Abstract
Experimental populations of model organisms provide valuable opportunities to unravel the genomic impact of selection in a controlled system. The Virginia body weight chicken lines represent a unique resource to investigate signatures of selection in a system where long-term, single-trait, bidirectional selection has been carried out for more than 60 generations. At 55 generations of divergent selection, earlier analyses of pooled genome resequencing data from these lines revealed that 14.2% of the genome showed extreme differentiation between the selected lines, contained within 395 genomic regions. Here, we report more detailed analyses of these data exploring the regions displaying within- and between-line genomic signatures of the bidirectional selection applied in these lines. Despite the strict selection regime for opposite extremes in body weight, this did not result in opposite genomic signatures between the lines. The lines often displayed a duality of the sweep signatures, where an extended region of homozygosity in one line, in contrast to mosaic pattern of heterozygosity in the other line. These haplotype mosaics consisted of short, distinct haploblocks of variable between-line divergence, likely the results of a complex demographic history involving bottlenecks, introgressions and moderate inbreeding. We demonstrate this using the example of complex haplotype mosaicism in the growth1 QTL. These mosaics represent the standing genetic variation available at the onset of selection in the founder population. Selection on standing genetic variation can thus result in different signatures depending on the intensity and direction of selection.
Keywords: Chicken, White Plymouth Rock, selective sweeps, body weight, quantitative trait
Initial thinking on how adaptive processes shape the genome was modeled by Smith and Haigh (1974), who demonstrated that a beneficial mutation favored by natural selection will increase in frequency within a population. Linkage disequilibrium in the flanking region of a selected allele will result in a characteristic valley of diversity around the selected variant, known as a hard selective sweep (Kaplan et al. 1989; Stephan et al. 1992). Aside from hard selective sweeps, selection on recessive variants, variants contributing to the standing genetic variation in a population, and partial sweeps (Hermisson and Pennings 2005; Teshima and Przeworski 2006) typically have weaker effects. Empirical studies, however, suggest that this type of selection is the most abundant mode of adaptation in recent evolution in both Drosophila melanogaster (Garud et al. 2015) and humans (Pritchard et al. 2010; Hernandez et al. 2011; Schrider and Kern 2017). Furthermore, polygenic adaptation describes how selection acts on standing genetic variation across the many loci contributing to a quantitative polygenic trait leading to a new phenotypic optimum by way of modest allele frequency changes across these loci (Pritchard and Di Rienzo 2010; Pritchard et al. 2010). Although this mode of adaptation would respond very rapidly to changes in the selective environment, it would not necessarily lead to fixation for any one variant (Pritchard et al. 2010).
Modes of adaptation are not mutually exclusive, and the genomic signature that results will be dependent upon factors such as the genetic architecture of the trait, standing genetic variation available within this architecture, effective sizes of standing haplotypes, and population demography. By exploiting population genetic signals, researchers are increasingly able to detect the underlying modes of selection, from initial sweep scans that identify valleys of low diversity resulting from hard sweeps, to various recent developments to detect and differentiate between soft and hard sweeps (Berg and Coop 2014; Garud et al. 2015; Schrider and Kern 2017). Whereas previous studies have attempted to uncover selection throughout human history (Coop et al. 2009; Schrider and Kern 2017), much can be learned from research with model organisms, such as selection in experimental populations of Drosophila (Burke et al. 2010) or mice (Chan et al. 2012). In particular, long-term selection experiments have well-defined population histories, likely have stronger selection signatures in the genome due to an isolation of the trait under selection, and allow breeding of crosses to test for adaptive trait associations to candidate sweeps. The Virginia body weight chicken lines, whose history of long-term, single-trait, bi-directional selection from a common founder White Plymouth Rock population has been well-characterized, affords us the opportunity to dissect the genomic selective-sweep signatures of strongly selected loci (Figure 1).
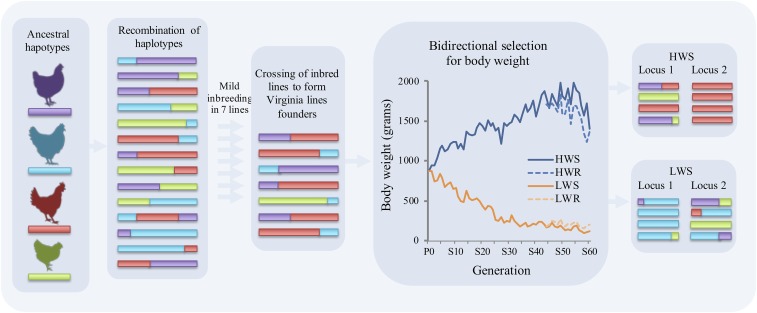
Figure 1.
Representation of the formation of genomic signatures in the Virginia body weight lines. Ancestral haplotypes that contributed to the formation of the White Plymouth Rock breed would have recombined over time. Haplotype variation was likely constrained in the seven partially inbred lines, which were crossed to form the founder population of the Virginia body weight lines. The bidirectional change in eight-week body weight in the Virginia body weight lines is presented in the graph from the parental (P0) across the selected generations (S1-S60) for the high weight selected (HWS; blue unbroken line) and low weight selected (LWS; orange unbroken line) body weight lines. Body weight within relaxed sublines are also shown for the high weight relaxed line (HWR; blue broken line) and low weight relaxed line (LWR; orange broken line) at corresponding selected generation. Haplotype frequency and ancestral haplotype origin are depicted at two hypothetical loci to the right. At locus 1, HWS continues to segregate with multiple haplotypes, whereas LWS is fixed for an extended blue haplotype. At locus 2, HWS is fixed for an extended red haplotype, whereas LWS continues to segregate with multiple haplotypes.
Founders for the Virginia lines were generated via crossing of seven partially inbred White Plymouth Rock lines, in effect constraining the standing diversity. From this base population in 1957, bidirectional selection for 8-week body weight was carried out to form the high (HWS) and low (LWS) Virginia body weight lines. The breeding regime structured to minimize inbreeding and minimize the stochastic fixation of alleles that could affect small breeding populations (Siegel 1962; Marquez et al. 2010). Relaxed sublines for both HWS and LWS were produced from selected generation 44 (high weight relaxed: HWR; low weight relaxed: LWR) (Dunnington et al. 2013). After 55 generations of divergent selection, a 15-fold difference in body weight exists between the lines (Jambui et al. 2017b). With its well-defined population history, bi-directional single trait selection regime, and well-defined polygenic architecture of the adaptive trait, the Virginia body weight lines represent an invaluable resource to investigate the genomic signatures of selection.
Previous research has demonstrated that standing genetic variations in many loci from the founder population of the Virginia lines contribute to the observable difference in body weight. Sweep scans using individual genotypes from a 60k SNP-chip have revealed numerous regions of differentiation between the selected lines (Johansson et al. 2010; Pettersson et al. 2013). These were explored for associations with body weight and a large number of them were found to have contributed to selection response using available standing genetic variation (Sheng et al. 2015). Using genotype data from these sweeps and earlier identified QTL, together with phenotypic data from an F15 intercross between the lines, 20 independent associations to 8-week body weight have been confirmed (Zan et al. 2017).
Recently, pooled genome resequencing was applied to several generations from these lines to investigate whether these data could reveal any additional features of the genomic impacts of bidirectional selection on body weight to those previously found using the 60k SNP-chip data. An initial evaluation of these data were integrated into a review of the knowledge gained from multiple lines of inquiry in the Virginia body weight lines, with a particular focus on identified QTL regions (Lillie et al. 2018). Here, performed is a more in-depth analysis of this pooled genome resequencing dataset to characterize the genomic signatures of highly differentiated regions across the whole genome between the bidirectionally-selected lines: the putative selective sweeps. As earlier shown for the QTL regions (Lillie et al. 2018), between-line differentiation was seldom due to complete fixation of different haplotypes in the lines. Instead, most selective sweeps resulted from extended runs of homozygosity in one line, contrasting to persistence of heterozygosity in the other. This duality of selection signatures and haplotype structures was illustrated by dissecting the complex differentiation in an earlier identified QTL on chromosome 1. Typically, this heterozygous region was comprised of multiple, distinct regions, with variable diversity, and between-line divergence. The directions of these relationships do not suggest any line bias, suggesting that the physiological plateau since generation 35 in the LWS due to a disruption of food-consumption and an inability to enter egg production at less than 1000 g (Siegel and Dunnington 1985; Siegel and Dunnington 1987; Jambui et al. 2017a) has not had a major influence on these patterns.
These empirical observations and our knowledge about the history of these populations imply that the duality of these signatures genome-wide reflect positive selection for one large-effect haplotype in one line while in the other line negative selection would remove this haplotype, allowing other haplotypes to continue to segregate. Mosaic haplotype structures within these segregating regions reflect standing variation in the founder population, likely resulting from ancestral haplotype recombination along a history of bottlenecking, inbreeding, and crossbreeding.
Materials and Methods
Virginia body weight chicken lines
All animal procedures were carried out by experienced handlers and in accordance with the Virginia Tech Animal Care Committee animal use protocols (IACUC-15-136). The Virginia body weight lines were formed from a founder population resulting from crossing seven lines originating in 1949 that had undergone mild inbreeding. Established in 1957, bidirectional selection for body weight at 8 weeks of age was initiated to produce the closed selected lines: high weight selected (HWS) and low weight selected (LWS) (Siegel 1962). Breeding focused on a response to selection, while attempting to minimize inbreeding (Marquez et al. 2010). Effective population sizes in the LWS and HWS lines have been estimated as 38.3 and 32.1, respectively (Marquez et al. 2010). Relaxed sublines for both HWS and LWS were produced from selected generation 44, and are referred to as high weight relaxed (HWR) and low weight relaxed (LWR) (Dunnington et al. 2013). All generations were hatched in the same incubators and reared in the same pens on the same diet. Pooled semen was used to produce each generation of relaxed lines.
Sequencing and genome alignments
The sequencing data used in this dataset was originally reported in (Lillie et al. 2018). In short, DNA for the genomic analyses was prepared from blood samples collected from 9-30 individuals from each line and pooled in equimolar ratios prior to library construction. Genome sequencing library construction and sequencing was carried out by SciLifeLab (Uppsala, Sweden) using two lanes on an Illumina Hiseq 2500. Reads were aligned to the Gallus gallus genome (Galgal5; INSDC Assembly GCA_000002315.3, Dec 2015) using BWA (Li and Durbin 2009). Genomes were sorted and duplicates were marked and removed with Picard (v1.92; http://picard.sourceforge.net). GATK (v3.3.0; McKenna et al. 2010) was used for realignment around indels. GATK UnifiedGenotyper was used to generate allele calls at all sites (option: emit all sites) and with ploidy = 30 (18 for LWS generation 50 as only 9 individuals went into this pool) to account for the pooled genome sample. Sites were filtered to only include those with >10 and <100 reads, wherefrom allele frequency, heterozygosity, and pairwise FST between all populations were calculated. Samtools (Li et al. 2009; v1.1; Li 2011) was used to generate mpileup files for PoPoolation2 (v1.201; Kofler et al. 2011), which was used to calculate FST over 1000 bp sliding windows with 50% overlaps between the population samples using the Karlsson et al. (2007) method, with minimum count 3, minimum coverage 10, maximum coverage 100, and minimum coverage fraction 1. Genome alignments were visualized in IGV (v2.3.52; Robinson et al. 2011; Thorvaldsdottir et al. 2013).
Differentiated regions
As reported in (Lillie et al. 2018), differentiated regions were identified by employing an empirical FST threshold of 0.953, representing the top 5% FST values in generation 55. Windows with FST values above this threshold were clustered into differentiated regions when they were less than 100 kb from one another. Clusters with less than 2 SNPs or less than 100 kb were removed from the dataset to retain only the stronger candidate regions. Mean and median heterozygosity were calculated for each line within each differentiated region. We used the Variant Effect Predictor (VEP) (McLaren et al. 2016) available from Ensembl (Aken et al. 2017) to investigate potential functionality of candidate alleles. Haplotype structure within regions of interest were visualized using adjusted allele frequencies (Lillie et al. 2017) (similar to the allele polarization step in the haplotype-block reconstruction approach used by Franssen et al. 2017). This approach adjusts allele frequencies within the sequenced lines to the generation of lowest haplotypic complexity, such that allele frequencies across all lines would be adjusted to 1-AF, for sites where allele frequency > 0.5 in the generation of lowest haplotypic complexity. In most cases, the generation of lowest haplotypic complexity contains one fixed extended haplotype within the region of interest, with raw allele frequencies equal to ∼0 or ∼1, generating adjusted allele frequencies equal to ∼0. These were then plotted using custom R scripts.
Data availability
Pooled genome data generated for this study are available via Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) under bioProject: PRJNA516366; bioSample: SAMN10787895; and accessions: SRR8480632-SRR8480641. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7674281.
Results
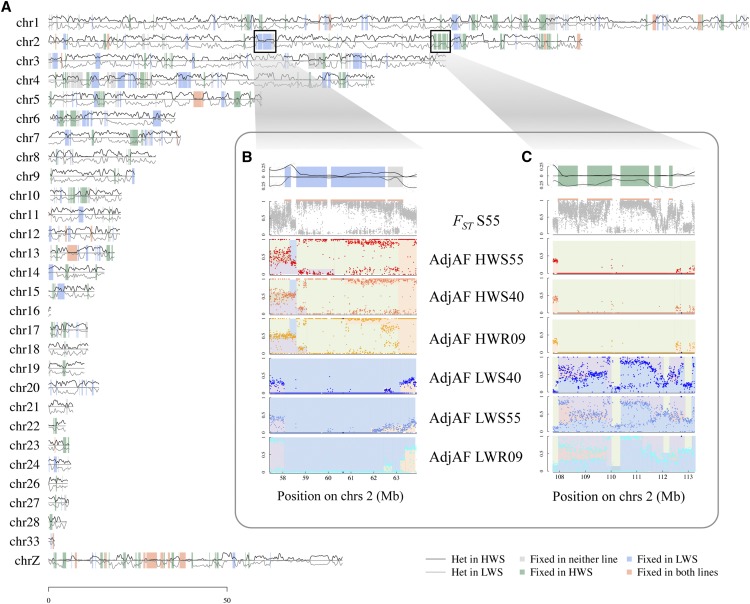
Alignment coverage of the reference genome after read alignment was between 91.22% and 91.44% across the sequenced pools. As reported earlier (Lillie et al. 2018), 395 differentiated regions between the HWS and LWS lines in selected generation 55 were identified from the clusters of high differentiation (FST > 0.953; Figure 2). These regions covered a total of 174.5 Mb, or 14.2% of the genome, which is an increase from the 244 differentiated regions identified in selected generation 40 (99.6 Mb or 8.1% of the genome; Figure S1).
Figure 2.
Heterozygosity across chromosomes of divergently selected Virginia body weight chicken lines. A. Heterozygosity in generation 55 for high weight selected (HWS; black line above x-axis) and low weight selected (LWS; gray line below x-axis) presented across the chicken chromosomes within the differentiated regions shaded using color code: gray where neither line is fixed (); green where only HWS is fixed (); blue where only LWS is fixed within the differentiated region; red where both lines are fixed. B. FST and heterozygosity patterns across chromosome 2:58-63 Mb for selected generations 55 and 40 and relaxed generation 9. C. FST and heterozygosity patterns across chromosome 2:108-113 Mb for selected generations 55 and 40 and relaxed generation 9. Panels within B and C insets: Panel 1: Detail of heterozygosity trace from chromosome map. Panel 2: Mean FST within 1 kb windows between HWS and LWS at generation 55 indicated with gray points; region of differentiation indicated with the orange line above FST plot; allele frequencies of SNP markers with association to body weight indicated with blue diamonds. Panel 3-8: Mean adjusted allele frequency (adjAF) of 5kb windows in HWS generation 55 (HWS55) / HWS generation 40 (HWS40) / high weight relaxed generation 9 (HWR9) / LWS generation 55 (HLWS55) / LWS generation 40 (LWS40) / low weight relaxed generation 9 (LWR9). Shaded colors within these plots have been used to highlight the runs of adjusted allele frequencies that contribute to different haploblocks.
Candidate selective sweeps are often polymorphic in one of the selected lines
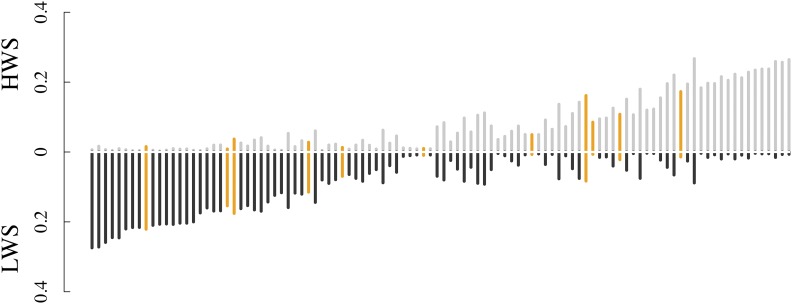
Compared to the genome-wide trend, there was a decline in heterozygosity at the extremes of high FST (S2 Figure). Few regions of differentiation showed fixation in both lines; rather, it was often the case that there was fixation across an extended region in one line, while many nucleotide positions in the region still segregated in the other (Figure 3; S3 Figure). Of the differentiated regions greater than 0.5 Mb in length, 34 regions (33%) were close to fixation in HWS (mean heterozygosity <= 0.1) while LWS continued to segregate (mean heterozygosity > 0.1), 33 regions (32%) were close to fixation in LWS (mean heterozygosity <= 0.1) while HWS continued to segregate (mean heterozygosity > 0.1), and 37 regions (35%) were close to fixation for alternative haplotypes in both lines (mean heterozygosity <= 0.1 in both). This demonstrated that, while one extended haplotype was fixed in one line, multiple haplotypes continue to segregate in the other. Fixation was as common in HWS as LWS, a trend that extended to regions with confirmed associations for the selected trait, 8-week body weight (Figure 3; S3 Figure) (Zan et al. 2017).
Figure 3.
Mean heterozygosity in HWS (above the x-axis; gray) and LWS (below the x-axis; black) at generation 55 within differentiated regions greater than 0.5 Mb in length. Differentiated regions overlapping known associations with 8-week body weight (Zan et al. 2017) are indicated in orange. Regions presented are sorted in order of increasing cumulative heterozygosity across both lines with increased heterozygosity in HWS.
Mosaic haplotypes in the growth1 QTL
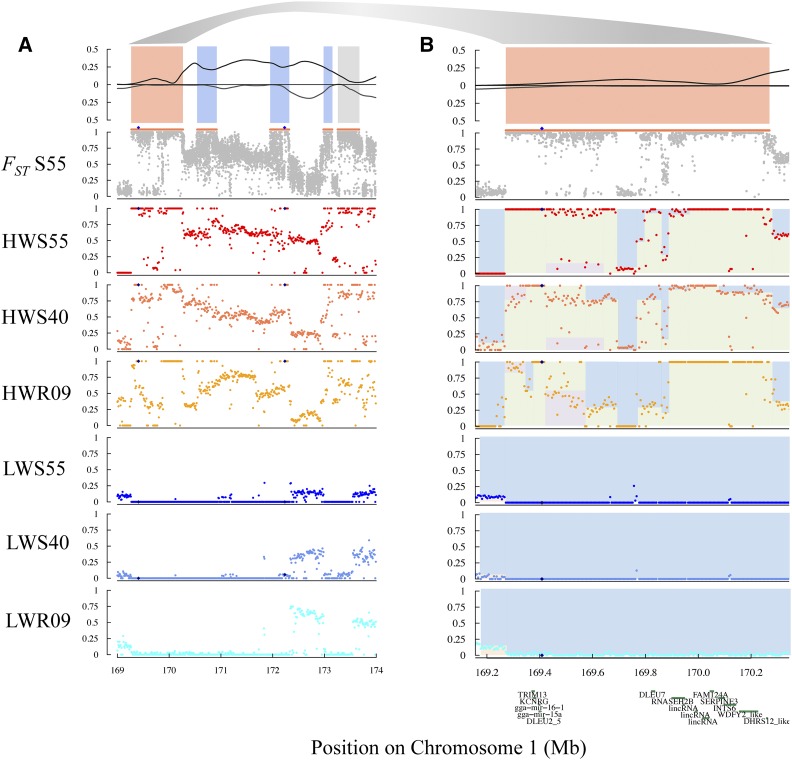
To illustrate the genomic mosaicism observed in many differentiated regions, we investigated the growth1 QTL in detail (Jacobsson et al. 2005; Zan et al. 2017). A long region of divergence was observed between 169.3 Mb and 173.7 Mb on chromosome 1 (in total 4.4 Mb), where a single extended haplotype was close to fixation in LWS by generation 40, whereas the pattern of polymorphism in the HWS suggest that multiple haplotypes segregate in this line (Figure 4). Approximately 28% of the nucleotide positions within this region were highly divergent between HWS and LWS at generation 55. In total, 10,148 from 36,934 sites had differences between the lines in allele frequency that were greater than 0.9. This level of divergence implies that the long haplotype fixed in LWS is not present in the HWS at generation 55 because it has been selected out.
Figure 4.
A. FST and heterozygosity patterns across chromosome 1:169-174 Mb for selected generations 55 and 40 and relaxed generation 9 of the Virginia body weight chicken lines. B. FST and heterozygosity patterns across chromosome 1:169.2-170.3 Mb for selected generations 55 and 40 and relaxed generation 9 with gene positions. Panel 1: Heterozygosity. Panel 2: Mean FST within 1 kb windows (gray points) in selected generation 55; defined region of differentiation indicated with orange line above FST plot; blue diamonds indicate allele frequencies of SNP markers with significant association to body weight from Zan et al. (2017) . Panel 3-8: Mean adjusted allele frequency of 5kb windows in HWS55 (panel 3), HWS40 (panel 4), HWR09 (panel 5), LWS55 (panel 6), LWS40 (panel 7), LWR09 (panel 8). Colors within the plots are used to highlight the runs of adjusted allele frequencies that contribute to different haploblocks.
We checked for the presence of a known insertion that has been previously associated with increased body weight in other chicken populations (Jia et al. 2016). By examining the soft-clipped reads (S4 Figure), we observed that this variant also segregated in our population within the short region fixed for divergent haplotypes in both LWS and HWS (approximately 50 kb in length; GGA1: 169,370,000-169,420,000). Contrary to the expectation that this insertion would dominate in the HWS line, it was instead present on the long haplotype fixed by generation 40 in LWS, and entirely absent in HWS. Furthermore, this insertion is linked to the LWS allele of SNP marker rs14916997, which was associated with low body weight (Zan et al. 2017).
To identify candidate linkages and thus other potential functional variants, the growth1 QTL was evaluated for further sub-haplotype fixations. In the nearby region (GGA1: 169,950,000-170,220,000), the HWS and LWS were also fixed by generation 55. Bounded by these two highly differentiated regions are 15 annotated genes (S1 Table), with 8 missense mutations (S2 Table). One missense mutation with predicted (SIFT) deleterious effect was present in HWS, located within the sixth exon of Ribonuclease H2 Subunit B gene (RNASEH2B), as well as the small (18 bp) deletion in LWS at GGA1: 169,407,811, which was absent from HWS.
Discussion
There was an overall increase (244 to 395) in the number of differentiated regions between the HWS and LWS from generation 40 to generation 55. This increase is consistent with the continuing phenotypic divergence between the lines resulting from the ongoing response to selection in the HWS (average 56-day body-weights 1264 g and 1506 g, respectively) contrasted to the plateau in the LWS at 142 g (Lillie et al. 2018).
Candidate selective sweeps were often polymorphic in one of the selected lines
Within highly differentiated candidate selective sweep regions, we observed that an extended haplotype was often fixed in one line while multiple haplotypes continued to segregate in the other, thus resembling a duality in these sweep signatures. These patterns are likely to be shaped by the intensity of selection on the locus, as well as the haplotype frequencies that were present in the base population. For example, haplotypes with equal but opposite effects present at the same frequencies at the onset of selection would be expected to result in relatively equal lengths of fixation (homozygosity) in both lines, and thus a high FST. This pattern was observed only a few times (Figure 3), and thus appears to have been a relatively rare event. More likely is that the haplotypes in the founder population had different effect sizes and were present at different frequencies when selection was initiated to develop the high and low body weight chicken lines. The long runs on homozygosity found only in one line within regions of differentiation therefore likely represent the haplotypes with the largest effect, with the signature possibly being amplified by being at a low frequency at the onset of selection. These patterns suggest that the effect size of functional variants, as well as founding haplotype frequencies, are likely to have influenced the selective signature in the genome of the Virginia body weight chicken lines. The lack of line-bias in these signatures suggest that the selection plateau reached in the LWS at about generation 35, that physiologically is due to a disrupted food-consumption in some birds and an inability to enter egg production at less than 1000 g (Siegel and Dunnington 1985; Siegel and Dunnington 1987; Jambui et al. 2017a), has not made any major impact on these patterns.
A challenge in selective sweep studies is to discriminate which differentiated regions are likely due to drift or selection. The influence of drift on these lines has been investigated in a previous study, which showed that the population size was sufficiently large to prevent genetic drift from overriding the effect of selection for the loci with the larger effects and that the probability of fixation for alternative haplotypes by drift was very low (Johansson et al. 2010). Many regions of differentiation are present both within and outside of those known to be associated with body-weight (Sheng et al. 2015; Lillie et al. 2018; Zan et al. 2017). Although it is known that the already associated regions only explain part of the variation in body-weight (Zan et al. 2017), the observation made here that fixation for alternative haplotypes is seldom the case even within regions of differentiation makes it difficult to quantify the expected influence of drift in shaping genomic signatures. By implementing a high FST cutoff, integrating association mapping results from previous studies in these lines, and comparing gene ontology and associations from other chicken populations, we have endeavored to build more confidence in differentiated regions than can be obtained separately using the approaches.
Mosaic haplotypes
Domestication of the chicken began roughly 7,000 years ago, predominately from red junglefowl, Gallus gallus (West and Zhou 1988; Sawai et al. 2010), but potentially also with contributions from Gallus sonneratii (gray) and Gallus layafetii (Sri Lankan) (Eriksson et al. 2008; Groeneveld et al. 2010; Tixier-Boichard et al. 2011). With subsequent distribution of chickens via human dispersal and trade, local breed formation would subject established chicken populations to genetic drift and selection to suit the environment and human-imposed selection for breed standards and production traits (Lyimo et al. 2014). Regardless of expectations that founder effects, bottlenecks, and selection would impact genetic diversity, nucleotide diversity and substitution rates were comparable for red jungle fowl and domestic chickens. This implies that domestication did not result overall in a substantial genome-wide loss of diversity in the species and had only a minor effect in an evolutionary context (Tixier-Boichard et al. 2011).
Thus, recombination of divergent haplotypes from European and Asian populations occurred while producing the WPR breed, forming the genetic substrate available for artificial selection for high and low body weight (Figure 1). During the several decades of WPR breeding that preceded the initiation of the Virginia body weight chicken lines, recombinant haplotypes are likely to have arisen, ultimately producing finely grained mosaics of the chromosomes entering the founders of the Virginia body weight lines. It is on this standing variation which the bi-directional selection experiment has acted (Figure 1). Short haplotype mosaics within the longer selective sweep regions were revealed when comparing the HWS and LWS genomes. The growth1 QTL region on chromosome 1 is a clear example, illustrating the haplotype mosaicism of sweep signatures observed across the genome.
Mosaic haplotypes in the growth1 QTL
The growth1 QTL was first defined in a microsatellite-based analysis of an F2 cross between the HWS and LWS, with a length of almost 110 cM (Jacobsson et al. 2005). This QTL was confirmed in SNP-based analyses of the F2 (Wahlberg et al. 2009) and fine mapped in the F2-F8 (Besnier et al. 2011; Brandt et al. 2016) and the F15 generation of the Advanced Intercross Line (Zan et al. 2017). In this latest study, two associations were found within growth1, with the SNP rs14916997 (GGA1: 169,408,309) having the strongest association (Zan et al. 2017). From the pooled genome data, it is evident that a major haplotype, between 169.3 Mb and 173.7 Mb (in total 4.4 Mb), is close to fixation in LWS (Figure 4). This haplotype was already close to fixation by generation 40, possibly indicating that selection has been relatively strong, and the resulting pattern resembles a hard sweep in LWS. Contrastingly, multiple haplotypes segregate in HWS, with a mixture of nucleotide positions that are divergently fixed when compared to LWS, intermixed with positions that segregate for both LWS and unique HWS alleles.
We observed a high level of divergence between HWS and LWS within this region, implying that the long haplotype fixed in LWS is not present in the HWS at generation 55 because it was selected out of this line. Highly dissimilar regions are intermixed with short, but continuous, stretches where the HWS and LWS haplotypes are nearly identical. Additionally, the boundaries between these shared and differentiated regions are distinct. They likely represent historical recombination events shared between one or more selected haplotypes, rather than being multiple classic hard sweeps, as these are expected to result in a gradual breakdown of population-wide linkage disequilibrium.
This interpretation is supported by other observations. First, for sharing of such interspersed haplotypes to be possible between the lines, formative recombination events must have occurred prior to the onset of bidirectional selection. Second, as several shared haplotype segments were sometimes observed in the divergent regions, such multiple events must have happened on the same haplotypes. Third, because the shared regions are often short (10s to a few 100 kb), the events are unlikely in a population with as small a population-size as the Virginia body weight chicken lines (effective population size, Ne ∼35) (Marquez et al. 2010). Finally, if the recombination events occurred during the selection experiment, selection on the recombinant haplotypes must have been strong in order to only retain them in the lines. Such is unlikely given the highly polygenic genetic architecture of 8-week body weight and the dilution of selection pressure across the many loci. Therefore, these haplotype mosaics most likely represent recombinant founder haplotypes resulting from their population history, including the formation of the WPR as a breed. Although recombination cannot be completely ruled out as a contributor to haplotype mosaicism, without individual-based sequencing, we cannot confirm or eliminate a role for recombination events in HWS and LWS.
Positive selection of this long haplotype to fixation in the LWS, coupled with negative selection for its removal from HWS, may serve as an explanation for why previous studies have seen a transgressive effect on 8-week body weight for SNP markers within this region. Zan et al. (2017) reported that while the HWS allele at the rs14916997 SNP marker (GGA1:169,408,309 bp) was associated with an increased 8-week body weight (additive genetic effect approximately 26 grams), the HWS allele at the nearby SNP marker rs316102705 (GGA1: 172,235,208 bp) was associated with a decrease in 8-week body weight (additive genetic effect of approximately -7 grams).
Notably, this region on chicken chromosome GGA1 appears often in association studies carried out in other populations, including comb traits (Shen et al. 2016), egg weight (Yi et al. 2015), feed intake (Yuan et al. 2015), abdominal fat percentage (Abasht and Lamont 2007; Sheng et al. 2013), shank metrics (Sheng et al. 2013), and growth and body weight at numerous life stages (Xie et al. 2012; Sheng et al. 2013; Abdalhag et al. 2015; Zhang et al. 2015). This may reflect a shared ancestral variant that has spread in domestic populations due to its beneficial effect, or that this is a gene rich region associated with many functionally important genes. Recently, an insertion (GGA1: 169,399,420) located upstream of the miR-15a-16 precursor was strongly associated with growth traits in an Xinghua & White Recessive Rock F2 cross, where presence of this variant results in an altered hairpin formation, reduction of miR-16 expression, and increased body weight, bone size, and muscle mass (Jia et al. 2016). Although this insertion was also present in multiple chicken breeds, and at high frequencies in broiler breeds (Jia et al. 2016), in our population, the insertion was present in LWS on the long, fixed haplotype, and was absence from the HWS, contrary to expectations that the insertion would substantially increase body weight.
This observation may be explained by alternative hypotheses; i.e., that this insertion: i) has an effect also in our lines but is linked to another polymorphism with a stronger opposite effect, ii) does not have an effect in our lines due to genetic background, iii) does not have an effect at all, suggesting that earlier reports revealed association between this polymorphism and the studied traits, rather than causation. Alternatively, as is often the case, the functional variant may lie outside coding regions. Nevertheless, further work within this QTL region will be required to fully characterized the functional variant responsible for the large difference in 8-week body weight in the Virginia body weight lines.
Conclusions
Over the course of long-term bidirectional selection, the Virginia body weight lines have experienced significant changes impacting behavioral, neurological, metabolic, and developmental processes (Dunnington and Siegel 1996). With a concerted effort to understand the fundamental genomics underlying these changes, we have employed linkage and association mapping, selective-sweep analyses using high-density SNP data and pooled genome sequencing across generations of the selected lines, relaxed lines, and a derived advanced intercross line (Wahlberg et al. 2009; Johansson et al. 2010; Pettersson and Carlborg 2010; Besnier et al. 2011; Pettersson et al. 2011; Ahsan et al. 2013; Pettersson et al. 2013; Sheng et al. 2015; Brandt et al. 2016). It has become clear that the difference in body weight between the selected lines relies on small to moderate effects from many loci, with the most recent analysis revealing 20 contributing loci (Zan et al. 2017).
Pooled genome resequencing revealed distinct hallmarks of selection in regions that were highly differentiated in the lines after 55 generations of long-term experimental selection. More often than not, the regions show the persistence of haplotypic diversity in one line, contrasted by fixation in the other, as demonstrated in the growth1 QTL region. Despite selection acting on the same pool of standing genetic variants, the genomic signatures of selection thus resemble classic hard sweeps in one line, contrasted to a mosaic pattern of divergence in the other. This haplotype mosaic emerges from recombination of multiple divergent founder haplotypes, probably shaped by historical bottlenecks, crossbreeding, and inbreeding, which forms the standing genetic variation available in this selection experiment.
ACKNOWLEDGMENTS
For help with genome sequencing, we thank Uppsala University SNP&SEQ Technology Platform (Science for Life Laboratory), a national infrastructure supported by the Swedish Research Council (VR-RFI) and the Knut and Alice Wallenberg Foundation. For computational resources, we thank the Uppsala Multidisciplinary Center for Advanced Computational Science and the associated Next generation sequencing Cluster and Storage (UPPNEX) project, funded by the Knut and Alice Wallenberg Foundation and the Swedish National Infrastructure for Computing (SNIC; project id: b2015010). This work was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (DNR 221-2013-450 to ÖC) and various sources at Virginia Tech. Lastly, individuals at Virginia Tech for a range of roles in producing and maintaining the chickens across more than half a century.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7674281.
Communicating editor: D. J. de Koning
Literature Cited
- Abasht B., Lamont S. J., 2007. Genome-wide association analysis reveals cryptic alleles as an important factor in heterosis for fatness in chicken F2 population. Anim. Genet. 38: 491–498. 10.1111/j.1365-2052.2007.01642.x [DOI] [PubMed] [Google Scholar]
- Abdalhag M. A., Zhang T., Fan Q. C., Zhang X. Q., Zhang G. X., et al. , 2015. Single nucleotide polymorphisms associated with growth traits in Jinghai yellow chickens. Genet. Mol. Res. 14: 16169–16177. 10.4238/2015.December.8.6 [DOI] [PubMed] [Google Scholar]
- Ahsan M., Li X., Lundberg A. E., Kierczak M., Siegel P. B., et al. , 2013. Identification of candidate genes and mutations in QTL regions for chicken growth using bioinformatic analysis of NGS and SNP-chip data. Front. Genet. 4: 226 10.3389/fgene.2013.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aken B. L., Achuthan P., Akanni W., Amode M. R., Bernsdorff F., et al. , 2017. Ensembl 2017. Nucleic Acids Res. 45: D635–D642. 10.1093/nar/gkw1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. J., Coop G., 2014. A population genetic signal of polygenic adaptation. PLoS Genet. 10: e1004412 10.1371/journal.pgen.1004412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnier F., Wahlberg P., Rönnegård L., Ek W., Andersson L., et al. , 2011. Fine mapping and replication of QTL in outbred chicken advanced intercross lines. Genet. Sel. Evol. 43: 3 10.1186/1297-9686-43-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt M., Ahsan M., Honaker C. F., Siegel P. B., Carlborg Ö., 2016. Imputation-based fine-mapping suggests that most QTL in an outbred chicken advanced intercross body weight line are due to multiple, linked loci. G3 (Bethesda). 10.1534/g3.116.036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R., et al. , 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467: 587–590. 10.1038/nature09352 [DOI] [PubMed] [Google Scholar]
- Chan Y. F., Jones F. C., McConnell E., Bryk J., Bunger L., et al. , 2012. Parallel selection mapping using artificially selected mice reveals body weight control loci. Curr. Biol. 22: 794–800. 10.1016/j.cub.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Coop G., Pickrell J. K., Novembre J., Kudaravalli S., Li J., et al. , 2009. The role of geography in human adaptation. PLoS Genet. 5: e1000500 10.1371/journal.pgen.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnington E. A., Honaker C. F., McGilliard M. L., Siegel P. B., 2013. Phenotypic responses of chickens to long-term, bidirectional selection for juvenile body weight-Historical perspective. Poult. Sci. 92: 1724–1734. 10.3382/ps.2013-03069 [DOI] [PubMed] [Google Scholar]
- Dunnington E. A., Siegel P. B., 1996. Long-term divergent selection for eight-week body weight in White Plymouth Rock chickens. Poult. Sci. 75: 1168–1179. 10.3382/ps.0751168 [DOI] [PubMed] [Google Scholar]
- Eriksson J., Larson G., Gunnarsson U., Bed’hom B., Tixier-Boichard M., et al. , 2008. Identification of the Yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4: e1000010 10.1371/journal.pgen.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen S. U., Barton N. H., Schlötterer C., 2017. Reconstruction of haplotype-blocks selected during experimental evolution. Mol. Biol. Evol. 34: 174–184. 10.1093/molbev/msw210 [DOI] [PubMed] [Google Scholar]
- Garud N. R., Messer P. W., Buzbas E. O., Petrov D. A., 2015. Recent selective sweeps in north american Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11: e1005004 10.1371/journal.pgen.1005004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld L. F., Lenstra J. A., Eding H., Toro M. A., Scherf B., et al. , 2010. Genetic diversity in farm animals - a review. Anim. Genet. 41: 6–31. 10.1111/j.1365-2052.2010.02038.x [DOI] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. 10.1534/genetics.104.036947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924. 10.1126/science.1198878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson L., Park H. B., Wahlberg P., Fredriksson R., Perez-Enciso M., et al. , 2005. Many QTLs with minor additive effects are associated with a large difference in growth between two selection lines in chickens. Genet. Res. 86: 115–125. 10.1017/S0016672305007767 [DOI] [PubMed] [Google Scholar]
- Jambui M., Honaker C. F., Siegel P. B., 2017a Correlated responses to long-term divergent selection for 8-week body weight in female White Plymouth Rock chickens: Sexual maturity. Poult. Sci. 96: 3844–3851. 10.3382/ps/pex224 [DOI] [PubMed] [Google Scholar]
- Jambui M., Honaker C. F., Siegel P. B., 2017b Selection for juvenile body weight in chickens: Standardizing for scaling. Poult. Sci. 96: 2562–2568. 10.3382/ps/pex080 [DOI] [PubMed] [Google Scholar]
- Jia X. Z., Lin H. R., Nie Q. H., Zhang X. Q., Lamont S. J., 2016. A short insertion mutation disrupts genesis of miR-16 and causes increased body weight in domesticated chicken. Sci. Rep. 6: 36433 10.1038/srep36433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Pettersson M. E., Siegel P. B., Carlborg Ö., 2010. Genome-wide effects of long-term divergent selection. PLoS Genet. 6: e1001188 10.1371/journal.pgen.1001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The hitchhiking effect revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson E. K., Baranowska I., Wade C. M., Salmon Hillbertz N. H. C., Zody M. C., et al. , 2007. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 39: 1321–1328. 10.1038/ng.2007.10 [DOI] [PubMed] [Google Scholar]
- Kofler R., Orozco-terWengel P., De Maio N., Pandey R. V., Nolte V., et al. , 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6: e15925 10.1371/journal.pone.0015925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie M., Sheng Z. Y., Honaker C. F., Andersson L., Siegel P. B., et al. , 2018. Genomic signatures of 60 years of bidirectional selection for 8-week body weight in chickens. Poult. Sci. 97: 781–790. 10.3382/ps/pex383 [DOI] [PubMed] [Google Scholar]
- Lillie M., Sheng Z. Y., Honaker C. F., Dorshorst B. J., Ashwell C. M., et al. , 2017. Genome-wide standing variation facilitates long-term response to bidirectional selection for antibody response in chickens. BMC Genomics 18: 99 10.1186/s12864-016-3414-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyimo C. M., Weigend A., Msoffe P. L., Eding H., Simianer H., et al. , 2014. Global diversity and genetic contributions of chicken populations from African, Asian and European regions. Anim. Genet. 45: 836–848. 10.1111/age.12230 [DOI] [PubMed] [Google Scholar]
- Marquez G. C., Siegel P. B., Lewis R. M., 2010. Genetic diversity and population structure in lines of chickens divergently selected for high and low 8-week body weight. Poult. Sci. 89: 2580–2588. 10.3382/ps.2010-01034 [DOI] [PubMed] [Google Scholar]
- Smith J. M., Haigh J., 1974. The Hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. 10.1017/S0016672300014634 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R., et al. , 2016. The Ensembl Variant Effect Predictor. Genome Biol. 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M., Besnier F., Siegel P. B., Carlborg Ö., 2011. Replication and explorations of high-order epistasis using a large advanced intercross line pedigree. PLoS Genet. 7: e1002180 10.1371/journal.pgen.1002180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M. E., Carlborg O., 2010. Dissecting the genetic architecture of complex traits and its impact on genetic improvement programs: lessons learnt from the Virginia chicken lines. Rev. Bras. Zootec. 39: 256–260. 10.1590/S1516-35982010001300028 [DOI] [Google Scholar]
- Pettersson M. E., Johansson A. M., Siegel P. B., Carlborg Ö., 2013. Dynamics of adaptive alleles in divergently selected body weight lines of chickens. G3 (Bethesda) 3: 2305–2312. 10.1534/g3.113.008375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Di Rienzo A., 2010. Adaptation - not by sweeps alone. Nat. Rev. Genet. 11: 665–667. 10.1038/nrg2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The Genetics of Human Adaptation: Hard Sweeps, Soft Sweeps, and Polygenic Adaptation. Curr. Biol. 20: R208–R215. 10.1016/j.cub.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H., Kim H. L., Kuno K., Suzuki S., Gotoh H., et al. , 2010. The origin and genetic variation of domestic chickens with special reference to junglefowls Gallus g. gallus and G. varius. PLoS One 5: e10639 10.1371/journal.pone.0010639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Kern A. D., 2017. Soft sweeps are the dominant mode of adaptation in the human genome. Mol. Biol. Evol. 34: 1863–1877. 10.1093/molbev/msx154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M. M., Qu L., Ma M., Dou T. C., Lu J., et al. , 2016. Genome-wide association studies for comb traits in chickens. PLoS One 11: e0159081 10.1371/journal.pone.0159081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z. Y., Pettersson M. E., Honaker C. F., Siegel P. B., Carlborg Ö., 2015. Standing genetic variation as a major contributor to adaptation in the Virginia chicken lines selection experiment. Genome Biol. 16: 219 10.1186/s13059-015-0785-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z., Pettersson M. E., Hu X., Luo C., Qu H., et al. , 2013. Genetic dissection of growth traits in a Chinese indigenous × commercial broiler chicken cross. BMC Genomics 14: 151 10.1186/1471-2164-14-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel P. B., 1962. Selection for body weight at 8 weeks of age. 1. Short term response and heritabilities. Poult. Sci. 41: 954–962. 10.3382/ps.0410954 [DOI] [Google Scholar]
- Siegel P. B., Dunnington E. A., 1985. Reproductive complications associated with selection for broiler growth, Poultry Genetics and Breeding, edited by Hill W. G., Mason J. M., Hewitt D. British Poultry Sci. Ltd. Longman Group, Harlow. [Google Scholar]
- Siegel P. B., Dunnington E. A., 1987. Selection for growth in chickens. Critical Reviews in Poultry Biology 1: 1–24. [Google Scholar]
- Stephan W., Wiehe T. H. E., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism - analytical results based on diffusion-theory. Theor. Popul. Biol. 41: 237–254. 10.1016/0040-5809(92)90045-U [DOI] [Google Scholar]
- Teshima K. M., Przeworski M., 2006. Directional positive selection on an allele of arbitrary dominance. Genetics 172: 713–718. 10.1534/genetics.105.044065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier-Boichard M., Bed’hom B., Rognon X., 2011. Chicken domestication: From archeology to genomics. C. R. Biol. 334: 197–204. 10.1016/j.crvi.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Wahlberg P., Carlborg Ö., Foglio M., Tordoir X., Syvanen A. C., et al. , 2009. Genetic analysis of an F(2) intercross between two chicken lines divergently selected for body-weight. BMC Genomics 10: 248 10.1186/1471-2164-10-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West B., Zhou B. X., 1988. Did chickens go north - New evidence for domestication. J. Archaeol. Sci. 15: 515–533. 10.1016/0305-4403(88)90080-5 [DOI] [Google Scholar]
- Xie L., Luo C. L., Zhang C. G., Zhang R., Tang J., et al. , 2012. Genome-wide association study identified a narrow chromosome 1 region associated with chicken growth traits. PLoS One 7: 9 10.1371/journal.pone.0030910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Shen M., Yuan J., Sun C., Duan Z., et al. , 2015. Genome-wide association study dissects genetic architecture underlying longitudinal egg weights in chickens. BMC Genomics 16: 746 10.1186/s12864-015-1945-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. W., Wang K. H., Yi G. Q., Ma M., Dou T. C., et al. , 2015. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet. Sel. Evol. 47: 82 10.1186/s12711-015-0161-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan Y., Sheng Z., Lillie M., Rönnegård L., Honaker C. F., et al. , 2017. Artificial selection response due to polygenic adaptation from a multilocus, multiallelic genetic architecture. Mol. Biol. Evol. 34: 2678–2689. 10.1093/molbev/msx194 [DOI] [PubMed] [Google Scholar]
- Zhang G. X., Fan Q. C., Wang J. Y., Zhang T., Xue Q., et al. , 2015. Genome-wide association study on reproductive traits in Jinghai Yellow Chicken. Anim. Reprod. Sci. 163: 30–34. 10.1016/j.anireprosci.2015.09.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Pooled genome data generated for this study are available via Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) under bioProject: PRJNA516366; bioSample: SAMN10787895; and accessions: SRR8480632-SRR8480641. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7674281.