Abstract
Glioblastoma (GBM) is the most common and lethal primary brain tumor in adults, with nearly 100% of patients ultimately succumbing to the disease. Median patient survival is 15 months, and no standard of care currently exists for recurrent cases. Glioma stem cells (GSCs), a rare and highly aggressive subpopulation of cells within these tumors, have recently emerged as drivers of tumor initiation and recurrence, and a growing body of evidence suggests that they must be completely eradicated to prevent relapse. Toward this goal, we have developed polyethylenimine-wrapped spherical nucleic acid nanoparticles (PEI– SNAs) targeting Gli1, a transcription factor within the Hedgehog signaling pathway that is crucial for the maintenance of GSCs. Here, we demonstrate that Gli1 PEI–SNAs bind scavenger receptors on GBM cells to undergo endocytosis in a caveolae/lipid raft/dynamin-dependent manner. They further achieve ~30% silencing of tumor-promoting Hedgehog pathway genes and downstream target genes that promote the aggressive, chemoresistant phenotype of GBM. This produces a 30% decrease in proliferation that correlates with a robust onset of GBM cell senescence as well as an ~60% decrease in metabolic activity with or without cotreatment with temozolomide (TMZ), the frontline chemotherapy for GBM. Most importantly, Gli1 PEI–SNAs impair the self-renewal capacity of GBM cells as indicated by a 30–40% reduction in the expression of stemness genes and further impair the formation of stem-like neurospheres. They also substantially improve neurosphere chemosensitivity as demonstrated by a 2-fold increase in the fraction of cells undergoing apoptosis in response to low doses of TMZ. These results underscore the potential for siRNA therapeutics targeting Gli1 to reduce GBM resistance to therapy and warrant further development of PEI–SNAs and Gli1-targeted therapies to alleviate drug resistance and recurrence for GBM patients.
Keywords: gene regulation, glioblastoma, Hedgehog signaling, polyethylenimine, spherical nucleic acids
INTRODUCTION
RNA interference (RNAi) therapeutics have received tremendous attention recently for their potential to revolutionize the management of diseases with a known genetic basis. In a clinical setting, RNAi offers the ability to silence the expression of genes that promote disease progression with greater potency and specificity than small molecule drugs. However, one unresolved challenge toward recognizing the clinical potential of RNAi is maximally delivering siRNA to the targeted disease site.1,2 This is because siRNA is highly susceptible to degradation by nucleases present ubiquitously in physiological conditions, is cleared rapidly from circulation, and cannot cross cellular membranes due its large size and negative charge. As a result, there is a need to develop carriers to protect siRNA and efficiently deliver it to sites of disease. One of the greatest challenges in developing clinically translatable siRNA carriers is achieving a balance between efficacy and toxicity; many strongly cationic carriers that are highly effective for intra-cellular siRNA delivery are also toxic due to their tendencies to destabilize cellular membranes and trigger immune responses.3–5 The reverse is also true: many materials with more favorable biocompatibility profiles are less effective as transfection agents.
Toward the goal of identifying carriers to maximize siRNA delivery efficacy and minimize toxicity, we have recently developed a hybrid delivery vehicle consisting of a spherical nucleic acid (SNA) core and a polyethylenimine (PEI) shell that provides greater cellular uptake and endosomal escape as compared to highly biocompatible SNAs and greater cytocompatibility and transfection efficiency as compared to PEI–siRNA polyplexes.6 SNAs, which consist of radially oriented, densely arranged siRNA stabilized on a gold nanoparticle core, exhibit a controlled siRNA architecture that imparts unique properties favoring siRNA delivery to biological systems. Most notably, SNAs are rapidly taken up by >50 cell types, provide steric and electrostatic hindrances against endonucleases, and do not induce an immune response in animal models.7–10 Further, their successful delivery of siRNA and miRNA to glioblastoma tumors has prompted the first clinical trial evaluating their use as therapeutics for glioblastoma and gliosarcoma.8,11,12 However, their tendency to accumulate within late endosomes limits their transfection efficiency.13 Simultaneously, polycationic materials such as PEI have been widely investigated as siRNA carriers for their ability to encapsulate and protect nucleic acids and rapidly enter cells. In particular, PEI is strongly cationic due to its high amine content, which enables PEI–siRNA polyplexes to overcome a major bottleneck to their intracellular delivery: achieving endosomal escape.4,5 However, toxicity common to polycationic materials has limited its clinical translation.14 Interestingly, we found that PEI-wrapped SNAs afford enhanced cellular uptake and endosomal escape to improve gene silencing efficacy while dramatically reducing the cytotoxicity of PEI,6 warranting continued investigation of these constructs as therapeutic gene regulatory agents for diseases with a genetic basis.
One devastating disease that might benefit from the continued development and application of RNAi therapeutics is glioblastoma multiforme (GBM). GBM is the most common and lethal neurological tumor in adults, representing nearly 50% of all malignant primary brain tumors.15 The three main treatment strategies, surgery, radiation, and chemotherapy, often fail to completely eradicate the disease, in part due to intrinsic or acquired resistance to therapy characteristic of GBM tumors. Consequently, tumor recurrence is inevitable, and nearly 100% of patients eventually succumb to disease.16,17 Recently, a growing body of work has identified a role of developmental pathways in GBM progression. One such pathway is the Hedgehog (Hh) signaling pathway.18–20 During development, Hh signaling plays a key regulatory role in tissue patterning and stem cell maintenance. It is subsequently inactivated in most differentiated adult tissues, during which the Ptch1 transmembrane receptor represses Smo, a G-protein coupled receptor (GPCR)-like protein. However, aberrant pathway activation is implicated in many cancers, including GBM. In GBM, this aberrant activation is most commonly initiated when extracellular Sonic Hh ligand binds to Ptch1, which relieves its suppression of Smo to drive an intracellular signaling cascade that ultimately results in the translocation of the Gli1 transcription factor to the nucleus, where it transcriptionally regulates the expression of genes that promote GBM progression. When activated in GBM, Hh/Gli1 signaling induces proliferation and survival signaling and also maintains an aggressive subpopulation of GBM cells called glioblastoma stem cells (GSCs).18,19,21 GSCs are pluripotent, highly tumorigenic cells that resist therapy and generate the bulk of GBM tumors. This is enabled by their ability to divide asymmetrically; they can divide to produce additional GSCs by self-renewal, or they can divide and differentiate into nontumorigenic progenitor and differentiated GBM cells.22,23 Further, GSCs cycle slowly and tend to reside within the hypoxic tumor core, rendering them highly refractory to radiation and chemotherapies that target rapidly dividing cells. As a result, traditional therapies fail to eliminate GSCs, which in turn drive tumor recurrence. Due to their highly aggressive characteristics, GSCs must be eradicated to achieve complete tumor regression.
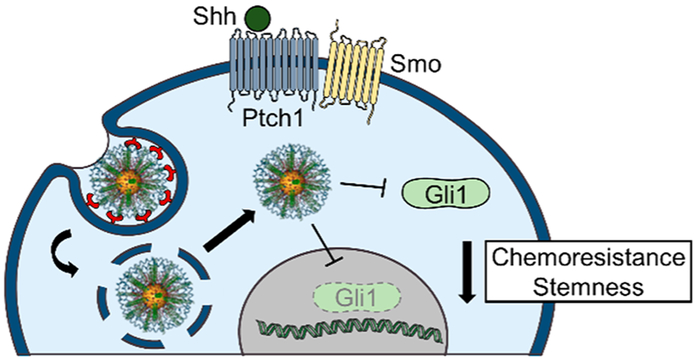
GSCs may be eliminated by targeting the developmental pathways that maintain them, such as Hh signaling. Therefore, we hypothesized that GBM progression could be halted by targeting Gli1 with RNAi therapeutics. Much research has previously been dedicated to targeting a diverse selection of genes toward the goal of reducing GBM chemoresistance, stemness, and ultimately disease progression. For example, nanocarriers delivering siRNA against genes that promote survival, drug resistance, and DNA repair such as Bcl2,24 Bcl2L12,8 survivin,25 cyclin D1,26 MDR1, EGFR and EGFRviii,27 and MGMT,28,29 have been previously demonstrated to improve GBM therapeutic response in preclinical testing. Importantly, many of these genes are known transcriptional targets of Gli1; therefore, we hypothesized that targeting Gli1 upstream of these genes might have broad effects toward halting GBM progression. In this work, we developed Gli1-targeted PEI–SNAs, characterized their cellular uptake and intracellular trafficking mechanisms, and demonstrated that they can reduce the chemoresistance and stemness of GBM cells (Figure 1). Our results demonstrate that PEI–SNAs bind cells via scavenger receptors and undergo both dynamin-dependent, caveolae-mediated endocytosis and macropinocytosis (Figure 2). Following endocytosis, the majority of PEI–SNAs are routed to late endosomes and lysosomes. Despite this, Gli1 PEI–SNAs can successfully reduce the expression of the Hh signaling components Gli1 and Smo by ~30%, and this corresponds to an ~30% reduction in Hh transcriptional target genes that promote GBM progression including CyclinD1, c-Myc, Bcl2, and ABCG2 (Figure 4). Gene regulation by Gli1 PEI–SNAs also mediates a 30% reduction in GBM cell proliferation (Figure 5A) and a distinguishable onset of cellular senescence (Figure 5B). Further, Gli1 PEI–SNAs reduce the metabolic activity of GBM cells by ~60% alone or in combination with TMZ (temozolomide, the frontline chemotherapy for GBM) (Figure 5C). Importantly, Gli1 PEI–SNAs impair the self-renewal capacity of GBM cells as indicated by a 30–40% reduction in the expression of stemness genes (Figure 6A,B) and by impairing the formation of neurospheres (Figure 6A,C,D). This translates to a substantial improvement in neurosphere chemosensitivity as demonstrated by a 2-fold increase in the fraction of cells undergoing apoptosis in response to low doses of TMZ (Figure 7). These findings underscore the potential for siRNA therapeutics targeting Gli1 to reduce GBM resistance to therapy and warrant the continued development of polycation–SNA hybrid siRNA carriers for potent gene regulation.
Figure 1.
Gli1-targeted PEI–SNAs were developed to suppress GBM-promoting Hedgehog signaling and mitigate the chemoresistance and stemness of GBM cells.
Figure 2.
Assessment of the uptake mechanism for PEI–SNAs, as determined by flow cytometry (top) or visualized by fluorescence microscopy (bottom). Data are average geometric means ± standard deviations. *p < 0.05 and **p < 0.005 relative to control cells by one-way ANOVA with post hoc Tukey. For fluorescence microscopy images, scale = 50 μm.
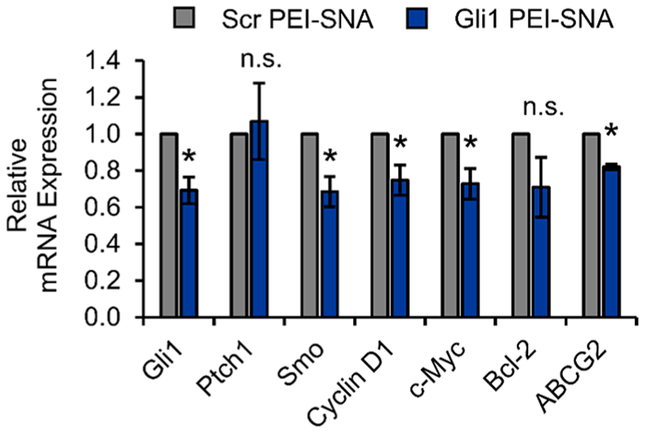
Figure 4.
Gli1 PEI–SNAs reduce the mRNA expression of Gli1 and downstream target genes by qPCR. Gene expression is normalized to that of GAPDH, and data shown are means ± SEM; *p < 0.05 relative to Scr PEI–SNA control by Student’s t-test.
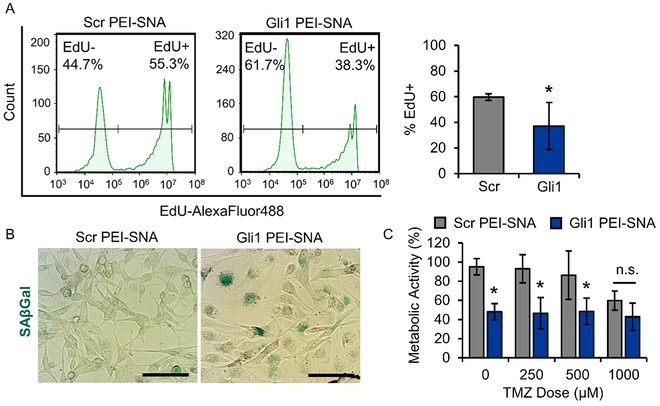
Figure 5.
Gli1 PEI–SNAs reduce proliferation and chemoresistance. (A) By EdU assay, Gli1 PEI–SNAs reduce U87 proliferation by ~30%. Flow cytometric histograms (left) and quantification of EdU+ cells (right). Data are means ± STDs, *p = 0.02. (B) SAβGal staining (teal) demonstrating that Gli1PEI–SNAs induce senescence in U87 cells. Scale = 100 μm. (C) By MTT assay, Gli1 PEI–SNAs reduce U87 metabolic activity alone and in combination with TMZ, *p < 0.01 relative to Scr PEI–SNA control with equivalent TMZ dose by one-way ANOVA with post hoc Tukey.
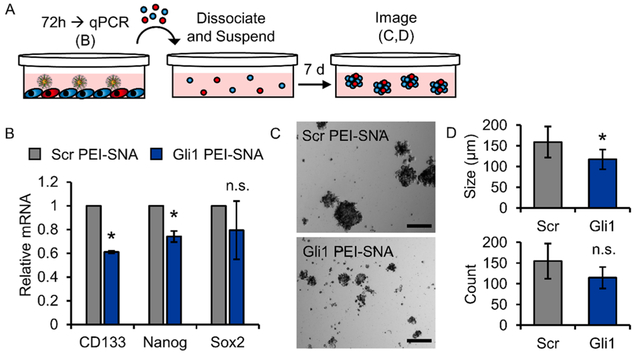
Figure 6.
Gli1 PEI–SNAs reduce stemness and impair self-renewal of U87 cells. (A) Schematic depicting the neurosphere culture model and experimental design; red cells illustrate GSCs. (B) qPCR showing expression of genes associated with stemness following exposure to PEI–SNAs. Gene expression is normalized to that of GAPDH. Data are means ± STDs; *p < 0.001 relative to Scr PEI–SNA. (C) Representative bright-field images of neurospheres cultured from U87 cells after exposure to PEI–SNAs. Scale = 200 μm. (D) Gli1 PEI–SNAs reduce the size and number of neurospheres formed, as measured from 25 tiled bright-field images per treatment group per experiment; *p = 0.03 by Student’s t-test.
Figure 7.
Gli1 PEI–SNAs potentiate neurosphere response to TMZ chemotherapy. (A) Confocal microscopy visualizing Gli1 PEI–SNA distribution into small (top) and large (bottom) neurospheres. Scale = 100 μm. (B) Flow cytometric histogram of Cy5–PEI–SNA uptake by cells grown as neurosperes. (C) Flow cytometric analysis of the mechanism by which Gli1 PEI–SNAs distribute throughout neurospheres. Data are geometric mean fluorescence intensity (GMFI) ± STD normalized to control cells, *p < 0.05 by one-way ANOVA with post hoc Tukey. (D) Experimental timeline for determining effect of cotreating neurospheres with Gli1 PEI–SNAs and TMZ. (E) Flow cytometric density plots of Annexin-V/PI apoptosis analysis of neurospheres cotreated with Gli1 PEI–SNAs and TMZ. (F) Summary of Annexin-V/PI apoptosis analysis. Data are means ± STDs from n = 2 replicates. *p < 0.05 by one-way ANOVA with post hoc Fisher’s least significant difference test.
EXPERIMENTAL SECTION
Nanoparticle Synthesis and Characterization.
Citrate-stabilized gold nanoparticles (AuNPs, 15 nm) were prepared using the Frens method30 and treated with 0.1% diethyl pyrocarbonate (DEPC) to inactivate RNases. SNAs were synthesized and characterized for siRNA loading as previously reported.31 Briefly, RNase-free AuNPs were suspended in 0.2% Tween-20 and 350 mM NaCl and subsequently functionalized with thiolated siRNA (1 nmol per mL of 10 nM AuNPs; Integrated DNA Technologies, Coralville, IA). The NaCl concentration was slowly increased to 500 mM and incubated overnight prior to passivation with 2 kDa methoxy-polyethylene glycol-thiol (mPEG-SH; Laysan Bio, Arab, AL) to increase stability. PEI–SNAs were synthesized by incubating purified SNAs suspended in water at 10 nM with 1 mg/mL 25 kDa branched PEI (Sigma-Aldrich, St. Louis, MO) for 15 min under sonication to prevent aggregation, and then, the PEI– SNAs were purified by centrifugation to remove unbound PEI. siRNA sequences used are as follows: Scr, 5′-UGAUAAGUCGUUGGUGCACdT-3′; Gli1, 5′-UUGGGAGUCAAAUUCCUGGCdT-3′. Using an OliGreen assay to measure siRNA loading,31 Scr-SNAs contained 53.3 ± 6.5 duplexes, and Gli1-SNAs contained 58.7 ± 11.2 duplexes. All loading was measured prior to coating SNAs with PEI.
Cell Culture and Stable Gene Expression.
U87-MG cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA), cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; VWR, Radnor, PA) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA), and maintained in a humidified incubator at 37 °C, 5% CO2. For neurosphere experiments, U87-MG cells were seeded as a single-cell suspension in low-adhesion plates cultured in NeuroCult NSA (STEMCELL Technologies, Vancouver, BC, Canada) medium supplemented with recombinant human epidermal growth factor (EGF, 20 ng/mL), recombinant human basic fibroblast growth factor (bFGF, 10 ng/mL), and heparin (2 μg/mL). To study the intracellular trafficking of PEI–SNAs, U87-MG cells were stably transformed with GFP-tagged endosomal markers using standard lentiviral transduction procedures. Briefly, Rab5-GFP (Addgene # 56530), Rab7-GFP (Addgene # 12605), Rab11-GFP (Addgene # 12674), or LAMP1-GFP (Addgene # 34831) were cloned into a lentiviral transfer vector (System Biosciences, Palo Alto, CA) by restriction cloning. Lentiviral particles were produced by triple-transfecting (TransIT-Lenti transfection reagent; Mirus Bio, Madison, WI) 293TN cells (System Biosciences, Palo Alto, CA) with either transfer vector and lentiviral packaging and envelope plasmids (Addgene #12260,12259). Lentivirus was harvested, filtered, and diluted in cell culture medium to transform U87-MG cells. Cells stably expressing the desired protein were selected with 1 mg/mL puromycin (VWR, Radnor, PA).
Assessment of Endocytosis Pathways.
Endocytosis pathways responsible for the cellular uptake of PEI–SNAs were investigated using flow cytometry and fluorescence microscopy. U87-MG cells were seeded in 24-well culture plates to 60–70% confluence and treated with inhibitors to various components of endocytic pathways according to the conditions detailed in Table S1. Treated cells were incubated with Cy5-labeled PEI–SNAs (40 nM siRNA) for 4 h and either trypsinized for flow cytometry analysis using a NovoCyte flow cytometer (ACEA Biosciences, San Diego, CA) or fixed in 4% formaldehyde, counterstained with DAPI and DyLightTM488-Phalloidin (Cell Signaling Technology, Danvers, MA), and mounted on slides using gelvatol mounting medium for fluorescence microscopy using a Zeiss AxioOb-server.Z1 microscope (Zeiss, Thornwood, NY). PEI–SNA distribution into neurospheres was conducted similarly by culturing neurospheres for 1 week prior to treatment with endocytosis inhibitors and PEI–SNAs and subsequently dissociating them for flow cytometry analysis or imaging them intact using a Zeiss LSM880 confocal microscope to collect z-stacks of whole spheres.
Confocal Microscopy and Image Analysis.
U87-MG cells stably expressing Rab5-GFP, Rab7-GFP, Rab11-GFP, or LAMP1-GFP were seeded in 35 mm glass bottom dishes to 60–70% confluence. Cells were incubated with Cy5-labeled PEI–SNAs for 24 h, counterstained with CellMask Orange (Thermo Fisher Scientific, Waltham, MA), and fed with FluoroBrite DMEM Media (Thermo Fisher Scientific, Waltham, MA). Cells were imaged live using a Zeiss LSM880 confocal microscope equipped with an incubated stage. Z-stacks were acquired to analyze PEI–SNA/endosomal colocalization throughout the entire volume of cells. Quantitative colocalization analysis was performed as previously described6 to calculate Mander’s colocalization coefficients32 for each endosomal marker. Briefly, image analysis was performed using data from three independent experiments. Regions of interest (ROIs) were identified by manually tracing individual cells based on the CellMask Orange channel. Both the Cy5 and GFP channels were median filtered with a 3-by-3-by-3 neighborhood and top-hat filtered using a 2 μm disk element. MCCs were calculated for each ROI in each image stack.
Gene Expression Analysis by Quantitative Real-Time Polymerase Chain Reaction (qPCR).
The gene regulation potency of Gli1 PEI–SNAs was evaluated using qPCR. U87-MG cells were seeded at 100 000 cells/well in a 12-well culture plate and grown overnight. Cells were incubated with PEI– SNAs (50 nM siRNA) for 24 h in complete medium, fed with fresh medium, and incubated a further 48 h at 37 °C, 5% CO2. RNA was isolated using an Isolate II RNA Mini Kit (Bioline, Taunton, MA), and qPCR was performed using SensiFAST SYBR One-Step Master Mix on a LightCycler 96 (Roche Diagnostics Corporation, Indianapolis, IN). Gene expression was normalized to that of GAPDH. Primer sequences are listed in Table S2.
Evaluation of Cell Proliferation, Senescence, and Viability.
Gli1 PEI–SNAs were evaluated for their ability to reduce GBM cell proliferation, senescence, and viability in combination with TMZ. To measure proliferation, U87-MG cells were treated with Gli1 PEI–SNAs as described for qPCR analysis and then assessed using an EdU assay (Thermo Fisher Scientific, Waltham, MA). Briefly, treated U87-MG cells were incubated with 10 μM EdU for 16 h, then trypsinized, fixed in 4% formaldehyde, and permeabilized with 0.05% saponin prior to staining according to the manufacturer’s protocol. EdU incorporation was detected by an AlexaFluor488-azide and measured by flow cytometry (Ex 488 nm/Em 530/30 nm) using a NovoCyte flow cytometer. Senescence was evaluated in cells treated with PEI–SNAs using a Senescence-Associated β-Galactosidase (SAβGal) Kit (Cell Signaling Technology, Danvers, MA) according to the manufacturer’s protocol. Stained cells were imaged using a Zeiss Axioobserver.Z1 microscope equipped with a color camera. U87-MG cell metabolic activity (taken to correlate with viability) was assessed following cotreatment with Gli1 PEI–SNAs and TMZ. Cells were seeded in 96-well plates at a density of 2500 cells/well and treated with Scr or Gli1 PEI–SNAs as described above. Treated cells were then exposed to TMZ concentrations ranging from 0–1000 μM for an additional 72 h and then evaluated using an MTT assay (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. Each treatment group was performed in triplicate, and separate cells treated with equivalent volumes of DMSO (used to reconstitute and store TMZ) were used to ensure that toxicity was due to TMZ treatment rather than the DMSO vehicle.
Assessment of Self-Renewal and Neurosphere TMZ Response.
A neurosphere culture model was used to assess the impact of Gli1 PEI–SNAs on the self-renewal capacity of U87-MG cells. First, U87-MG cells were seeded in standard adherent culture at a density of 25 000 cells/mL in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were treated with Gli1 PEI– SNAs or Scr PEI–SNAs for 24 h at 37 °C, 5% CO2, fed with fresh medium, and incubated a further 48 h at 37 °C, 5% CO2. Cells were then trypsinized and seeded in suspension at a density of 10 000 cells/mL to grow as neurospheres as described above for 1 week at 37 °C, 5% CO2. After 7 days, the entirety of each well was imaged using a Zeiss AxioObserver.Z1 microscope equipped with automated stage control. Images were stitched using Zeiss Efficient Navigation software (ZEN 2.0; Zeiss) and exported for analysis, and then, spheres were counted and measured in ImageJ. The sphere size reported is the diameter of the projected area imaged by bright-field microscopy.
To assess the effect of Gli1 PEI–SNAs on neurosphere response to TMZ, we used a similar neurosphere model. Cells were pretreated with PEI–SNAs in adherent culture and then seeded as neurospheres as described above. At 72 h postseeding of the neurospheres, cells were exposed to PEI– SNAs for an additional 24 h and then resuspended in fresh medium containing TMZ. Spheres were incubated in TMZ for 72 h and subsequently assessed for apoptosis using an Annexin-V–FITC/PI assay (Cayman Chemical Company, Ann Arbor, MI). Annexin-V–FITC/PI staining was analyzed on a NovoCyte flow cytometer.
Statistical Analysis.
All experiments were performed in triplicate, and data represent means ± standard deviations from three independent replicates unless otherwise indicated. Groups with significant differences were identified using oneway ANOVA with a post hoc Tukey test (or Student’s t-test when only two groups were compared), and differences were considered significant at p < 0.05. Statistical tests were performed in MATLAB software (MathWorks, Natick, MA), and flow cytometry data was analyzed using NovoExpress software (ACEA Biosciences, San Diego, CA).
RESULTS AND DISCUSSION
Evaluation of PEI–SNA Endocytosis Mechanism.
To begin our evaluation of Gli1 PEI–SNAs, we were interested in understanding the mechanism by which PEI–SNAs are taken up by cells. Importantly, the mechanism of endocytosis can determine the intracellular fate of the siRNA cargo, which must reach the cytosol to facilitate gene silencing. Based on previous studies that have separately demonstrated that both PEI-based polyplexes33 and SNAs34 undergo clathrin-independent, caveolae-mediated endocytosis, we expected to observe similar results. We further anticipated that PEI–SNAs would bind to cells via class A scavenger receptors, which has been previously reported for SNAs.34 For our studies, we used chemical inhibitors to different endocytosis mechanisms (Table S1) to determine which pathways were necessary for cellular uptake of PEI–SNAs fluorescently labeled with Cy5-siRNA. Cells were subsequently analyzed by flow cytometry and fluorescence microscopy to identify changes in both net cellular association and in cellular localization of PEI–SNAs. As expected, we found that fucoidan (FCD), a potent scavenger receptor inhibitor, significantly decreases the net cellular uptake of PEI–SNAs by up to 75%, and little association of PEI–SNAs with cells was detected by fluorescence microscopy (Figure 2). Similarly, we found that methyl-β-cyclodextrin (MβCD), a caveolae/lipid raft inhibitor, but not chlorpromazine (CPZ), a clathrin inhibitor, significantly reduces the uptake of PEI–SNAs by up to 50% (Figure 2). While cargo internalized by clathrin or caveolae-dependent mechanisms can ultimately be routed to lysosomes, caveolae can also fuse with intermediate vesicles called caveosomes, which do not acidify and can avoid lysosomal trafficking in some cases.35,36 Interestingly, one study found that polyplexes taken up by clathrin-mediated endocytosis are routed to lysosomes for degradation, but polyplexes taken up by caveolae-dependent mechanisms were more likely to evade lysosomes and induce efficient transfection.33 We additionally found that the cellular uptake of PEI–SNAs occurs in a dynamin-dependent manner, indicated by a significant 50% reduction in PEI–SNA uptake in cells treated with dynasore (Figure 2). Cytochalasin D, which disrupts actin polymerization to inhibit phagocytosis, does not significantly reduce PEI–SNA uptake (Figure 2).
Evaluation of PEI–SNA Intracellular Trafficking.
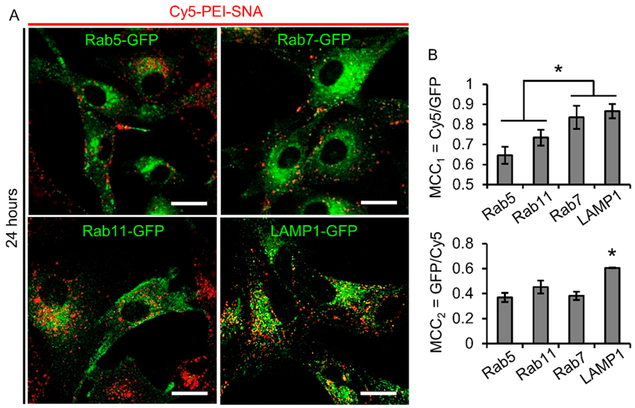
Having demonstrated that PEI–SNAs undergo both dynamin-dependent, caveolae-mediated endocytosis and macro-pinocytosis, we next sought to understand the consequences of this uptake mechanism on the intracellular trafficking of PEI– SNAs. While our previous research demonstrates that PEI– SNAs are visible in early endosomes within 1 h and undergo reduced lysosomal accumulation relative to SNAs and PEI– siRNA polyplexes,6 we were further interested in determining whether PEI–SNAs accumulate within other endosomal compartments, including early endosomes, late endosomes, recycling endosomes, and lysosomes. In these studies, we used U87-MG cells engineered to stably express fluorescently tagged endosomal compartments, including Rab5-GFP, Rab7-GFP, Rab11-GFP, and LAMP1-GFP, which label early endosomes, late endosomes, recycling endosomes, and lysosomes, respectively, to determine the subcellular localization of PEI–SNAs by confocal microscopy. Cells were incubated with Cy5–PEI–SNAs for 24 h and imaged by confocal microscopy to capture z-stacks containing the entire volume of the cells. To quantitatively assess colocalization between PEI– SNAs and endosomal compartments, Manders’ colocalization coefficients (MCCs) were calculated for each endosomal compartment. MCCs measure the fractional overlap of fluorescent signals32 and range from 0–1, where MCC = 0 indicates that no colocalization is present, and MCC = 1 indicates that the two signals colocalize perfectly. Here, we have calculated both the fractional overlap of Cy5–PEI–SNAs and GFP-endosomes (MCC1) to evaluate the fraction of siRNA signal within endosomes as well as the fractional overlap of GFP-endosomes and Cy5–PEI–SNAs (MCC2) to evaluate the fraction of endosomes containing siRNA.
We found that Cy5–PEI–SNAs colocalize to the greatest extent with LAMP1-GFP (MCC1 = 0.87 ± 0.04) and Rab7-GFP (MCC1 = 0.84 ± 0.04), which was significantly greater than colocalization with Rab11-GFP (MCC1 = 0.73 ± 0.06) or Rab5-GFP (MCC1 = 0.65 ±0.04, Figure 3 A,B). Additionally, LAMP1-GFP colocalizes with Cy5–PEI–SNAs (MCC2 = 0.61 ± 0.002) to a significantly greater extent than other endosomal markers (MCC2 all ~0.45, Figure 3A,B). Therefore, we conclude that PEI–SNAs accumulate to the greatest extent within Rab7+ late endosomes and LAMP1+ lysosomes. This is consistent with previous studies, which demonstrate that avoiding retention within the endolysosomal network remains a challenge for RNAi therapeutics.6,37,38 These results demonstrate that future research should seek to reduce the lysosomal entrapment of these constructs possibly through the incorporation of cell penetrating peptides or ionizable lipids to provide additional mechanisms of achieving endosomal escape. Additionally, the relatively strong colocalization of PEI–SNAs with Rab5+ early endosomes (MCC > 0.5) suggests that PEI– SNAs are continually endocytosed through 24 h incubation. Unexpectedly, we also observed relatively high colocalization with Rab11+ recycling endosomes, suggesting a mechanism by which PEI–SNAs may be exocytosed. While it is unusual for nanoparticles to exhibit colocalization with Rab11+ vesicles,39 previous studies investigating the intracellular fate of SNAs have demonstrated that while the gold core is retained within lysosomes, the oligonucleotide shell is gradually cleared from cells over a period of 24 h, likely due to nuclease degradation and oligonucleotide exocytosis.13 A similar process may explain our results, which show that Cy5-siRNA can colocalize with Rab11+ recycling endosomes.
Figure 3.
Intracellular trafficking of PEI–SNAs. (A) Confocal microscopy was used to visualize Cy5–PEI–SNA localization to endocytic compartments after 24 h incubation with cells. Endocytic compartments were labeled by stably expressing GFP-tagged markers for early endosomes (Rab5+), recycling endosomes (Rab11+), late endosomes (Rab7+), or lysosomes (LAMP1+). Scale bar = 20 μm. (B) Results from quantitative colocalization analysis to calculate the fractional overlap of Cy5–PEI–SNAs with GFP-endosomal markers and vice versa. MCC = Manders’ colocalization coefficient; *p < 0.05 by one-way ANOVA with post hoc Tukey.
Gli1 PEI–SNAs Regulate the Expression of Hh Signaling Components and Downstream Target Genes.
Next, we evaluated the gene regulation potency of Gli1 PEI–SNAs using qPCR to measure mRNA expression of Hh signaling components and downstream target genes known to promote GBM progression. Gli1 PEI–SNAs significantly reduce Gli1 expression by 30% relative to PEI–SNAs carrying a scrambled control siRNA sequence (Scr PEI–SNAs; Figure 4). We also observed a significant 30% decrease in the expression of the GPCR-like protein Smo (Figure 4), which is also considered an oncogene that can induce aberrant Hh signaling to further drive cancer progression.40 Mutations to Smo are common to many cancers, and therapeutics targeting Smo often fail in the clinic, because acquired Smo mutations render tumor cells refractory to Smo-targeted therapy.41 Our results demonstrate that Gli1-targeted RNAi can reduce the expression of both oncogenes. Interestingly, we did not observe significant differences in the expression of the transmembrane receptor Ptch1. Ptch1 normally inhibits Smo to suppress Hh activity and is thus considered the tumor suppressor of the Hh pathway, so we were encouraged that Gli1 PEI–SNAs did not reduce the expression of this gene. We also observed ~30% decreases in the pro-GBM Gli1 transcriptional target genes cyclin D1, c-Myc, Bcl-2, and ABCG2 (Figure 4). Cyclin D1 promotes cell cycle progression through the G1/S transition, and its overexpression in GBM correlates with poor prognosis. Further, silencing cyclin D1 inhibits proliferation, induces apoptosis, and reduces the invasive capacity of GBM cells.26 While c-Myc also promotes proliferation in numerous cancers, it has been further recognized as a key regulator of glioma cancer stem cells.22,42 Bcl-2 opposes apoptosis and contributes substantially to GBM therapy resistance, and inhibiting Bcl-2 can enhance the response of glioma cells to TMZ.43 ABCG2 is an ABC transporter known for its drug efflux activity to promote cell survival and also regulates self-renewal and the expression of stemness genes in glioma cells.44 Taken together, these results demonstrate that Gli1 PEI–SNAs can downregulate multiple genes that contribute to GBM progression, chemoresistance, and stemness, and we expect the observed gene regulation to exert tumor suppressive effects.
Gli1 PEI–SNAs Slow Proliferation, Induce Senescence, and Reduce Chemoresistance of GBM Cells.
To test whether the gene regulation capacity of Gli1 PEI–SNAs is sufficient to induce a tumor suppressive response against GBM cells, we investigated the impact of Gli1 PEI–SNAs on proliferation, senescence, and response to TMZ. Using a 5-ethynyl-2′-deoxyuridine (EdU) assay to measure proliferation, we found that Gli1 PEI–SNAs significantly reduce the fraction of proliferative (EdU+) U87-MG cells by ~30% relative to cells treated with Scr PEI–SNAs (Figure 5A). This is consistent with our measured decreases in cyclin D1 and c-Myc expression (Figure 4) and with previous reports demonstrating that suppressing Gli1 reduces GBM proliferation.18,19 In parallel with our observed decrease in proliferation, we recently reported that silencing Gli1 can induce senescence in PTEN-deficient U87-MG cells.18 To determine whether Gli1 PEI–SNAs could also elicit this effect, we employed a senescence-associated β-galactosidase (SAβ-Gal) assay to visually identify cells undergoing senescence. SAβGal staining demonstrated that U87-MG cells treated with Gli1 PEI–SNAs broadly undergo senescence, as indicated by teal SAβGal staining, while cells treated with Scr PEI–SNAs do not (Figure 5B). During senescence, cells enter a stable state of cell cycle arrest but remain metabolically active.45 While much remains unknown regarding the anticancer implications of senescence, a growing body of research has demonstrated that senescence can induce tumor suppressive effects and even compensate for apoptosis in some contexts, such as when expression of the tumor suppressor PTEN is lost.46 Interestingly, senescent cells acquire a senescence-associated secretory phenotype (SASP), which can trigger immune responses to either promote tumor progression or promote immune clearance of tumor cells.47,48 In aggregate, our data suggests that Gli1 PEI–SNA-mediated senescence exerts tumor suppressive effects, though future research should continue to investigate the effects of the SASP induced by Gli1 silencing on GBM progression.
Next, we were interested in investigating the effects of Gli1 PEI–SNAs on GBM cell sensitivity to the frontline chemo-therapy, TMZ. One way we could envision using such a Gli1-targeted therapy is to first administer our Gli1 RNAi therapeutic to downregulate cellular resistance mechanisms and subsequently treat with TMZ. To model this schedule, we first treated U87-MG cells with Gli1 PEI–SNAs, incubated the cells for 72 h, and then treated cells with TMZ doses ranging from 0–1000 μM for an additional 72 h, after which we measured cellular metabolic activity using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Our results demonstrate that Gli1 PEI–SNAs significantly reduce U87-MG metabolic activity by ~60% alone and in combination with low doses of TMZ (Figure 5C), suggesting that Gli1 PEI–SNAs might reduce the required dose of TMZ to achieve a therapeutic effect. Notably, the observed decrease in metabolic activity appears to be largely due to Gli1 PEI– SNAs alone rather than TMZ, which may reflect the fact that U87-MG cells grown in adherent culture are highly refractory to TMZ, as shown by our own previous studies and studies conducted by others.18,50 This is supported by the observation that TMZ failed to reduce metabolic activity in cells treated with Scr PEI–SNAs except at the highest dose tested (1000 μM), which is 20-fold higher than the maximum clinically feasible TMZ dose at the tumor site.49 At this dose of TMZ, Scr PEI–SNAs reduced metabolic activity by ~40%, while we observed a similar but slightly larger decrease of ~57% in cells treated with Gli1 PEI–SNAs. In addition to supporting prior knowledge that adherent-cultured U87-MG cells are TMZ-insensitive, our results are also consistent with our finding that Gli1 PEI–SNAs induce senescence in adherent-cultured U87-MG cells. Because TMZ is an alkylating agent and therefore exhibits selective toxicity to actively dividing cells, the onset of senescence could explain why TMZ is ineffective toward reducing metabolic activity in this context. The results from these intriguing experiments prompted us to next evaluate combination therapy using TMZ and Gli1 PEI–SNAs in GBM neurospheres, which exhibit greater sensitivity to clinically relevant TMZ doses.50
Gli1 PEI–SNAs Reduce Stemness and Impair Self-Renewal of GBM Cells.
We were next interested in whether Gli1 PEI–SNAs could reverse the stemness of GBM cells to impair the self-renewal capacity of GSCs. First, we determined the extent to which Gli1 PEI–SNAs could reduce the expression of stemness genes in adherent-cultured cells (Figure 6A,B). By qPCR, we found that Gli1 PEI–SNAs significantly reduce the expression of CD133 and NANOG by 38 and 25%, respectively (Figure 6B). We also observed a slight but insignificant decrease in Sox2 expression (Figure 6B). To determine whether these changes are sufficient to impair self-renewal, we used a sphere-formation assay, in which U87-MG cells are seeded in suspension to form multicellular neuro-spheres that exhibit increased stemness relative to adherent-cultured cells.18 In these experiments, U87-MG cells were pretreated with PEI–SNAs for 72 h and then dissociated and suspended in serum-free medium supplemented with growth factors to form neurospheres over a period of 7 days (Figure 6A). Bright-field images show that neurospheres grown from cells pretreated with Gli1 PEI–SNAs are significantly smaller than those grown from cells pretreated with Scr PEI–SNAs, exhibiting an ~40 μm decrease in average projected diameter (Figure 6C,D). Further, we found that treatment with Gli1 PEI–SNAs reduced the total number of neurospheres formed by 26% (Figure 6D), though this result was not statistically significant. Notably, we detected this reduction in size and number of neurospheres 10 days after Gli1 PEI–SNA treatment, whereas previous work demonstrates that a single transfection with SNAs can silence gene expression for up to 96 h.8 Here, we demonstrate that PEI–SNAs are a useful vehicle for achieving sustained effects of Gli1-targeted therapy. Consistent with our other results, this suggests that Gli1 PEI– SNAs can impair the stemness of GBM cells to reduce the formation of neurospheres through self-renewal. Our findings are corroborated by prior studies, which reported that either transient transfection of siGli1 or treatment with a Smo inhibitor into GBM neurospheres significantly hinder neuro-sphere growth.19,21 Similar results have been obtained using the pharmacological Gli inhibitor, GANT61, encapsulated within PLGA nanoparticles. PLGA–GANT61 reduced tumor-sphere formation in colon and breast cancer cell lines.51 The ability of Hh inhibitors to decrease the number of cancer stem cells (CSCs) has been demonstrated in vivo as well; one study reported that PLGA–PEG nanoparticles delivering another pharmacological Gli inhibitor, HPI-1, reduced the number of ALDH+ CSCs in a murine orthotopic pancreatic cancer xenograft.52 In totality, these results suggest that Gli1 PEI– SNAs can impair the self-renewal capacity of U87-MG cells and may be useful to eliminate the aggressive GSC subpopulation.
Gli1 PEI–SNAs Potentiate the Neurosphere Response to TMZ Chemotherapy.
Finally, we were interested in whether treating neurospheres with Gli1 PEI–SNAs could improve neurosphere response to TMZ. We first confirmed that PEI–SNAs could enter neurospheres by confocal microscopy and flow cytometry. Confocal microscopy demonstrates that Cy5-labeled PEI–SNAs can easily penetrate small neurospheres (<100 μm) but accumulate to the greatest extent within the periphery of larger neurospheres (Figure 7A) within 24 h. Flow cytometry analysis of neurospheres dissociated after treatment demonstrates that Cy5-labeled PEI–SNAs can enter 91.3% of cells grown as neurospheres (Figure 7B). Taken together with our microscopy data, this suggests that PEI–SNAs can penetrate neurospheres greater than 100 μm in diameter but show greater accumulation near the periphery than in the center. This is consistent with previous work demonstrating that SNAs can successfully transfect human tumor neurospheres,8 so we were encouraged that our PEI–SNAs retain this property. We were further interested in examining the mechanism by which PEI–SNAs penetrate neurospheres. We found that the scavenger receptor inhibitor FCD reduces the uptake of PEI–SNAs by neuro-sphere-cultured cells by 43%, and the caveolae/lipid raft inhibitor MβCD reduces uptake by 64% (Figure 7C), demonstrating that these are mechanisms that are required for PEI–SNA distribution through neurospheres.
To investigate the capacity for Gli1 PEI–SNAs to potentiate the neurosphere response to TMZ, we treated cells according to the timeline in Figure 7D. Adherent-cultured cells were pretreated with PEI–SNAs for 72 h, seeded as neurospheres, and incubated a further 72 h. Spheres were treated with a second PEI–SNA pulse for 24 h, resuspended in TMZ-containing medium, and incubated for 72 h. Spheres were dissociated and analyzed for apoptosis using an Annexin-V– FITC/propidium iodide (PI) assay. We found that spheres primed with Gli1 PEI–SNAs prior to low-dose TMZ treatment (50 μM) exhibited a nearly 2-fold increase in the fraction of apoptotic cells relative to cells primed with Scr PEI–SNAs (Figure 7E,F). Gli1 PEI–SNAs also increased the fraction of apoptotic cells in response to 100 and 200 μM TMZ by 62 and 50%, respectively. This is consistent with previous research, which demonstrated that the pharmacological Hedgehog pathway inhibitor cyclopamine can increase the fraction of Caspase3+ apoptotic GBM stem cell cultures in combination with TMZ.19 An additional report found that cyclopamine could potentiate the cytotoxic effects of TMZ in CD133+ glioma stem cells.53 However, the clinical translation of cyclopamine has been deterred by severe toxicity and rapid clearance from circulation.54 Further, cyclopamine targets Smo, upstream of Gli1 in the Hedgehog signaling pathway, and a growing body of work has demonstrated that cancer cells frequently acquire resistance to Smo antagonists, potentially limiting their utility.55 Among Gli1 inhibitors, furthest along in development is GANT61, which, despite its potency in preclinical models, is poorly stable at physiological conditions.55 Further, the ability of GANT61 to cross the blood– brain barrier remains poorly understood. Because SNAs have previously demonstrated sufficient stability in physiological conditions and can accumulate within glioma xenografts and improve tumor response to TMZ,8,11,28 we are hopeful that a Gli1-targeted construct might behave similarly. Based on our results and the results of others, we hypothesize that Gli1 PEI– SNAs may also accumulate within GBM tumors in vivo to exert antitumor effects that also extend to GSCs, though this remains to be evaluated in future work.
CONCLUSIONS
In this work, we have demonstrated that Gli1 PEI–SNAs can improve the response of GBM cells and aggressive GBM neurospheres to TMZ chemotherapy. We found that Gli1 PEI–SNAs bind scavenger receptors on GBM cells to undergo endocytosis in a caveolae/lipid raft/dynamin-dependent manner and that this leads to trafficking through the classical endolysosomal pathway. Despite many of the nanoparticles accumulating within late endosomes and lysosomes, Gli1 PEI– SNAs could silence the expression of genes associated with the Hedgehog signaling pathway that promote the aggressive, chemoresistant phenotype of GBM. Further, this leads to a decrease in proliferation that correlates with an onset of GBM cell senescence as well as a decrease in metabolic activity with or without TMZ cotreatment. Importantly, Gli1 PEI–SNAs reduced the growth of stem-like neurospheres and sensitized neurospheres to low doses of TMZ chemotherapy. These results warrant further development of PEI–SNAs and Gli1-targeted therapies to alleviate drug resistance and recurrence for GBM patients.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Institutes of General Medical Sciences and the National Institutes of Health under a Maximizing Investigators’ Research Award (MIRA) (grant number R35-GM119659). Microscopy access was supported by grants from the NIH-NIGMS (P20 GM103446) including a shared instrumentation grant (S10 OD016361), the NSF (IIA-1301765), and the State of Delaware. J.R.M. received support from the Department of Defense through a National Defense Science and Engineering Graduate Fellowship. S.A.I. received support from the University of Delaware Summer Scholars program. The content is solely the responsibility of the authors and does not necessarily represent the views of the funding agencies.
ABBREVIATIONS
- GBM
glioblastoma
- PEI
polyethylenimine
- SNA
spherical nucleic acid
- TMZ
temozolomide
- RNAi
RNA interference
- Hh
Hedgehog
- GPCR
G-protein coupled receptor
- GSC
glioblastoma stem cell
- MβCD
methyl-β-cyclodextrin
- CPZ
chlorpromazine
- FCD
fucoidan
- CytD
cytochalasin D
- MCC
Manders’ colocalization coefficient
- EdU
5-ethynyl-2′-deoxyuridine
- SAβGal
senescence-associated β-galactosidase
- SASP
senescence-associated secretory phenotype
- qPCR
quantitative polymerase chain reaction
- PI
propidium iodide
- AuNP
gold nanoparticle
- DEPC
diethyl pyrocarbonate
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.molpharmaceut.8b00707.
Table of conditions used for uptake inhibitor experiments; table of primers used for qPCR (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Whitehead KA; Langer R; Anderson DG Knocking down Barriers: Advances in SiRNA Delivery. Nat. Rev. Drug Discovery 2009, 8 (2), 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WCW Analysis of Nanoparticle Delivery to Tumours. Nat. Rev. Mater 2016, 1 (5), 16014. [Google Scholar]
- (3).Mitra M; Kandalam M; Rangasamy J; Shankar B; Maheswari UK; Swaminathan S; Krishnakumar S Novel Epithelial Cell Adhesion Molecule Antibody Conjugated Polyethyleneimine-Capped Gold Nanoparticles for Enhanced and Targeted Small Interfering RNA Delivery to Retinoblastoma Cells. Mol. Vis 2013, 19, 1029–1038. [PMC free article] [PubMed] [Google Scholar]
- (4).Hall A; Lächelt U; Bartek J; Wagner E; Moghimi SM Polyplex Evolution: Understanding Biology, Optimizing Performance. Mol. Ther 2017, 25 (7), 1476–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Neuberg P; Kichler A Recent Developments in Nucleic Acid Delivery with Polyethylenimines. Adv. Genet 2014, 88, 263–288. [DOI] [PubMed] [Google Scholar]
- (6).Melamed JR; Kreuzberger NL; Goyal R; Day ES Spherical Nucleic Acid Architecture Can Improve the Efficacy of Polycation-Mediated SiRNA Delivery. Mol. Ther.–Nucleic Acids 2018, 12 (9), 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cutler JI; Auyeung E; Mirkin C a. Spherical Nucleic Acids. J. Am. Chem. Soc 2012, 134 (3), 1376–1391. [DOI] [PubMed] [Google Scholar]
- (8).Jensen SA; Day ES; Ko CH; Hurley LA; Luciano JP; Kouri FM; Merkel TJ; Luthi AJ; Patel PC; Cutler JI; et al. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Sci. Transl. Med 2013, 5 (209), 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Massich MD; Giljohann DA; Seferos DS; Ludlow LE; Horvath CM; Mirkin CA Regulating Immune Response Using Polyvalent Nucleic Acid - Gold Nanoparticle Conjugates. Mol. Pharmaceutics 2009, 6 (6), 1934–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kapadia CH; Melamed JR; Day ES Spherical Nucleic Acid Nanoparticles: Therapeutic Potential. BioDrugs 2018, 32 (4), 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kouri FM; Hurley LA; Day ES; Hua Y; Merkel TJ; Queisser A; Peng C; Ritner C; Hao L; Daniel WL; et al. MiR-182 Integrates Apoptosis, Growth and Differentiation Programs in Glioblastoma. Genes Dev. 2015, 29 (7), 732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Northwestern University. NU-0129 in Treating Patients with Recurrent Glioblastoma or Gliosarcoma Undergoing Surgery. https://clinicaltrials.gov/ct2/show/NCT03020017.
- (13).Wu XA; Choi CHJ; Zhang C; Hao L; Mirkin CA Intracellular Fate of Spherical Nucleic Acid Nanoparticle Conjugates. J. Am. Chem. Soc 2014, 136, 7726–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet 2014, 15 (8), 541–555. [DOI] [PubMed] [Google Scholar]
- (15).Ostrom QT; Gittleman H; Liao P; Rouse C; Chen Y; Dowling J; Wolinsky Y; Kruchko C; Barnholtz-Sloan J CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro. Oncol 2014, 16 (suppl 4), iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Weller M; Cloughesy T; Perry JR; Wick W Standards of Care for Treatment of Recurrent Glioblastoma-Are We There Yet? Neuro. Oncol 2013, 15 (1), 4–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Seystahl K; Wick W; Weller M Therapeutic Options in Recurrent Glioblastoma – an Update. Crit. Rev. Oncol. Hematol 2016, 99, 389–408. [DOI] [PubMed] [Google Scholar]
- (18).Melamed JR; Morgan JT; Ioele SA; Gleghorn JP; Sims-mourtada J; Day ES Investigating the Role of Hedgehog/ GLI1 Signaling in Glioblastoma Cell Response to temozolomide. Oncotarget 2018, 9 (43), 27000–27015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Clement V; Sanchez P; de Tribolet N; Radovanovic I; Ruiz i Altaba A HEDGEHOG-GLI1 Signaling Regulates Human Glioma Growth, Cancer Stem Cell Self-Renewal, and Tumorigenicity. Curr. Biol 2007, 17 (2), 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cui D; Xu Q; Wang K; Che X Gli1 Is a Potential Target for Alleviating Multidrug Resistance of Gliomas. J. Neurol. Sci 2010, 288 (1–2), 156–166. [DOI] [PubMed] [Google Scholar]
- (21).Bar EE; Chaudhry A; Lin A; Fan X; Schreck K; Matsui W; Piccirillo S; Vescovi AL; DiMeco F; Olivi A; et al. Cyclopamine-Mediated Hedgehog Pathway Inhibition Depletes Stem-like Cancer Cells in Glioblastoma. Stem Cells 2007, 25 (10), 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bradshaw A; Wickremsekera A; Tan ST; Peng L; Davis PF; Itinteang T Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front. Surg 2016, 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lathia J; Mack S; Mulkearns-Hubert EE; Valentim CLL; Rich JN Cancer Stem Cells in Glioblastoma. Genes Dev. 2015, 29 (12), 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).George J; Banik NL; Ray SK Combination of Taxol and Bcl-2 SiRNA Induces Apoptosis in Human Glioblastoma Cells and Inhibits Invasion, Angiogenesis and Tumour Growth. J. Cell. Mol. Med 2009, 13 (10), 4205–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liu Y; Miao C; Wang Z; He X; Shen W Survivin Small Interfering RNA Suppresses Glioblastoma Growth by Inducing Cellular Apoptosis. Neural Regen Res. 2012, 7 (12), 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wang J; Wang Q; Cui Y; Liu ZY; Zhao W; Wang CL; Dong Y; Hou L; Hu G; Luo C; et al. Knockdown of Cyclin D1 Inhibits Proliferation, Induces Apoptosis, and Attenuates the Invasive Capacity of Human Glioblastoma Cells. J. Neuro-Oncol 2012, 106 (3), 473–484. [DOI] [PubMed] [Google Scholar]
- (27).Padfield E; Ellis HP; Kurian KM Current Therapeutic Advances Targeting EGFR and EGFRvIII in Glioblastoma. Front. Oncol 2015, 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sita TL; Kouri FM; Hurley LA; Merkel TJ; Chalastanis A; May JL; Ghelfi ST; Cole LE; Cayton TC; Barnaby SN; et al. Dual Bioluminescence and Near-Infrared Fluorescence Monitoring to Evaluate Spherical Nucleic Acid Nano-conjugate Activity in Vivo. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (16), 4129–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kato T; Natsume A; Toda H; Iwamizu H; Sugita T; Hachisu R; Watanabe R; Yuki K; Motomura K; Bankiewicz K; et al. Efficient Delivery of Liposome-Mediated MGMT-SiRNA Reinforces the Cytotoxity of Temozolomide in GBM-Initiating Cells. Gene Ther. 2010, 17 (11), 1363–1371. [DOI] [PubMed] [Google Scholar]
- (30).Frens G Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nature, Phys. Sci 1973, 241 (105), 20–22. [Google Scholar]
- (31).Melamed J; Riley R; Valcourt D; Billingsley M; Kreuzberger N; Day E Quantification of SiRNA Duplexes Bound to Gold Nanoparticle Surfaces. Methods Mol. Biol 2017, 1570, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Manders E; Verbeek F; Aten J Measurement of Co-Localization of Objects in Dual-Colour Confocal Images. J. Microsc 1993, 169 (3), 375–382. [DOI] [PubMed] [Google Scholar]
- (33).Rejman J; Bragonzi A; Conese M Role of Clathrin-and Caveolae-Mediated Endocytosis in Gene Transfer Mediated by Lipoand Polyplexes. Mol. Ther 2005, 12 (3), 468–474. [DOI] [PubMed] [Google Scholar]
- (34).Choi CHJ; Hao L; Narayan SP; Auyeung E; Mirkin C a. Mechanism for the Endocytosis of Spherical Nucleic Acid Nanoparticle Conjugates. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (19), 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Pelkmans L; Kartenbeck J; Helenius A Caveolar Endocytosis of Simian Virus 40 Reveals a New Two-Step Vesicular-Transport Pathway to the ER. Nat. Cell Biol 2001, 3 (5), 473–483. [DOI] [PubMed] [Google Scholar]
- (36).Munsell EV; Ross NL; Sullivan MO Journey to the Center of the Cell: Current Nanocarrier Design Strategies Targeting Biopharmaceuticals to the Cytoplasm and Nucleus. Curr. Pharm. Des 2016, 22 (9), 1227–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wittrup A; Ai A; Liu X; Hamar P; Trifonova R; Charisse K; Manoharan M; Kirchhausen T; Lieberman J Visualizing Lipid-Formulated SiRNA Release from Endosomes and Target Gene Knockdown. Nat. Biotechnol 2015, 33 (8), 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Gilleron J; Querbes W; Zeigerer A; Borodovsky A; Marsico G; Schubert U; Manygoats K; Seifert S; Andree C; Stöter M; et al. Image-Based Analysis of Lipid Nanoparticle-Mediated SiRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol 2013, 31 (7), 638–646. [DOI] [PubMed] [Google Scholar]
- (39).Sandin P; Fitzpatrick LW; Simpson JC; Dawson KA High-Speed Imaging of Rab Family Small GTpases Reveals Rare Events in Nanoparticle Trafficking in Living Cells. ACS Nano 2012, 6 (2), 1513–1521. [DOI] [PubMed] [Google Scholar]
- (40).Ng JMY; Curran T The Hedgehog’s Tale: Developing Strategies for Targeting Cancer. Nat. Rev. Cancer 2011, 11 (7), 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Metcalfe C; De Sauvage FJ Hedgehog Fights Back: Mechanisms of Acquired Resistance against Smoothened Antagonists. Cancer Res. 2011, 71 (15), 5057–5061. [DOI] [PubMed] [Google Scholar]
- (42).Wang J; Wang H; Li Z; Wu Q; Lathia JD; McLendon RE; Hjelmeland AB; Rich JN C-Myc Is Required for Maintenance of Glioma Cancer Stem Cells. PLoS One 2008, 3 (11), e3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Voss V; Senft C; Lang V; Ronellenfitsch MW; Steinbach JP; Seifert V; Kogel D The Pan-Bcl-2 Inhibitor (–)-Gossypol Triggers Autophagic Cell Death in Malignant Glioma. Mol. Cancer Res. 2010, 8 (7), 1002–1016. [DOI] [PubMed] [Google Scholar]
- (44).Wee B; Pietras A; Ozawa T; Bazzoli E; Podlaha O; Antczak C; Westermark B; Nelander S; Uhrbom L; Forsberg-Nilsson K; et al. ABCG2 Regulates Self-Renewal and Stem Cell Marker Expression but Not Tumorigenicity or Radiation Resistance of Glioma. Sci. Rep 2016, 6 (1), 25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Campisi J; D’Adda Di Fagagna F Cellular Senescence: When Bad Things Happen to Good Cells. Nat. Rev. Mol. Cell Biol 2007, 8 (9), 729–740. [DOI] [PubMed] [Google Scholar]
- (46).Lee J-J; Kim BC; Park M-J; Lee Y-S; Kim Y-N; Lee BL; Lee J-S PTEN Status Switches Cell Fate between Premature Senescence and Apoptosis in Glioma Exposed to Ionizing Radiation. Cell Death Differ. 2011, 18 (4), 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Coppé J-P; Desprez P-Y; Krtolica A; Campisi J The Senescence-Associated Secretory Phenotype: The Dark Side of Tumor Suppression. Annu. Rev. Pathol.: Mech. Dis 2010, 5 (1), 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Childs BG; Baker DJ; Kirkland JL; Campisi J; van Deursen JM Senescence and Apoptosis: Dueling or Complementary Cell Fates? EMBO Rep. 2014, 15 (11), 1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Biswas NK; Chandra V; Sarkar-Roy N; Das T; Bhattacharya RN; Tripathy LN; Basu SK; Kumar S; Das S; Chatterjee A; et al. Variant Allele Frequency Enrichment Analysis in Vitro Reveals Sonic Hedgehog Pathway to Impede Sustained Temozolomide Response in GBM. Sci. Rep 2015, 5 (1), 7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Mihaliak AM; Gilbert CA; Li L; Daou MC; Moser RP; Reeves A; Cochran BH; Ross AH Clinically Relevant Doses of Chemotherapy Agents Reversibly Block Formation of Glioblastoma Neurospheres. Cancer Lett. 2010, 296 (2), 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Borah A; Palaninathan V; Girija AR; Balasubramanian S; Rochani AK; Maekawa T; Kumar DS Poly-Lactic-Co-Glycolic Acid Nanoformulation of Small Molecule Antagonist GANT61 for Cancer Annihilation. NanoWorld J 2017, 3 (1), 1–10. [Google Scholar]
- (52).Chenna V; Hu C; Pramanik D; Aftab BT; Karikari C; Campbell NR; Hong S-M; Zhao M; Rudek M. a.; Khan SR; et al. A Polymeric Nanoparticle Encapsulated Small-Molecule Inhibitor of Hedgehog Signaling (NanoHHI) Bypasses Secondary Mutational Resistance to Smoothened Antagonists. Mol. Cancer Ther 2012, 11 (1), 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ulasov IV; Nandi S; Dey M; Sonabend AM; Lesniak MS Inhibition of Sonic Hedgehog and Notch Pathways Enhances Sensitivity of CD133(+) Glioma Stem Cells to Temozolomide Therapy. Mol. Med 2011, 17 (1–2), 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Rimkus T; Carpenter R; Qasem S; Chan M; Lo H-W Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers 2016, 8 (2), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Infante P; Alfonsi R; Botta B; Mori M; Di Marcotullio L Targeting GLI Factors to Inhibit the Hedgehog Pathway. Trends Pharmacol. Sci 2015, 36 (8), 547–558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.