Abstract
Hypercholesterolemia, the driving force of atherosclerosis, accelerates the expansion and mobilization of hematopoietic stem and progenitor cells (HSPCs). The molecular determinants connecting hypercholesterolemia with hematopoiesis are unclear. Here we report that a somite-derived pro-hematopoietic cue, AIBP, orchestrates HSPC emergence from the hemogenic endothelium, a type of specialized endothelium manifesting hematopoietic potential. Mechanistically, AIBP-mediated cholesterol efflux activates endothelial Srebp2, the master transcription factor for cholesterol biosynthesis, which in turn transactivates Notch and promotes HSPC emergence. Srebp2 inhibition impairs hypercholesterolemia-induced HSPC expansion. Srebp2 activation and Notch upregulation are associated with HSPC expansion in hypercholesterolemic human subjects. Genome-wide ChIP-seq, RNA-seq, and ATAC-seq indicate that Srebp2 trans-regulates Notch pathway genes required for hematopoiesis. Our studies outline an AIBP-regulated Srebp2-dependent paradigm for HSPC emergence in development and HPSC expansion in atherosclerotic cardiovascular disease.
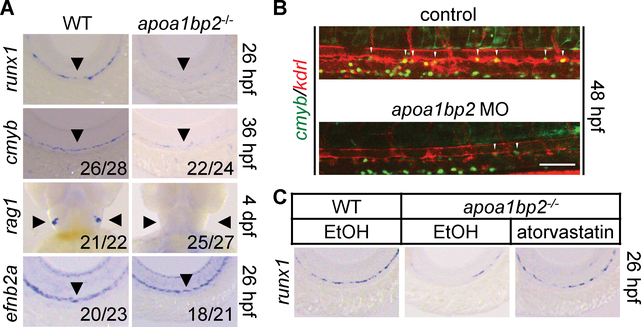
HSPCs maintain hematopoietic output by generating the whole spectrum of blood cell lineages in vertebrate animals. Previous studies demonstrate that blood vessels play an essential role in HSPC specification in development (1–4). During embryogenesis, HSPCs are emerged from a rare population of endothelial cells (ECs) residing on the floor of the dorsal aorta (DA) (1–4). Our earlier studies show that apoA-I binding protein 2 (Aibp2, aka Yjefn3) regulates angiogenesis from the DA (5). Since HSCs arise from the ventral DA (1–3), we investigated the role of Aibp2 in hematopoiesis. We generated apoa1bp2−/− zebrafish (fig. S1A–E), which appeared morphologically normal (fig. S2). The expression of HSC marker genes runx1 and cmyb in the ventral DA, and rag1 that marks HSC-derived T-lymphocytes in the thymus, were significantly reduced in apoa1bp2−/− animals (Fig. 1A and fig. S3A). Aibp2 depletion had no observable effect on DA specification as revealed by unaffected arterial efnb2a expression (6) (Fig. 1A). Morpholino antisense oligos (MO)-mediated Aibp2 knockdown (fig. S4A–B) or antibody-mediated extracellular Aibp2 neutralization (fig. S4C–D) reproduced Aibp2 knockout effect on hematopoiesis. Consistently, Aibp2 deficiency reduced the number of cmyb+kdrl+ cells, which mark nascent HSCs in the ventral DA between 28 and 60 hours post-fertilization (hpf) (Fig. 1B and fig. S3B). The results were validated using FACS analysis of cmyb+kdrl+ cells (fig. S3C&D). Although blood flow regulates hematopoiesis (7), it appeared normal in Aibp2-deficient gata1:DsRed zebrafish (movies. S1&2). These data suggest that Aibp2 governs HSC ontogeny in a direct and non-cell autonomous fashion.
Fig. 1. Effect of Aibp2 and cholesterol on HSC emergence.
A. Whole-mount in situ hybridization (WISH) analysis of runx1, cmyb, rag1, and efnb2a expression. B. HSC emergence in control or Aibp2-deficient cmyb:GFP; kdrl:mCherry zebrafish. C. WISH analysis of runx1 in animals with the indicated treatments. Ethanol: EtOH. Arrowheads in A indicate DA or thymus, and in B show cmyb+kdrl+ HSCs. Scale bar, 100 μm.
The expression of primitive hematopoiesis genes gata-1 and l-plastin at 24 hpf was normal in apoa1bp2 morphants whereas the marker of HSC-derived leukocytes (l-plastin+) at 4 days post-fertilization (dpf) was reduced (fig. S5). We also examined the integrity of non-hematopoietic tissues by surveying the expression of associated marker genes. Development of the pronephros (cdh17), somite (desma), and sclerotome (nkx3.1) in the trunk (fig. S6A), sonic hedgehog (shh) signaling (shha and vegfa), and arterial (dll4) and venous vasculature (ephb4) development showed no apparent changes in the absence of Aibp2 (fig. S6B). Pan-endothelial markers fli1 and kdrl were increased in Aibp2-deficient animals (5). These results suggest that Aibp2 plays a direct role in HSC specification.
Our previous study showed increased cholesterol content in Aibp2-deficient embryos (5). To determine the effect of cholesterol on HSC emergence, apoa1bp2 knockouts or morphants were treated with a cholesterol-lowering drug, atorvastatin. Atorvastatin treatment restored largely runx1 expression (Fig. 1C and fig. S7A–E) and reduced free cholesterol levels in Aibp2-deficient animals (fig. S7B&E). Furthermore, atorvastatin expanded the cmyb+kdrl+ cells in the DA floor (fig. S7F&G). These results indicate that an effective cholesterol metabolism program orchestrates HSC emergence.
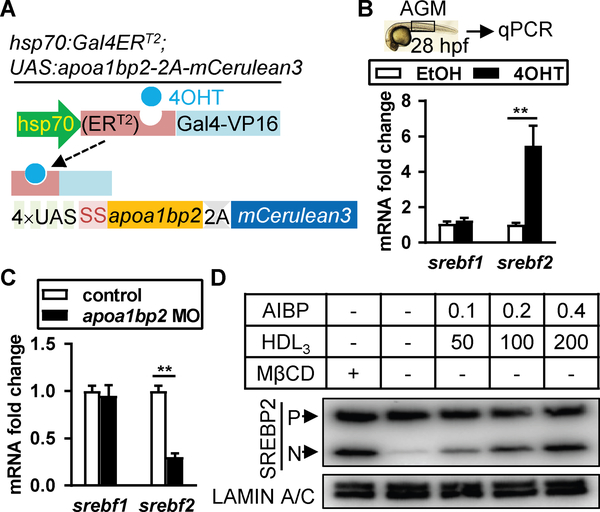
Cholesterol synthesis requires the master transcription factor Srebp2 (8), which is produced as an endoplasmic reticulum (ER)-bound precursor. Cholesterol depletion activates Srebp2 via two-step proteolytic cleavages, which releases its N-terminal transcriptional activation domain into the nucleus dictating the expression of genes for cholesterol biosynthesis such as Hmgcr, Srebf2, and cholesterol uptake Ldlr (8). Zebrafish genes srebf1 and srebf2 encode Srebp1 and Srebp2, respectively. Srebp1 is primarily responsible for fatty acid synthesis (8). We generated the double transgenic animal hsp70:Gal4ERT2; UAS:apoa1bp2–2A-mCerulean3, which expressed untagged Aibp2 upon heat shock with the addition of 4-hydroxy-tamoxifen (4OHT) (Fig. 2A and fig. S8A&B). Srebp2 binds its own promoter and upregulates its mRNA expression (8), measurement of which mirrors its transcriptional activity. Aibp2 deficiency reduced, and 4OHT-induced Aibp2 overexpression increased, srebf2 expression. Whereas srebf1 expression was not changed (Fig. 2B&C and fig. S8C). These results suggest that Aibp2 regulates Srebp2 activity.
Fig. 2. Effect of Aibp2 on Srebp2 activity in ECs.
A. DNA constructs used to make the transgenic zebrafish with heat shock-induced Aibp2 expression. SS: secretion signal. B-C. qRT-PCR analyses of srebf1 and srebf2 in the AGM regions of Aibp2 overexpression (B) or knockdown zebrafish (C). D. Immunoblots of Srebp2 in HUVECs incubated with or without AIBP/HDL3 (μg/ml) for 4 hours. **, p<0.01. P: Srebp2 precursor; N: nuclear Srebp2.
We also explored the hypercholesterolemia effect on embryonic hematopoiesis. High cholesterol diet (HCD)-fed adult female cmyb:GFP zebrafish produced embryos with significantly higher cholesterol content (fig. S9A&B) and greater srebf2 expression (fig. S9C). The embryos produced by the HCD-fed females showed more cmyb+kdrl+ HSCs compared to the embryos produced by females fed a control diet (fig. S9D&E). Similarly, hypercholesterolemic female mice produce E11.5 embryos with increased frequency of c-Kit+CD144+CD45.2− and RUNX1-enriched hemogenic endothelial cells (HECs) and hematopoietic precursors (fig. S9F–K). The data suggest that plasma cholesterol content regulates the developmental HSC program.
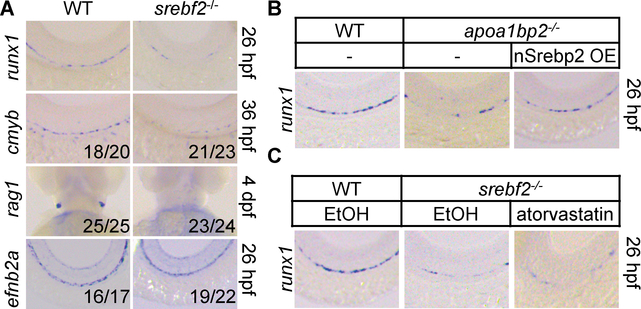
Cellular cholesterol homeostasis is sustained by LDL cholesterol uptake, Srebp2-mediated cholesterol synthesis, and HDL-mediated cholesterol efflux. Since cholesterol pools in the plasma membrane and ER are interconnected (9), we next probed the effect of cholesterol efflux on Srebp2 activation. Cholesterol sequestrant Methyl-β-cyclodextrin (MβCD) robustly activated SREBP2 in human umbilical vein ECs (HUVECs) (fig. S8D&E). AIBP augments the capacity of HDL to accept cholesterol (5), and their combinatorial treatment dose-dependently activated SREBP2 (Fig. 2D and fig. S8F). These results suggest that Aibp2-mediated cholesterol efflux activates SREBP2. We hypothesized that Srebp2 mediates Aibp2 effect on hematopoiesis. We thus generated the srebf2−/− zebrafish (fig. S10A–D). Srebp2 disruption markedly decreased runx1, cmyb, and rag1 expression (Fig. 3A and fig. S11A) but did not influence efnb2a expression (Fig. 3A). Similarly, Srebp2 knockdown disrupted HSC emergence but showed no effect on DA specification, and the hematopoiesis defect was rescued by srebf2 overexpression (fig. S12A&B). Srebp2 knockdown significantly reduced the cmyb+kdrl+ HSCs (fig. S12C&D), but had no effect on the formation of adjacent supporting tissues, shh signaling, and arterial and venous vessel specification (fig. S6A&B), while mildly increased the expression of pan-endothelial markers (fig. S6C). Srebp2 depletion specifically reduced the expression of Srebp2 but not Srebp1 downstream target genes (fig. S12E). These phenotypes support our hypothesis that Aibp2, through Srebp2 activation, controls HSC emergence. To test this, we created transgenic animal kdrl:Gal4ERT2; UAS:Flag-nSrebp2–2A-mCerulean3, in which the addition of 4OHT induced EC-specific transcriptionally active nuclear Srebp2 (nSrebp2) expression (10) (fig. S11B&C). Nuclear srebf2 mRNA injection into Aibp2 knockouts (Fig. 3B and fig. S13A) or 4OHT treatment of Aibp2-deficient kdrl:Gal4ERT2; UAS:Flag-nSrebp2–2A-mCerulean3 animals rescued impaired HSC emergence (fig. S11D&E). Atorvastatin, which activates Srebp2 (11), markedly augmented srebf2 but not srebf1 expression (fig. S11F). Atorvastatin treatment augmented HSC emergence (fig. S13C&D), which was abolished by Srebp2 disruption (Fig. 3C and fig. S13B–D). Atorvastatin-enhanced HSC emergence is not due to HSC hyper-proliferation since similar numbers of Brdu-positive cmyb+ cells in DA were found in control and atorvastatin-treated animals at 30 and 36 hpf (fig. S14A&B). Collectively, these findings suggest that Srebp2 acts downstream of Aibp2 to orchestrate HSC specification.
Fig. 3. Effect of Srebp2 on HSC emergence.
A. WISH analysis of the indicated genes in WT or srebf2−/− zebrafish. B and C. WISH analysis of runx1 expression in the DA. nSrebp2 OE: nuclear Srebp2 overexpression.
The key role of Notch in HSC specification prompted us to explore the role of Srebp2 in Notch signaling (12). We employed a Notch reporter zebrafish tp1:d2GFP, which expresses an EGFP variant with shortened half-life under the control of tandem Notch responsive elements (13). Ablation of Aibp2 substantially reduced tp1+kdrl+ HSPCs, which can be reversed by Aibp2 overexpression (fig. S15A&B). Similarly, Srebp2 depletion decreased, whereas enforced nSrebp2 expression restored tp1+kdrl+ HSPCs, in the ventral DA (fig. S15A&B). Furthermore, nSrebp2 overexpression rescued HSPC emergence in apoa1bp2 morphants (fig. S15A&B), indicating that Srebp2 mediates the Aibp2 effect on Notch signaling.
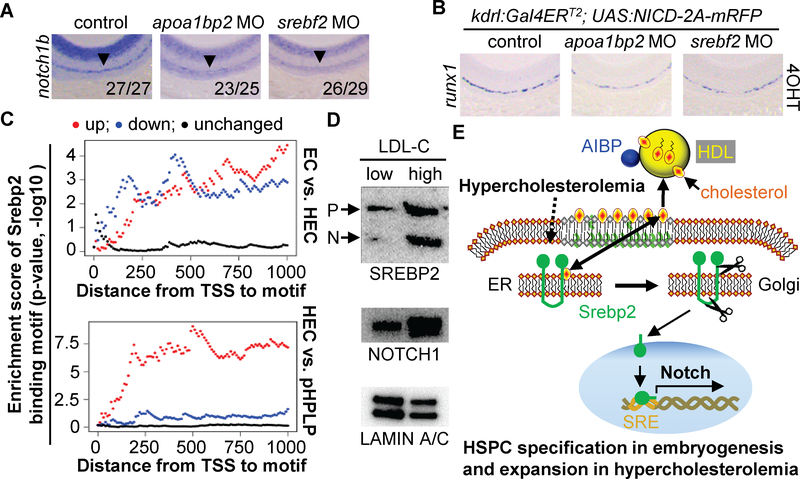
Aibp2 or Srebp2 deficiency markedly reduced the expression of notch1b but not notch1a, notch2, or notch3 in the DA (Fig. 4A and fig. S15C), suggesting that attenuated Notch signaling caused impaired HSC emergence in apoa1bp2 or srebf2 morphants. To investigate this possibility, transgenic animals kdrl:Gal4ERT2; UAS:NICD-2A-mRFP that selectively express 4OHT-inducible endothelial NICD were created (fig. S15D). Indeed, NICD expression restored runx1 mRNA expression in apoa1bp2- or srebf2-deficient animals (Fig. 4B and fig. S15E&F). Our findings agree with other findings that notch1 is intrinsically required for HSC fate (14–16). The notch1b promoter contains putative Srebp2 binding motifs, which was validated by chromatin immunoprecipitation (ChIP) and qPCR of the targeted region (fig. S16A&B). By analyzing a mouse Srebp2 ChIP-seq data (17), we found a prominent Srebp2 binding peak in the promoters of Notch1 (fig. S16C), and validated the Srebp2 binding (fig. S17B).
Fig. 4. Effect of AIBP-regulated Srebp2 activity on Notch signaling.
A and B. WISH analysis of notch1b and runx1. The numerator indicates number of zebrafish with the representative phenotype, and denominator indicates the total number of animals assessed. C. Srebp2 binding motif enrichment in differentially expressed gene groups. TSS: translation start site. D. Immunoblotting of SREBP2 and NOTCH1 in the HSPCs isolated from low LDL-C (1.826 ± 0.089 mM; n=5) and high LDL-C (4.796 ± 0.454 mM; n=5) subjects. LAMIN A/C serves as the loading control. E. Working model. Bilateral cholesterol transport occurs between the ER and plasma membrane. AIBP-accelerated cholesterol efflux to HDL or hypercholesterolemia activates Srebp2, which transactivates Notch for hematopoiesis.
Furthermore, we performed a bioinformatics scan of the whole mouse genome using the putative Srebp2 binding motif, which is enriched at the center of Srebp2 ChIP-seq peaks (fig. S16D). Our results indicate that Srebp2 binding motif is highly enriched in the promoters of genes for cholesterol metabolism and Notch signaling (fig. S16E, table S1 and S2), suggesting that this motif is highly conserved. For example, the Srebp2 binding motif and ChIP-Seq peak are present in the promoters of Srebp2-regulated cholesterol biosynthesis genes Srebf2, Hmgcr, and Ldlr (fig. S17A), all of which were experimentally verified in murine ECs with Srebp2 overexpression (fig. S17B&C). Furthermore, Srebp2 ChIP-seq results (17) also validated Srebp2-mediated regulation of Notch signaling and cholesterol metabolism (fig. S18A&B).
To further investigate the role of Srebp2 in hematopoiesis, we compared gene expression profiles in paired murine ECs, a Ly6a-GFP+ population that contains HECs, and pre-HSCs and progenitors with lymphoid potential (pHPLPs) (18). Compared to ECs, 752 genes are upregulated and 569 genes are downregulated in HECs, whereas compared to HECs, 752 genes are increased and 977 genes are decreased in pHPLPs (fig. S19A). Notch pathway genes are significantly enriched in the upregulated genes of HECs compared to ECs or pHPLPs (table S3 and fig. S19B). Except for Scap, most Srebp2regulated cholesterol metabolism genes were repressed in HECs (table S3). SCAP is a protein chaperone of Srebp2, and is retained in the ER membrane by sterol-induced interaction with ER resident protein INSIG1/2 (8). Scap increased up to 4-fold in HECs compared with ECs (table S3). Srebp2 binding motif or ChIP peak is markedly enriched in the promoters of upregulated genes, but to a less extent in the promoters of downregulated genes (Fig. 4C and fig. S18C). Consistent with this, our ATAC-seq results unveiled that the Srebp2 binding motif and ChIP-seq peak are indeed located within active transcription-associated open chromatin regions of HECs, with 42% binding motifs (fig. S16F&G) and 79% ChIP-seq peaks (fig. S18D&E) overlapping the ATAC-seq peaks in HECs. Thus, our systemic bioinformatics analyses independently validate our findings that Srebp2 is a critical regulator of the Notch pathway.
We further explored the effect of hypercholesterolemia on adult hematopoiesis. As reported (19, 20), Western diet (WD) feeding augmented HSPC frequency in Ldlr−/− mice, and Srebp2 suppression by betulin abolished WD-induced augmentation of HSPC frequency (fig. S20A–C). To relate our findings to human disease, we assessed the circulating CD34+CD45+ HSPCs in healthy volunteers. We found that LDL cholesterol levels are correlated with HSPC frequency (fig. S20D), and that Srebp2 and Notch are activated/upregulated in HSPCs isolated from hypercholesterolemic subjects (Fig. 4D and table S4). Collectively, our data document a conserved Srebp2-dependent mechanism that regulates HSPC maintenance in hypercholesterolemia.
Accumulating studies indicate that Srebp2 has moonlighting activities (17, 21). We show that the somite-derived pro-hematopoietic Aibp2 controls hematopoiesis by targeting Srebp2-regulated cholesterol metabolism and Notch signaling (Fig. 4E). In murine HECs, only the cholesterogenic gene Scap but not others is significantly upregulated. Given that SCAP gain-of-function increases sterol-independent Srebp2 bioavailability (22), its upregulation may contribute to increased Srebp2 activation in HECs. Possibly, the Srebp2 function in HECs is shifted more towards Notch activation than cholesterol regulation. Our findings also corroborate the essential role of somite in providing proper Notch signaling for HSC specification, e.g., Wnt16-induced Dlc/Dld presented by the sclerotome regulates Notch1b activity in the migrating HSC precursors (23, 24).
Hypercholesterolemia is the driving force for atherosclerosis that underlies heart attacks and strokes. Hypercholesterolemia activates endothelial Srebp2 (8, 21). Srebp2 activation and Notch1 upregulation are detected in circulating HSPCs of hypercholesterolemic human subjects. Possibly, the Srebp2-regulated Notch1 signaling also orchestrates HSPC homeostasis in hypercholesterolemia. It appears that both AIBP-mediated cholesterol efflux and hypercholesterolemia converge on endothelial Srebp2 activation. Taken together, we have uncovered a cholesterol metabolism pathway governing HSPC emergence in development as well as HSPC expansion in hypercholesterolemia. These insights may have relevance for hematological and cardiovascular disorders.
Supplementary Material
Acknowledgements:
We thank the Brown and Goldstein lab (UTSW), T Osborne (SBP), S Gerety (WTSI), and B Link (MCW) for providing antibodies, DNA construct, or transgenic zebrafish. We thank Dr. C Liu (UCSD) and the H Wu’s lab (BCM) for technical support. The project was supported by grants (HL114734, HL132155, 16BGIA27790081, 18TPA34250009) to L.F., (HL04880, DK49216) to L.Z., (DK092365, DK111599, RP110776) to P.W., (HL135737) to Y.I.M., (RR160083, CA204468) to W.L., (HL133254) to J.P.C. & K.C., (GM125632, RP120348, RP170002) to K.C., and AHA (17POST33410671) to X.Y.
Footnotes
Competing interests: Y.I.M. and L.F. are inventors on patent application (US20160115211A1 and WO2014193822A1). L.Z. holds stock and is a consultant and on the SAB of FATE Therapeutics, Scholar Rock, Camp4 and Celularity.
Data and materials availability: The ATAC-seq data of this study are deposited in NCBI GEO (GSE122204).
References and Notes
- 1.Bertrand JY et al. , Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boisset JC et al. , In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Kissa K, Herbomel P, Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Nguyen PD et al. , Haematopoietic stem cell induction by somite-derived endothelial cells controlled by meox1. Nature 512, 314–318 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Fang L et al. , Control of angiogenesis by AIBP-mediated cholesterol efflux. Nature 498, 118–122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawson ND et al. , Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development 128, 3675–3683 (2001). [DOI] [PubMed] [Google Scholar]
- 7.North TE et al. , Hematopoietic stem cell development is dependent on blood flow. Cell 137, 736–748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown MS, Goldstein JL, The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A, Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin S-W, Beis D, Mitchell T, Chen J-N, Stainier DY, Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Brown MS, Goldstein JL, A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. P Natl Acad of Sci USA 96, 11041–11048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butko E, Pouget C, Traver D, Complex regulation of HSC emergence by the Notch signaling pathway. Dev. Biol 409, 129–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark BS et al. , Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis. Development 139, 15991610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadland BK et al. , A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood 104, 3097–3105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumano K et al. , Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 18, 699–711 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Kim AD et al. , Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 33, 2363–2373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo YK et al. , Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 13, 367–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solaimani Kartalaei P et al. , Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med 212, 93–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy AJ et al. , ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J. Clin. Invest 121, 4138–4149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westerterp M et al. , Regulation of hematopoietic stem and progenitor cell mobilization by cholesterol efflux pathways. Cell stem cell 11, 195–206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z et al. , Oxidative Stress Activates Endothelial Innate Immunity via Sterol Regulatory Element Binding Protein 2 (SREBP2) Transactivation of MiRNA-92a. Circulation, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton JD et al. , Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U. S. A 100, 12027–12032 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clements WK et al. , A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 474, 220–224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi I et al. , Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 512, 319–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.