Abstract
Breast cancer is the most commonly diagnosed cancer and the second leading cause of cancer death in women. Radioresistance remains one of the most critical barriers in radiation therapy for breast cancer. In this study, we employed a parallel-reaction monitoring (PRM)-based targeted proteomic method to examine the reprogramming of the heat shock proteome during the development of radioresistance in breast cancer. In particular, we investigated the differential expression of heat shock proteins (HSPs) in two pairs of matched parental/radioresistant breast cancer cell lines. We were able to quantify 43 and 42 HSPs in the MCF-7 and MDA-MB-231 pairs of cell lines, respectively. By analyzing the commonly altered proteins, we found that several members of the HSP70 and HSP40 subfamilies of HSPs exhibited substantially altered expression upon development of radioresistance. Moreover, the expression of HSPB8 is markedly elevated in the radioresistant lines relative to the parental MCF-7 and MDA-MB-231 cells. Together, our PRM-based targeted proteomics method revealed the reprogramming of the heat shock proteome during the development of radioresistance in breast cancer cells and offered potential targets for sensitizing breast cancer cells toward radiation therapy.
Graphical Abstract

INTRODUCTION
Breast cancer is the most common malignancy among women and the second leading cause of cancer deaths in women in the United States.1 Locoregional treatment of breast cancer with radiation therapy has evolved over the past several decades,2 which has been a critical component of the multimodal management of invasive breast cancer.3 However, development of radioresistance becomes one of the most critical obstacles in cancer radiotherapy.4,5 Therefore, identifying molecular targets responsible for the radioresistant phenotype of breast cancer cells is critical for enhancing the efficacy of anti-cancer therapy and for reducing the mortality of breast cancer patients.
Heat shock proteins (HSPs) are molecular chaperones which bear important functions in protein folding/unfolding, cell cycle regulation, and cellular protection against stress.6–8 HSPs have been frequently shown to affect disease progression. For instance, HSP90, a molecular chaperone for protein folding and client protein stabilization,9 is overexpressed in many types of tumors and known to be associated with breast cancer progression.10 Furthermore, some heat shock proteins, e.g., HSP257 and HSP27,8 were found to promote the development of radioresistance in cancer cells. We reason that a systematic assessment of how HSPs modulate the development of radioresistance may lead to novel molecular targets for sensitizing cancer cells to radiation therapy.
We recently developed a parallel-reaction monitoring (PRM)-based targeted proteomics method to examine the expression of HSPs at the entire proteome scale.11 The method was successfully employed to reveal the differentially expressed HSPs during melanoma metastasis and led to the identification of DNAJB4 as a novel suppressor for melanoma metastasis.11 Here, we employed the PRM method to investigate comprehensively the implications of HSPs in radioresistance of breast cancer by profiling the differential expression of HSPs in two pairs of matched wild-type (WT)/radioresistant breast cancer cells (i.e., MCF-7 and MDA-MB-231 cells, and their radioresistant counterparts).12,13 We quantified the expressions of 43 and 42 unique HSPs in the MCF-7 and MDA-MB-231 pairs, respectively. Our results support that HSP70, HSP40, and HSPB8 proteins play important roles in radioresistance of breast cancer cells.
MATERIALS AND METHODS
Generation of Radioresistant Breast Cancer Cell Lines.
MCF-7 cells were treated with a total of 60 Gy of γ rays to generate the radioresistant clone (clone 6, referred as C6) and further exposed to an additional term of 30 fractions of γ rays.12 Similarly, the radioresistant clone (clone 5, referred as C5) of MDA-MB-231 cells was generated by exposing the cells to a total dose of 30 Gy of ionizing radiation.13 The radioresistant phenotype of the MCF-C6 and MDA-MB-231-C5 cells was characterized by clonogenic survival assay, as reported previously.12,13
Cell Culture.
MCF-7 WT/radioresistant and MDA-MB-231 WT/radioresistant cells were cultured in Dulbecco’s modified eagle medium (DMEM). Culture media were supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and penicillin (100 IU/mL). The cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. Approximately 2 × 107 cells were harvested, washed with cold PBS for three times, and lysed by incubating on ice for 30 min with CelLytic M cell lysis reagent (Sigma) containing 1% protease inhibitor cocktail. The cell lysates were centrifuged at 9000g at 4 °C for 30 min, and the resulting supernatants were collected. For SILAC labeling experiments, the cells were cultured in SILAC medium containing unlabeled lysine and arginine, or [13C6,15N2]-lysine and [13C6]-arginine, for at least five cell doublings.
Tryptic Digestion of Whole-Cell Protein Lysates.
The whole-cell lysates prepared from MCF-7 or MDA-MB-231 breast cancer cells and their radioresistant counterparts were combined at 1:1 ratio, incubated with 8 M urea for protein denaturation, and then treated with dithiothreitol and iodoacetamide for cysteine reduction and alkylation, respectively. The proteins were subsequently digested with modified MS-grade trypsin (Pierce) at an enzyme/substrate ratio of 1:100 in 50 mM NH4HCO3 (pH 8.5) at 37 °C overnight. The peptide mixture was then dried in a Speed-vac, desalted with OMIX C18 pipet tips (Agilent Technologies), and analyzed by LC-MS and MS/MS in the PRM mode.
LC-PRM Analysis.
All LC-PRM experiments were carried out on a Q Exactive Plus quadrupole-Orbitrap mass spectrometer coupled with an EASY-nLC 1200 system (Thermo Fishier Scientific). The samples were automatically loaded onto a 4 cm trapping column (150 μm i.d.) which was packed with ReproSil-Pur 120 C18-AQ resin (5 μm in particle size and 120 Å in pore size, Dr. Maisch GmbH HPLC) at a flow rate of 3 μL/min. The trapping column was coupled to a 20 cm fused silica analytical column (PicoTip Emitter, New Objective, 75 μm i.d.) packed with ReproSil-Pur 120 C18-AQ resin (3 μm in particle size and 120 Å in pore size, Dr. Maisch GmbH HPLC). The peptides were then separated using a 140 min linear gradient of 9−38% acetonitrile in 0.1% formic acid and at flow rate of 300 nL/min. The spray voltage was 1.8 kV. Peptide ions were collisionally activated in the HCD cell at a normalized collision energy of 29 to yield MS/MS, which were recorded in the Orbitrap analyzer at a resolution of 17 500 with an AGC target of 1 × 105.
The linear predictor of empirical retention time (RT) from normalized RT (iRT)14 for targeted peptides of HSPs was determined by the linear regression of RTs of BSA standard peptides obtained for the current chromatography setup.11,15,16 This RT−iRT linear relationship was redefined between every eight LC-MS/MS runs by injecting another BSA tryptic digestion mixture. The targeted precursor ions were monitored in scheduled PRM mode with an 8 min retention time window.
All raw files were processed using Skyline (version 3.5) for the generation of extracted-ion chromatograms and for peak integration.17 A total of 2−5 unique peptides with the highest abundances of precursor ions were selected for the quantification of each HSP. The 6 most abundant ions found in MS/MS acquired from shotgun proteomic analysis were chosen for peptide identification and quantification in PRM mode, where a mass accuracy of 20 ppm or better was imposed for fragment ions during the identification of peptides in the Skyline platform. The targeted peptides were first manually checked to ensure that the chromatographic profiles of multiple fragment ions derived from the light and heavy forms of the same peptide could be overlaid. The data were then processed to ensure that the distribution of the relative intensities of multiple transitions associated with the same precursor ion is correlated with the theoretical distribution in the MS/MS acquired from shotgun proteomic analysis. The sum of peak areas from all transitions of light or heavy peptides was used for quantification.
TCGA Data, GEO Data, and Signaling Network Analysis.
Clinical data and mRNA expression profiles for the Molecular Taxonomy of Breast Cancer International Consortium (META-BRIC)18 cohort which contains 1982 breast cancer samples were obtained from cBioPortal (http://www.cbioportal.org/data_sets.jsp)19 and analyzed via its web-based Python API. The patients in the METABRIC cohort were stratified based on with/without radiotherapy, and we further stratified for those patients who received radiotherapy based on median mRNA expressions of genes encoding HSPs. Kaplan−Meier survival curves were generated to assess the association between the mRNA expression of the HSP genes and overall survival (OS) of breast cancer patients. p-values were generated using log rank (Mantel-Cox) test, and a p-value <0.05 is considered significant.
GSE datasets were derived from Gene Expression Omnibus (GEO) database. Wild-type animals in the GSE42742 dataset and breast cancer patients in the GSE40640 dataset were employed for analysis. Signaling network analysis was performed using the online tool GeneMANIA (https://genemania.org/).20
Western Blot.
Cells were cultured in 6-well plates and lysed at 50−70% confluency following the aforementioned procedures. The concentrations of the resulting protein lysates were determined by using the Bradford Assay (Bio-Rad). The whole-cell lysate for each sample (10 μg) was denatured by boiling in Laemmli loading buffer and resolved by using SDS-PAGE. Subsequently, the proteins were transferred onto a nitrocellulose membrane at 4 °C overnight. The resulting membrane was blocked with PBS-T (PBS with 0.1% Tween 20) containing 5% milk (Bio-Rad) at 4 °C for 6 h. The membrane was subsequently incubated with primary antibody at 4 °C overnight and then with secondary antibody at room temperature for 1 h. After thorough washing with PBS-T, the HRP signal was detected using Pierce ECL Western blotting substrate (Thermo).
Human HSPB8 antibody (Cell Signaling, no. 3059, 1:2000 dilution) was employed as the primary antibody for Western blot analysis. Horseradish peroxidase-conjugated anti-rabbit IgG was used as the secondary antibody. Membranes were also probed with anti-actin antibody (Cell Signaling, no. 4967, 1:10000 dilution) to verify equal protein loading.
Real-Time PCR.
Cells were seeded in 6-well plates at 50−70% confluence level. Total RNA was extracted from cells using TRI Reagent (Sigma). Approximately 3 μg of RNA was reverse transcribed by employing M-MLV reverse transcriptase (Promega) and an oligo(dT)18 primer. After a 60 min incubation at 42 °C, the reverse transcriptase was deactivated by heating at 75 °C for 5 min. Quantitative real-time PCR was performed using iQ SYBR Green Supermix kit (Bio-Rad) on a Bio-Rad iCycler system (Bio-Rad), and the running conditions were at 95 °C for 3 min and 45 cycles at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 45 s. The comparative cycle threshold (Ct) method (ΔΔCt) was used for the relative quantification of gene expression,21 and the primer sequences are listed in Table S1.
RESULTS AND DISCUSSION
Quantitative Assessment of Differential Expression of Heat Shock Proteins in Paired Radioresistant and Parental Breast Cancer Cells.
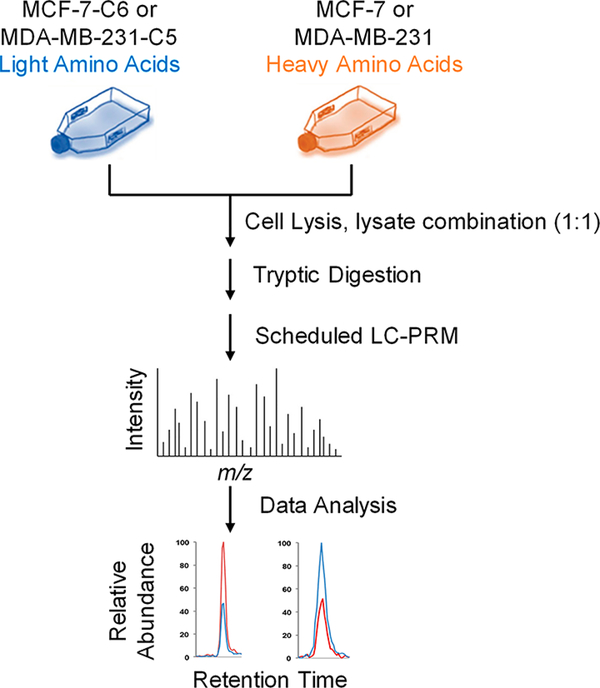
To explore the potential roles of HSPs in developing radioresistance in breast cancer cells, we utilized our recently developed PRM-based targeted proteomic method,11 together with stable isotope labeling by amino acids in cell culture (SILAC)22 (Figure 1), to examine the differential expression of HSPs in two pairs of breast cancer cells, i.e., parental (WT) MCF-7 and the radioresistant C6 clone, and WT MDA-MB-231 and the radio-resistant C5 clone.
Figure 1.
General workflow for the scheduled LC-PRM analysis in the quantitative assessment of the human heat shock proteome. Shown is a flowchart of the forward SILAC labeling and LC-PRM experiment.
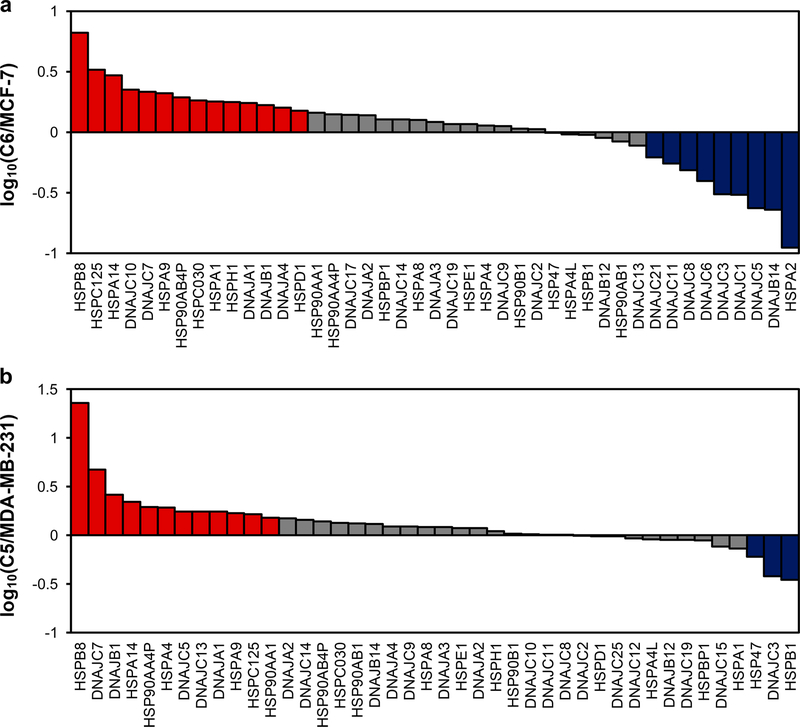
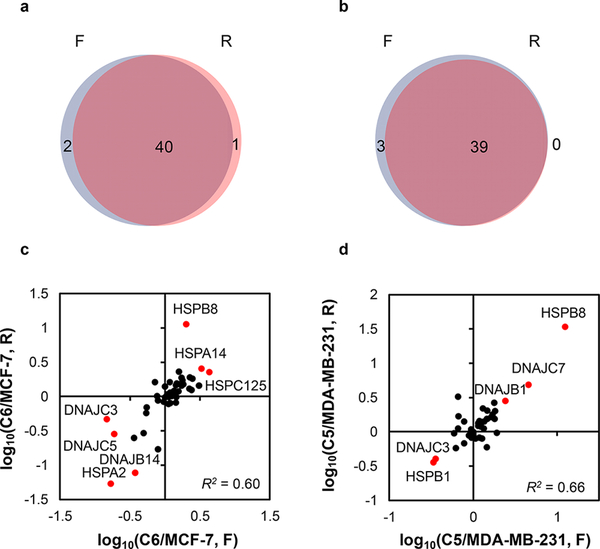
Approximately 50% of the human HSPs (Figure 2 and Table S2) were quantified with the PRM-based targeted proteomic method. Specifically, 43 and 42 unique HSPs were quantified in the MCF-7 WT/C6 and MDA-MB-231 WT/C5 paired cells, respectively. All PRM transitions (4−6) used for each tryptic peptide derived from HSPs displayed the same retention time, with the dot product (dotp) value23 being >0.7 (Figure S1). In addition, more than 90% of the quantified proteins appeared in both forward and reverse SILAC labeling experiments (Figure 3a,b). Furthermore, the ratios of the quantified peptides obtained from forward and reverse SILAC labeling experiments exhibited an excellent linear fit (Figure 3c,d). The results, therefore, confirmed the excellent reproducibility of our PRM method.
Figure 2.
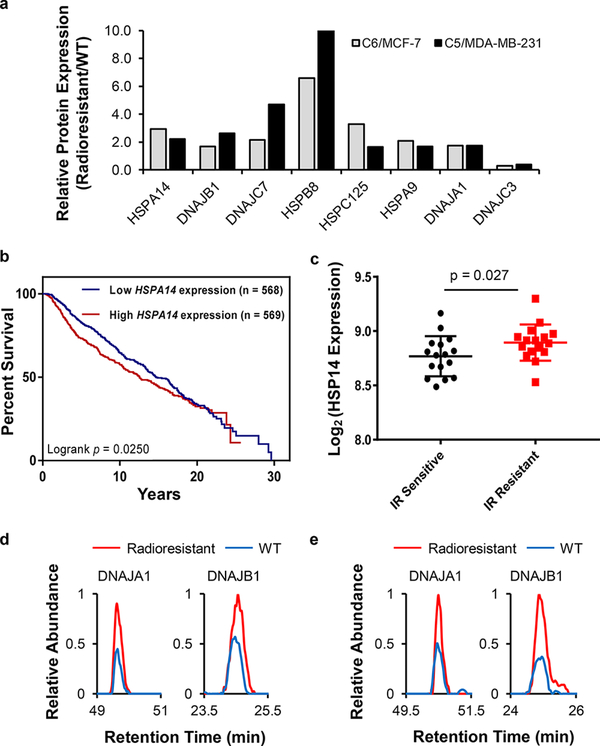
Alterations in expression levels of heat shock proteins in WT/radioresistant MCF-7 (a) and MDA-MB-231 (b) cells. The data represent the average ratios obtained from two biological replicates (i.e., one forward and one reverse SILAC labeling experiments). The red and blue bars designate those HSPs that were up- and down-regulated, respectively, by at least 1.5-fold in the radioresistant over the corresponding parental breast cancer cells.
Figure 3.
Analytical performance of the PRM method. Venn diagrams showing the overlap between quantified heat shock proteins obtained from forward and reverse SILAC labeling experiments in paired MCF-7 WT/C6 (a) and MDA-MB-231 WT/C5 (b) cells. Scatter plots displaying the correlation between the ratios obtained from forward and reverse SILAC labeling experiments in paired MCF-7 WT/C6 (c) and MDA-MB-231 WT/C5 (d) cells.
Members of HSP70 and HSP40 Proteins Are Altered upon the Development of Radioresistance in Breast Cancer Cells.
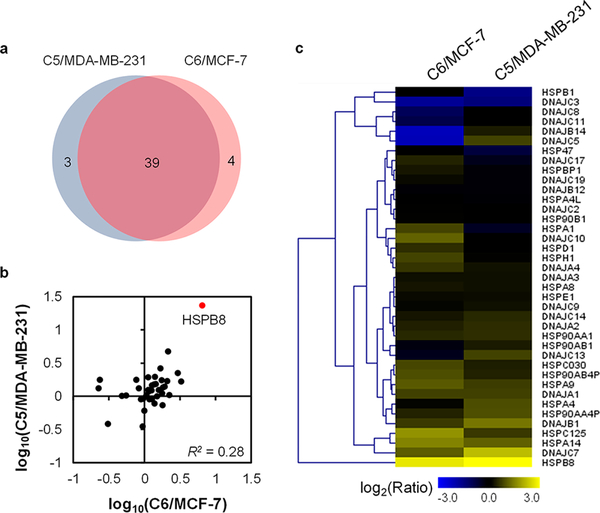
Figure 4 shows that 39 HSPs were commonly quantified for the two pairs of radioresistant/parental breast cancer cells. In addition, the relative expression of HSPs between the two pairs of matched breast cancer cells exhibited similar profiles (Figure 4b,c). Moreover, 8 HSPs (i.e., HSPA9, HSPA14, HSPB8, HSPC125, DNAJA1, DNAJB1, DNAJC3, and DNAJC7) were commonly altered in both lines of radioresistant cells relative to the corresponding parental lines (Figures 4c and 5a). Furthermore, the relative mRNA expression of genes encoding HSPA9, HSPA14, and HSPC125 were positively correlated with the overall survival of breast cancer patients undergoing radiation therapy, with higher mRNA levels of these genes being associated with poorer overall survival (Figures 5b and S2a,b). Along this line, both HSPA9 and HSPA14 belong to the HSP70 subfamily, which was previously shown to be associated with radioresistance in human glioma spheroids.24 In addition, interrogation of several Gene Expression Omnibus (GEO) datasets indicated that HSP70 was also correlated with radiation sensitivity of breast cancer patients. The expression of HSPA9 was increased for the mouse mammary glands after ionizing radiation (IR) (GSE42742, n = 30, Figure S2c),25 while elevated expression of HSP14 was observed in IR-resistant breast cancer patients relative to IR-sensitive patients (GSE40640, n = 32, Figure 5c).26,27 This result indicates that HSPA9 and HSPA14 may promote the development of radioresistance in breast cancer patients.
Figure 4.
Comparison of the quantified HSPs in the two pairs of breast cancer cells. (a) Venn diagram displaying the overlap between quantified HSPs from the MCF-7 WT/C6 pair and the MDA-MB-231 WT/C5 pair. (b) Correlation between the expression ratios of HSPs obtained for radioresistant/parental cells. (c) Heatmap showing the differences in expression of HSPs in the two pairs of WT/radioresistant breast cancer cell lines. Genes were clustered according to Euclidean distance. The data represent the mean of the results obtained from one forward and one reverse SILAC labeling results, and Table S2 lists the ratios obtained from individual measurements.
Figure 5.
Changes in the expression of HSP70 and HSP40 proteins during the development of radioresistance in breast cancer cells. (a) Bar graph showing the commonly up- or down-regulated HSPs in the two lines of radioresistant breast cancer cells relative to the corresponding parental lines (Table S2, n = 2). (b) Patient survival correlates with HSPA14 expression in the METABRIC cohort with radiotherapy treatment. p-value was calculated by using the log-rank test. (c) Differential expression of HSPA14 gene in lymphocytes of IR-sensitive or normal responding patients in the GSE40640 dataset (n = 32, 16 for IR-sensitive and 16 for IR-resistant). The error bar represents standard deviation. p-value was calculated using unpaired two-tailed Student’s t-test. Representative PRM traces showing the relative quantification results of DNAJA1 and DNAJB1 proteins in the MCF-7 WT/C6 (d) and MDA-MB-231 WT/C5 (e) cells.
Aside from the 3 HSPs whose mRNA expression levels are closely associated with patient survival, 4 out of 8 commonly changed HSPs (DNAJA1, DNAJB1, DNAJC3, and DNAJC7) belong to the HSP40 subfamily, and 3 were up-regulated in the radioresistant cell lines (DNAJA1, DNAJB1, and DNAJC7). On the grounds that HSP70 and HSP40 function together as a chaperone complex,28,29 HSP40 may assist HSP70 in promoting the development of radioresistance. In this regard, DNAJA1 (a.k.a. HDJ2) and DNAJB1 (a.k.a. HDJ1), which are known to be co-chaperones of HSP70,30 were up-regulated by ~2-fold in the two radioresistant breast cancer cell lines relative to the corresponding parental lines (Figure 5d,e). In addition, DNAJA1 was involved with radiosensitivity of glioblastoma multiforme cells,31,32 and DNAJB1 was shown to sensitize lung cancer cells to gefitinib through regulating the EGFR signaling pathway.33 Along this line, the EGFR signaling pathway also plays an important role in radioresistance of tumor cells.34,35 Together, the altered expression of these HSP70 and HSP40 proteins may contribute to radioresistance in breast cancer cells.
HSPB8 Is Up-Regulated in Radioresistant Breast Cancer Cell Lines.
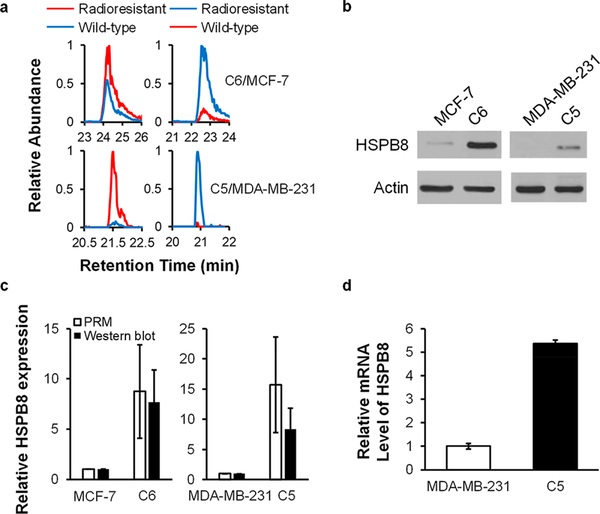
HSPB8 exhibits a pronouncedly increased expression (by ~10-fold) in the radioresistant lines of both MCF-7 and MDA-MB-231 cells (Figures 5a and 6a), and we validated the up-regulation of HSPB8 by Western blot analysis (Figure 6b,c). Meanwhile, the mRNA level of HSPB8 was also elevated in the radioresistant line (Figure 6d), indicating that the augmented expression of HSPB8 occurs through a transcriptional mechanism.
Figure 6.
HSPB8 is up-regulated in radioresistant breast cancer cell lines. (a) PRM traces for the relative quantification of HSPB8. (b) Western blot for the validation of the relative expression levels of HSPB8 protein in WT/radioresistant breast cancer cells. (c) Quantitative comparisons of the ratios of HSPB8 protein in WT/radioresistant breast cancer cells obtained from PRM (n = 2, one forward and one reverse SILAC labelings) and Western blot analyses (n = 3). (d) Relative mRNA levels of HSPB8 gene in MDA-MB-231 WT/C5 cells (n = 3).
HSPB8 is a member of small HSPs mediating the clearance of misfolded proteins;36 thus, it can modulate the proliferation and migration of breast cancer cells, including the MCF-7 and MDA-MB-231 cells used in the present study.37 HSPB8 was also found to promote drug resistance under many circumstances. For instance, it is involved in the development of bortezomib resistance in multiple myeloma cells,38 irinotecan resistance in colorectal cancer cells,39 and tamoxifen resistance in breast cancer cells.40 Furthermore, HSPB8 coordinates with HSP70-BAG3 and functions as a chaperone complex,41 which is modulated by NF-κB protein.42 Others have shown that NF-κB could promote the development of radioresistance in both MCF-7 and MDA-MB-231 cells.13 Thus, NF-κB may regulate the HSPB8-HSP70-BAG3 complex in breast cancer cells and modulate their radiosensitivity. Since the expression of HSPB8 is uniformly and markedly increased in both lines of radioresistant breast cancer cells, HSPB8 could be critical in conferring the development of radioresistance in breast cancer cells.
Signaling Network of HSPs in the Development of Radioresistance.
Since we have discovered that HSPs may play important roles in radioresistance development, we next explored the potential signaling networks involving those HSPs. Previously, we found that a group of kinases were overexpressed in radioresistant breast cancer cells, including checkpoint kinase 1 (CHK1), cyclin-dependent kinases 1 and 2 (CDK1 and CDK2), and the catalytic subunit of DNA-dependent protein kinase.43 These altered kinome profiles of radioresistant MCF-7/C6 cells suggest the contributions of those kinases involved in cell cycle progression and DNA repair to tumor-adaptive radioresistance. For instance, CHK1 was previously shown to promote the development of radioresistance.44,45 In addition, signaling network analysis (Figure S3) suggested that the altered HSPs (HSPC125, a.k.a. NDUFAF4, DNAJB1, and HSPB8) interact with the previously identified kinases (CHK1, CDK1, and CDK2). Therefore, these kinases may coordinate with the currently identified cluster of HSPs to form a prosurvival network that confers radioresistance in breast cancer.
CONCLUSIONS
In this study, we employed our recently developed PRM-based targeted proteomic method to examine the reprogramming of the human heat shock proteome during the development of radioresistance in breast cancer cells. We investigated the differential expression of heat shock proteins (HSPs) in two pairs of matched parental/radioresistant breast cancer cell lines. We were able to quantify 43 and 42 HSPs in the MCF-7 pair and the MDA-MB-231 pair, respectively. By comparing the commonly perturbed HSPs, we found that altered expressions of several members of the HSP70 and HSP40 protein subfamilies are accompanied with the development of radioresistance. In addition, HSPB8 is a target of NF-κB, which is a transcription factor known to promote radioresistance and is present at markedly elevated levels in both lines of radioresistant breast cancer cells; thus, HSPB8 could be critical in the development of radioresistance in breast cancer. In summary, our PRM-based targeted proteomic method constitutes a unique tool to examine the reprogramming of heat shock proteome and reveals potential targets to sensitize cancer cells toward radiation therapy.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (R01 CA210072), and M.H. was supported by a T32 Institutional Training Grant (ES018827).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.8b00330.
All the raw files for LC-PRM analyses of heat shock proteins for the paired radioresistant breast cancer cells were deposited into PeptideAtlas with the identifier number PASS01261 (http://www.peptideatlas.org/PASS/PASS01261).
Figures S1−S3 and Table S1, showing representative extracted-ion chromatograms, TCGA data, and signaling network analysis results (PDF)
Table S2, an Excel file containing the processed LC-PRM data (XLSX)
REFERENCES
- (1).Power EJ, Chin ML, and Haq MM (2018) Breast cancer incidence and risk reduction in the hispanic population. Cureus 10, No. e2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Krug D, Baumann R, Budach W, Dunst J, Feyer P, Fietkau R, Haase W, Harms W, Piroth MD, Sautter-Bihl M-L, Sedlmayer F, Souchon R, Wenz F, and Sauer R (2017) Current controversies in radiotherapy for breast cancer. Radiat. Oncol 12, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jagsi R (2014) Progress and controversies: Radiation therapy for invasive breast cancer. Ca-Cancer J. Clin. 64, 135–152. [DOI] [PubMed] [Google Scholar]
- (4).Kuwahara Y, Roudkenar MH, Urushihara Y, Saito Y, Tomita K, Roushandeh AM, Sato T, Kurimasa A, and Fukumoto M (2017) Clinically relevant radioresistant cell line: a simple model to understand cancer radioresistance. Med. Mol. Morphol 50, 195–204. [DOI] [PubMed] [Google Scholar]
- (5).Duru N, Fan M, Candas D, Menaa C, Liu H-C, Nantajit D, Wen Y, Xiao K, Eldridge A, Chromy BA, Li S, Spitz DR, Lam KS, Wicha MS, and Li JJ (2012) HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin. Cancer Res 18, 6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Feder ME, and Hofmann GE (1999) Heat shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol 61, 243–282. [DOI] [PubMed] [Google Scholar]
- (7).Baek S-H, Min J-N, Park E-M, Han M-Y, Lee Y-S, Lee YJ, and Park Y-M (2000) Role of small heat shock protein HSP25 in radioresistance and glutathione-redox cycle. J. Cell. Physiol 183, 100–107. [DOI] [PubMed] [Google Scholar]
- (8).Zhang B, Qu J-Q, Xiao L, Yi H, Zhang P-F, Li M-Y, Hu R, Wan X-X, He Q-Y, Li J-H, Ye X, Xiao Z-Q, and Feng X-P (2012) Identification of heat shock protein 27 as a radioresistancerelated protein in nasopharyngeal carcinoma cells. J. Cancer Res. Clin. Oncol 138, 2117–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Trepel J, Mollapour M, Giaccone G, and Neckers L (2010) Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, and Kluger HM (2007) High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 67, 2932. [DOI] [PubMed] [Google Scholar]
- (11).Miao W, Li L, and Wang Y (2018) A targeted proteomic approach for heat shock proteins reveals DNAJB4 as a suppressor for melanoma metastasis. Anal. Chem 90, 6835–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ahmed KM, Dong S, Fan M, and Li JJ (2006) Nuclear factor-κB p65 inhibits mitogen-activated protein kinase signaling pathway in radioresistant breast cancer cells. Mol. Cancer Res 4, 945. [DOI] [PubMed] [Google Scholar]
- (13).Cao N, Li S, Wang Z, Ahmed KM, Degnan ME, Fan M, Dynlacht JR, and Li JJ (2009) NF-κB-mediated HER2 overexpression in radiation-adaptive resistance. Radiat. Res. 171, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, and Rinner O (2012) Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Miao W, Xiao Y, Guo L, Jiang X, Huang M, and Wang Y (2016) A high-throughput targeted proteomic approach for comprehensive profiling of methylglyoxal-induced perturbations of the human kinome. Anal. Chem. 88, 9773–9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Miao W, Li L, and Wang Y (2018) Identification of Helicase Proteins as Clients for HSP90. Anal. Chem. 90, 11751–11755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Caldas C, Aparicio S, et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, and Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signaling 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, and Morris Q (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Livak KJ, and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- (22).Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, and Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386. [DOI] [PubMed] [Google Scholar]
- (23).de Graaf EL, Altelaar AF, van Breukelen B, Mohammed S, and Heck AJ (2011) Improving SRM assay development: a global comparison between triple quadrupole, ion trap, and higher energy CID peptide fragmentation spectra. J. Proteome Res 10, 4334–4341. [DOI] [PubMed] [Google Scholar]
- (24).Fedrigo CA, Grivicich I, Schunemann DP, Chemale IM, Santos D. d., Jacovas T, Boschetti PS, Jotz GP, Filho AB, and da Rocha AB (2011) Radioresistance of human glioma spheroids and expression of HSP70, p53 and EGFr. Radiat. Oncol. 6, 156–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Nguyen DH, Fredlund E, Zhao W, Perou CM, Balmain A, Mao J-H, and Barcellos-Hoff MH (2013) Murine microenvironment metaprofiles associate with human cancer etiology and intrinsic subtypes. Clin. Cancer Res 19, 1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Greve B, Bölling T, Amler S, Rössler U, Gomolka M, Mayer C, Popanda O, Dreffke K, Rickinger A, Fritz E, Eckardt-Schupp F, Sauerland C, Braselmann H, Sauter W, Illig T, Riesenbeck D, Könemann S, Willich N, Mörtl S, Eich HT, and Schmezer P (2012) Evaluation of different biomarkers to predict individual radiosensitivity in an inter-laboratory comparison−lessons for future studies. PLoS One 7, No. e47185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mayer C, Popanda O, Greve B, Fritz E, Illig T, Eckardt-Schupp F, Gomolka M, Benner A, and Schmezer P (2011) A radiation-induced gene expression signature as a tool to predict acute radiotherapy-induced adverse side effects. Cancer Lett. 302, 20–28. [DOI] [PubMed] [Google Scholar]
- (28).Alderson TR, Kim JH, and Markley JL (2016) Dynamical structures of Hsp70 and Hsp70−Hsp40 complexes. Structure 24, 1014–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Vjestica A, Zhang D, Liu J, and Oliferenko S (2013) Hsp70-Hsp40 chaperone complex functions in controlling polarized growth by repressing Hsf1-driven heat stress-associated transcription. PLoS Genet. 9, No. e1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Terada K, and Mori M (2000) Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J. Biol. Chem 275, 24728–24734. [DOI] [PubMed] [Google Scholar]
- (31).Wang C-C, Liao Y-P, Mischel PS, Iwamoto KS, Cacalano NA, and McBride WH (2006) HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 66, 6756. [DOI] [PubMed] [Google Scholar]
- (32).Wang C-C, Mischel P, and McBride W (2005) HDJ-2 is an important target of farnesyltransferase inhibitor, R115777, induced radiosensitization. Cancer Res. 65, 390. [DOI] [PubMed] [Google Scholar]
- (33).Park S-Y, Choi H-K, Seo JS, Yoo J-Y, Jeong J-W, Choi Y, Choi K-C, and Yoon H-G (2015) DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim. Biophys. Acta, Mol. Cell Res 1853, 2722–2730. [DOI] [PubMed] [Google Scholar]
- (34).Dittmann K, Mayer C, Paasch A, Huber S, Fehrenbacher B, Schaller M, and Rodemann HP (2015) Nuclear EGFR renders cells radio-resistant by binding mRNA species and triggering a metabolic switch to increase lactate production. Radiother. Oncol. 116, 431–437. [DOI] [PubMed] [Google Scholar]
- (35).Sturla L-M, Alexander MS, Valerie K, and Mikkelsen RB (2006) A novel mechanism for ionizing radiation-induced radioresistance of tumor cells involving SHP2 and nuclear localization of EGFR. Cancer Res. 66, 1029. [Google Scholar]
- (36).Crippa V, D’Agostino VG, Cristofani R, Rusmini P, Cicardi ME, Messi E, Loffredo R, Pancher M, Piccolella M, Galbiati M, Meroni M, Cereda C, Carra S, Provenzani A, and Poletti A (2016) Transcriptional induction of the heat shock protein B8 mediates the clearance of misfolded proteins responsible for motor neuron diseases. Sci. Rep 6, 22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Piccolella M, Crippa V, Cristofani R, Rusmini P, Galbiati M, Cicardi ME, Meroni M, Ferri N, Morelli FF, Carra S, Messi E, and Poletti A (2017) The small heat shock protein B8 (HSPB8) modulates proliferation and migration of breast cancer cells. Oncotarget 8, 10400–10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hamouda A, Belhacene N, Puissant A, Colosetti P, Robért G, Jacquel A, Mari B, Auberger P, and Luciano F (2014) The small heat shock protein B8 (HSPB8) confers resistance to bortezomib by promoting autophagic removal of misfolded proteins in multiple myeloma cells. Oncotarget 5, 6252–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Choi DH, Ha JS, Lee WH, Song JK, Kim GY, Park JH, Cha HJ, Lee BJ, and Park JW (2007) Heat shock protein 27 is associated with irinotecan resistance in human colorectal cancer cells. FEBS Lett. 581, 1649–1656. [DOI] [PubMed] [Google Scholar]
- (40).Gonzalez-Malerva L, Park J, Zou L, Hu Y, Moradpour Z, Pearlberg J, Sawyer J, Stevens H, Harlow E, and LaBaer J (2011) High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc. Natl. Acad. Sci. U. S. A 108, 2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, Pansarasa O, Cereda C, Poletti A, Alberti S, and Carra S (2016) A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol. Cell 63, 796–810. [DOI] [PubMed] [Google Scholar]
- (42).Nivon M, Abou-Samra M, Richet E, Guyot B, Arrigo A-P, and Kretz-Remy C (2012) NF-κB regulates protein quality control after heat stress through modulation of the BAG3−HspB8 complex. J. Cell Sci 125, 1141. [DOI] [PubMed] [Google Scholar]
- (43).Guo L, Xiao Y, Fan M, Li JJ, and Wang Y (2015) Profiling global kinome signatures of the radioresistant MCF-7/C6 breast cancer cells using MRM-based targeted proteomics. J. Proteome Res. 14, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wang W-J, Wu S-P, Liu J-B, Shi Y-S, Huang X, Zhang Q-B, and Yao K-T (2013) MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 73, 1219. [DOI] [PubMed] [Google Scholar]
- (45).Zhang Y, Lai J, Du Z, Gao J, Yang S, Gorityala S, Xiong X, Deng O, Ma Z, Yan C, Susana G, Xu Y, and Zhang J (2016) Targeting radioresistant breast cancer cells by single agent CHK1 inhibitor via enhancing replication stress. Oncotarget 7, 34688–34702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.