Abstract
Background
Cyanobacteria produce hydrocarbons corresponding to diesel fuels by means of aldehyde-deformylating oxygenase (ADO). ADO catalyzes a difficult and unusual reaction in the conversion of aldehydes to hydrocarbons and has been widely used for biofuel production in metabolic engineering; however, its activity is low. A comparison of the amino acid sequences of highly active and less active ADOs will elucidate non-conserved residues that are essential for improving the hydrocarbon-producing activity of ADOs.
Results
Here, we measured the activities of ADOs from 10 representative cyanobacterial strains by expressing each of them in Escherichia coli and quantifying the hydrocarbon yield and amount of soluble ADO. We demonstrated that the activity was highest for the ADO from Synechococcus elongatus PCC 7942 (7942ADO). In contrast, the ADO from Gloeobacter violaceus PCC 7421 (7421ADO) had low activity but yielded high amounts of soluble protein, resulting in a high production level of hydrocarbons. By introducing 37 single amino acid substitutions at the non-conserved residues of the less active ADO (7421ADO) to make its sequence more similar to that of the highly active ADO (7942ADO), we found 20 mutations that improved the activity of 7421ADO. In addition, 13 other mutations increased the amount of soluble ADO while maintaining more than 80% of wild-type activity. Correlation analysis showed a solubility-activity trade-off in ADO, in which activity was negatively correlated with solubility.
Conclusions
We succeeded in identifying non-conserved residues that are essential for improving ADO activity. Our results may be useful for generating combinatorial mutants of ADO that have both higher activity and higher amounts of the soluble protein in vivo, thereby producing higher yields of biohydrocarbons.
Electronic supplementary material
The online version of this article (10.1186/s13068-019-1409-8) contains supplementary material, which is available to authorized users.
Keywords: Aldehyde-deformylating oxygenase, Biohydrocarbons, Cyanobacteria, Protein engineering
Background
The biosynthesis of hydrocarbons has provided a remarkable means of producing substitutes for petroleum-based fuels [1]. Biohydrocarbons are produced by various organisms, including bacteria, cyanobacteria, yeasts, plants, insects, birds, and green algae [2, 3]. Cyanobacteria-derived hydrocarbons have gained attention as a renewable energy source that can help limit global warming [4]. Moreover, the cyanobacterial biosynthesis of hydrocarbons is attractive because the growth rate of cyanobacteria is rapid, and their genetic manipulation is well established [5–10].
Cyanobacteria can convert fatty acyl–acyl carrier protein (ACP) and fatty acyl-coenzyme A (CoA), which are intermediates of fatty acid metabolism, into hydrocarbons that are 13–17 carbons in length, corresponding to diesel fuels, through two-step reactions [11]. The reactions are catalyzed by two enzymes, namely, acyl-ACP reductase (AAR) and aldehyde-deformylating oxygenase (ADO) [11, 12]. AAR reduces fatty acyl-ACPs or fatty acyl-CoAs to fatty aldehydes using NADPH [11, 13, 14]. The aldehyde products are delivered to ADO via the binding of AAR to ADO [14]. Subsequently, ADO converts saturated or monounsaturated fatty aldehydes (Cn) into corresponding alkanes or alkenes (Cn−1) and formate, respectively [11, 12]. Escherichia coli coexpressing cyanobacterial AAR and ADO can produce and secrete hydrocarbons [11]. Therefore, AAR and ADO are key enzymes for the biosynthesis of hydrocarbons. Thus, AAR and ADO have been applied for the production of biohydrocarbons in several organisms [10], including cyanobacteria [15–20], E. coli [11, 21–25], yeasts [26–28], chemoautotrophic eubacteria [29], and filamentous fungi [30]. In particular, ADO has been extensively used with other enzymes because ADO can catalyze a difficult and unusual reaction in the conversion of aldehydes to hydrocarbons [10, 23, 27, 31–35].
ADO is a member of a ferritin-like non-heme diiron family of enzymes, including methane monooxygenase, ribonucleotide reductase, and acyl-ACP desaturase [36]. ADO has eight α-helices, and its active site consists of two histidine and four carboxylate residues, with two iron atoms accommodated in an antiparallel four-helix bundle [37–40]. The substrate binding site is composed of a hydrophobic channel that terminates at the diiron center. An external reducing system composed of ferredoxin (Fd), ferredoxin NADP+ reductase (FNR), and NADPH is required for ADO activity [41]. Although ADO has been widely used for biofuel production by the metabolic engineering of cyanobacteria and other organisms, it has very low activity. Its maximum turnover number is ~ 1 min−1 [42], and the rate-limiting step in the biosynthesis of hydrocarbons in E. coli coexpressing AAR and ADO is the reaction catalyzed by ADO [10, 24]. There have been a few attempts to improve the hydrocarbon-producing activity of ADO by introducing amino acid substitutions based on the crystal structures of cyanobacterial ADOs [11, 22, 39]. These studies have revealed residues that are essential for maintaining the hydrocarbon-producing activity of ADO. Such residues include C83 located adjacent to the substrate binding site and those located near the diiron center [the residue numbering of ADO from Prochlorococcus marinus MIT 9313 (9313ADO) is used throughout this article]. In addition, several mutations at residues that compose the substrate binding channel have been reported to change the substrate specificity of ADO [33, 34, 43]. However, most of the residues at the mutation sites in these studies are highly conserved among ADOs from various cyanobacteria, and there have been few reports on mutations at non-conserved residues. Because the amino acid residues of an enzyme are generally conserved to maintain its activity [44–46] and because functionally important residues are generally less mutationally tolerant than residues with less stringent functional constraints [46–48], mutations that improve enzymatic activity are expected to be introduced mainly at non-conserved residues. Thus, to improve the hydrocarbon-producing activity of ADO, a survey of amino acid substitutions at non-conserved sites is necessary. A comparison of the amino acid sequences of highly active and less active ADOs will help to elucidate the mutations at non-conserved residues that are essential for improving hydrocarbon-producing activity.
To compare the activity of the ADO mutants, a method that can accurately measure ADO activity must be employed. However, accurate measurements of ADO activity in vitro have not been possible because the natural substrates of ADO, namely, medium- and long-chain (C10–C18) aldehydes, have low solubility and form micelles in solution [14]. Consequently, the dissociation reaction of an aldehyde molecule from micelles is the rate-determining step in the overall reaction and obscures the catalytic reaction of ADO [43, 49–51]. Instead, ADO activity for medium- and long-chain aldehydes has been measured in vivo by quantifying the hydrocarbon production yield [11, 16, 22, 52–54]. Several studies have shown that ADO from Nostoc punctiforme PCC 73102 (73102ADO) or Synechococcus elongatus PCC 7942 (7942ADO) produces the highest yield of hydrocarbons when ADO is expressed in E. coli, yeast, or cyanobacteria [11, 28, 35, 53]. In contrast, other studies have reported that ADOs from Thermosynechococcus elongatus BP-1 (TeADO), Cyanothece sp. PCC 7425 (7425ADO), and Planktothrix agardhii NIVA-CYA 126/8 (PaADO) produce hydrocarbons in amounts that are higher than or comparable to those produced by 73102ADO and 7942ADO [28, 52, 54], which contradicts previous reports. The inconsistency of these results may be attributed to the methods used for evaluating ADO activity. All of these studies compared the yield of hydrocarbons without considering the amount of the soluble form of ADO inside the cells. However, because the hydrocarbon production yield is directly related to both ADO activity and the amount of the soluble form of ADO, it is necessary to measure the amount of soluble ADO to accurately evaluate ADO activity.
Previously, we established a method for accurate measurements of ADO activity [22]. In this method, both AAR and ADO are coexpressed in E. coli, and fatty aldehyde substrates are supplied from AAR to ADO. ADO activity was calculated as the amount of hydrocarbon produced [quantified using gas chromatography–mass spectrometry (GC–MS)] divided by the amount of the soluble form of ADO [quantified using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or western blotting]. The same method has been successfully applied to measure the activity of AAR, and we found that AAR from Synechococcus elongatus PCC 7942 (7942AAR) had the highest activity among the examined AARs [24]. Thus, our method can be applied to accurately determine the activity of various ADOs and to identify residues that are essential for increasing ADO activity. In this method, factors affecting the hydrocarbon-producing activity of ADO include the catalytic efficiency of ADO, the efficiency of substrate delivery from AAR to ADO by their interactions, and the efficiency of coupling between ADO and the Fd/FNR/NADPH reducing system. Thus, this method can identify ADO residues that are essential for improving hydrocarbon-producing activity in terms of these factors.
Here, using this method, we compared the hydrocarbon-producing activity of ADOs from 10 representative cyanobacterial strains. 7942AAR, which has the highest activity among the various AARs [24], was used as a supplier of aldehyde substrates. We found that 7942ADO has the highest activity. In contrast, ADO from Gloeobacter violaceus PCC 7421 (7421ADO) has low activity but yields high amounts of the soluble protein, resulting in a high production level of hydrocarbons. By constructing 37 mutants of 7421ADO, in which non-conserved residues were substituted into the amino acids used in 7942ADO, we identified non-conserved residues that are responsible for the high activity of ADO, as well as mutations that increase the amount of soluble ADO in E. coli. Our results may be useful for generating combinatorial mutants of ADO that have both higher activity and higher amounts of the soluble protein in vivo, thereby producing a higher yield of biohydrocarbons.
Results
Selection of 10 representative ADOs
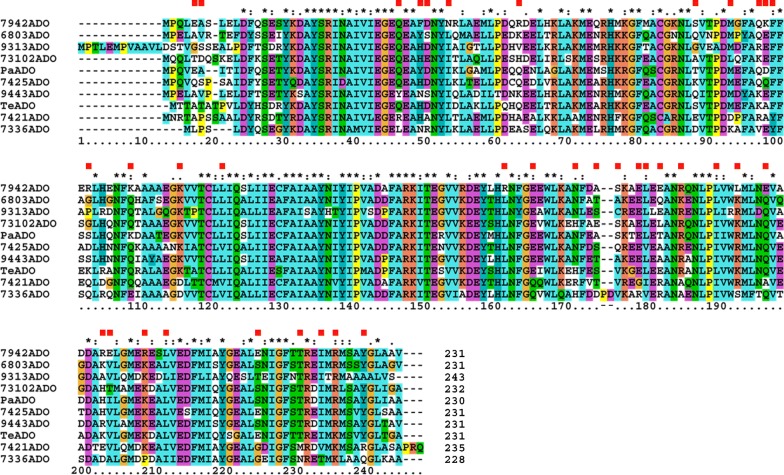
Previously, we compared the aldehyde-producing activity of AARs from eight representative cyanobacteria [24]. Here, we compared the activity of the ADOs from the same eight representative cyanobacteria: 73102ADO, 7421ADO, 7942ADO, 9313ADO, and TeADO and ADOs from Synechocystis sp. PCC 6803 (6803ADO), Synechococcus sp. PCC 7336 (7336ADO), and Microcystis aeruginosa PCC 9443 (9443ADO) [24]. In addition, we also used 7425ADO and PaADO because these ADOs have been reported to produce high amounts of hydrocarbons [28, 52]. Thus, a total of 10 ADOs were selected for the activity assays. Multiple sequence alignments of the 10 ADOs are shown in Fig. 1. The amino acid sequence identities among them are, on average, 69% (Additional file 1: Table S1).
Fig. 1.
Amino acid sequences of 10 representative ADOs. The asterisk denotes completely conserved residues, and the colon and dot denote partially conserved residues. Red squares indicate the mutation sites of the 7421ADO mutants used in the present study. The amino acid sequences of ADOs are shown in descending order of activity. The residue numbering of ADO from 9313ADO is shown at the bottom of the sequence alignments
A phylogenetic tree of cyanobacterial ADOs was constructed based on the ADO amino acid sequences (Additional file 2: Figure S1). There are two large groups (groups 1 and 2) and one small group (group 3) of ADO sequences in the phylogenetic tree, which is similar to that constructed using AAR amino acid sequences [24]. Group 1 contains 93 ADO sequences, which are mainly from freshwater cyanobacteria, including Nostoc punctiforme PCC 73102, Synechococcus elongatus PCC 7942, Planktothrix agardhii NIVA-CYA 126/8, Synechocystis sp. PCC 6803, Microcystis aeruginosa PCC 9443, Cyanothece sp. PCC 7425, and Thermosynechococcus elongatus BP-1. Group 2 contains 31 ADO sequences, which are mainly from marine cyanobacteria, including Prochlorococcus marinus MIT 9313. Group 3 contains 10 ADO sequences from both marine and freshwater cyanobacteria, such as Synechococcus sp. PCC 7336 and Gloeobacter violaceus PCC 7421, respectively. The amino acid sequences of the ADOs from Group 3 are distinct from those categorized into groups 1 and 2 (Additional file 1: Table S1).
ADO activity
ADO activity towards the C16 and C18 aldehydes was measured with an in vivo activity assay that we previously developed [22, 24] because the in vitro activity measurement was precluded by the low solubility of the substrates (see above). Thus, we coexpressed 7942AAR, having the highest activity among the various AARs [24], with one of the 10 selected ADOs in E. coli to produce hydrocarbons. In this method, ADOs that exhibit higher hydrocarbon-producing activity should have higher catalytic efficiency, higher affinity for 7942AAR, and/or more efficient coupling with an external reducing system. A GC–MS profile of the extract from the E. coli cell culture coexpressing AAR and ADO showed predominant amounts of heptadecene and pentadecane and a small amount of heptadecane (Additional file 3: Figure S2), consistent with previous studies [22, 24]. The predominant production of heptadecene (alkene) is probably related to the fact that most acyl-ACPs are desaturated during fatty acid synthesis in E. coli [55].
A control experiment showed that fatty aldehydes and corresponding fatty alcohols can be extracted using ethyl acetate (Additional file 4: Figure S3a, b). Thus, the GC–MS profiles of the extracts from the E. coli cell cultures coexpressing AAR and ADO showed peaks of fatty aldehydes and fatty alcohols, in addition to those of alkanes/alkenes (Additional file 4: Figure S3d). However, because the peaks for aldehydes and alcohols had extremely low intensities and were partially overlapped with those from E. coli cells (Additional file 4: Figure S3c, d), it was not possible to accurately quantify their amounts and therefore to estimate ADO activity using them.
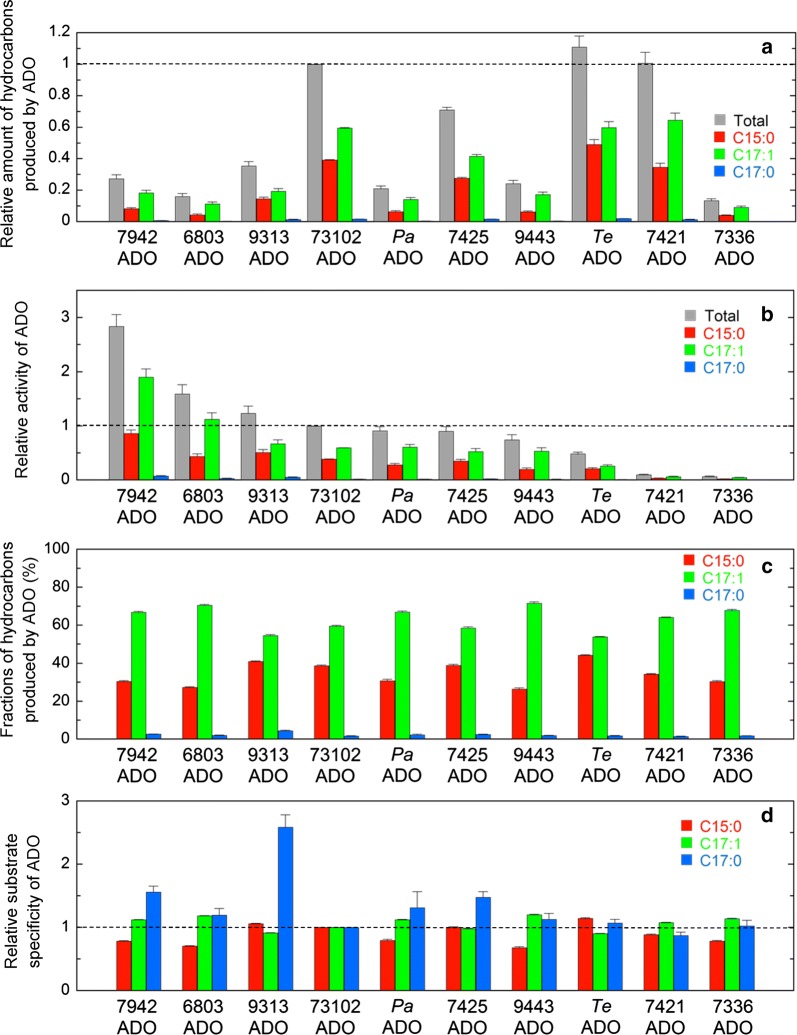
Figure 2a shows the amounts of pentadecane, heptadecene, and heptadecane and their total amounts in E. coli cell cultures coexpressing ADO from one of 10 representative cyanobacteria and 7942AAR. Because 73102ADO had been previously considered to have the highest activity [11], the amounts of hydrocarbons relative to those produced by 73102ADO are shown in Fig. 2. The actual amounts of produced hydrocarbons are shown in Additional file 5: Table S2. The total amounts of hydrocarbons produced in E. coli were the highest for TeADO, followed by, in descending order, 7421ADO, 73102ADO, 7425ADO, 9313ADO, 7942ADO, 9443ADO, PaADO, 6803ADO, and 7336ADO. The amounts of hydrocarbons produced by 6803ADO and 7336ADO were less than 20% of that produced by TeADO.
Fig. 2.
Activity and substrate specificity of 10 representative ADOs. a The amounts of hydrocarbons produced in E. coli coexpressing ADO and 7942AAR. The amounts of pentadecane (C15:0), heptadecene (C17:1), and heptadecane (C17:0) and their combined total amount are shown in red, green, blue, and gray, respectively. The values relative to the total amount of hydrocarbon produced in E. coli coexpressing 73102ADO and 7942AAR are shown. b The hydrocarbon-producing activity of ADO relative to that of 73102ADO. The values are expressed as the amount of hydrocarbon divided by the amount of the soluble form of ADO. c Fractions of pentadecane, heptadecene, and heptadecane relative to the total amount of hydrocarbon produced in E. coli. d The substrate specificity of ADO relative to that of 73102ADO. In all panels, the data are shown in descending order of activity. A horizontal dotted line shows the value for 73102ADO. The measurements were carried out more than three times, and the means ± standard errors are shown
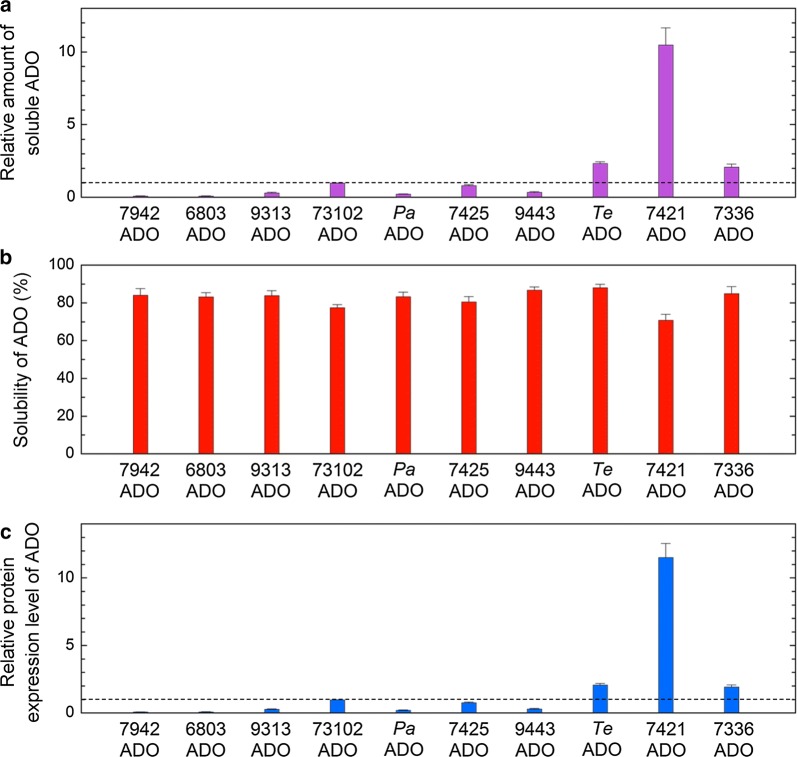
Factors affecting the amounts of hydrocarbons produced in E. coli are ADO activity and the amount of the soluble form of ADO. Therefore, the total amount of hydrocarbon in the E. coli cell culture, normalized to the amount of the soluble form of ADO, was used as an index of ADO activity. The amount of the soluble form of ADO was quantified by western blotting (Fig. 3a; Additional file 6: Figure S4) because western blotting and SDS-PAGE have been successfully used to quantify the amounts of soluble and insoluble forms of proteins [56–58]. The 7425ADO protein migrated more slowly than the other ADOs. This may be because the 7425ADO protein has the highest number of negative charges at pH 8.8 among the ADOs used in this study (see Additional file 7: Table S3), as abnormally slow migration by SDS-PAGE has been previously reported for proteins that have many negative charges [59, 60]. The western blotting results showed that there were large differences in the amount of the soluble form among the various ADOs. We found that the amount of the soluble protein was high for 7421ADO, TeADO, and 7336ADO but low for PaADO, 6803ADO, and 7942ADO (Fig. 3a; Additional file 6: Figure S4). Notably, the variations in the amounts of soluble ADOs were not due to handling errors associated with western blotting because the reproducibility of the data was confirmed by triplicate experiments, as shown in Additional file 6: Figure S4a; the error bars for quantifications of the amounts of soluble ADO and AAR are shown in Fig. 4 and Additional file 6: Figure S4b, respectively.
Fig. 3.
Solubility and protein expression levels of 10 representative ADOs quantified by western blotting. a The amount of the soluble form of ADO normalized to that of 73102ADO. b The solubility (%) of ADO calculated as the ratio of the amount of the soluble form to the total amount of the soluble and insoluble forms of ADO. c The protein expression level of ADO, calculated as the total amount of the soluble and insoluble forms, is normalized to that of 73102ADO. In all panels, the data are shown in descending order of activity. A horizontal dotted line shows the value for 73102ADO. The measurements were carried out more than three times, and the means ± standard errors are shown
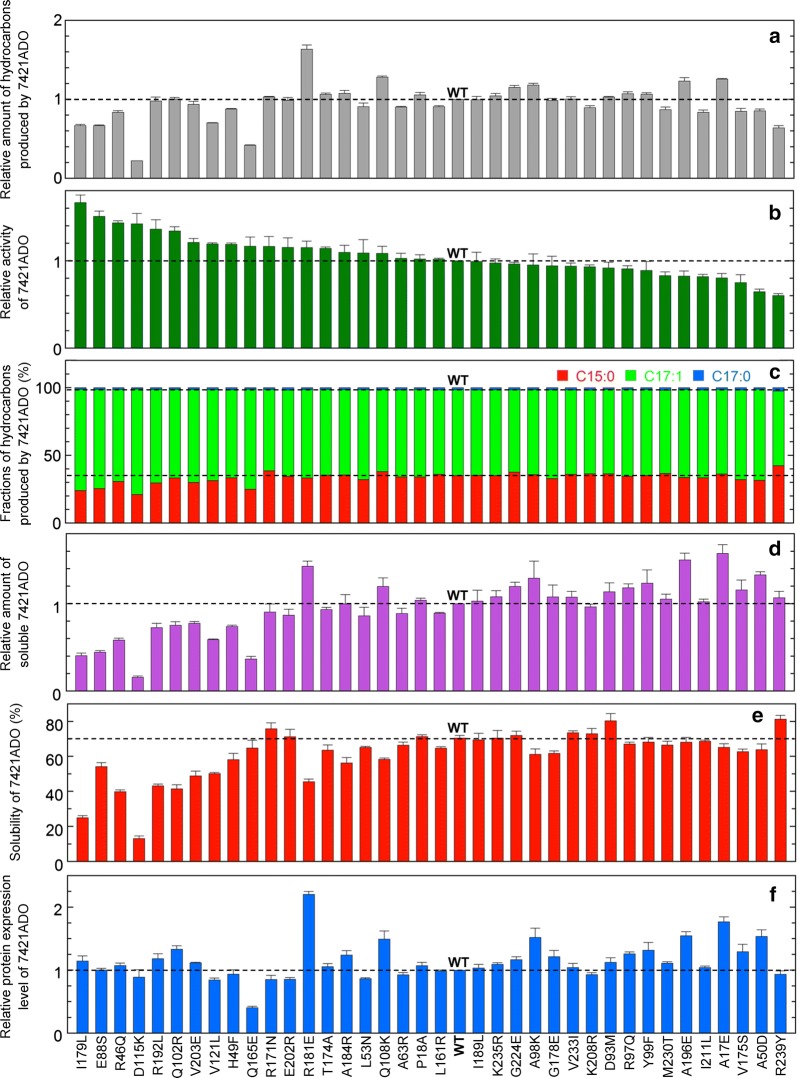
Fig. 4.
Activity and solubility of mutants of 7421ADO. a The amount of total hydrocarbon produced in E. coli coexpressing 7942AAR and a mutant of 7421ADO relative to that of wild-type (WT) 7421ADO. b The hydrocarbon-producing activity of a 7421ADO mutant relative to that of the WT. c Fractions of pentadecane, heptadecene, and heptadecane relative to the total amount of hydrocarbon produced by a 7421ADO mutant. d The amount of the soluble form of a 7421ADO mutant relative to that of the WT. e Solubility (%) of ADO. f The protein expression level of 7421ADO relative to that of the WT. In all panels, the data are shown in descending order of activity. A horizontal dotted line shows the value for the WT. The measurements were carried out more than three times, and the means ± standard errors are shown
Figure 2b shows the hydrocarbon-producing activity of ADO expressed as the total amount of hydrocarbons in the E. coli cell culture (Fig. 2a) divided by the amount of the soluble form of ADO (Fig. 3a). The activities relative to that of 73102ADO are shown. The hydrocarbon-producing activity was the highest for 7942ADO, followed by, in descending order, 6803ADO, 9313ADO, 73102ADO, PaADO, 7425ADO, 9443ADO, TeADO, 7421ADO, and 7336ADO (Fig. 2b). Although the amount of the soluble form of ADO for 7421ADO, TeADO, and 7336ADO was high, their activities were low.
Additional file 6: Figure S4b shows that the amount of soluble AAR ranged from 80 to 175% when the value of E. coli coexpressing 7942AAR and 73102ADO was used as a reference. However, this variation does not affect the measurement of ADO activities. We have previously shown that the rate-limiting step in the biosynthesis of hydrocarbons in E. coli coexpressing 7942AAR and 73102ADO is the reaction catalyzed by ADO [24]. Moreover, when the amount of the soluble AAR protein was reduced to 1/3 (33%), the amount of hydrocarbon produced by 73102ADO was decreased to 1/2 [24]. This result indicates that when the amount of soluble AAR is reduced to 2/3 (66%), the efficiency of substrate supply by AAR becomes the same as that of hydrocarbon production by 73102ADO. Thus, if the amount of soluble AAR is larger than 80%, aldehyde substrates that are sufficient for a hydrocarbon production level that is 1.2-fold higher than that of 73102ADO are supplied. Figure 2a shows that the levels of hydrocarbons produced by all ADOs were less than 1.2-fold that of 73102ADO. Therefore, in the present study, the aldehyde supply from AAR was sufficient for estimating ADO activities, and normalizing the amount of hydrocarbon to the amount of soluble AAR was unnecessary when estimating ADO activity.
Solubility and protein expression level
The solubility and protein expression levels of the ADOs in E. coli were quantified by western blotting (Fig. 3b, c). Here, the protein expression level was determined as the total amount of the soluble and insoluble forms of ADO, while solubility was estimated as the ratio of the amount of the soluble form to the total amount of the soluble and insoluble forms of ADO. We found that solubility was highest for TeADO (88%), followed by, in descending order, 9443ADO, 7336ADO, 7942ADO, 9313ADO, PaADO, 6803ADO, 7425ADO, 73102ADO, and 7421ADO (Fig. 3b). The high solubility of TeADO may be explained by the fact that proteins derived from thermophilic bacteria have high stability [61]. In contrast, the solubility of 7421ADO was lowest (70%). This may be due to the significantly high expression level of the 7421ADO protein (Fig. 3c). Nonetheless, the amount of the soluble form of 7421ADO, which corresponds to the product of the protein expression level and solubility, was highest among the ADOs (Fig. 3a).
Substrate specificity
All 10 ADOs examined in this study showed that their major product is heptadecene (Fig. 2c), indicating that these ADOs have a similar substrate specificity. However, there were slight differences in the substrate specificities of the 10 ADOs (Fig. 2d). 9443ADO and 6803ADO had relatively higher substrate specificities for the 18-carbon fatty aldehyde than the other ADOs because the amount of pentadecane was less than 30% of the total amount of produced hydrocarbon (Fig. 2c). In contrast, the amount of pentadecane was more than 40% of the total amount of hydrocarbon produced with TeADO and 9313ADO, which are derived from freshwater and marine cyanobacteria, respectively (Fig. 2c), indicating that these ADOs had a relatively higher substrate specificity for the 16-carbon fatty aldehyde. Among the three types of detected hydrocarbons, heptadecane exhibited the lowest production (Fig. 2a). Nevertheless, the level of heptadecane produced by 9313ADO was ~ 2.5 times higher than that produced by 73102ADO (Fig. 2d).
Correlation analysis
Correlation analysis of activity, solubility, the protein expression level, the amount of hydrocarbon, and the amount of soluble ADO measured for the 10 representative ADOs was performed (Additional file 8: Figure S5). Strong correlations were not observed, except for a correlation between the protein expression level and the amount of soluble ADO. However, clear correlations between ADO properties were observed when the data from the 7421ADO mutants were used (see below).
We also carried out correlation analyses between the above properties and the amino acid sequence identities. To determine whether the activity was higher for the ADOs with amino acid sequences that were more similar to that of the most active ADO, we plotted the ADO activities against the sequence identities with 7942ADO, which exhibited the highest activity (Additional file 9: Figure S6a). There appears to be a positive correlation, but when the data point for 7942ADO is omitted, the correlation disappears, indicating that the activity level of ADO is not determined by the overall similarity of amino acid sequences. Similarly, solubility, the relative protein expression level, the relative amount of hydrocarbon, and the relative amount of soluble ADO do not correlate well with the sequence identities of TeADO, 7421ADO, TeADO, and 7421ADO, respectively, which have the highest number of corresponding properties (Additional file 9: Figure S6b–e). These results indicate that local differences in amino acid sequences, which cannot be inferred from the phylogenetic tree of ADOs, determine these ADO properties, suggesting that a limited number of non-conserved residues determine the activity level of an ADO.
Mutational analysis of non-conserved residues of 7421ADO
As shown above, hydrocarbon-producing activity is diverse among the various ADOs. This difference can be ascribed to the residues not being completely conserved among ADOs. To identify the non-conserved residues that are essential for ADO activity, we introduced mutations at the non-conserved residues of the less active ADO (7421ADO) to make its amino acid sequence more similar to that of the highly active ADO (7942ADO). The sites for mutation were selected using the multiple sequence alignment shown in Fig. 1, in which the amino acid sequences of the ADOs are shown in descending order of activity. Eighty-four ADO residues are completely conserved. Among the non-conserved residues, including partially conserved residues, both 7421ADO and 7942ADO have the same amino acids at 56 positions, which were excluded from the group of candidates for mutational studies. In addition, positions excluded from the group of candidates were those with the same amino acids as the highly active 7942ADO or the less active 7336ADO. Thus, we selected 37 positions in 7421ADO and replaced them with the corresponding residues in 7942ADO one at a time (Fig. 1). We then coexpressed each of the 37 mutants of 7421ADO with 7942AAR in E. coli and measured the amounts of produced hydrocarbons and soluble ADO, as well as the activity, substrate specificity, solubility, and protein expression level of the ADO (Fig. 4).
As expected, mutations affected the hydrocarbon-producing activity of 7421ADO. Twenty out of 37 mutations improved ADO activity (Fig. 4b). Among them, four mutations increased the activity of all hydrocarbons by more than 40% (I179L, E88S, R46Q, and D115K mutations, in descending order of activity). Six other mutations increased the activity by more than 17% (R192L, Q102R, V203E, V121L, H49F, and Q165E). However, the amount of the soluble form of ADO was decreased in the 10 mutants (Fig. 4d). These results are consistent with the fact that 7942ADO has high activity but yields low amounts of the soluble protein (Figs. 2, 3). Ten other mutations increased the activity by up to 17% but maintained more than 85% of the amount of soluble ADO (R171N, E202R, R181E, T174A, A184R, L53N, Q108K, A63R, P18A, and L161R) (Fig. 4b, d).
In contrast, 17 mutations in 7421ADO decreased the activity while increasing or maintaining the amount of soluble ADO (Fig. 4b, d). Among them, 13 mutations had more than 80% wild-type activity while increasing the amount of soluble protein (I189L, K235R, G224E, A98K, G178E, V233I, D93M, R97Q, Y99F, M230T, A196E, I211L, and A17E). However, three mutations (R239Y, A50D, and V175S) decreased the activity by more than 20%.
Substrate specificity was almost unchanged by the mutations, although the fraction of pentadecane was slightly decreased for the mutants having the 10 highest activities (Fig. 4c). The results are consistent with the fact that all representative ADOs have similar specificity for the substrates (Fig. 2c).
In addition to their effects on activity, the mutations also affected the amounts of hydrocarbons produced by ADO. Among the 37 mutants examined, the R181E mutant showed the highest yield of total hydrocarbon, which was 60% higher than that of the wild-type 7421ADO (Fig. 4a). Subsequently, a more than 20% increase in hydrocarbon production was observed for the Q108K, A17E, A196E, A98K, and G224E mutants of 7421ADO, in descending order. However, the activities of these mutants, especially those of A17E and A196E, both of which had activities of ~ 0.8-fold that of the wild type (Fig. 4b), did not increase by much, and the amount of the soluble form of ADO was increased by more than 20% (Fig. 4d). In particular, A17E showed the highest amount of soluble ADO among all mutants (57% increase compared with that of the wild type), followed by A196E and R181E.
The amount of the soluble form of ADO depends on both the solubility and protein expression level of ADO. A decrease in solubility was observed for the mutants with high activities (Fig. 4e). For some mutants, the protein expression level was increased by more than 50%, especially for R181E (2.2-fold greater than that of the wild type) (Fig. 4f). This increase in the protein expression level resulted in the higher amount of soluble ADO (Fig. 4d).
Correlation analysis among the properties of 7421ADO mutants
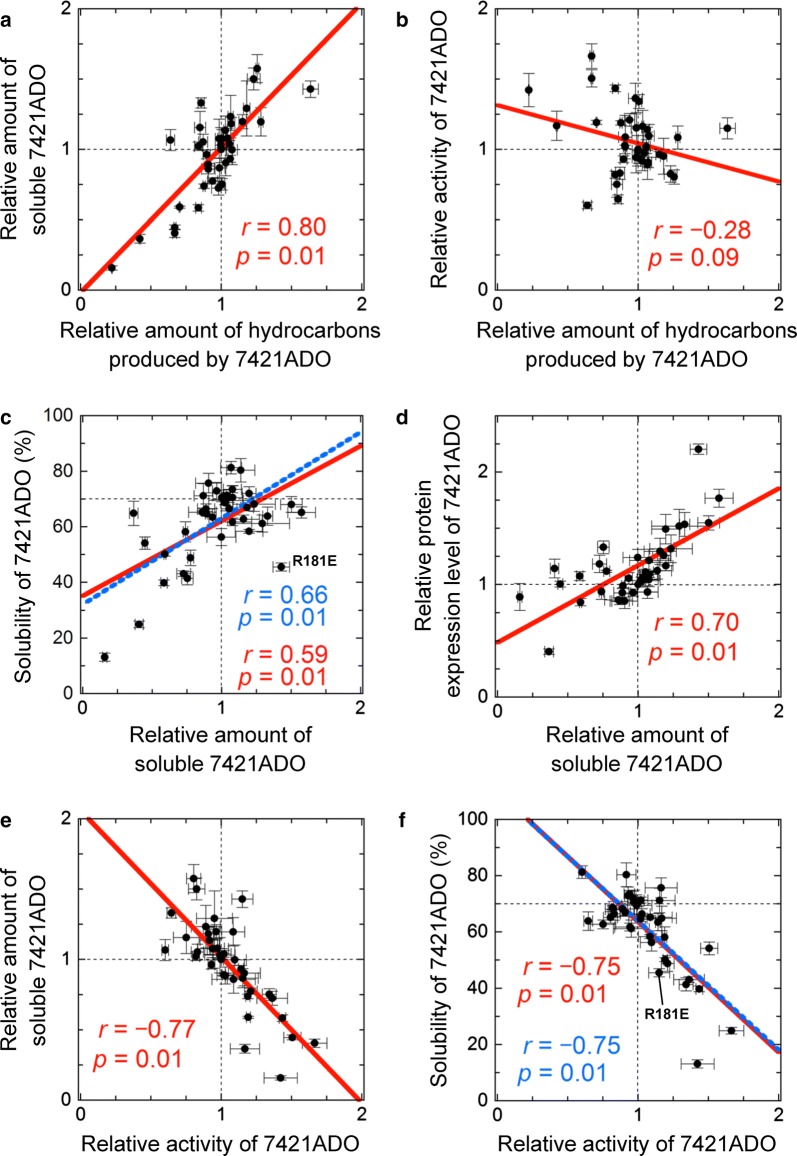
We performed a correlation analysis among the properties measured for the 37 mutants of 7421ADO (Fig. 5; Additional file 10: Figure S7). The amounts of total hydrocarbons produced in E. coli cells coexpressing AAR and ADO should be directly related to both the activity of the ADO and the amount of the soluble form of ADO. We found that there was a good positive correlation between the amount of total hydrocarbon and the amount of the soluble form of ADO (Fig. 5a). In contrast, a clear correlation was not observed between the amount of hydrocarbon and activity (Fig. 5b).
Fig. 5.
Correlation analysis of the mutants of 7421ADO. a, b The relative amount of total hydrocarbon produced in E. coli coexpressing 7942AAR and a mutant of 7421ADO plotted against the relative amount of soluble ADO (a) and relative activity for total hydrocarbons (b). c, d The relative amount of soluble 7421ADO plotted against the solubility (c) and relative protein expression level of 7421ADO (d). e, f The relative activity for total hydrocarbons of 7421ADO plotted against the relative amount of soluble 7421ADO (e) and solubility (f). In each panel, the red continuous line indicates a linear regression, and the corresponding correlation coefficient, r, and p value are shown. In c, f, the data point for R181E is highlighted. The blue dotted line indicates a linear regression obtained without using the data for R181E
The amount of the soluble form of ADO should be directly related to both the solubility and protein expression level of the ADO. In fact, the amount of the soluble form was positively correlated with both the solubility and protein expression level of the ADO (Fig. 5c, d). In contrast, the activity of the ADO was negatively correlated with both the amount of soluble ADO and solubility (Fig. 5e, f) but not with the protein expression level of the ADO (Additional file 10: Figure S7c).
The correlation between the amount of the soluble form and solubility (Fig. 5c) may seem to contradict the results for the R181E mutant, which showed higher amounts of the soluble protein but had lower solubility than the wild-type 7421ADO. However, this may be due to the twofold higher expression level of the protein relative to wild-type 7421ADO expression, and the solubility of R181E may not be correctly estimated. When this data point is omitted from Fig. 5c, a more significant correlation is observed between solubility and the amount of soluble ADO. Similarly, the correlation between the solubility and the amount of hydrocarbon produced by the ADO becomes more significant when the data point for R181E is omitted (Additional file 10: Figure S7a).
Discussion
Comparison of various ADOs
Comparing the activities of ADOs from various cyanobacteria is important when selecting an ADO that is suitable for hydrocarbon production. However, there have been no studies in which an accurate comparison of ADO activity has been performed by normalizing hydrocarbon production to the amount of the soluble form of ADO. Here, we measured both the hydrocarbon production yield and the amount of the soluble form of ADO and accurately evaluated ADO activity. We demonstrated that the hydrocarbon-producing activity is highest for 7942ADO (Fig. 2). However, since the amount of the soluble protein was low for 7942ADO, the total amount of hydrocarbon produced in E. coli expressing 7942ADO together with 7942AAR was moderate among the 10 ADOs examined.
In contrast, 7421ADO, TeADO, and 73102ADO had low activity but showed both high amounts of the soluble ADO protein and high yields of hydrocarbon production. In particular, the high amount of the soluble protein for 7421ADO compensated for its low activity and resulted in high amounts of produced hydrocarbons. Similarly, TeADO yielded the second highest amount of soluble protein (Fig. 3a) and the highest amount of hydrocarbon (Fig. 2a). In general, enzymes from thermophilic bacteria have high optimal temperatures and low activity at 25–37 °C due to the slower dynamics of the enzymes [62]. Thus, TeADO may be useful when hydrocarbon production is conducted at higher temperatures. Finally, consistent with previous reports [11, 50, 53], 73102ADO produced a high amount of hydrocarbon. However, our results also indicate that this was not due to the high activity of the enzyme but to both a moderate activity and a moderate expression level of the 73102ADO protein.
Aldehydes produced by AAR have been reported to be efficiently delivered to ADO by the binding of AAR to ADO [14]. It is possible that AAR and ADO that are derived from the same strain interact with each other more efficiently than those derived from different strains, resulting in a higher production of hydrocarbons. In accordance with this, the combination of AAR and ADO, both of which are derived from Synechococcus elongatus PCC 7942, showed the highest activity (Fig. 2b). However, when 73102ADO was coexpressed with one of 12 representative AARs, the highest AAR activity was observed for 7942AAR, but not for AAR from Nostoc punctiforme PCC 73102 [24]. These results suggest that AAR and ADO derived from the same strain do not always interact more tightly than those derived from different strains.
Unexpectedly, we observed a variation in the amount of soluble 7942AAR protein depending on the coexpressed ADO (Additional file 6: Figure S4). It may be possible that the interaction between AAR and ADO stabilizes the AAR protein, resulting in an increase in the amount of soluble AAR. If this is the case, the present results might suggest that 9313ADO, with which the highest amount of soluble 7942AAR was coexpressed, binds 7942AAR more tightly than other ADOs and stabilizes it, leading to high accumulation of AAR.
Mutational analysis of ADO
In the present mutational analysis of 7421ADO, we identified 20 non-conserved residues that increased ADO activity. However, the mutants that increased the activity showed a decrease in hydrocarbon production due to reductions in their solubility and, therefore, the amount of soluble ADO. There was a good negative correlation between activity and solubility (Fig. 5f). It has been reported that there is a trade-off between the stability and activity of an enzyme [63]. Because a protein exerts its function through dynamic motions, an enzyme with high activity shows enhanced structural flexibility [64], which results in a decrease in stability, leading to aggregation and lower solubility.
We also identified mutations that increased hydrocarbon production by introducing single substitutions at non-conserved residues (i.e., R181E, Q108K, A17E, A196E, A98K, and G224E). The highest and second highest hydrocarbon production levels were observed for the R181E and Q108K mutants, which increased both the activity by 15% and 9% and the amount of soluble protein by 43% and 20%, respectively, compared with those of the wild type. Other mutants that produced 10% or more hydrocarbon than the wild type had more soluble ADO than and similar activity to the wild type. These results suggest that mutations that increase both the activity and the amount of the soluble protein are the most effective in increasing hydrocarbon production and that the mutations that increase either the activity or the amount of the soluble protein while maintaining the amount of the soluble protein or activity, respectively, are the second most effective.
In addition to R181E and Q108K, there were eight mutations that improved ADO activity while maintaining more than 85% of the amount of soluble ADO (Fig. 4). Moreover, 13 other mutations increased the amount of soluble ADO while maintaining more than 80% of wild-type activity. In particular, A17E and A196E increased the amount of soluble ADO by more than 50% compared with that of the wild type. A combination of these mutations may enable us to generate 7421ADO mutants that have higher activity and yield higher amounts of the soluble protein in vivo, leading to higher production of hydrocarbons.
In addition to the mutations at non-conserved residues, appropriate mutations at conserved residues might improve the activity and solubility of ADO. Previous studies have shown that although mutations at conserved residues decrease enzymatic activity in many cases [46–48], conserved residues located far from the active site of a given protein may provide targets for substitution to improve protein properties [39, 65].
Mapping the mutation sites onto the ADO structure
To understand the effects of mutations on ADO from the viewpoint of the three-dimensional structure, we mapped the mutation sites onto the structure of 7421ADO. Although X-ray crystal structures have been determined for 9313ADO [37, 43], 7942ADO [38], and 6803ADO [39], the structure of 7421ADO has not been solved. Thus, the structure of 7421ADO was constructed by homology modeling using the I-TASSER server [66]. The mutation sites of 20 mutants that increased activity, 3 mutants that decreased activity by more than 20%, and 2 mutants that increased the amount of soluble ADO by more than 50% compared with the wild type were mapped onto the ADO structure (Fig. 6). All of these mutation sites, except for E88, can be categorized into the following three regions.
Fig. 6.
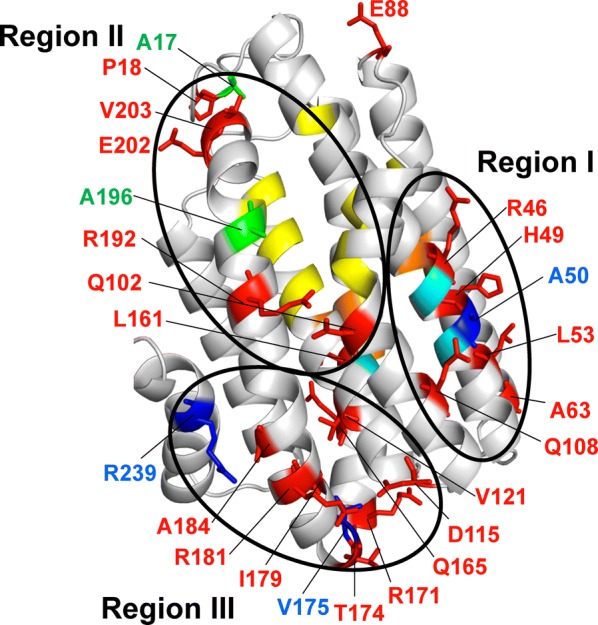
Map of the mutation sites onto the structure of 7421ADO. The structure was built by homology modeling using I-TASSER [66]. The substrate binding site (yellow), iron binding sites (orange), and proton supply channel to the active center (cyan) are shown in the structure [10]. The mutation sites of 20 mutants that increased activity, 3 mutants that decreased activity by more than 20%, and 2 mutants that increased the amount of soluble ADO by more than 50% are indicated by red, blue, and green sticks, respectively. The three regions are indicated by black circles
Region I, including R46, H49, A50, L53, A63, and Q108, is close to the residues involved in the putative proton relay pathway (composed of E47, N51, E108, and E123) from the solvent to the diiron center [37] (Fig. 6). In particular, R46 is also close to the iron binding sites corresponding to the catalytic center of ADO. Thus, activity enhancement by the mutations in region I may be due to perturbation of the active site residues.
Region II, including A17, P18, Q102, L161, R192, A196, E202, and V203, is located close to the substrate-entry/product-exit site of ADO [37] (Fig. 6). Therefore, mutations in this region may affect the open/close dynamics of the substrate-entry site, leading to enhanced activity. Consistent with this, previous studies have shown that the W178R mutation introduced in region II of 7942ADO enhanced ADO activity by removing the Trp residue that points towards the substrate and may interfere with substrate entry [39]. On the other hand, the increase in the amount of soluble ADO by the A17E and A196E mutations may be attributed to the stabilization of ADO that occurs by replacing exposed hydrophobic residues with hydrophilic residues.
Finally, region III, including D115, V121, Q165, R171, T174, V175, I179, R181, A184, and R239, is located close to the turns between α-helices that are far from the active site and the substrate binding site (Fig. 6). The I179L mutation that best improved activity is involved in this region. Similarly, E88, which is not included in either region, is located at a turn between the α-helices. Residues on loops between helices and/or strands have been shown to be important in determining the catalytic reactions of enzymes [67]. Hinge-like motions in the loop regions of enzymes have been suggested to facilitate large-scale, slower motions that produce catalytically competent states [68]. Thus, the introduction of mutations in region III may enhance activity and stability by modulating the structural dynamics of ADO, especially the open/close dynamics of the substrate-entry site near region III [69, 70]. In support of this, Shakeel et al. [70] recently reported that the A161E mutation in 73102ADO (corresponding to V173E in 7421ADO), which is introduced in region III, improves ADO activity at high temperatures [70].
In addition, it may be possible that mutations in ADO affect the interactions with AAR and/or the reducing system composed of Fd and FNR and then improve/reduce the activity and solubility of ADO. Future studies on the ADO residues that are essential for binding with AAR and Fd/FNR will clarify the reason for the mutational effects observed in the present study.
Taken together, the present results suggest that residues in regions I, II, and III are good targets for introducing mutations to improve ADO activity. In addition, stabilization of the ADO protein via removal of exposed hydrophobic residues may improve the solubility of ADO. A combination of these mutations will increase hydrocarbon production by ADO.
Substrate specificity
Our previous studies showed that there is a difference in substrate specificity between AARs from marine and freshwater cyanobacteria, suggesting that the type of habitat of the host cyanobacteria may determine the substrate specificity of AAR [24]. In contrast, the present results indicate that the substrate specificity of ADO is not affected by the type of habitat of the host cyanobacteria (Fig. 2), suggesting that the residues essential for determining the substrate specificity of ADO are conserved among ADOs. This suggestion is supported by three lines of evidence. First, the substrate specificity of 7421ADO was not remarkably changed by any of the mutations at its non-conserved residues (Fig. 4). Second, the residues that constitute the substrate binding channel are reported to be conserved among ADOs from several cyanobacteria [38]. Third, mutations at conserved residues can change the substrate specificity of the ADO [33, 43]. Therefore, these results may suggest that both the activity and substrate specificity of ADO can be regulated separately by introducing mutations at non-conserved and conserved residues, respectively.
Conclusions
In this study, we compared the hydrocarbon-producing activities of 10 representative ADOs and demonstrated that 7942ADO had the highest activity. By creating 37 mutants of the less active 7421ADO, in which a non-conserved residue was replaced by the corresponding residue in the highly active 7942ADO, we identified 20 non-conserved residues that are responsible for the high activity of ADO. Among them, the Q108K and R181E mutations increased both the activity and the amount of the soluble form of ADO, and eight mutations increased the ADO activity while maintaining more than 85% of the amount of soluble ADO. We also identified 13 other mutations that increased the amount of soluble ADO while maintaining more than 80% of the wild-type activity. Our results may be useful for generating combinatorial mutants of ADO that have both higher activity and higher amounts of the soluble protein in vivo, thereby producing higher yields of biohydrocarbons.
Methods
Plasmids
The DNA fragments encoding 6803ADO, 73102ADO, 7336ADO, 7421ADO, 7942ADO, 9443ADO, and TeADO were prepared by overlap extension polymerase chain reaction. The codons were optimized for high expression in E. coli [71]. The codon-optimized DNA fragments coding for 9313ADO, 7425ADO, and PaADO were constructed by Eurofin Operon Biotechnologies (Tokyo, Japan). For the coexpression of AAR and ADO in E. coli, the pETDuet-1 coexpression vector (Merck Millipore, Darmstadt, Germany) was used, into which the codon-optimized DNA fragments of AAR and ADO were cloned via the NcoI and EcoRI restriction sites and the NdeI and AvrII restriction sites, respectively [22]. Both the AAR and ADO genes were preceded by a T7 promoter, lac operator, and ribosome binding site. 7942AAR was used for coexpression because it is known to have high aldehyde-producing activity [24]. All AAR and ADO proteins had a C-terminal extension of Gly-Ser-Ser-Gly and a 6 × His-tag. The ESPRESSO server, which estimates protein expression (http://cblab.meiyaku.jp/ESPRESSO/) [72], predicted that all the AAR and ADO constructs could be expressed in E. coli. The mutants of 7421ADO were constructed according to the protocols of the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA).
Hydrocarbon production in E. coli coexpressing AAR and ADO
Alkanes and alkenes were produced in E. coli coexpressing AAR and ADO, as previously described [22, 24]. Briefly, E. coli BL21(DE3)pLysS competent cells were transformed with the pETDuet-1 coexpression vector, containing both AAR and ADO genes, and inoculated onto a 2 × YT agarose plate containing carbenicillin (50 μg/mL) and chloramphenicol (34 μg/mL). The colonies on the plate were seeded into M9 medium containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) and pre-cultured at 37 °C overnight. The preculture was then seeded at an optical density of 0.1 at 600 nm into M9 medium containing 100 µM ammonium iron (II) sulfate and 1 mM isopropyl β-d-1-thiogalactoside. The culture was incubated in a 96 deep-well plate at 37 °C for 16 h. The cell culture was sonicated, and 500 µL of the cell lysate was centrifuged to separate the supernatant and pellet fractions, which were used to quantify the amounts of the soluble and insoluble forms of the AAR and ADO proteins by western blotting (for 10 representative ADOs) and SDS-PAGE (for the 7421ADO mutants). Next, 800 µL of the cell lysate was mixed with an equal amount of ethyl acetate by vortexing. The organic phase was separated from the aqueous phase by centrifugation and was subjected to GC–MS analysis using a Shimadzu gas chromatograph mass spectrometer GCMS-QP2010 Ultra (Shimadzu, Kyoto, Japan), as described elsewhere [22, 24].
The total amounts of hydrocarbons [pentadecane (C15:0), heptadecene (C17:1), and heptadecane (C17:0)] produced in the culture were normalized to the amount of the soluble form of the ADO protein expressed in E. coli, which was quantified using western blotting or SDS-PAGE. After normalization, the total amounts of hydrocarbons were compared with those of 73102ADO (for 10 representative ADOs) or the wild-type 7421ADO (for 7421ADO mutants), and the ratio was used as an index of hydrocarbon-producing activity of ADO. The measurements were carried out more than three times, and the means ± standard errors are shown.
Western blotting
The amounts of the soluble and insoluble forms of ADO proteins were measured by SDS-PAGE or western blotting as previously described [22, 24]. Briefly, supernatant and pellet samples, prepared from the same amounts of cell lysates, were analyzed by SDS-PAGE using 12% gels. The proteins were electrotransferred onto a polyvinylidene difluoride membrane (Millipore), and the membrane was blocked with 100 mL of phosphate-buffered saline containing 5% skim milk. The membrane was incubated with an anti-His-tag antibody conjugated with horseradish peroxidase (MBL, Japan). The AAR and ADO proteins, both having a His-tag at the C-terminus, were visualized by color reactions with 5 mL of 3,3′,5,5′-tetramethylbenzidine solution (ATTO, Tokyo, Japan). Gel images of western blotting were acquired using a Gel Doc EZ imager (Bio-Rad, Hercules, CA, USA). Quantification of the bands of AAR and ADO was performed using ImageLab software (Bio-Rad).
Analysis of amino acid sequences
A total of 134 orthologs of ADO were found on the BLAST server [73] using the amino acid sequence of 73102ADO as a query. The multiple sequence alignment of the 134 ADO amino acid sequences was carried out on the BLAST server. The phylogenetic tree of cyanobacteria based on the ADO amino acid sequences was drawn using NJplot [74]. The multiple sequence alignment of 10 representative ADOs was carried out using the Clustal X software [75].
The habitats of 134 cyanobacterial species were examined using MicrobeDB (http://microbedb.jp/), CyanoBase (http://genome.microbedb.jp/cyanobase/) [76], and the Pasteur Culture Collection of Cyanobacteria (PCC; http://cyanobacteria.web.pasteur.fr/). Ninety-three and 31 species are freshwater and marine cyanobacteria, respectively, and 10 species can live in both freshwater and marine environments.
Additional files
Additional file 1: Table S1. Amino acid sequence identities (%) among the ADO sequences used in the present study.
Additional file 2: Figure S1. Phylogenetic tree of cyanobacterial ADOs based on ADO amino acid sequences. ADO sequence groups 1, 2, and 3 are shown in hatched yellow, cyan, and magenta, respectively. Host strains of ADOs used in the present study are indicated in red. Vertical bars show habitats of derived cyanobacteria: marine (blue), freshwater (red), and both marine and freshwater environments (green). The scale of the phylogenetic tree is shown in the upper right corner.
Additional file 3: Figure S2. GC–MS profiles of extracts from E. coli cell cultures coexpressing 7942AAR and ADO from one of 10 representative cyanobacteria. a Whole GC–MS profiles. Pentadecane (C15:0), heptadecene (C17:1), and heptadecane (C17:0) were eluted at retention times of 14.15, 16.32, and 16.54 min, respectively. The region in the black square is expanded and shown in Additional file 4: Figure S3c. b Peaks of pentadecane. c Peaks of heptadecene and heptadecane. Color codes for various ADOs are shown in panel (a).
Additional file 4: Figure S3. GC–MS profiles of fatty aldehydes and corresponding fatty alcohols. a, b GC–MS profiles of fatty aldehyde and alcohol standards, including hexadecanal (C16:0 aldehyde), hexadecanol (C16:0 alcohol), octadecanal (C18:0 aldehyde), and octadecanol (C18:0 alcohol), which were eluted at retention times of 17.69, 18.35, 19.74 and 20.32 min, respectively. The concentrations of the standards were 50 mg/L (a) and 10 mg/L (b). c, d GC–MS profiles of the extracts from E. coli cell culture expressing neither AAR nor ADO (c) and from E. coli coexpressing 7942AAR and 73102ADO (d). In panel (d), peaks 1, 2, and 3 correspond to hexadecanal, hexadecanol, and octadecenal, respectively, while these peaks are not present in panel (c).
Additional file 5: Table S2. The absolute amounts of hydrocarbons (in milligrams) produced in E. coli coexpressing ADO and 7942AAR per liter of E. coli cell culture using M9 medium.
Additional file 6: Figure S4. Solubility and protein expression levels of ADO and 7942AAR in E. coli. a Western blotting analysis of the supernatant and pellet fractions of the E. coli cell culture lysates. Images of triplicate experiments are shown. The E. coli cell culture coexpressing ADO and 7942AAR was sonicated and centrifuged to separate the supernatant (S) and pellet (P) fractions. M denotes the molecular weight markers. The bands for AAR (38.6 kDa) and ADO (26.5–28.3 kDa) are indicated by arrows. The results for E. coli transformed with the pETDuet-1 vector containing neither the AAR nor ADO gene are shown in lanes 17 and 18 as a control. In lanes 9 and 10, the 7425ADO protein migrated more slowly than the other ADOs (see text for details). b The amount of the soluble form of 7942AAR relative to that observed in E. coli coexpressing 73102ADO.
Additional file 7: Table S3. Charge and molecular weight of each ADO used in the present study.
Additional file 8: Figure S5. Correlation analysis among various properties of 10 representative ADOs. The relative amount of total hydrocarbon plotted against relative activity (a), relative amount of soluble ADO (b), solubility (c), and relative protein expression level of ADO (d). The relative activity of ADO plotted against the relative amount of soluble ADO (e), the relative protein expression level (f), and solubility (g). The relative amount of soluble ADO plotted against the relative protein expression level (h) and solubility (i). Solubility plotted against the relative protein expression level (j). In each panel, the red continuous line indicates a linear regression. The corresponding correlation coefficients, r, and the p values are shown.
Additional file 9: Figure S6. Correlation analysis of various properties of ADO with amino acid sequence identities. a The relative activity of ADO plotted against sequence identity (%) with the amino acid sequence of 7942ADO. b, c The solubility (b) and protein expression level (c) of ADO plotted against sequence identity (%) with the amino acid sequences of TeADO and 7421ADO, respectively. d The amount of total hydrocarbon produced in E. coli coexpressing 7942AAR and ADO from one of 10 representative cyanobacteria plotted against sequence identity (%) with the amino acid sequence of TeADO. e The relative amount of soluble ADO plotted against sequence identity (%) with the amino acid sequence of 7421ADO. In each panel, the red continuous line and blue dotted line respectively indicate a linear regression obtained using all data and that obtained without the data point for the highest value. The corresponding correlation coefficients, r, and the p values are shown.
Additional file 10: Figure S7. Correlation analysis among various properties of the 7421ADO mutants. a, b The relative amounts of hydrocarbons plotted against the solubility (a) and relative protein expression level of 7421ADO (b). c The relative activity of 7421ADO plotted against the relative protein expression level. d The solubility of ADO plotted against the relative protein expression level. In (a, d), the data point for R181E is highlighted. The blue dotted line indicates a linear regression obtained without using the data for R181E.
Authors’ contributions
HK and MA designed the study. HK, YH, and MA performed the experiments. HK, YH, and MA analyzed the data. HK, YH, and MA wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Data availability
The datasets supporting the conclusions of this article are included within the article and its additional files.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by Grant-in-Aid for JSPS Research Fellow (HK) and Grants-in-Aid for Scientific Research (YH, MA) from the Ministry of Education, Culture, Sports, Science and Technology, Japan and the Institute for Fermentation, Osaka (MA). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- 6803ADO
ADO from Synechocystis sp. PCC 6803
- 73102ADO
ADO from Nostoc punctiforme PCC 73102
- 7336ADO
ADO from Synechococcus sp. PCC 7336
- 7421ADO
ADO from Gloeobacter violaceus PCC 7421
- 7425ADO
ADO from Cyanothece sp. PCC 7425
- 7942AAR
AAR from Synechococcus elongatus PCC 7942
- 7942ADO
ADO from Synechococcus elongatus PCC 7942
- 9313ADO
ADO from Prochlorococcus marinus MIT 9313
- 9443ADO
ADO from Microcystis aeruginosa PCC 9443
- AAR
acyl-ACP reductase
- ACP
acyl carrier protein
- ADO
aldehyde-deformylating oxygenase
- CoA
coenzyme A
- Fd
ferredoxin
- FNR
ferredoxin NADP+ reductase
- PaADO
ADO from Planktothrix agardhii NIVA-CYA 126/8
- GC-MS
gas chromatography-mass spectrometry
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TeADO
ADO from Thermosynechococcus elongatus BP-1
- WT
wild type
Contributor Information
Hisashi Kudo, Email: hisashikudo23@gmail.com.
Yuuki Hayashi, Email: hayashi@bio.c.u-tokyo.ac.jp.
Munehito Arai, Phone: +81-3-5454-6751, Email: arai@bio.c.u-tokyo.ac.jp.
References
- 1.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 2.Kolattukudy PE. Biosynthesis of surface lipids. Biosynthesis of long-chain hydrocarbons and waxy esters is discussed. Science. 1968;159:498–505. doi: 10.1126/science.159.3814.498. [DOI] [PubMed] [Google Scholar]
- 3.Marsh EN, Waugh MW. Aldehyde decarbonylases: enigmatic enzymes of hydrocarbon biosynthesis. ACS Catal. 2013;3:2515–2521. doi: 10.1021/cs400637t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angermayr SA, Hellingwerf KJ, Lindblad P, de Mattos MJ. Energy biotechnology with cyanobacteria. Curr Opin Biotechnol. 2009;20:257–263. doi: 10.1016/j.copbio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Wang J, Zhang W, Meldrum DR. Application of synthetic biology in cyanobacteria and algae. Front Microbiol. 2012;3:344. doi: 10.3389/fmicb.2012.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kallio P, Pasztor A, Akhtar MK, Jones PR. Renewable jet fuel. Curr Opin Biotechnol. 2014;26:50–55. doi: 10.1016/j.copbio.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Kang MK, Nielsen J. Biobased production of alkanes and alkenes through metabolic engineering of microorganisms. J Ind Microbiol Biotechnol. 2017;44:613–622. doi: 10.1007/s10295-016-1814-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie M, Wang W, Zhang W, Chen L, Lu X. Versatility of hydrocarbon production in cyanobacteria. Appl Microbiol Biotechnol. 2017;101:905–919. doi: 10.1007/s00253-016-8064-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Zhu K. Microbial production of alka(e)ne biofuels. Curr Opin Biotechnol. 2017;50:11–18. doi: 10.1016/j.copbio.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Arai M, Hayashi Y, Kudo H. Cyanobacterial enzymes for bioalkane production. In: Zhang W, Song X, editors. Synthetic biology of Cyanobacteria. Singapore: Springer Singapore; 2018. pp. 119–154. [DOI] [PubMed] [Google Scholar]
- 11.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 12.Warui DM, Li N, Norgaard H, Krebs C, Bollinger JM, Jr, Booker SJ. Detection of formate, rather than carbon monoxide, as the stoichiometric coproduct in conversion of fatty aldehydes to alkanes by a cyanobacterial aldehyde decarbonylase. J Am Chem Soc. 2011;133:3316–3319. doi: 10.1021/ja111607x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin F, Das D, Lin XN, Marsh EN. Aldehyde-forming fatty acyl-CoA reductase from cyanobacteria: expression, purification and characterization of the recombinant enzyme. FEBS J. 2013;280:4773–4781. doi: 10.1111/febs.12443. [DOI] [PubMed] [Google Scholar]
- 14.Warui DM, Pandelia ME, Rajakovich LJ, Krebs C, Bollinger JM, Jr, Booker SJ. Efficient delivery of long-chain fatty aldehydes from the Nostoc punctiforme acyl–acyl carrier protein reductase to its cognate aldehyde-deformylating oxygenase. Biochemistry. 2015;54:1006–1015. doi: 10.1021/bi500847u. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q, Wang W, Zhao H, Lu X. Effects of fatty acid activation on photosynthetic production of fatty acid-based biofuels in Synechocystis sp. PCC6803. Biotechnol Biofuels. 2012;5:17. doi: 10.1186/1754-6834-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Liu X, Lu X. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol Biofuels. 2013;6:69. doi: 10.1186/1754-6834-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peramuna A, Morton R, Summers ML. Enhancing alkane production in cyanobacterial lipid droplets: a model platform for industrially relevant compound production. Life. 2015;5:1111–1126. doi: 10.3390/life5021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kageyama H, Waditee-Sirisattha R, Sirisattha S, Tanaka Y, Mahakhant A, Takabe T. Improved alkane production in nitrogen-fixing and halotolerant cyanobacteria via abiotic stresses and genetic manipulation of alkane synthetic genes. Curr Microbiol. 2015;71:115–120. doi: 10.1007/s00284-015-0833-7. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida S, Takahashi M, Ikeda A, Fukuda H, Kitazaki C, Asayama M. Overproduction and easy recovery of biofuels from engineered cyanobacteria, autolyzing multicellular cells. J Biochem. 2015;157:519–527. doi: 10.1093/jb/mvv011. [DOI] [PubMed] [Google Scholar]
- 20.Yoshino T, Liang Y, Arai D, Maeda Y, Honda T, Muto M, Kakunaka N, Tanaka T. Alkane production by the marine cyanobacterium Synechococcus sp. NKBG15041c possessing the a-olefin biosynthesis pathway. Appl Microbiol Biotechnol. 2015;99:1521–1529. doi: 10.1007/s00253-014-6286-2. [DOI] [PubMed] [Google Scholar]
- 21.Harger M, Zheng L, Moon A, Ager C, An JH, Choe C, Lai YL, Mo B, Zong D, Smith MD, et al. Expanding the product profile of a microbial alkane biosynthetic pathway. ACS Synth Biol. 2013;2:59–62. doi: 10.1021/sb300061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi Y, Yasugi F, Arai M. Role of cysteine residues in the structure, stability, and alkane producing activity of cyanobacterial aldehyde deformylating oxygenase. PLoS ONE. 2015;10:e0122217. doi: 10.1371/journal.pone.0122217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao YX, Xiao WH, Zhang JL, Xie ZX, Ding MZ, Yuan YJ. Heterologous biosynthesis and manipulation of alkanes in Escherichia coli. Metab Eng. 2016;38:19–28. doi: 10.1016/j.ymben.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Kudo H, Nawa R, Hayashi Y, Arai M. Comparison of aldehyde-producing activities of cyanobacterial acyl-(acyl carrier protein) reductases. Biotechnol Biofuels. 2016;9:234. doi: 10.1186/s13068-016-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song X, Yu H, Zhu K. Improving alkane synthesis in Escherichia coli via metabolic engineering. Appl Microbiol Biotechnol. 2016;100:757–767. doi: 10.1007/s00253-015-7026-y. [DOI] [PubMed] [Google Scholar]
- 26.Zhou YJ, Buijs NA, Zhu Z, Qin J, Siewers V, Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foo JL, Susanto AV, Keasling JD, Leong SS, Chang MW. Whole-cell biocatalytic and de novo production of alkanes from free fatty acids in Saccharomyces cerevisiae. Biotechnol Bioeng. 2017;114:232–237. doi: 10.1002/bit.25920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Zhou YJ, Kang MK, Krivoruchko A, Buijs NA, Nielsen J. Enabling the synthesis of medium chain alkanes and 1-alkenes in yeast. Metab Eng. 2017;44:81–88. doi: 10.1016/j.ymben.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Crepin L, Lombard E, Guillouet SE. Metabolic engineering of Cupriavidus necator for heterotrophic and autotrophic alka(e)ne production. Metab Eng. 2016;37:92–101. doi: 10.1016/j.ymben.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Sinha M, Weyda I, Sorensen A, Bruno KS, Ahring BK. Alkane biosynthesis by Aspergillus carbonarius ITEM 5010 through heterologous expression of Synechococcus elongatus acyl-ACP/CoA reductase and aldehyde deformylating oxygenase genes. AMB Express. 2017;7:18. doi: 10.1186/s13568-016-0321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallio P, Pasztor A, Thiel K, Akhtar MK, Jones PR. An engineered pathway for the biosynthesis of renewable propane. Nat Commun. 2014;5:4731. doi: 10.1038/ncomms5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menon N, Pasztor A, Menon BR, Kallio P, Fisher K, Akhtar MK, Leys D, Jones PR, Scrutton NS. A microbial platform for renewable propane synthesis based on a fermentative butanol pathway. Biotechnol Biofuels. 2015;8:61. doi: 10.1186/s13068-015-0231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao L, Li JJ, Jia C, Li M, Lu X. Structure-oriented substrate specificity engineering of aldehyde-deformylating oxygenase towards aldehydes carbon chain length. Biotechnol Biofuels. 2016;9:185. doi: 10.1186/s13068-016-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Liang Y, Wu W, Tan X, Lu X. Microbial synthesis of propane by engineering valine pathway and aldehyde-deformylating oxygenase. Biotechnol Biofuels. 2016;9:80. doi: 10.1186/s13068-016-0496-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patrikainen P, Carbonell V, Thiel K, Aro EM, Kallio P. Comparison of orthologous cyanobacterial aldehyde deformylating oxygenases in the production of volatile C3–C7 alkanes in engineered E. coli. Metab Eng Commun. 2017;5:9–18. doi: 10.1016/j.meteno.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Chang WC, Warui DM, Booker SJ, Krebs C, Bollinger JM., Jr Evidence for only oxygenative cleavage of aldehydes to alk(a/e)nes and formate by cyanobacterial aldehyde decarbonylases. Biochemistry. 2012;51:7908–7916. doi: 10.1021/bi300912n. [DOI] [PubMed] [Google Scholar]
- 37.Buer BC, Paul B, Das D, Stuckey JA, Marsh EN. Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound. ACS Chem Biol. 2014;9:2584–2593. doi: 10.1021/cb500343j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia C, Li M, Li J, Zhang J, Zhang H, Cao P, Pan X, Lu X, Chang W. Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases. Protein Cell. 2015;6:55–67. doi: 10.1007/s13238-014-0108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Bao L, Jia C, Li M, Li JJ, Lu X. Identification of residues important for the activity of aldehyde-deformylating oxygenase through investigation into the structure–activity relationship. BMC Biotechnol. 2017;17:31. doi: 10.1186/s12896-017-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park AK, Kim IS, Jeon BW, Roh SJ, Ryu MY, Baek HR, Jo SW, Kim YS, Park H, Lee JH, et al. Crystal structures of aldehyde deformylating oxygenase from Limnothrix sp. KNUA012 and Oscillatoria sp. KNUA011. Biochem Biophys Res Commun. 2016;477:395–400. doi: 10.1016/j.bbrc.2016.06.090. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Lu X, Li JJ. Conversion of fatty aldehydes into alk (a/e)nes by in vitro reconstituted cyanobacterial aldehyde-deformylating oxygenase with the cognate electron transfer system. Biotechnol Biofuels. 2013;6:86. doi: 10.1186/1754-6834-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andre C, Kim SW, Yu XH, Shanklin J. Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2. Proc Natl Acad Sci USA. 2013;110:3191–3196. doi: 10.1073/pnas.1218769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khara B, Menon N, Levy C, Mansell D, Das D, Marsh EN, Leys D, Scrutton NS. Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase. ChemBioChem. 2013;14:1204–1208. doi: 10.1002/cbic.201300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worth CL, Gong S, Blundell TL. Structural and functional constraints in the evolution of protein families. Nat Rev Mol Cell Biol. 2009;10:709–720. doi: 10.1038/nrm2762. [DOI] [PubMed] [Google Scholar]
- 45.Gong S, Worth CL, Bickerton GR, Lee S, Tanramluk D, Blundell TL. Structural and functional restraints in the evolution of protein families and superfamilies. Biochem Soc Trans. 2009;37:727–733. doi: 10.1042/BST0370727. [DOI] [PubMed] [Google Scholar]
- 46.Echave J, Spielman SJ, Wilke CO. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 2016;17:109–121. doi: 10.1038/nrg.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abriata LA, Palzkill T, Dal Peraro M. How structural and physicochemical determinants shape sequence constraints in a functional enzyme. PLoS ONE. 2015;10:e0118684. doi: 10.1371/journal.pone.0118684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das D, Eser BE, Han J, Sciore A, Marsh EN. Oxygen-independent decarbonylation of aldehydes by cyanobacterial aldehyde decarbonylase: a new reaction of diiron enzymes. Angew Chem Int Ed Engl. 2011;50:7148–7152. doi: 10.1002/anie.201101552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eser BE, Das D, Han J, Jones PR, Marsh EN. Oxygen-independent alkane formation by non-heme iron-dependent cyanobacterial aldehyde decarbonylase: investigation of kinetics and requirement for an external electron donor. Biochemistry. 2011;50:10743–10750. doi: 10.1021/bi2012417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das D, Ellington B, Paul B, Marsh EN. Mechanistic insights from reaction of a-oxiranyl-aldehydes with cyanobacterial aldehyde deformylating oxygenase. ACS Chem Biol. 2014;9:570–577. doi: 10.1021/cb400772q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coursolle D, Lian J, Shanklin J, Zhao H. Production of long chain alcohols and alkanes upon coexpression of an acyl-ACP reductase and aldehyde-deformylating oxygenase with a bacterial type-I fatty acid synthase in E. coli. Mol Biosyst. 2015;11:2464–2472. doi: 10.1039/C5MB00268K. [DOI] [PubMed] [Google Scholar]
- 53.Cao YX, Xiao WH, Liu D, Zhang JL, Ding MZ, Yuan YJ. Biosynthesis of odd-chain fatty alcohols in Escherichia coli. Metab Eng. 2015;29:113–123. doi: 10.1016/j.ymben.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Kang MK, Zhou YJ, Buijs NA, Nielsen J. Functional screening of aldehyde decarbonylases for long-chain alkane production by Saccharomyces cerevisiae. Microb Cell Fact. 2017;16:74. doi: 10.1186/s12934-017-0683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bloch K. Enzymic synthesis of monounsaturated fatty acids. Acc Chem Res. 1969;2:193–202. doi: 10.1021/ar50019a001. [DOI] [Google Scholar]
- 56.Niwa T, Ying BW, Saito K, Jin W, Takada S, Ueda T, Taguchi H. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc Natl Acad Sci USA. 2009;106:4201–4206. doi: 10.1073/pnas.0811922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor SC, Posch A. The design of a quantitative western blot experiment. Biomed Res Int. 2014;2014:361590. doi: 10.1155/2014/361590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bass JJ, Wilkinson DJ, Rankin D, Phillips BE, Szewczyk NJ, Smith K, Atherton PJ. An overview of technical considerations for Western blotting applications to physiological research. Scand J Med Sci Sports. 2017;27:4–25. doi: 10.1111/sms.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graceffa P, Jancso A, Mabuchi K. Modification of acidic residues normalizes sodium dodecyl sulfate-polyacrylamide gel electrophoresis of caldesmon and other proteins that migrate anomalously. Arch Biochem Biophys. 1992;297:46–51. doi: 10.1016/0003-9861(92)90639-E. [DOI] [PubMed] [Google Scholar]
- 60.Alves VS, Pimenta DC, Sattlegger E, Castilho BA. Biophysical characterization of Gir2, a highly acidic protein of Saccharomyces cerevisiae with anomalous electrophoretic behavior. Biochem Biophys Res Commun. 2004;314:229–234. doi: 10.1016/j.bbrc.2003.12.086. [DOI] [PubMed] [Google Scholar]
- 61.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic–mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 63.Arai M. Unified understanding of folding and binding mechanisms of globular and intrinsically disordered proteins. Biophys Rev. 2018;10:163–181. doi: 10.1007/s12551-017-0346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iwakura M, Maki K, Takahashi H, Takenawa T, Yokota A, Katayanagi K, Kamiyama T, Gekko K. Evolutional design of a hyperactive cysteine- and methionine-free mutant of Escherichia coli dihydrofolate reductase. J Biol Chem. 2006;281:13234–13246. doi: 10.1074/jbc.M508823200. [DOI] [PubMed] [Google Scholar]
- 65.Suemori A. Conserved and non-conserved residues and their role in the structure and function of p-hydroxybenzoate hydroxylase. Protein Eng Des Sel. 2013;26:479–488. doi: 10.1093/protein/gzt026. [DOI] [PubMed] [Google Scholar]
- 66.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malabanan MM, Amyes TL, Richard JP. A role for flexible loops in enzyme catalysis. Curr Opin Struct Biol. 2010;20:702–710. doi: 10.1016/j.sbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 69.Yu H, Yan Y, Zhang C, Dalby PA. Two strategies to engineer flexible loops for improved enzyme thermostability. Sci Rep. 2017;7:41212. doi: 10.1038/srep41212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shakeel T, Gupta M, Fatma Z, Kumar R, Kumar R, Singh R, Sharma M, Jade D, Gupta D, Fatma T, et al. A consensus-guided approach yields a heat-stable alkane-producing enzyme and identifies residues promoting thermostability. J Biol Chem. 2018;293:9148–9161. doi: 10.1074/jbc.RA117.000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirose S, Noguchi T. ESPRESSO: a system for estimating protein expression and solubility in protein expression systems. Proteomics. 2013;13:1444–1456. doi: 10.1002/pmic.201200175. [DOI] [PubMed] [Google Scholar]
- 73.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 75.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 76.Fujisawa T, Narikawa R, Maeda SI, Watanabe S, Kanesaki Y, Kobayashi K, Nomata J, Hanaoka M, Watanabe M, Ehira S, et al. CyanoBase: a large-scale update on its 20th anniversary. Nucleic Acids Res. 2017;45:D551–D554. doi: 10.1093/nar/gkw1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Amino acid sequence identities (%) among the ADO sequences used in the present study.
Additional file 2: Figure S1. Phylogenetic tree of cyanobacterial ADOs based on ADO amino acid sequences. ADO sequence groups 1, 2, and 3 are shown in hatched yellow, cyan, and magenta, respectively. Host strains of ADOs used in the present study are indicated in red. Vertical bars show habitats of derived cyanobacteria: marine (blue), freshwater (red), and both marine and freshwater environments (green). The scale of the phylogenetic tree is shown in the upper right corner.
Additional file 3: Figure S2. GC–MS profiles of extracts from E. coli cell cultures coexpressing 7942AAR and ADO from one of 10 representative cyanobacteria. a Whole GC–MS profiles. Pentadecane (C15:0), heptadecene (C17:1), and heptadecane (C17:0) were eluted at retention times of 14.15, 16.32, and 16.54 min, respectively. The region in the black square is expanded and shown in Additional file 4: Figure S3c. b Peaks of pentadecane. c Peaks of heptadecene and heptadecane. Color codes for various ADOs are shown in panel (a).
Additional file 4: Figure S3. GC–MS profiles of fatty aldehydes and corresponding fatty alcohols. a, b GC–MS profiles of fatty aldehyde and alcohol standards, including hexadecanal (C16:0 aldehyde), hexadecanol (C16:0 alcohol), octadecanal (C18:0 aldehyde), and octadecanol (C18:0 alcohol), which were eluted at retention times of 17.69, 18.35, 19.74 and 20.32 min, respectively. The concentrations of the standards were 50 mg/L (a) and 10 mg/L (b). c, d GC–MS profiles of the extracts from E. coli cell culture expressing neither AAR nor ADO (c) and from E. coli coexpressing 7942AAR and 73102ADO (d). In panel (d), peaks 1, 2, and 3 correspond to hexadecanal, hexadecanol, and octadecenal, respectively, while these peaks are not present in panel (c).
Additional file 5: Table S2. The absolute amounts of hydrocarbons (in milligrams) produced in E. coli coexpressing ADO and 7942AAR per liter of E. coli cell culture using M9 medium.
Additional file 6: Figure S4. Solubility and protein expression levels of ADO and 7942AAR in E. coli. a Western blotting analysis of the supernatant and pellet fractions of the E. coli cell culture lysates. Images of triplicate experiments are shown. The E. coli cell culture coexpressing ADO and 7942AAR was sonicated and centrifuged to separate the supernatant (S) and pellet (P) fractions. M denotes the molecular weight markers. The bands for AAR (38.6 kDa) and ADO (26.5–28.3 kDa) are indicated by arrows. The results for E. coli transformed with the pETDuet-1 vector containing neither the AAR nor ADO gene are shown in lanes 17 and 18 as a control. In lanes 9 and 10, the 7425ADO protein migrated more slowly than the other ADOs (see text for details). b The amount of the soluble form of 7942AAR relative to that observed in E. coli coexpressing 73102ADO.
Additional file 7: Table S3. Charge and molecular weight of each ADO used in the present study.
Additional file 8: Figure S5. Correlation analysis among various properties of 10 representative ADOs. The relative amount of total hydrocarbon plotted against relative activity (a), relative amount of soluble ADO (b), solubility (c), and relative protein expression level of ADO (d). The relative activity of ADO plotted against the relative amount of soluble ADO (e), the relative protein expression level (f), and solubility (g). The relative amount of soluble ADO plotted against the relative protein expression level (h) and solubility (i). Solubility plotted against the relative protein expression level (j). In each panel, the red continuous line indicates a linear regression. The corresponding correlation coefficients, r, and the p values are shown.
Additional file 9: Figure S6. Correlation analysis of various properties of ADO with amino acid sequence identities. a The relative activity of ADO plotted against sequence identity (%) with the amino acid sequence of 7942ADO. b, c The solubility (b) and protein expression level (c) of ADO plotted against sequence identity (%) with the amino acid sequences of TeADO and 7421ADO, respectively. d The amount of total hydrocarbon produced in E. coli coexpressing 7942AAR and ADO from one of 10 representative cyanobacteria plotted against sequence identity (%) with the amino acid sequence of TeADO. e The relative amount of soluble ADO plotted against sequence identity (%) with the amino acid sequence of 7421ADO. In each panel, the red continuous line and blue dotted line respectively indicate a linear regression obtained using all data and that obtained without the data point for the highest value. The corresponding correlation coefficients, r, and the p values are shown.
Additional file 10: Figure S7. Correlation analysis among various properties of the 7421ADO mutants. a, b The relative amounts of hydrocarbons plotted against the solubility (a) and relative protein expression level of 7421ADO (b). c The relative activity of 7421ADO plotted against the relative protein expression level. d The solubility of ADO plotted against the relative protein expression level. In (a, d), the data point for R181E is highlighted. The blue dotted line indicates a linear regression obtained without using the data for R181E.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.