Abstract
Little is known about chromosome segregation in polyploid prokaryotes. In this study, whether stringent or variable chromosome segregation occurs in polyploid thermophilic bacterium Thermus thermophilus was analyzed. A stable heterozygous strain (HL01) containing two antibiotic resistance markers at one gene locus was generated. The inheritance of the two alleles in the progeny of the heterozygous strain was then followed. During incubation without selection pressure, the fraction of heterozygous cells decreased and that of homozygous cells increased, while the relative abundance of each allele in the whole population remained constant, suggesting chromosome segregation had experienced random event. Consistently, in comparison with Bacillus subtilis in which the sister chromosomes were segregated equally, the ratios of DNA content in two daughter cells of T. thermophilus had a broader distribution and a larger standard deviation, indicating that the DNA content in the two daughter cells was not always identical. Further, the protein homologs (i.e., ParA and MreB) which have been suggested to be involved in bacterial chromosome partitioning did not actively participate in the chromosome segregation in T. thermophilus. Therefore, it seems that protein-based chromosome segregation machineries are less critical for the polyploid T. thermophilus, and chromosome segregation in this bacterium are not stringently controlled but tend to be variable, and random segregation can occur.
Keywords: polyploid prokaryotes, Thermus thermophilus, random chromosome segregation, ParA and MreB homologs
An increasing number of bacterial and archaeal species are recognized as being oligo- or polyploid, e.g., containing on average more than one chromosomal copy per cell. Vastly different levels of the number of genome equivalents per cell have been measured in microorganisms belonging to such distinct groups as the Archaea, Cyanobacteria and Proteobacteria (Breuert et al. 2006; Schneider et al. 2007; Hildenbrand et al. 2011; Pecoraro et al. 2011). The reported genome copy numbers in these polyploid microorganisms range from two to several hundred, and there are extreme cases with several thousand copies, like in the giant endosymbiont bacterium Epulopiscium sp. (Mendell et al. 2008). It has been previously shown that a certain strain of T. thermophilus, a member of the deep-branching Deinococcus-Thermus group, which has been established as a model thermophilic organism, is also polyploid, carrying four to five genome copies per cell under slow growth condition, these genome copies are assumed to be genetically identical (Henne et al. 2004; Ohtani et al. 2010). Almost 40 years ago, the same ploidy level has been determined for the related radiation-resistant bacterium Deinococcus radiodurans (Hansen 1978). Several possible evolutionary advantages of having more than one genome copy have been suggested. For example, genome redundancy is associated with higher resistance against DNA damaging agents and permits the accumulation of otherwise deleterious recessive mutations (Comai 2005; Pecoraro et al. 2011; Soppa 2013, 2014). An important question is how polyploid microorganisms partition the multiple genome copies to daughter cells. The mechanism of genome partitioning has been extensively studied in model bacteria like Escherichia coli (Lampo et al. 2015; Cass et al. 2016), Bacillus subtilis (Kloosterman et al. 2016; Wang et al. 2017) and Caulobacter crescentus (Taylor et al. 2017), which are monoploid (only very slowly growing E. coli cells are truly monoploid). In a stark contrast, very few literatures could be found describing chromosomal dynamics in bacterial cells with more than two complete chromosome copies. Basically, chromosome partitioning in polyploid bacteria could be either stringent or variable (e.g., random). There is an additional layer of complexity in a dividing polyploid cell – if, for some reason, the cell carries two different alleles at the same locus (heterozygous), there would be different genetic outcomes in the progeny depending on the basic mode of chromosome partitioning. In a stringent mode, each chromosomal copy is actively passed on to the recipient cells via various partitioning mechanisms. Such a partitioning mode would lead to progeny which is genetically identical to the parents. If the chromosomes are divided randomly in the daughter cells, similar to high-copy-number plasmids, these cells can receive any combination and/or different numbers of the parental genomes. An important consequence of this mode would be that, if the parental cell was heterozygous, the daughter cells can be genetically different from the parent due to allele segregation. Such a mode has been proposed for polyploid euryarchaeota Methanococcus jannaschii (Malandrin et al. 1999), and polyploid cyanobacteria Anabaena sp. 7120 (Hu et al. 2007) and Synechocystis sp. (Schneider et al. 2007) based on DNA content analysis in dividing cells.
In T. thermophilus, it is not known whether the chromosome copies segregate into daughter cells via a stringent or variable mode. In contrast to some polyploid cyanobacteria, the genomes of so far sequenced members of the Thermales order (including T. thermophilus) contain homologs of the actin-like MreB protein as well as presumably complete chromosomal ParAB - parS systems, which have been suggested to be involved in active genome partitioning in many bacterial species. For instance, in E. coli it has been shown that MreB is important for both chromosomal bulk DNA and origin segregation (Kruse et al. 2003, 2005, 2006; Madabhushi and Marians 2009). Similarly, depletion of mreB in B. subtilis and C. crescentus leads to considerable defects in chromosome segregation, where replication origins fail to localize correctly (Soufo and Graumann 2003; Gitai et al. 2004, 2005; Shebelut et al. 2009; Sundararajan and Goley 2017). Disruptions of parAB in the chromosomal ParAB - parS systems also cause defects in chromosome segregation in many bacteria, e.g., in B. subtilis (Ireton et al. 1994; Sharpe and Errington 1996; Lewis and Errington 1997; Lee and Grossman 2006; Murray and Errington 2008; Sullivan et al. 2009; Gruber and Errington 2009; Scholefield et al. 2011), C. crescentus (Mohl and Gober 1997; Mohl et al. 2001; Toro et al. 2008; Ptacin et al. 2010; Shebelut et al. 2010), Streptomyces coelicolor (Kim et al. 2000; Jakimowicz et al. 2002), Vibrio cholera (Fogel and Waldor 2006; Yamaichi et al. 2007), Pseudomonas aeruginosa (Lasocki et al. 2007), Corynebacterium glutamicum (Donovan et al. 2010), Streptococcus pneumonia (Minnen et al. 2011) and Mycobacterium smegmatis (Ginda et al. 2013). In this manuscript, the mode of chromosome segregation in T. thermophilus HB27 was investigated. A heterozygous strain in which two antibiotic resistance alleles could be stably maintained in the presence of selection pressure was initially generated. The inheritance of the two alleles in the absence of selection was then followed in the daughter cells by molecular and genetic techniques. The results from these experiments suggested that in T. thermophilus, partitioning of the chromosomal copies would experience random event. Analysis of dividing nucleoids in T. thermophilus and B. subtilis by fluorescence microscopy further confirmed that the chromosome segregation occurred less stringently in T. thermophilus than in B. subtilis. In addition, through generating ΔmreBΔparA double deletion mutant followed by phenotype analysis with respect to chromosome segregation, it was found that in T. thermophilus both the ParA and MreB homologs were not actively participated in chromosomal bulk DNA segregation. The data from these experiments strongly suggest that the protein-based chromosome segregation machineries are less important for the polyploid T. thermophilus cells, and the chromosome segregation in this bacterium is likely non-stringent or often irregular.
Materials And Methods
Bacterial strains and growth conditions
E. coli DH5α was used as a host strain for plasmid constructions and was grown at 37° in LB medium (contains 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl). T. thermophilus HB27 wild type (DSM 7039) and derivative strains were grown at 70° or 60° in TB medium. TB medium contained 8 g/L trypticase peptone (BD Biosciences, Heidelberg, Germany), 4 g/L yeast extract, and 3 g/L NaCl, and had a pH of 7.5. B. subtilis 168 (DSM 402) was grown in LB medium at 30°. The growth media were supplemented with ampicillin (100 µg/mL for E. coli), kanamycin (50 µg/mL for E. coli and 20 µg/mL for T. thermophilus), bleomycin (“Bleocin”, Calbiochem, 15 µg/mL for E. coli and 3 µg/mL for T. thermophilus) or X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucopyranoside, 100 µg/mL) when necessary.
Determination of chromosome copy numbers
The real-time quantitative PCR was essentially performed based on the method described (Breuert et al. 2006; Li et al. 2015). Specifically, two loci (TTC1610 (near oriC) and TTC0574 (closer to terC) on the chromosome were chosen as the investigation targets. Approximately 1 kbp standard fragments of the two loci were PCR amplified using T. thermophilus genomic DNA as a template. The PCR products were purified from agarose gels followed by photometrical measurement to determine the concentrations (Nanodrop 2000). The molecular concentrations of the standard fragments were calculated using the online software “oligo calc” (www.basic.northwestern.edu/biotools). A series of dilutions of the standard fragments containing defined numbers of molecules were used in the real time PCR analyses. Standard curves were then generated by plotting the Ct values against the corresponding molecular concentrations in the reaction (molecules/μL). Quantified cell numbers (determined both by spectrophotometry and Neubauer counting chamber) from exponentially growing cultures were harvested and lysed by cell lysis buffer, the cell lysis efficiency was determined by cell counting. The cell lysates were then dialyzed, and aliquots from the dilution series of the cell lysates were used as templates in the real time PCR reactions. For real time PCR reactions, fragments of 139 bp (TTC1610) and 119 bp (TTC0574) were amplified, and three independent repeats were performed. Eventually, the chromosomal copy numbers per cell were calculated based on the created standard curves and the cell density.
Plasmid construction
All plasmids and strains used are listed in Table 1 and all primers used are listed in Table 2. For construction of pUC-Δ42, the flanking regions about 1 kbp of the bgl gene (TTP0042, encoding β-glucosidase) were respectively PCR amplified from T. thermophilus HB27, using primers that permitted to generate sufficient overlaps between each other; the two flanking regions were then cloned into XbaI digested pUC18 vector via Gibson Assembly method (New England Biolabs) (Gibson et al. 2009). The plasmids pCT3FK-2 and pJ-ΔpyrE::blm were used in the generation of the stable heterozygous strain HL01. pCT3FK (Angelov et al. 2009) in which a kanamycin resistance marker (kat) was sandwiched by two flanking regions of pyrE gene (TTC1380) was mutagenized (Change-IT Multiple Mutation Site Directed Mutagenesis Kit, Affymetrix, USA) to introduce two ZraI sites in the flanking sequences, followed by restriction and re-ligation to excise 521 bp of flanking sequences, while maintain the native pyrE flanking regions intact, this gave pCT3FK-2. For generation of pJ-ΔpyrE::blm, a 2.3 kbp pyrFE region was PCR amplified and cloned in pJET1.2 giving pJ-pyrFE; two NdeI sites were introduced to pJ-pyrFE by the same mutagenesis tool at positions -3 and +551 relative to the ORF of the pyrE gene. The mutagenized pJ-pyrFE was then digested with NdeI to get rid of the pyrE gene, and ligated with a bleomycin resistance marker blm (sequence data from Brouns et al. 2005). The plasmid pMK-parB-sgfp allowing expression of ParB-sGFP in T. thermophilus was constructed in a former study (Li et al. 2015). The plasmids pUC-ΔmreB::kat and pUC-ΔparA::blm were gene deletion vectors for generation of double knockout mutant ΔmreB::kat/ΔparA::blm in T. thermophilus HB27. The two flanking regions (about 1 kbp) of the parA (TTC1605) and mreB (TTC1464) genes, and the blm/kat cassette were respectively amplified by PCR, using T. thermophilus HB27 genomic DNA and plasmids containing the blm/kat cassettes (de Grado et al. 1999; Brouns et al. 2005) as the templates. Gibson assembly reaction was used to assemble the three PCR fragments into XbaI-digested pUC18 respectively, as they contained sufficient overlaps between each other.
Table 1. Strains and plasmids used in this study.
| Name | Descriptiona | Reference |
|---|---|---|
| Plasmids | ||
| pUC-Δ42 | clean deletion vector for generating homozygous and heterozygous Δbgl in T. thermophilus HB27, ori pUC, AmpR | this study |
| pCT3FK-2 | allele exchange vector for generating ΔpyrE::kat and HL01, ori pBR322, KmR | this study |
| pJ-pyrFE | intermediate vector for generating pJ-ΔpyrE::blm, ori pJET, AmpR | this study |
| pJ-ΔpyrE::blm | allele exchange vector for generating ΔpyrE::blm and HL01, ori pJET, AmpR, BlmR | this study |
| pMK-parB-sgfp | pMK18 derived vector, allowing expression of parB-sGFP in T. thermophilus, Tth (repA), Ec (oriE), KmR | Li et al. 2015 |
| pUC-ΔparA::blm | allele exchange vector for generating ΔparA::blm in T. thermophilus HB27, ori pUC, AmpR, BlmR | this study |
| pUC-ΔmreB::kat | allele exchange vector for generating ΔmreB::kat in T. thermophilus HB27, ori pUC, AmpR, KmR | this study |
| Strains | ||
| HB27 | T. thermophilus wild type | DSM 7039 |
| HB27 wt/Δ-bgl | T. thermophilus HB27 heterozygous derivative with bgl clean deleted | this study |
| HB27 ΔpyrE::kat | T. thermophilus HB27 homozygous derivative with pyrE replaced by kat | this study |
| HB27 ΔpyrE::blm | T. thermophilus HB27 homozygous derivative with pyrE replaced by blm | this study |
| HL01 | T. thermophilus HB27 heterozygous derivative with pyrE replaced by kat and blm simultaneously | this study |
| HB27/ParB-sGFP | T. thermophilus HB27 derivative permitting expression of ParB-sGFP | Li et al. 2015 |
| ΔmreB::kat/ΔparA::blm | T. thermophilus HB27 derivative with parA replaced by blm and mreB replaced by kat | this study |
| 168 | B. subtilis wild-type strain | DSM 402 |
Ec (oriE), replication origin for E. coli; Tth (repA), replication origin for T. thermophilus.
Table 2. Primers used in this study.
| Name | Sequences (5′-3′)a | Usage |
|---|---|---|
| 1610-F | TCAAGGAGAAGGGCTACAG | generating standard fragment for qPCR in the TTC1610 region |
| 1610-R | CCTTGTAGCTCACGGAAAC | |
| 1610-F-1 | ACGCCATCCTGGTCAAGGTG | primers of qPCR reactions for detecting TTC1610 copy numbers |
| 1610-R-1 | AGGTCGGCGATGAAGCTGTC | |
| 0574-F | CCGGCAGGTAGACGTCAAAG | generating standard fragment for qPCR in the TTC0574 region |
| 0574-R | TGAGCCGGAGGGAGTTTGAG | |
| 0574-F-1 | GTGACCACCACGCTTTCGGG | primers of qPCR reactions for detecting TTC0574 copy numbers |
| 0574-R-1 | TTAGGCCGCCAGGATCAGTACG | |
| bglF1-F | tgcatgcctgcaggtcgactAGACCATCCCCCAGGAGCTC | amplifying bgl upstream flanking region for pUC-Δ42 |
| bglF1-R | cctctggcggggcacttagCTCGGTCATAGGCGTTTCTC | |
| bglF2-F | gagaaacgcctatgaccgagCTAAGTGCCCCGCCAGAGG | amplifying bgl downstream flanking region for pUC-Δ42 |
| bglF2-R | gctcggtacccggggatcctGCCAGAACCAGAACGAAAAG | |
| dbgl-F | GCCGTCTACATCTTCCTCAC | PCR determination of bgl deletion |
| dbgl-R | TACCTTCCCGAGGACATCAC | |
| pCT3FKm-Zra-F | ACCTGCAGACGTCCAAGCTTGGCACTGGCCGTCGTTTTACAA | site-directed mutagenesis of pCT3FK |
| pCT3FKm-Zra-R | TTTCAGCAGACGTCGTTTCCTTTCTTTTCAGAGGGTATTTTA | |
| pyr.F | CCGAGCCCTTGGCCCATATC | cloning of the pyrFE region |
| pyr.R | CAGGACCGCCACCCTCATA | |
| pyrEm-Nde.F | GAGGAAGCGCATATGAGACCTCCTCC | site-directed mutagenesis of pJ-pyrFE |
| pyrEm-Nde.R | CAGGACGTCCATATGCCCCTACTCTAC | |
| pyrF.R | GGACCCTCCCGGTACCTTTC | combing with pyr.F used for probe generation for Southern blot |
| pyr.R2 | GCTTTCCAGGTTGACGGTAAGC | combing with pyr.F used for PCR detection of pyrE gene replacements |
| mreB-1-F | catgcctgcaggtcgactAAACGGGACCGATTCCTC | amplifying mreB downstream flanking region for pUC-ΔmreB::kat |
| mreB-1-R | agagcgcccaatacgcaaaccGAGCTCGCCTCGGACATCTAC | |
| mreB-2-F | cttggaggagaaacgccTGCCGATGTCTTCGCCTTTAAGC | amplifying mreB upstream flanking region for pUC-ΔmreB::kat, and Southern blot probe template generation for mreB deletion detection |
| mreB-2-R | cggtacccggggatcctGGGTGGACCTCATCATTGAC | |
| kat-F | cggtttgcgtattgggcgctctTCCCCGGGAGTATAACAGAAACC | amplifying kat for pUC-ΔmreB::kat |
| kat-R | ggcgtttctcctccaagAATTCCGTTCAAAATGGTATG | |
| dmreB-F | TTGAGGATCTCCCGGATGTC | PCR determination of mreB deletion |
| dmreB-R | ATCGCGAGCCGCATTGAGAA | |
| parA-F1-F | tgcatgcctgcaggtcgactTCCTCCAAGGAGCGGTACTG | amplifying parA upstream flanking region for pUC-ΔparA::blm |
| parA-F1-R | gttatactcccggggatcccCCTGGCAGAGGAGGTGATGG | |
| parA-F2-F | gactgatctagaggatccccGCCCTTAGCATAACGGATAC | amplifying parA downstream flanking region for pUC-ΔparA::blm |
| parA-F2-R | cggtacccggggatcctCAAGTACGCGGGCTACATTG | |
| blm-F | GGGATCCCCGGGAGTATAAC | amplifying blm for pUC-ΔparA::blm |
| blm-R | GGGGATCCTCTAGATCAGTC | |
| probe-F | CCTCGGCTTCCTCAAGCTCTTC | Southern blot probe template generation for parA deletion detection |
| probe-R | CCGAAGAGGACGCGCACCGC | |
| dparA-F | TAGCGCCTTTCCCCCGCCAC | PCR determination of parA deletion |
| dparA-R | ACCTGGTGGTGTTGGAGAAG |
Enzyme restriction sites are in bold and sequences that create the overlaps for the Gibson assembly reactions are in lowercase.
Determination of the genetic outcomes of the heterozygous strain wt/Δ-bgl
The gene clean deletion vector pUC-Δ42 was linearized by HindIII and transformed into T. thermophilus HB27 based on the protocol described by de Grado et al. (1999). The transformation reaction was appropriately diluted and plated on TB agar plates supplemented with X-Gluc. After incubation at 70° for 2 d, colonies that exhibited intermediate color were selected for genotype analysis. The correct heterozygous strain (wt/Δ-bgl) was then grown in TB medium for 24 h and the culture was restreaked on TB-X-Gluc plates. The phenotype of the progeny was discriminated by colony color, and the genotype was determined by PCR.
Generation of homozygous T. thermophilus gene deletion mutants
The allele exchange vectors pCT3FK-2 and pJ-ΔpyrE::blm were linearized by ScaI and HindIII respectively, then transformed into T. thermophilus HB27 in the same manner. The transformation reactions were respectively streaked on TB agar plates supplemented with kanamycin or bleomycin. Several colonies were randomly selected, and PCR (using primers flanking the pyrE gene) was used to sort out the colonies which only contained the mutant allele. This resulted in two homozygous strains, i.e., ΔpyrE::kat and ΔpyrE::blm, which carried either the kat or the blm resistance marker at the pyrE gene locus. For generation of ΔmreB::kat/ΔparA::blm mutant, single ΔmreB::kat mutant was initially created. The pUC-ΔmreB::kat vector was linearized by HindIII and transformed into wild-type T. thermophilus cells followed by plating on agar plates containing kanamycin. Several transformants were exposed to PCR analysis to determine mutants containing homozygous ΔmreB::kat allele. Linearized pUC-ΔparA::blm was sequentially transformed into the correct ΔmreB::kat strain, transformants were selected on agar plates containing both antibiotics. The homozygous double deletion mutant ΔmreB::kat/ΔparA::blm was prescreened by PCR and further confirmed by Southern blot.
Construction of a stable heterozygous strain and kinetics of allele segregation
The heterozygous strain HL01 was obtained by transforming linearized pJ-ΔpyrE::blm to the homozygous ΔpyrE::kat strain and selecting on TB agar plates supplemented with both kanamycin and bleomycin. The heterozygous strain was initially verified by PCR as well, and further confirmed by Southern blot and DNA sequencing of the respective genomic regions. The allele segregation experiment was performed by growing cells of HL01 in 5 mL of TB medium supplemented with both antibiotics for 12 h, followed by washing the cells three times in 1 × PBS buffer and inoculation in 50 mL antibiotic-free TB medium (time point 0). Growth was continued for 120 h at 70° with agitation. Samples (2 mL) were taken at various time points as indicated in the results to determine the fraction of each phenotype by spreading the cells on TB plates (without antibiotics) and re-streaking the colonies on plates supplemented with kanamycin or bleomycin. The same colony was parallelly streaked on TB plate to determine the growth. The relative amount of each allele of the samples was estimated by Southern blot followed by band intensity measurements (Image J, NIH, USA).
Fluorescence microscopy
To measure the relative DNA content in two dividing cells by fluorescence microscopy, the T. thermophilus strains (HB27, HB27/ParB-sGFP, and HB27 ΔmreB::kat/ΔparA::blm) and B. subtilis 168 cells were grown to exponential phase. Cells from 0.5 mL cultures were washed once in 1 × PBS buffer, fixed by ethanol, and resuspended in the same volume of PBS buffer. When necessary, the DNA-specific dye DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) and CFS (6-carboxyfluorescein) which stains the cell membrane were added to the cell suspension with final concentrations of 0.2 µg/mL and 10 µg/mL respectively, followed by incubating at room temperature for 30 min. The residual dyes were removed by washing the cells three times with 1 × PBS buffer. After mounting the cells on glass slides, fluorescence microscopy was carried out (Zeiss Axio Imager M1). The filter “DAPI” was used to observe DAPI-stained DNA signal, and the filter “AF488” was used to observe CFS-stained membrane signal and ParB-sGFP signal. The acquired micrographic images were analyzed with the AxioVision software (Carl Zeiss, Germany) and Image J (NIH, USA).
Data availability
The author affirm that all plasmids are available upon request, and all data necessary for confirming the results of the article are included within the article, figures and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7746878.
Results
Real-time PCR determination of the chromosomal copy number of T. thermophilus
The T. thermophilus strain HB8 has been reported to contain four to five copies of chromosomes per cell under slow growth condition (Ohtani et al. 2010). For the T. thermophilus type strain HB27, the chromosomal copy numbers are not clearly reported. Using real-time PCR method, the chromosomal copy numbers of T. thermophilus HB27 during exponential growth in rich medium was determined. This real-time PCR method was developed by Breuert et al. (2006), and has been used to determine the exact genome copy numbers in the cell populations of various archaeal and bacterial species (Hildenbrand et al. 2011; Pecoraro et al. 2011; Griese et al. 2011; Lange et al. 2011; Spaans et al. 2015; Zerulla et al. 2016), emphasizing the reliablity of this method. With the aim of accuracy, two loci (TTC1610 (near oriC) and TTC0574 (relatively closer to terC) on the chromosome was chosen as the investigation targets. The copy numbers of the two loci determined by the real-time PCR method are summarized in Table 3, which clearly showed that multiple sets of chromosomes existed per cell during the exponential growth phase in T. thermophilus HB27, and this is consistent with the study performed in the strain HB8 (Ohtani et al. 2010).
Table 3. Chromosome copy numbers in T. thermophilus HB27.
| Gene locus | Real-time PCRa | Fluorescent microscopyb |
|---|---|---|
| TTC1610 (oriC region) | 7.87 ± 0.73 | 7.53 ± 2.71 |
| TTC0574 (relatively closer to terC) | 7.01 ± 1.55 | / |
real-time PCR method was used to measure the chromosome copy numbers.
Fluorescent microscopy was used to count ParB-sGFP fluorescent foci numbers per cell reflecting the copy numbers of the chromosomal origin region (100 cells were counted).
Heterozygous T. thermophilus HB27 cells and their genetic outcomes
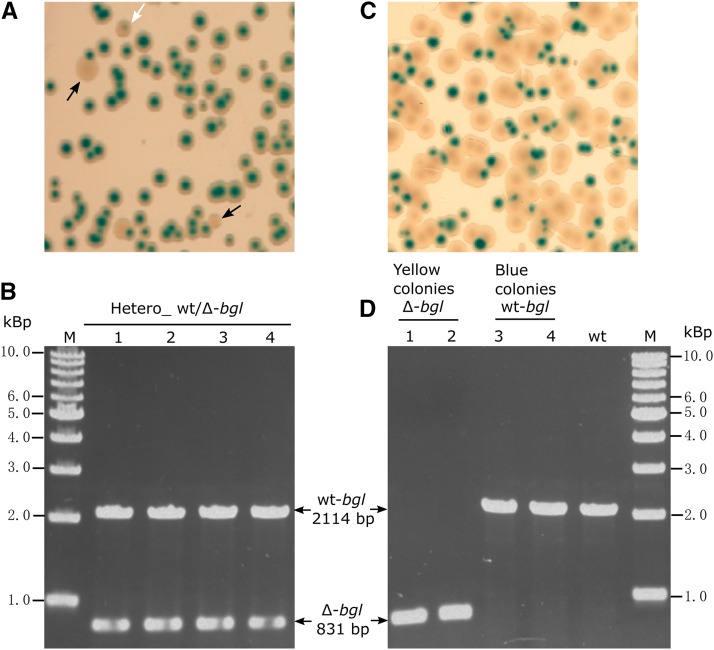
Since T. thermophilus HB27 is polyploid, it is conceivable that under certain condition, heterozygous cells containing two different alleles at one gene locus would appear. In practice, this kind of heterozygous cells was previously noticed during studies of T. thermophilus HB27 when generating deletion mutants of non-essential genes. Take bgl gene for example, wild-type T. thermophilus HB27 cells express β-glucosidase (Bgl). It is easy to distinguish Bgl- colonies (yellow) from Bgl+ colonies (blue) on agar plates supplemented with indicator substrate, i.e., X-Gluc (Ohta et al. 2006). Interestingly, when creating Δbgl mutant through direct double-crossover homologous recombination method, although apparent homozygous Δbgl mutant (yellow color) could be easily detected on TB X-Gluc plates (Figure 1A), transformants which projected intermediate phenotype (the color was between yellow and blue) could also be found (Figure 1A). PCR analysis showed that these “intermediate” colonies were carrying both knockout and wild-type alleles simultaneously at the bgl locus (Figure 1B). However, they were genetically unstable, since after growing in liquid medium without selection followed by re-streaking on agar plates with X-Gluc, colonies showing homozygous for the knockout or wide-type bgl allele were observed (Figure 1C, D). This result indicated that T. thermophilus cells could form heterozygous state carrying different alleles at one locus, and without selection pressure, the alleles could segregate resulting in different genetic outcomes in the progeny.
Figure 1.
Heterozygous wt/Δ-bgl cells and their genetic outcomes. (A) Phenotype of homozygous and heterozygous Δbgl transformants on TB-X-Gluc plate. Black arrows: yellow colonies (homozygous Δbgl); white arrow: colony with intermediate color (heterozygous wt/Δ-bgl). (B) Genotypes of the transformants showing intermediate phenotype were determined by PCR (using primers binding the flanking regions of bgl). (C, D) The genetic outcomes of the heterozygous wt/Δ-bgl strain. The phenotype (C) and genotype (D) of the progeny of the heterozygous strain after growing without selection and re-streaking on TB-X-Gluc plate. wt-bgl: genotype of the blue colonies from (C), Δ-bgl: genotype of the yellow colonies from (C).
Construction of a stable heterozygous strain
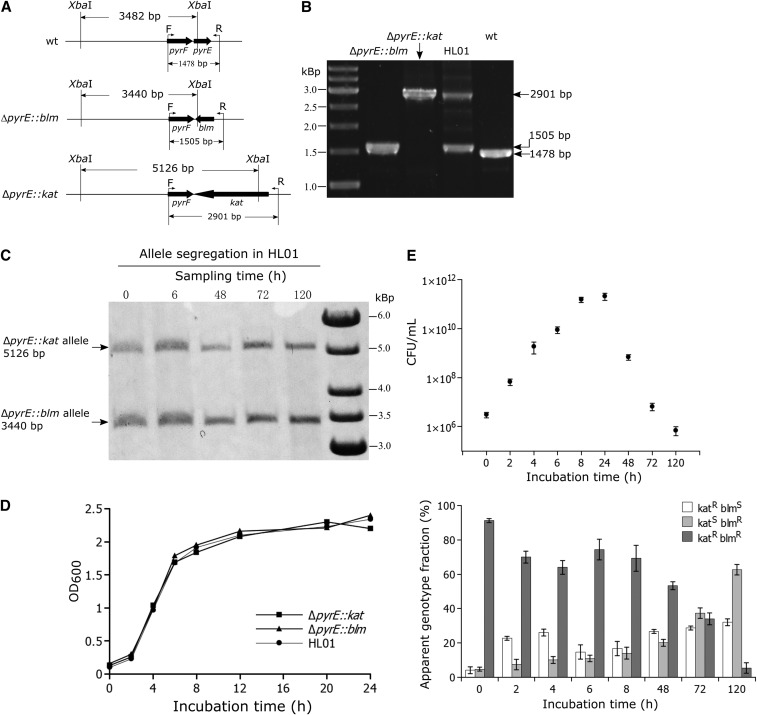
In order to investigate what mechanism had drived the allele segregation phenomenon, a stable heterozygous strain was constructed by inserting two different selection markers at the same chromosomal locus (pyrE). To this end, initially, two homozygous strains (ΔpyrE::kat and ΔpyrE::blm) were created by replacing the pyrE gene with a kanamycin and a bleomycin resistant cassette, respectively (Figure 2A). Numerous examples have shown that homozygous gene deletion mutant could be obtained in T. thermophilus in spite of its polyploid genomic background (Angelov et al. 2013; Leis et al. 2014; Li et al. 2015; Ohtani et al. 2015; Wang et al. 2016). In the experience, when a non-essential gene was targeted by a vector containing an antibiotic resistant marker sandwiched by two homology arms of that target region, near 90% of the resultant transformants were homozygous deletion mutant of that gene. In the generation of ΔpyrE::kat and ΔpyrE::blm, eight transformants were respectively chosen for PCR determination of the deletion mutants, and only one transformant was found to contain the wild-type pyrE allele (Figure S1). The possibility of obtaining homozygous gene deletion mutants with high frequency in the polyploid T. thermophilus genome, again suggested that after the mutant allele has been introduced into the specific genomic region, allele segregation would lead to homozygous mutant cells. Thereafter, the homozygous ΔpyrE::kat strain was transformed with linearized pJ-ΔpyrE::blm plasmid. Selection on TB agar plates supplemented with both antibiotics allowed to generate the heterozygous strain HL01. PCR, Southern blot and DNA sequencing was used to verify the genotype of the heterozygous strain, which excluded the possibility of vector integration and unequivocally showed that in HL01 the pyrE gene was completely replaced by both the kat and blm resistance cassettes and no wild-type pyrE allele was detectable (Figure 2A, B, C). The heterozygous state of the HL01strain could be stably maintained in medium supplemented with both kanamycin and bleomycin. The cell growth curves showed that there were no growth rate difference among the two homozygous strains (ΔpyrE::kat and ΔpyrE::blm) and the heterozygous strain HL01 (Figure 2D).
Figure 2.
Genotype confirmation and allele segregation kinetics of the HL01 strain. (A) Schematic drawings of the genotypes of the ΔpyrE::blm and ΔpyrE::kat strains. XbaI site was found in pyrE, blm and kat respectively. (B) PCR confirmation of the ΔpyrE::blm, ΔpyrE::kat and HL01 strains using primers F and R (the positions are shown in A). The predicted sizes of the PCR products were 1478 bp for the wild type, 2901 bp for ΔpyrE::kat, 1505 bp for ΔpyrE::blm, and both 2901 bp and 1505 bp for HL01. (C, E) Apparent genotype and allele abundance measurements of the heterozygous strain HL01 grown in liquid TB medium at 70° in the absence of selection. (C) The changes in the relative abundance of the two alleles (kat and blm) were acquired by measuring the intensity of the bands from the Southern blot. For the Southern blot, a 450 bp biotin-labeled fragment of the pyrF gene was used as the probe for hybridization; genomic DNA was prepared from samples taken at the indicated time points and was digested with XbaI. The in silico predicted sizes are 5126 bp and 3440 bp for the ΔpyrE::kat and ΔpyrE::blm alleles, respectively. One representative of three independent analyses is shown. (E) The viable counts were determined from the antibiotic-free plates (upper panel). The changes in the fraction of each phenotype (KatRBlmR – dark-gray bar, KatRBlmS – white bar and KatSBlmR – light-gray bar) in the population were followed by spreading the samples on antibiotic-free plates and restreaking 50 colonies for each time point on plates containing kanamycin or bleomycin (lower panel). Shown are mean and standard deviation from the three independent experiments. (D) The cell growth curves of the ΔpyrE::blm, ΔpyrE::kat and HL01 strains.
Allele segregation kinetics in the heterozygous strain HL01 in the absence of selection
To detect and quantify allele segregation in the heterozygous strain, HL01 was continuously grown in the absence of selection pressure for 120 h (without dilution or regrowth from stationary phase). Samples were withdrawn at different time points and were used to determine: i) the drug resistance phenotype of 50 individual colonies per time point and ii) the relative frequency of the two alleles in the samples, using Southern blot followed by band intensity measurement. The drug resistance profile implies the genotype of the tested colony (homozygous for either the kat or the blm allele, or heterozygous), while changes in the relative abundance of each allele in the whole population could be detected by Southern blot. In this experiment, it was observed that during growth in the absence of selection pressure the fraction of heterozygous cells (as determined by the resistance phenotype) decreased and that of homozygous cells increased (Figure 2E). The Southern blot analysis showed that, despite the significant changes in the observed genotypes, the relative amount of kat/blm allele in the whole population kept almost constant throughout the experiment (Figure 2C). Apart from random chromosome partitioning, another mechanism, which is gene conversion (Pecoraro et al. 2011; Lange et al. 2011), could serve to explain the allele segregation phenomenon as well. However, in addition to changing the genotype frequency, gene conversion also leads to changes in the allele abundance in the whole population (see discussion). Since the relative amount of blm/kat allele did not change, gene conversion was ruled out. Taken together, the data from this experiment indicated that random chromosome segregation is the most likely mechanism leading to allele segregation in the heterozygous cells when they were grown in the absence of antibiotic selection pressure.
Distribution of DNA content in two dividing daughter cells
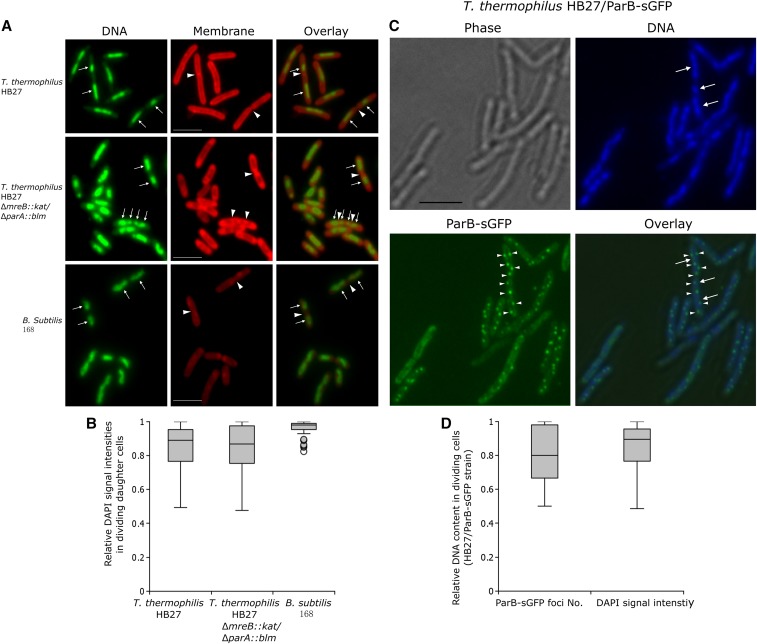
In bacterial parAB – parS systems, ParB can bind its cis-acting element parS (Lin and Grossman 1998). In T. thermophilus, it has been previously shown that the chromosomal parS site is located near the chromosomal oriC, and that ParB-sGFP fusion proteins could bind parS forming fluorescent foci (Li et al. 2015). The fluorescent foci numbers per cell to some extent represent the copy numbers of parS, i.e., of the chromosomal oriC region. Based on these observations, and to further confirm that the chromosome partitioning mode in T. thermophilus is non-stingent and often variable, the DNA content in dividing daughter cells was measured based on the signal intensities of DAPI-stained nucleoids, as well as on the numbers of fluorescent foci formed by ParB-sGFP/parS protein-DNA complexes. The two methods have been used also in studying of random chromosome segregation pattern in cyanobacterial species (Schneider et al. 2007; Hu et al. 2007). Specifically, exponentially growing cells of T. thermophilus HB27 were stained with DAPI (a DNA stain) and 6-carboxyfluorescein (CFS, a membrane stain), and the relative DNA staining intensities of 100 dividing cells (cells in which the nucleoids were separated and the division septum was visible) were measured. B. subtilis 168 was stained and analyzed in the same way and was used as a control in which stringent chromosome partitioning has been shown. Representative images from these measurements are shown in Figure 3A. The fluorescence signals of the two daughter nucleoids in dividing T. thermophilus cells (Figure 3A, top row; Figure S2) revealed unequal intensities of the nucleoids in contrast to B. subtilis cells, in which an almost equal distribution of the DAPI signal in the two daughter cells was observed (Figure 3A, bottom row). Further, it seems that in the T. thermophilus dividing cells with unequal amount of sister nucleoids, septa were also not always formed exactly at mid-cell positions (Figure 3A, top row; Figure S2). Figure 3B shows the relative DNA content, determined by DAPI signal intensity, in 100 pairs of daughter nucleoids in dividing cells of T. thermophilus and in B. subtilis. In the B. subtilis cells, the signal ratios distributed from 0.82 to one (mean 0.96, SD 0.04), while in the T. thermophilus cells, the relative ratios had a much broader distribution (from 0.49 to one with a mean of 0.84 and a SD of 0.15). In the polyploid Synechocystis sp., a similar analysis has been performed (Schneider et al. 2007), in which the relative DNA contents in the B. subtilis dividing cells varied from 0.76 to one, with a mean value of 0.94 and a SD of 0.05, emphasizing the reproducibility of this method. The above analysis suggested that unequal distribution of the genetic material occurred in the dividing T. thermophilus cells. It seems that although chromosome could segregate irregularly, the frequencies of anucleate cells were actually low, only 1.25% cells were found to be anucleate based on the fluorescent microscopic analysis. The phenomenon was also observed in cyanobacterium Anabaena sp. PCC 7120 which has been suggested to exert random chromosome segregation as well (Hu et al. 2007). This may be due to the fact that the daughter cells of these polyploid bacteria are likely to get at least one chromosome by chance, and subsequent chromosome replication may compensate for variance created during cell division.
Figure 3.
Relative DNA contents in dividing cells of T. thermophilus HB27, HB27/ParB-sGFP, ΔmreB::kat/ΔparA::blm, and B. subtlis 168. (A) Representative images of exponentially growing and dividing T. thermophilus and B. subtlis cells stained with 6-carboxyfluorescein (membrane) and DAPI (DNA) used in the analysis of the DNA intensity scatter. The white arrows show examples of separated nucleoids in cells where the formations of septa are visible (triangles), bars indicate 2 µm. (B) Box plot of the distribution of DAPI intensity in 100 randomly selected dividing T. thermophilus HB27, ΔmreB::kat/ΔparA::blm and B. subtilis cells. The fluorescence intensity ratio in the two daughter cells was calculated by dividing the intensity value of the cell with lower fluorescence signal by the one with a higher one. The whiskers represent the minimum and the maximum of all the data, circles show outliers. (C) The T. thermophilus chromosomal parAB – parS system was used to determine relative DNA contents (i.e., relative numbers of the oriC region) in dividing cells. Representative images show cells with different numbers of ParB-sGFP/parS fluorescent foci in the two daughter nucleoids (white triangles: fluorescent foci, white arrows: sister nucleoids, bar indicates 2 µm). (D) Box plot of the distributions of relative ParB-sGFP/parS fluorescent foci numbers and relative DAPI signal intensities in 100 dividing HB27/parB-sGFP cells. The ratio was achieved by dividing the lower value by the higher one.
The above results were further verified by using the T. thermophilus parAB – parS system. For this, a C-terminal sGFP fusion of the chromosomally encoded ParB protein was constructed (Li et al. 2015), and the ParB-sGFP fusion protein was expressed from the replicative plasmid pMK18 (de Grado et al. 1999). When expressed in T. thermophilus HB27, well-defined fluorescent foci could be observed (Figure 3C). The fluorescent foci numbers represent the copy numbers of parS sites, as has been shown previously (Li et al. 2015). In the example image Figure 3C, five to 12 fluorescent foci could be observed in the growing cells with dividing nucleoids, indicating five to 12 copies of parS (and of oriC region) existed in one cell, again confirming the polyploidy of T. thermophilus (Table 3). The pairwise distribution of fluorescent foci numbers in 100 pairs of daughter cells was almost identical to the distribution of the relative DAPI signals (Figure 3D), with a mean of 0.83 and a SD of 0.16. This result further confirmed that random chromosome segregation could occur in the polyploid T. thermophilus cells.
Inactivation of parA and mreB in T. thermophilus HB27
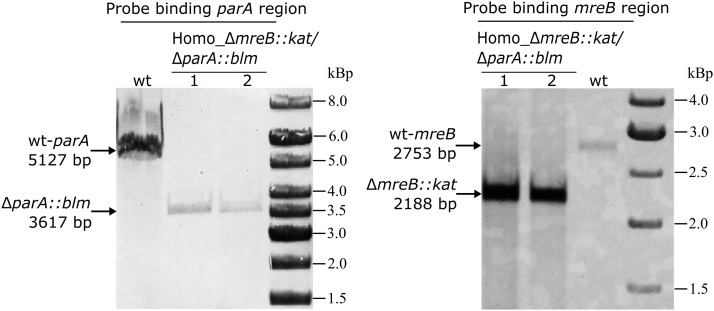
The Par system and actin-like protein MreB have been shown to participate in active chromosome partitioning in many bacterial species. By searching out the complete genome of T. thermophilus HB27 (Henne et al. 2004), only one copy of chromosomally encoded ParA and MreB homolog could be identified. And given their important role in other bacteria, their function with respect to chromosome segregation was investigated in this species. The mreB and parA genes were deleted by replacing them with the kat and blm resistance markers respectively (a schematic design see Figure S3A). Initially, homozygous ΔmreB::kat was generated (Figure S3B). The suicide vector pUC-ΔparA::blm was then transformed into the correct ΔmreB::kat strain, and selected on agar plates with both antibiotics (see materials and methods). Seven colonies were randomly selected and exposed to PCR prescreen, this was to determine transformants which contained homozygous ΔparA::blm allele in the ΔmreB::kat background. Six colonies were found to be homozygous ΔmreB::kat/ΔparA::blm mutants (Figure S3C). Southern blot analysis further confirmed that the double deletion mutant had correct insertion of kat and blm at the mreB and parA loci respectively, and no wild-type alleles were left (Figure 4). To investigate potential chromosome segregation defects in the mutant, exponentially growing cells were collected, stained by DAPI and CFS and analyzed by fluorescence microscopy. In dividing ΔmreB::kat/ΔparA::blm cells, the replicated nucleoid could segregate normally to the daughter cells (a representative image is shown in Figure 3A, middle row). Furthermore, compared with the wild type, no considerable increase in the frequency of anucleate or DNA-less cells in ΔmreB::kat/ΔparA::blm was observed (1.25% in the wild type and 1.50% in the mutant, 400 cells were analyzed), suggesting that the deletion of parA and mreB in T. thermophilus has no effect on the chromosomal bulk nucleoid segregation. The relative DNA content in 100 pairs of dividing cells, measured by DAPI signal intensities, were plotted as well. The relative DNA content in the dividing daughter cells of ΔmreB::kat/ΔparA::blm distributed from 0.48 to one, with a mean of 0.85 and a SD of 0.13. This data are almost indistinguishable from that of the wild type strain (Figure 3B). Taken together, the results suggested that in T. thermophilus, ParA and MreB do not actively participate in the chromosomal bulk DNA segregation process. A similar observation has been reported for the cyanobacterium Anabaena sp. PCC 7120, which is also polyploid (Hu et al. 2007), whereas in haploid bacteria such as B. subtilis and C. cresentus, ParA and MreB have been shown to be involved in chromosomal bulk DNA and origin segregations (Ireton et al. 1994; Lewis and Errington 1997; Mohl and Gober 1997; Soufo and Graumann 2003; Gitai et al. 2004, 2005; Shebelut et al. 2009, 2010; Scholefield et al. 2011).
Figure 4.
Generation of the ΔmreB::kat/ΔparA::blm double deletion mutant in T. thermophilus. The parA and mreB genes were replaced by blm and kat cassette respectively, genotype confirmation of ΔmreB::kat/ΔparA::blm was performed by Southern blot, using probes binding the mreB and parA regions for hybridization respectively. The genomic DNAs were digested with BamHI for confirmation of parA deletion, and were digested with BglII for confirmation of mreB deletion. The in silico predicted sizes are respectively indicated with arrows on the left side of each image.
Discussion
Allele segregation in heterozygous T. thermophilus cells
This work showed that the T. thermophilus strain HB27 contains on average seven to eight chromosomal copies per cell when grown in rich medium. This number was estimated using two independent methods, e.g., a real time PCR-based method and a fluorescence microscopy method, based on counting the numbers of fluorescent ParB-sGFP/parS foci per cell. The estimation of the chromosomal copy number is in good agreement with the one reported in another T. thermophilus strain, HB8 (Ohtani et al. 2010). One of several possible explanations for polyploidy is that this strategy favors the survival of bacteria living under harsh conditions. For example, due to multiple genome copies, D. radiodurans is able to rapidly reassemble the chromosomes destroyed by desiccation and ionizing radiation (Zahradka et al. 2006; Slade et al. 2009). T. thermophilus which grows at temperatures up to 85° may also suffer frequent DNA damage, and polyploidy may facilitate DNA repair by homologous recombination.
An interesting phenomenon observed repeatedly in T. thermophilus is the allele segregation in heterozygous cells containing two different alleles in one locus (Figure 1). It was considered that this could be the result of either random chromosome segregation or gene conversion. While both processes can result in homogeneity in the progeny, their intrinsic mechanisms are different. Gene conversion is essentially a non-reciprocal exchange of information between homologous chromosomes as well as between repeated sequences on the same chromosome (Paulsson et al. 2017). In principle, for a heterozygous cell containing both mutant and wild-type allele at one locus, gene conversion can conditionally occur in both directions, e.g., if the mutant allele is converted against the wild-type allele, the abundance (copies) of the mutant allele will be eliminated and that of the wild-type allele will be increased, and the reverse is also true (Khakhlova and Bock 2006; Soppa 2011). In polyploid archaea, allele equalization among the multiple genome copies via gene conversion has been repeatedly demonstrated (Hildenbrand et al. 2011; Lange et al. 2011; Guschinskaya et al. 2016). For instance, Lange et al. (2011) showed that in Haloferax volcanii, the heterozygous strain leuB/ΔleuB::trpA would convert to homozygous leuB or ΔleuB::trpA strain. In the presence of tryptophan, ΔleuB::trpA allele was converted to leuB allele, resulting in the increase of the fraction of homozygous leuB strain, in the meanwhile, the abundance of the ΔleuB::trpA allele was eliminated and that of the leuB allele was accordingly increased in the whole population; in the presence of leucine, the conversion was occurred in the opposite direction. On the other hand, if random chromosome segregation occurred in polyploid cells carrying two different alleles at one gene locus, daughter cells tended to receive different combination and/or numbers of the replicated parental chromosomes, which would also lead to change in the genotype frequency of the progeny, however, the relative allele abundance would be always maintained. To be able to discriminate which mechanism had led to the observed allele segregation in the heterozygous T. thermophilus cells, a stable heterozygous strain (HL01) containing two different markers (kat and blm) at the pyrE locus was generated (Figure 2B, C). When this strain was grown in the absence of antibiotic selection, a gradual segregation of the two markers was observed (Figure 2E). In this experiment the fraction of homozygous cells increased significantly, while at the same time the relative abundance of kat and blm in the population remained constant (Figure 2C, E). This indicates that the allele segregation was probably triggered by chromosome random segregation but not by gene conversion (see above). Interestingly, during the allele segregation kinetics, the fraction of the heterozygous strain decreased more steady in the stationary phase than in the exponential phase (e.g., the fraction was increased at the time point 6 h and 8h compared with 4 h). This may be explained by the high frequency of natural transformation in the exponential growth phase of T. thermophilus HB27. Although cell divisions proceeded faster in the exponential phase, DNA exchange among the cells (due to the uptake by natural transformation of free DNA released by dead cells) might also occur (César et al. 2011; Alvarez et al. 2011). Such an exchange would counteract allele segregation and would affect the frequency of the apparent genotypes of the two alleles. Indeed, addition of EDTA, which is known to impede DNA uptake, to the allele segregation reactions resulted in a more rapid segregation of the two alleles in the exponential phase (2 h - 8 h) (Figure S4). Another mechanism contributing to the steady allele segregation in the late stationary phase may be reductive cell division (Kolter et al. 1993; Navarro Llorens et al. 2010). Such reductive cell division has been shown to cause decrease in the average genome copy numbers in many polyploid bacteria and archaea, as shown by the extreme example, the copy numbers of cyanobacterium Synechocystis PCC 6803 could decrease from 218 copies in exponential growth phase to 58 copies in stationary growth phase (Griese et al. 2011). In T. thermophilus, the average chromosome copy number per cell was four to five under slow growth condition (Ohtani et al. 2010), while that was seven to eight in fast-growing culture, indicating that the value is also conditionally regulated. Therefore, it is conceivable that in this experiment, the decrease of average chromosome copies in the stationarily-growing cells would actually favor the daughter cells receive the same type of parental chromosome by random partitioning.
Chromosome random partitioning in polyploid bacterial cells
The complete segregation of bacterial chromosomes undergoes three separated steps: separation of the replicated origin regions, segregation of the replicated bulk chromosome, and finally resolution of the termini at the division septum (Wang and Rudner 2014; Badrinarayanan et al. 2015). Nevertheless, the molecular mechanisms involved in the chromosome segregation in non-model bacteria are only just beginning to be elucidated. Most sequenced bacterial genomes (with the exception of E. coli) contain a chromosomally encoded par locus, which is consisted of three components: two trans-acting proteins (ParA and ParB) and a cis-acting site (parS). The ParAB – parS system is believed to drive active chromosome segregation in many bacteria, including origin and/or bulk DNA segregations. Deletion of par genes is lethal in some bacteria, e.g., in C. crescentus (Mohl and Gober 1997; Mohl et al. 2001), V. cholera (parAB2) (Yamaichi et al. 2007) and Myxococcus xanthus (Iniesta 2014). In C. crescentus, the ParA retraction has been shown to mediate the latter part of ParB translocation, thus is crucial for the final stages of chromosome segregation (Shebelut et al. 2010). Deletion of the parAB2 genes located on chromosome II of V. cholera could lead to localization and segregation defects of the chromosome II, and yield cells with only chromosome I (Yamaichi et al. 2007). In some other species, par mutants are viable but exhibit various severity defects of chromosome bulk DNA or origin segregation in the vegetative and/or sporulation phase of growth, e.g., in B. subtilis, S. coelicolor, V. cholera (parAB1 of chromosome I), P. aeruginosa, C. glutamicum, S. pneumoniae, and M. smegmatis (Ireton et al. 1994; Sharpe and Errington 1996; Lewis and Errington 1997; Kim et al. 2000; Jakimowicz et al. 2002; Fogel and Waldor 2006; Lasocki et al. 2007; Murray and Errington 2008; Sullivan et al. 2009; Gruber and Errington 2009; Donovan et al. 2010; Scholefield et al. 2011; Minnen et al. 2011; Ginda et al. 2013). The bacterial actin-like protein MreB has also been suggested to be involved in chromosome segregation in addition to its role in cell morphology determination (Sundararajan and Goley 2017). In B. subtilis, Soufo and Graumann (2003) showed that depletion of mreB led to the appearance of 25% anucleate cells, and depletion of the other two mreB homologs (mbl or mreBH) also caused a considerable increase in the fraction of anucleate cells. In E. coli, the initial studies truly showed that MreB is necessary for origin and bulk nucleoid segregation; when the E. coli cells were treated with A22 which is a new antibiotic specifically targeting MreB, chromosome segregation was inhibited, resulting in cells containing large confluent bodies of nucleoids (Kruse et al. 2003, 2006). In C. cresentus, alterations in MreB expression also cause defects in chromosome segregation (Gitai et al. 2004, 2005). The A22 treated C. cresentus cells experienced growth condition-specific defects in segregation of the chromosomal origin-proximal regions (Gitai et al. 2005; Shebelut et al. 2009). In this work, the T. thermophilus parA and mreB gene were simultaneously deleted by replacement with antibiotic-resistant genes. The results showed that the parA and mreB deletion mutant had a similar chromosome partitioning pattern as the wild type (Figure 3A, B), indicating no apparent chromosome segregation defects occurred in the ΔmreBΔparA muant. Thus, both the Par and MreB systems are not involved in the chromosomal bulk DNA segregation in T. thermophilus. A similar result was found in the polyploid cyanobacterium Anabaena sp. PCC 7120, in which the lack of mreB did not lead to any DNA segregation defects in the two dividing daughter cells (Hu et al. 2007). Still, it is of interest what the function is of the parABS and mreB loci in T. thermophilus when they are not critical for cell growth and chromosomal DNA segregation. The parABS loci are located extremely approching oriC (appoximately 6 kb distance) (Li et al. 2015), and ParB could specifically bind parS forming nucleoprotein complexes spreading around the origin region, indicating that parABS are actually functional. Thus, parABS may contribute to other cell functions, such as serve to be transcriptional regulators. Recent studies have shown that analogous to their counterparts on the plasmids, the chromosomal Par proteins can also regulate transcriptions of other genes located around parS sites (Bartosik et al. 2014; Baek et al. 2014; Attaiech et al. 2015). For example, in P. aeruginosa, deletion of parB leads to global transcriptional changes affecting more than 1000 genes (Bartosik et al. 2014). Thus, it will prove interesting to test whether this is the situation for the T. thermophilus Par system in the future. Further, although MreB did not participate in the chromosome segregation, it was found to be a cell shape determinant in T. thermophilus. The cells of the ΔmreB::kat/ΔparA::blm double gene deletion mutant and ΔmreB::kat single gene deletion mutant have become much thicker and rounder compared with those of the wild-type strain (Figure 3A, middle row). The average ratio of cell length/diameter was 4.24 ± 0.21 in ΔmreB::kat, while that in the wild type was 8.71 ± 0.86, in addition, a certain number of the mutant cells have become drastically abnormal (not shown).
Taken together, it seems that the specific protein-based chromosome segregation machineries, e.g., ParAB – parS system and/or MreB are critical for certain bacteria containing a single chromosome during most of the cell cycles, as they ensure that the daughter cells receive an equal number of the parental chromosome during separation of sister chromosomes. Alternatively, bacteria normally containing multiple chromosomes per cell may not require an active segregation mechanism, since the frequency of daughter cells receiving none of the parental chromosomes is actually low (Graumann 2010). This is analogous to high-copy-number plasmid systems, which typically lack an active segregation mechanism (Ebersbach and Gerdes 2005).
An intriguing question is how the polyploid bacteria segregate their chromosome copies to the daughter cells. The allele segregation kinetics experiment in the heterozygous T. thermophilus strain HL01 suggested that the chromosome segregation mode in T. thermophilus is variable, and random segregation could occur. This suggestion was strengthened by monitoring the relative DNA content in dividing cells via DAPI-staining and by in vivo monitoring of ParB-sGFP/parS protein-DNA complexes (Figure 3). Similar conclusions were drawn for the polyploid cyanobacteria Anabaena sp. PCC 7120 (Hu et al. 2007) and Synechocystis sp. (Schneider et al. 2007), and the polyploid euryarchaeota M. jannaschii (Malandrin et al. 1999). In these prokaryotes, it would be possible that nonprotein-based chromosome segregation mechanisms, e.g., physical forces from extrusion of DNA from replication forks may help push it toward opposite poles (Jun and Mulder 2006; Jun and Wright 2010; Fisher et al. 2013; Badrinarayanan et al. 2015), thereby random chromosome segregation occur frequently. The observation that septa were not always formed exactly at the middle position of T. thermophilus cells could actually support the proposal. Further, it is worthy to note that the relaxation of DNA segregation control may not be a general trait for all polyploid bacteria or archaea. It has been shown that chromosomes were segregated precisely in the polyploid archaeon, Halobacterium salinarum (Breuert et al. 2006) as well as in the polyploid cyanobacterium, Synechococcus elongates (Jain et al. 2012).
Acknowledgments
The plasmids pCT3FK and pMK18 were kindly provided by Prof. Wolfgang Liebl and Dr. Angel Angelov from Technical University of Munich. This work was supported by the National Natural Science Foundation of China (grant number 31700059), Natural Science Foundation of Shannxi province (grant number 2018JQ3037) and Young Talent Fund of University Association for Science and Technology in Shaanxi, China (grant number 20180210).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7746878.
Communicating editor: S. Jaspersen
Literature Cited
- Alvarez L., Bricio C., Gómez M. J., Berenguer J., 2011. Lateral transfer of the denitrification pathway genes among Thermus thermophilus strains. Appl. Environ. Microbiol. 77: 1352–1358. 10.1128/AEM.02048-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov A., Li H., Geissler A., Leis B., Liebl W., 2013. Toxicity of indoxyl derivative accumulation in bacteria and its use as a new counterselection principle. Syst. Appl. Microbiol. 36: 585–592. 10.1016/j.syapm.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Angelov A., Mientus M., Liebl S., Liebl W., 2009. A two-host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst. Appl. Microbiol. 32: 177–185. 10.1016/j.syapm.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Attaiech L., Minnen A., Kjos M., Gruber S., Veening J. W., 2015. The ParB-parS chromosome segregation system modulates competence development in Streptococcus pneumoniae. MBio 6: e00662 10.1128/mBio.00662-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A., Le T. B., Laub M. T., 2015. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 31: 171–199. 10.1146/annurev-cellbio-100814-125211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J. H., Rajagopala S. V., Chattoraj D. K., 2014. Chromosome segregation proteins of Vibrio cholerae as transcription regulators. MBio 5: e01061-14 10.1128/mBio.01061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik A. A., Glabski K., Jecz P., Mikulska S., Fogtman A., et al. , 2014. Transcriptional profiling of ParA and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS One 9: e87276 10.1371/journal.pone.0087276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuert S., Allers T., Spohn G., Soppa J., 2006. Regulated polyploidy in halophilic archaea. PLoS One 1: e92 10.1371/journal.pone.0000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns S. J., Wu H., Akerboom J., Turnbull A. P., de Vos W. M., et al. , 2005. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 280: 11422–11431. 10.1074/jbc.M413623200 [DOI] [PubMed] [Google Scholar]
- Cass J. A., Kuwada N. J., Traxler B., Wiggins P. A., 2016. Escherichia coli chromosomal loci segregate from midcell with universal dynamics. Biophys. J. 110: 2597–2609. 10.1016/j.bpj.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- César C. E., Álvarez L., Bricio C., van Heerden E., Littauer D., et al. , 2011. Unconventional lateral gene transfer in extreme thermophilic bacteria. Int. Microbiol. 14: 187–199. [DOI] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. 10.1038/nrg1711 [DOI] [PubMed] [Google Scholar]
- de Grado M., Castan P., Berenguer J., 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42: 241–245. 10.1006/plas.1999.1427 [DOI] [PubMed] [Google Scholar]
- Donovan C., Schwaiger A., Krämer R., Bramkamp M., 2010. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J. Bacteriol. 192: 3441–3451. 10.1128/JB.00214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G., Gerdes K., 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39: 453–479. 10.1146/annurev.genet.38.072902.091252 [DOI] [PubMed] [Google Scholar]
- Fisher J. K., Bourniquel A., Witz G., Weiner B., Prentiss M., et al. , 2013. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153: 882–895. 10.1016/j.cell.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel M. A., Waldor M. K., 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20: 3269–3282. 10.1101/gad.1496506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., 3rd, Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Ginda K., Bezulska M., Zió-lkiewicz M., Dziadek J., Zakrzewska-Czerwińska J., et al. , 2013. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol. Microbiol. 87: 998–1012. 10.1111/mmi.12146 [DOI] [PubMed] [Google Scholar]
- Gitai Z., Dye N. A., Reisenauer A., Wachi M., Shapiro L., 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120: 329–341. 10.1016/j.cell.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Gitai Z., Dye N., Shapiro L., 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101: 8643–8648. 10.1073/pnas.0402638101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P. L., 2010. The chromosome segregation machinery in bacteria, pp. 31–48 in Bacterial Chromatin, edited by Dame R. T., Dorman C. J. Springer, Dordrecht: 10.1007/978-90-481-3473-1_3 [DOI] [Google Scholar]
- Griese M., Lange C., Soppa J., 2011. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323: 124–131. 10.1111/j.1574-6968.2011.02368.x [DOI] [PubMed] [Google Scholar]
- Gruber S., Errington J., 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137: 685–696. 10.1016/j.cell.2009.02.035 [DOI] [PubMed] [Google Scholar]
- Guschinskaya N., Brunel R., Tourte M., Lipscomb G. L., Adams M. W., et al. , 2016. Random mutagenesis of the hyperthermophilic archaeon Pyrococcus furiosus using in vitro mariner transposition and natural transformation. Sci. Rep. 6: 36711 10.1038/srep36711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. T., 1978. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J. Bacteriol. 134: 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A., Brüggemann H., Raasch C., Wiezer A., Hartsch T., et al. , 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22: 547–553. 10.1038/nbt956 [DOI] [PubMed] [Google Scholar]
- Hildenbrand C., Stock T., Lange C., Rother M., Soppa J., 2011. Genome copy numbers and gene conversion in methanogenic archaea. J. Bacteriol. 193: 734–743. 10.1128/JB.01016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Yang G., Zhao W., Zhang Y., Zhao J., 2007. MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 63: 1640–1652. 10.1111/j.1365-2958.2007.05618.x [DOI] [PubMed] [Google Scholar]
- Iniesta A. A., 2014. ParABS system in chromosome partitioning in the bacterium Myxococcus xanthus. PLoS One 9: e86897 10.1371/journal.pone.0086897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K., N. W. 4th Gunther, and A. D. Grossman, 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176: 5320–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain I. H., Vijayan V., O’Shea E. K., 2012. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc. Natl. Acad. Sci. USA 109: 13638–13643. 10.1073/pnas.1211144109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D., Chater K., Zakrzewska-Czerwínska J., 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 45: 1365–1377. 10.1046/j.1365-2958.2002.03102.x [DOI] [PubMed] [Google Scholar]
- Jun S., Wright A., 2010. Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol. 8: 600–607. 10.1038/nrmicro2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S., Mulder B., 2006. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. USA 103: 12388–12393. 10.1073/pnas.0605305103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhlova O., Bock R., 2006. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46: 85–94. 10.1111/j.1365-313X.2006.02673.x [DOI] [PubMed] [Google Scholar]
- Kim H. J., Calcutt M. J., Schmidt F. J., Chater K. F., 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182: 1313–1320. 10.1128/JB.182.5.1313-1320.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman T. G., Lenarcic R., Willis C. R., Roberts D. M., Hamoen L. W., et al. , 2016. Complex polar machinery required for proper chromosome segregation in vegetative and sporulating cells of Bacillus subtilis. Mol. Microbiol. 101: 333–350. 10.1111/mmi.13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Siegele D. A., Tormo A., 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47: 855–874. 10.1146/annurev.mi.47.100193.004231 [DOI] [PubMed] [Google Scholar]
- Kruse T., Blagoev B., Løbner-Olesen A., Wachi M., Sasaki K., et al. , 2006. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 20: 113–124. 10.1101/gad.366606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Bork-Jensen J., Gerdes K., 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55: 78–89. 10.1111/j.1365-2958.2004.04367.x [DOI] [PubMed] [Google Scholar]
- Kruse T., Møller-Jensen J., Løbner-Olesen A., Gerdes K., 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22: 5283–5292. 10.1093/emboj/cdg504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampo T. J., Kuwada N. J., Wiggins P. A., Spakowitz A. J., 2015. Physical modeling of chromosome segregation in Escherichia coli reveals impact of force and DNA relaxation. Biophys. J. 108: 146–153. 10.1016/j.bpj.2014.10.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Zerulla K., Breuert S., Soppa J., 2011. Gene conversion results in the equalization of genome copies in the polyploid haloarchaeon Haloferax volcanii. Mol. Microbiol. 80: 666–677. 10.1111/j.1365-2958.2011.07600.x [DOI] [PubMed] [Google Scholar]
- Lasocki K., Bartosik A. A., Mierzejewska J., Thomas C. M., Jagura-Burdzy G., 2007. Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J. Bacteriol. 189: 5762–5772. 10.1128/JB.00371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. S., Grossman A. D., 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60: 853–869. 10.1111/j.1365-2958.2006.05140.x [DOI] [PubMed] [Google Scholar]
- Leis B., Angelov A., Li H., Liebl W., 2014. Genetic analysis of lipolytic activities in Thermus thermophilus HB27. J. Biotechnol. 191: 150–157. 10.1016/j.jbiotec.2014.07.448 [DOI] [PubMed] [Google Scholar]
- Lewis P. J., Errington J., 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol. 25: 945–954. 10.1111/j.1365-2958.1997.mmi530.x [DOI] [PubMed] [Google Scholar]
- Li H., Angelov A., Pham V. T., Leis B., Liebl W., 2015. Characterization of chromosomal and megaplasmid partitioning loci in Thermus thermophilus HB27. BMC Genomics 16: 317 10.1186/s12864-015-1523-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. C., Grossman A. D., 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685. 10.1016/S0092-8674(00)81135-6 [DOI] [PubMed] [Google Scholar]
- Madabhushi R., Marians K. J., 2009. Actin homolog MreB affects chromosome segregation by regulating topoisomerase IV in Escherichia coli. Mol. Cell 33: 171–180. 10.1016/j.molcel.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malandrin L., Huber H., Bernander R., 1999. Nucleoid structure and partition in Methanococcus jannaschii: an archaeon with multiple copies of the chromosome. Genetics 152: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. E., Clements K. D., Choat J. H., Angert E. R., 2008. Extreme polyploidy in a large bacterium. Proc. Natl. Acad. Sci. USA 105: 6730–6734. 10.1073/pnas.0707522105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnen A., Attaiech L., Thon M., Gruber S., Veening J. W., 2011. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 81: 676–688. 10.1111/j.1365-2958.2011.07722.x [DOI] [PubMed] [Google Scholar]
- Mohl D. A., Gober J. W., 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88: 675–684. 10.1016/S0092-8674(00)81910-8 [DOI] [PubMed] [Google Scholar]
- Mohl D. A., Easter J. Jr., Gober J. W., 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42: 741–755. 10.1046/j.1365-2958.2001.02643.x [DOI] [PubMed] [Google Scholar]
- Murray H., Errington J., 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135: 74–84. 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- Navarro Llorens J. M., Tormo A., Martínez-García E., 2010. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34: 476–495. 10.1111/j.1574-6976.2010.00213.x [DOI] [PubMed] [Google Scholar]
- Ohta T., Tokishita S., Imazuka R., Mori I., Okamura J., 2006. beta-Glucosidase as a reporter for the gene expression studies in Thermus thermophilus and constitutive expression of DNA repair genes. Mutagenesis 21: 255–260. 10.1093/mutage/gel025 [DOI] [PubMed] [Google Scholar]
- Ohtani N., Tomita M., Itaya M., 2010. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J. Bacteriol. 192: 5499–5505. 10.1128/JB.00662-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N., Tomita M., Itaya M., 2015. Curing the megaplasmid pTT27 from Thermus thermophilus HB27 and maintaining exogenous plasmids in the plasmid-free strain. Appl. Environ. Microbiol. 82: 1537–1548. 10.1128/AEM.03603-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J., El Karoui M., Lindell M., Hughes D., 2017. The processive kinetics of gene conversion in bacteria. Mol. Microbiol. 104: 752–760. 10.1111/mmi.13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecoraro V., Zerulla K., Lange C., Soppa J., 2011. Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-) oligoploid and polyploid species. PLoS One 6: e16392 10.1371/journal.pone.0016392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin J. L., Lee S. F., Garner E. C., Toro E., Eckart M., et al. , 2010. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12: 791–798. 10.1038/ncb2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D., Fuhrmann E., Scholz I., Hess W. R., Graumann P. L., 2007. Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 8: 39 10.1186/1471-2121-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G., Whiting R., Errington J., Murray H., 2011. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol. Microbiol. 79: 1089–1100. 10.1111/j.1365-2958.2010.07507.x [DOI] [PubMed] [Google Scholar]
- Sharpe M. E., Errington J., 1996. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol. 21: 501–509. 10.1111/j.1365-2958.1996.tb02559.x [DOI] [PubMed] [Google Scholar]
- Shebelut C. W., Guberman J. M., van Teeffelen S., Yakhnina A. A., Gitai Z., 2010. Caulobacter chromosome segregation is an ordered multistep process. Proc. Natl. Acad. Sci. USA 107: 14194–14198. 10.1073/pnas.1005274107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebelut C. W., Jensen R. B., Gitai Z., 2009. Growth conditions regulate the requirements for Caulobacter chromosome segregation. J. Bacteriol. 191: 1097–1100. 10.1128/JB.00862-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D., Lindner A. B., Paul G., Radman M., 2009. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136: 1044–1055. 10.1016/j.cell.2009.01.018 [DOI] [PubMed] [Google Scholar]
- Soppa J., 2011. Ploidy and gene conversion in Archaea. Biochem. Soc. Trans. 39: 150–154. 10.1042/BST0390150 [DOI] [PubMed] [Google Scholar]
- Soppa J., 2013. Evolutionary advantages of polyploidy in halophilic archaea. Biochem. Soc. Trans. 41: 339–343. 10.1042/BST20120315 [DOI] [PubMed] [Google Scholar]
- Soppa J., 2014. Polyploidy in archaea and bacteria: about desiccation resistance, giant cell size, long-term survival, enforcement by a eukaryotic host and additional aspects. J. Mol. Microbiol. Biotechnol. 24: 409–419. 10.1159/000368855 [DOI] [PubMed] [Google Scholar]
- Soufo H. J., Graumann P. L., 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13: 1916–1920. 10.1016/j.cub.2003.10.024 [DOI] [PubMed] [Google Scholar]
- Spaans S. K., van der Oost J., Kengen S. W., 2015. The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles 19: 741–750. 10.1007/s00792-015-0750-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. L., Marquis K. A., Rudner D. Z., 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137: 697–707. 10.1016/j.cell.2009.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan K., Goley E. D., 2017. Cytoskeletal proteins in Caulobacter crescentus: spatial orchestrators of cell cycle progression, development, and cell shape. Subcell. Biochem. 84: 103–137. 10.1007/978-3-319-53047-5_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. A., Panis G., Viollier P. H., Marczynski G. T., 2017. A novel nucleoid-associated protein coordinates chromosome replication and chromosome partition. Nucleic Acids Res. 45: 8916–8929. 10.1093/nar/gkx596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro E., Hong S. H., McAdams H. H., Shapiro L., 2008. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. USA 105: 15435–15440. 10.1073/pnas.0807448105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hoffmann J., Watzlawick H., Altenbuchner J., 2016. Markerless gene deletion with cytosine deaminase in Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 82: 1249–1255. 10.1128/AEM.03524-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rudner D. Z., 2014. Spatial organization of bacterial chromosomes. Curr. Opin. Microbiol. 22: 66–72. 10.1016/j.mib.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Brandão H. B., Le T. B., Laub M. T., Rudner D. Z., 2017. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355: 524–527. 10.1126/science.aai8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y., Fogel M. A., Waldor M. K., 2007. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 104: 630–635. 10.1073/pnas.0608341104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka K., Slade D., Bailone A., Sommer S., Averbeck D., et al. , 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443: 569–573. [DOI] [PubMed] [Google Scholar]
- Zerulla K., Ludt K., Soppa J., 2016. The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology 162: 730–739. 10.1099/mic.0.000264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The author affirm that all plasmids are available upon request, and all data necessary for confirming the results of the article are included within the article, figures and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7746878.