Immunopathology of schistosomiasis. Immunol Cell Biol. 2007;85:148–154. doi: 10.1038/sj.icb.7100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashton PD, Harrop R, Shah B, Wilson R. The schistosome egg: development and secretions. Parasitology. 2001;122:329–338. doi: 10.1017/S0031182001007351. [DOI] [PubMed] [Google Scholar]
- 9.Jones MK, Bong SH, Green KM, Holmes P, Duke M, Loukas A, et al. Correlative and dynamic imaging of the hatching biology of Schistosoma japonicum from eggs prepared by high pressure freezing. PLoS Negl Trop Dis. 2008;2:e334. doi: 10.1371/journal.pntd.0000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meevissen MH, Balog CI, Koeleman CA, Doenhoff MJ, Schramm G, Haas H, et al. Targeted glycoproteomic analysis reveals that kappa-5 is a major, uniquely glycosylated component of Schistosoma mansoni egg antigens. Mol Cell Proteomics. 2011;10(M110):005710. doi: 10.1074/mcp.M110.005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdulla MH, Lim KC, McKerrow JH, Caffrey CR. Proteomic identification of IPSE/alpha-1 as a major hepatotoxin secreted by Schistosoma mansoni eggs. PLoS Negl Trop Dis. 2011;5:e1368. doi: 10.1371/journal.pntd.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson BJ, Newland SA, Gibbs SE, Tourlomousis P, Santos P, Patel MN, et al. The Schistosoma mansoni T2 ribonuclease omega-1 modulates inflammasome-dependent IL-1beta secretion in macrophages. Int J Parasitol. 2015;45:809–813. doi: 10.1016/j.ijpara.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Pearce EJM, Kane C, Sun J, Taylor J, McKee AS, Cervi L. Th2 response polarization during infection with the helminth parasite Schistosoma mansoni. Immunol Rev. 2004;201:117–126. doi: 10.1111/j.0105-2896.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- 14.Everts B, Perona-Wright G, Smits HH, Hokke CH, van der Ham AJ, Fitzsimmons CM, et al. Omega-1, a glycoprotein secreted by Schistosoma mansoni eggs, drives Th2 responses. J Exp Med. 2009;206:1673–1680. doi: 10.1084/jem.20082460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alger HM, Sayed AA, Stadecker MJ, Williams DL. Molecular and enzymatic characterisation of Schistosoma mansoni thioredoxin. Int J Parasitol. 2002;32:1285–1292. doi: 10.1016/S0020-7519(02)00108-X. [DOI] [PubMed] [Google Scholar]
- 16.Asahi H, Stadecker MJ. Analysis of egg antigens inducing hepatic lesions in schistosome infection. Parasitol Int. 2003;52:361–367. doi: 10.1016/S1383-5769(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhou XH, Wu JY, Huang XQ, Kunnon SP, Zhu XQ, Chen XG. Identification and characterization of Schistosoma japonicum Sjp40, a potential antigen candidate for the early diagnosis of schistosomiasis. Diagn Microbiol Infect Dis. 2010;67:337–345. doi: 10.1016/j.diagmicrobio.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 18.DeWalick S, Tielens AG, van Hellemond JJ. Schistosoma mansoni: the egg, biosynthesis of the shell and interaction with the host. Exp Parasitol. 2012;132:7–13. doi: 10.1016/j.exppara.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 19.DeWalick S, Hensbergen PJ, Bexkens ML, Grosserichter-Wagener C, Hokke CH, Deelder AM, et al. Binding of von Willebrand factor and plasma proteins to the eggshell of Schistosoma mansoni. Int J Parasitol. 2014;44:263–268. doi: 10.1016/j.ijpara.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Rychter JW, Van Nassauw L, Brown JK, Van Marck E, Knight PA, Miller HR, et al. Impairment of intestinal barrier and secretory function as well as egg excretion during intestinal schistosomiasis occur independently of mouse mast cell protease-1. Parasite Immunol. 2010;32:221–231. doi: 10.1111/j.1365-3024.2009.01182.x. [DOI] [PubMed] [Google Scholar]
- 21.Pearce EJM. Priming of the immune response by schistosome eggs. Parasite Immunol. 2005;27:265–270. doi: 10.1111/j.1365-3024.2005.00765.x. [DOI] [PubMed] [Google Scholar]
- 22.Lejoly-Boisseau H, Appriou M, Seigneur M, Pruvost A, Tribouley-Duret J, Tribouley J. Schistosoma mansoni: in vitro adhesion of parasite eggs to the vascular endothelium Subsequent inhibition by a monoclonal antibody directed to a carbohydrate epitope. Exp Parasitol. 1999;91:20–29. doi: 10.1006/expr.1999.4348. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Lu J, Hu W, Wang SY, Cui SJ, Chi M, et al. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2006;2:e29. doi: 10.1371/journal.ppat.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeWalick S, Bexkens ML, van Balkom BW, Wu YP, Smit CH, Hokke CH, et al. The proteome of the insoluble Schistosoma mansoni eggshell skeleton. Int J Parasitol. 2011;41:523–532. doi: 10.1016/j.ijpara.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Mathieson W, Wilson RA. A comparative proteomic study of the undeveloped and developed Schistosoma mansoni egg and its contents: the miracidium, hatch fluid and secretions. Int J Parasitol. 2010;40:617–628. doi: 10.1016/j.ijpara.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Cass CL, Johnson JR, Califf LL, Xu T, Hernandez HJ, Stadecker MJ, et al. Proteomic analysis of Schistosoma mansoni egg secretions. Mol Biochem Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albonico M, Ame SM, Vercruysse J, Levecke B. Comparison of the Kato-Katz thick smear and McMaster egg counting techniques for monitoring drug efficacy against soil-transmitted helminths in schoolchildren on Pemba Island, Tanzania. Trans R Soc Trop Med Hyg. 2012;106:199–201. doi: 10.1016/j.trstmh.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Levecke B, Brooker SJ, Knopp S, Steinmann P, Sousa-Figueiredo JC, Stothard JR, et al. Effect of sampling and diagnostic effort on the assessment of schistosomiasis and soil-transmitted helminthiasis and drug efficacy: a meta-analysis of six drug efficacy trials and one epidemiological survey. Parasitology. 2014;141:1826–1840. doi: 10.1017/S0031182013002266. [DOI] [PubMed] [Google Scholar]
- 29.Silva-Moraes V, Ferreira JM, Coelho PM, Grenfell RF. Biomarkers for schistosomiasis: towards an integrative view of the search for an effective diagnosis. Acta Trop. 2014;132:75–79. doi: 10.1016/j.actatropica.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Dalton JP, Day SR, Drew AC, Brindley PJ. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology. 1997;115:29–32. doi: 10.1017/S0031182097001091. [DOI] [PubMed] [Google Scholar]
- 31.Mulvenna J, Hamilton B, Nagaraj SH, Smyth D, Loukas A, Gorman JJ. Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol Cell Proteomics. 2009;8:109–121. doi: 10.1074/mcp.M800206-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Winiewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 33.Guo T, Kouvonen P, Koh CC, Gillet LC, Wolski WE, Röst HL, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nature Med. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blattmann P, Heusel M, Aebersold R. SWATH2stats: An R/Bioconductor package to process and convert quantitative SWATH-MS proteomics data for downstream analysis tools. PLoS One. 2016;11:e0153160. doi: 10.1371/journal.pone.0153160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 36.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom Rev. 2014;33:452–470. doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 37.Curwen RS, Ashton PD, Johnston DA, Wilson RA. The Schistosoma mansoni soluble proteome: a comparison across four life-cycle stages. Mol Biochem Parasitol. 2004;138:57–66. doi: 10.1016/j.molbiopara.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Wang T, Zhao M, Rotgans BA, Strong A, Liang D, Ni G, et al. Proteomic analysis of the Schistosoma mansoni miracidium. PLoS One. 2016;11:e0147247. doi: 10.1371/journal.pone.0147247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao X, Fu Z, Zhang M, Han Y, Han Q, Lu K, et al. Excretory/secretory proteome of 14-day schistosomula, Schistosoma japonicum. J Proteomics. 2016;130:221–230. doi: 10.1016/j.jprot.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Cao X, Fu Z, Zhang M, Han Y, Han H, Han Q, et al. iTRAQ-based comparative proteomic analysis of excretory-excretory proteins of schistosomulo and adult worms of Schistosoma japonicum. J Proteomics. 2016;138:30–39. doi: 10.1016/j.jprot.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Sotillo J, Pearson M, Becker L, Mulvenna J, Loukas A. A quantitative proteomic analysis of the tegumental proteins from Schistosma mansoni schistosomula reveals novel potential therapeutic targets. Int J Parasitol. 2015;45:505–516. doi: 10.1016/j.ijpara.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Wynn TA, Thompson RW, Cheever AW, Mentink-Kane MM. Immunopathogenesis of schistosomiasis. Immunol Rev. 2004;201:156–167. doi: 10.1111/j.0105-2896.2004.00176.x. [DOI] [PubMed] [Google Scholar]
- 43.Abouel-Nour MF, Lotfy M, Attallah AM, Doughty BL. Schistosoma mansoni major egg antigen Smp40: molecular modeling and potential immunoreactivity for anti-pathology vaccine development. Mem Inst Oswaldo Cruz. 2006;101:365–372. doi: 10.1590/S0074-02762006000400004. [DOI] [PubMed] [Google Scholar]
- 44.Lima LM, Santos SB, Oliveira RR, Cardoso LS, Oliveira SC, Góes AM, et al. Schistosoma antigens down modulate the in vitro inflammatory response in individuals infected with human T cell lymphotropic virus type 1. Neuroimmunomodulation. 2013;20:233–238. doi: 10.1159/000348700. [DOI] [PubMed] [Google Scholar]
- 45.Kaisar MMM, Ritter M, Del Fresno C, Jónasdóttir HS, van der Ham AJ, Pelgrom LR, et al. Dectin-1/2-induced autocrine PGE2 signalling licenses dendritic cells to prime Th2 responses. PLoS Biol. 2018;16:e2005504. doi: 10.1371/journal.pbio.2005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.VonLichtenberg F, Erickson DG, Sadun EH. Comparative histopathology of schistosome granulomas in the hamster. Am J Pathol. 1973;72:149–178. [PMC free article] [PubMed] [Google Scholar]
- 47.Hirata M, Hara T, Kage M, Fukuma T, Sendo F. Neutropenia augments experimentally induced Schistosoma japonicum egg granuloma formation in CBA mice, but not in C57BL/6 mice. Parasite Immunol. 2002;24:479–488. doi: 10.1046/j.1365-3024.2002.00491.x. [DOI] [PubMed] [Google Scholar]
- 48.Hirata M, Fukuma T. Cytokine regulation in experimentally-induced Schistosoma japonicum egg granuloma formation. Parasitol Int. 2003;52:341–349. doi: 10.1016/S1383-5769(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 49.Smith P, Fallon RE, Mangan NE, Walsh CM, Saraiva M, Sayers JR, et al. Schistosoma mansoni secretes a chemokine binding protein with antiinflammatory activity. J Exp Med. 2005;202:1319–1325. doi: 10.1084/jem.20050955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng L, Hu Y, Wang Y, Huang X, Xu Y, Shen Y, et al. Recruitment of neutrophils mediated by Vγ2 γδ T cells deteriorates liver fibrosis induced by Schistosoma japonicum infection in C57BL/6 Mice. Infect Immun. 2017;85:e01020. doi: 10.1128/IAI.01020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuah C, Jones MK, Burke ML, Owen HC, Anthony BJ, McManus DP, et al. Spatial and temporal transcriptomics of Schistosoma japonicum-induced hepatic granuloma formation reveals novel roles for neutrophils. J Leukoc Biol. 2013;94:353–365. doi: 10.1189/jlb.1212653. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y, Zheng H, Chen Y, Zhang L, Wang K, Guo J, et al. Schistosoma japonicum genome sequencing and functional analysis consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.González AY, Sulbarán GS, Ballen DE, Cesari IM. Immunocapture of circulatoring Schistosoma mansoni cathepsin B antigen (Sm31) by anti-Sm31 polyclonal antibodies. Parasitol Int. 2016;65:191–195. doi: 10.1016/j.parint.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Dvořák J, Fajtová P, Ulrychová L, Leontovyč A, Rojo-Arreola L, Suzuki BM, et al. Excretion/secretion products from Schistosoma mansoni adults, eggs and schistosomula have unique peptidase specificity profiles. Biochimie. 2016;122:99–109. doi: 10.1016/j.biochi.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojo JU, Melkus MW, Kottapalli KR, Okiya OE, Sudduth J, Zhang W, et al. Smp80-based schistosomiasis vaccine mediated epistatic interactions identified potential immune signatures for vaccine efficacy in mice and baboons. PLoS One. 2017;12:e0171677. doi: 10.1371/journal.pone.0171677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doenhoff MJ, Stanley RG, Pryce D, Curtis RC, Parry H, Griffiths K, et al. Identification of a fibrinolytic enzyme in Schistosoma mansoni eggs and modulated blood fibrinogen metabolism in S. mansoni-infected mice. Parasitology. 2003;126:231–234. doi: 10.1017/S0031182002002809. [DOI] [PubMed] [Google Scholar]
- 57.Molehin AJ, Gobert GN, Driguez P, McManus DP. Characterisation of a secretory serine protease inhibitor (SjB6) from Schistosoma japonicum. Parasit Vectors. 2014;7:330. doi: 10.1186/1756-3305-7-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Wang S, Zhan B, He W, Chu L, Qiu D, et al. Therapeutic effect of Schistosoma japonicum cystatin on bacterial sepsis in mice. Parasit Vectors. 2017;10:222. doi: 10.1186/s13071-017-2162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang X, Liu J, Yue Y, Chen W, Song M, Zhan X, et al. Cloning, expression and characterisation of type II cystatin from Schistosoma japonicum, which could regulate macrophage activation. Parasitol Res. 2014;113:3985–3992. doi: 10.1007/s00436-014-4064-9. [DOI] [PubMed] [Google Scholar]
- 60.Wang S, Xie Y, Yang X, Wang X, Yan K, Zhong Z, et al. Therapeutic potential of recombinant cystatin from Schistosoma japonicum in TNBS-induced experimental colitis of mice. Parasit Vectors. 2016;9:6. doi: 10.1186/s13071-015-1288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L, He B, Hou W, He L. Cysteine protease inhibitor of Schistosoma japonicum - a parasite-derived negative immunoregulatory factor. Parasitol Res. 2017;116:901–908. doi: 10.1007/s00436-016-5363-0. [DOI] [PubMed] [Google Scholar]
- 62.Katagiri K, Hattori M, Minato N, Kinashi T. Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Cell Biol. 2002;22:1001. doi: 10.1128/MCB.22.4.1001-1015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Yang Z, Li Y, Yu F, Brindley PJ, McManus DP, et al. Reconstruction and in silico analysis of the MAPK signaling pathways in the human blood fluke, Schistosoma japonicum. FEBS. 2006;580:36177. doi: 10.1016/j.febslet.2006.05.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. S. japonicum mature eggs isolation. Eggs isolated from mice feces after six weeks of infection.
Additional file 2: Figure S2. S. japonicum immature eggs isolation. a S. japonicum female worm in the media laying eggs. b S. japonicum immature eggs isolated after 24 h incubation.
Additional file 3: Figure S3. S. japonicum egg secretory proteins production. a S. japonicum liver eggs incubated in a well with RPMI media (time 0 h). White arrow indicates immature eggs, black arrow indicates mature eggs. b S. japonicum liver eggs incubated in a well with RPMI media, at 37 °C, 5% CO2 (time 3 h). c S. japonicum miracidia. After incubation in RPMI media the eggs were induced to hatch by being placed in water and exposed to direct light for 1 h.
Additional file 4: Table S1. List of proteins identified in the whole eggs and those shared with ESP from S. japonicum eggs.
Additional file 5: Table S2. Proteins identified with significant variation in S. japonicum immature eggs compared with mature eggs, listed in descending order of variation.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012835, available at http://www.proteomexchange.org/submission/index.html.