Abstract
The root-knot nematode (RKN) species Meloidogyne incognita and M. javanica cause substantial root system damage and suppress yield of susceptible cowpea cultivars. The narrow-based genetic resistance conferred by the Rk gene, present in some commercial cultivars, is not effective against Rk-virulent populations found in several cowpea production areas. The dynamics of virulence within RKN populations require a broadening of the genetic base of resistance in elite cowpea cultivars. As part of this goal, F and F populations from the cross CB46-Null (susceptible) x FN-2-9-04 (resistant) were phenotyped for M. javanica induced root-galling (RG) and egg-mass production (EM) in controlled growth chamber and greenhouse infection assays. In addition, F families of the same cross were phenotyped for RG on field sites infested with Rk-avirulent M. incognita and M. javanica. The response of F to RG and EM indicated that resistance to RKN in FN-2-9-04 is partially dominant, as supported by the degree of dominance in the F and F populations. Two QTL associated with both RG and EM resistance were detected on chromosomes Vu01 and Vu04. The QTL on Vu01 was most effective against aggressive M. javanica, whereas both QTL were effective against avirulent M. incognita. Allelism tests with CB46 x FN-2-9-04 progeny indicated that these parents share the same RKN resistance locus on Vu04, but the strong, broad-based resistance in FN-2-9-04 is conferred by the additive effect of the novel resistance QTL on Vu01. This novel resistance in FN-2-9-04 is an important resource for broadening RKN resistance in elite cowpea cultivars.
Keywords: Meloidogyne spp., Quantitative Trait Loci, Vigna unguiculata
Root-knot nematode (RKN) species, particularly Meloidogyne incognita and M. javanica, cause substantial damage to root systems and suppress yield of susceptible cowpea (Vigna unguiculata L. Walp) cultivars by impairing water and nutrient uptake, and the partitioning and translocation of photo-assimilates (Bird and Loveys 1975; McClure 1977; Taylor and Sasser 1978; Williamson and Hussey 1996; Sikora et al. 2005). Host-plant resistance is an important strategy to mitigate the impact of nematode infestation (Hall and Frate 1996; Roberts 1992; Ehlers et al. 2000b; Castagnone-Sereno 2002; National Research Council 2006), both in Africa where access to agronomic inputs including nematicides is limited (Sasser 1980; Luc et al. 2005), and in developed agriculture where resistant varieties are the best option economically (Ehlers et al. 2000b).
Narrow-based resistance conferred by a single dominant gene Rk has provided protection against RKN in cowpea agricultural systems worldwide (Amosu and Franckowiak 1974; Singh and Reddy 1986; Helms et al. 1991; Fery et al. 1994; Roberts et al. 1995; Roberts et al. 1996; Roberts et al. 1997; Ehlers and Hall 1997; Ehlers et al. 2009). The resistance conferred by gene Rk is highly effective against avirulent forms of RKN populations (Roberts et al. 1995; Hall and Frate 1996; Roberts et al. 1997; Ehlers et al. 2000a,b; Roberts et al. 2013), but Rk-virulent and aggressive forms of common RKN species have been identified (Swanson and Van Gundy 1984; Roberts et al. 1995; Hall and Frate 1996; Roberts et al. 1997; Petrillo et al. 2006). Selection for virulence to Rk (Roberts et al. 1997; Petrillo and Roberts 2005; Petrillo et al. 2006) has prompted efforts to broaden the genetic base of resistance in elite cowpea cultivars (Hall and Frate 1996; Roberts et al. 1996; Roberts et al. 1997; Ehlers et al. 2000b; Roberts et al. 2013). The threat imposed by virulence in RKN populations led to the discovery of new resistance genes, Rk - with a dominant effect (Roberts et al. 1996; Roberts et al. 1997; Ehlers et al. 2000b) and rk - with recessive and additive effect (Roberts et al. 1996; Ehlers et al. 2000a), to broaden the genetic base of resistance, and advanced breeding materials with a combination of these resistance genes have shown promising performance under RKN infestation (Roberts et al. 1996; Roberts et al. 1997; Ehlers et al. 2000b; Ehlers et al. 2002). For example, the additive effect of gene Rk in breeding line IT84S-2049 (which also carries gene Rk) contributes substantially to an enhanced resistance to Rk-virulent populations of M. incognita and to M. javanica compared to gene Rk alone (Roberts et al. 1996; Roberts et al. 1997; Roberts et al. 2005). The rk locus modifies slightly the resistance provided by gene Rk under Rk-virulent RKN isolates (Ehlers et al. 2000b), and it is present in cowpea cv. CB27 (Ehlers et al. 2000a). Although the contribution of these resistance genes is known, their individual action is not clearly understood. However, these examples of resistance gene combinations have shown that broad-based genetic resistance can be developed through effective gene pyramiding of independent sets of resistance genes from distinct genetic sources (Ehlers et al. 2002).
The Rk locus has been mapped on chromosome Vu04 (Huynh et al. 2016), the previous cowpea linkage group 11 of the cowpea consensus genetic map (Lucas et al. 2011; Muñoz-Amatriaín et al. 2017). This genomic region and flanking markers associated with RKN resistance within this region are important resources for introgressing this resistance into elite cowpea cultivars. Also, markers flanking the resistance in this genomic region can be utilized as a reference to decipher the genetic relationship between the resistance conferred by gene Rk and potential novel sources of resistance to RKN. A broad-based resistance to RKN has been identified through a series of field, greenhouse and seedling growth pouch tests in a cowpea accession FN-2-9-04 from Mozambique (Ndeve et al. 2018). This accession carries higher levels of resistance to avirulent M. incognita and M. javanica than that conferred by the Rk gene alone. The performance of FN-2-9-04 under M. javanica infestation was contrasted to cowpea breeding lines and cowpea cultivars carrying sets of RKN resistance genes, including RkRk/, RkRk/, RkRk//gg and IT84S-2049 which indicated that the RKN resistance in accession FN-2-9-04 is unique. Therefore, to characterize the resistance in FN-2-9-04, genetic analyses were conducted to determine its genomic architecture and localization through genetic linkage analysis and QTL mapping.
Materials and Methods
Plant materials
Four F, three F and one F populations (Table 1) were developed under greenhouse conditions at the University of California Riverside (UCR). Accession FN-2-9-04 was crossed with CB46-Null, CB46, Ecute and INIA-41. A single F seed from each of the crosses CB46-Null x FN-2-9-04, CB46 x FN-2-9-04 and INIA-41 x FN-2-9-04 was grown to derive three independent F populations, and 150 F lines of population CB46-Null x FN-2-9-04 were advanced to generate 150 F families (Table 1). Four F populations (CB46-Null x FN-2-9-04, CB46 x FN-2-9-04, INIA-41 x FN-2-9-04, Ecute x FN-2-9-04) and subsets of their F populations were phenotyped for root-galling and egg-mass production in greenhouse and seedling growth-pouch screens, respectively, following infection with nematode isolates listed in Table 1. Five to ten seeds per F population were also screened in each test. The subsets of F populations and F families (Table 1) also were phenotyped for root-galling in field experiments.
Table 1. Cowpea populations used for inheritance studies and QTL mapping, their size, phenotyping conditions, target trait, nematode isolate used and year of testing.
| Exp a | Population | Size | Environment | Trait | Nematode isolate | Year |
|---|---|---|---|---|---|---|
| 1 | bCB46-Null/FN-2-9-04 (F) | 163 | SGP-UCR | EM | M.j | 2015 |
| 2 | bCB46/FN-2-9-04 (F) | 172 | SGP-UCR | EM | M.j | 2015 |
| 3 | bINIA-41/FN-2-9-04 (F) | 126 | GH-UCR | RG | M.j | 2015 |
| 4 | bCB46-Null/FN-2-9-04 (F) | 177 | GH-UCR | RG | M.j | 2015 |
| 5 | bCB46/FN-2-9-04 (F) | 197 | GH-UCR | RG | M.j | 2015 |
| 6 | CB46/ FN-2-9-04 (F) | 400 | CVARS | RG | Avr-M.i | 2015 |
| 7 | CB46/FN-2-9-04 (F) | 162 | KARE | RG | Avr-M.i | 2015 |
| 8 | CB46-Null/FN-2-9-04 (F) | 150 | SCREC | RG | M.j | 2016 |
| 9 | CB46-Null/FN-2-9-04 (F) | 150 | SCREC | RG | Avr-M.i | 2016 |
Exp. = experiment; SGP = seedling growth-pouches; GH = greenhouse; RG = root-galling; EM = egg masses; Avr-M.i = avirulent M. incognita and M.j – M. javanica Project 811; UCR = University of California Riverside; CVARS = University of California Coachella Valley Agricultural Research Station; KARE = University of California Kearney Agricultural Research and Extension Center; bExperiment included the F plus Ecute x FN-2-9-04 F plants.
CB46 is a California blackeye cultivar carrying gene Rk (Helms et al. 1991), and the CB46-Null genotype is a near-isogenic breeding line (NIL) derived from CB46. This breeding line has the CB46 background, but it is susceptible (minus Rk via backcrossing) (Huynh et al. 2016). Ecute and INIA-41 are landraces and FN-2-9-04 is an accession from Mozambique. FN-2-9-04 is resistant to both the avirulent M. incognita isolates and M. javanica isolate used in this study, whereas CB46-Null, CB46, Ecute and INIA-41 are all susceptible to M. javanica. In addition, CB46-Null and Ecute are susceptible to the avirulent M. incognita isolates (Beltran and Project 77), whereas INIA-41 is resistant.
Root-knot nematode isolates
Four RKN isolates were used to phenotype plant materials for response to infection. Three M. incognita isolates, Beltran, Project 77 and an equivalent isolate indigenous to CVARS are avirulent to the Rk gene, with little or no galling and EM production on root systems of plants carrying gene Rk (Roberts et al. 1995; Roberts et al. 1996; Roberts et al. 1997), whereas M. javanica isolate Project 811 is an aggressive isolate due to its enhanced parasitic ability (Ehlers et al. 2000b; Ehlers et al. 2009), inducing galling and reproducing successfully on roots of plants carrying Rk (Thomason and McKinney 1960; Roberts et al. 1997; Ehlers et al. 2009).
Resistance phenotyping: egg-mass production
The F and F populations (Table 1) plus parental genotypes were phenotyped for M. javanica EM production in seedling growth-pouches according to Ehlers et al. (2000b) and Atamian et al. (2012). Briefly, a single seed of each F and F was planted per plastic pouch, and the plants were grown in a controlled environment chamber with day/night temperatures set at 28/22 oC under 16 h day-length. Plants were inoculated two weeks after germination with 1500 freshly hatched second-stage juveniles (J) of M. javanica. Two days after inoculation, plants were supplied daily with fertilizer for 3-5 days using half-strength Hoagland’s solution (Hoagland and Arnon 1950). Thirty-five days after inoculation, the pouches were irrigated with erioglaucine dye (Sigma Chemical Co., St. Louis, MO, USA) to stain egg-masses, which were counted under 10X magnification.
Resistance phenotyping: root-galling
Phenotyping for resistance to root-galling was conducted under greenhouse and field conditions in 2015 and 2016 (Table 1). In the greenhouse, the F and F populations and parental genotypes phenotyped for response to M. javanica egg-mass production in seedling growth-pouches (in growth chamber conditions) were then transplanted into 4L pots containing UC mix 3 soil and maintained at 28/22 oC day/night temperatures. After 21 days, each plant was inoculated with 10 ml of M. javanica egg suspension in water adjusted to 1000 eggs/ml. All greenhouse-grown plants were irrigated twice per day by drip-irrigation for about 90 days to allow seed production, and F seeds were collected from each F plant. After seed collection, the plant tops were cut at 2 – 3 cm above the soil line, and the roots were washed and scored for root-galling response under 10X magnification, using a 0 - 9 gall index (GI) modified from Bridge and Page (1980): 0 = no galls on root system; 1 = very few, small galls and hard to see; 5 = generally large galls can be seen on the root system and the taproot slightly galled, with galls of different sizes; 9 = large galls on the root system, and most lateral roots lost.
Field experiments were conducted in 2015 and 2016 at three sites (Table 1). At CVARS and KARE, 400 and 162 CB46 x FN-2-9-04 F lines, respectively, were phenotyped for root-galling response to avirulent M. incognita (isolate Project 77 at KARE and an equivalent to it at CVARS). In 2016 at SCREC parental genotypes, F and F populations were phenotyped for root-galling response in separate fields infested with avirulent M. incognita isolate Beltran or M. javanica (Table 3). In both experiments (Exps. 8 and 9), F families with 25 – 30 plants/family were planted in single plots. The M. javanica isolate used in the pot and seedling growth-pouch screens was the same isolate used to infest field sites. For both F and F generations, 25 - 30 seeds were planted on a 1.5 m-long single row plot, and 60 days after plant emergence plant tops were cut at 2 – 3 cm above the soil line, and the root systems dug and evaluated for root-galling using the same root-galling index described for the pot tests (Bridge and Page 1980).
Table 3. Best fit segregation ratios (resistant:susceptible) in 119 and 141 F plants from crosses CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04, respectively, determined using SNP marker loci at the two nematode resistance QTL regions.
| Genotypes (Observed)a | ||||||||
|---|---|---|---|---|---|---|---|---|
| F Population | BB + AB | AA | Exp | P-value | Trait | Vu | Isolate | |
| 96 | 23 | 13:3b | 0.002 | 0.95-0.99 | RG | 1 | ||
| CB46-NullxFN-2-9-04 | 93 | 26 | 13:3b | 0.56 | 0.25-0.50 | RG | 4 | Avr-M.i |
| CB46-NullxFN-2-9-04 | 97 | 22 | 13:3b | 0.002 | 0.95-0.99 | RG | 1 | M.j |
| CB46-NullxFN-2-9-04 | 98 | 21 | 13:3b | 0.04 | 0.75-0.90 | EM | 1 | M.j |
| CB46xFN-2-9-04 | 111 | 30 | 13:3b | 0.44 | 0.50-0.75 | RG | 1 | Mj |
| CB46xFN-2-9-04 | 109 | 32 | 13:3b | 1.19 | 0.25-0.50 | EM | 1 | Mj |
BB = alleles from resistant parent, AB = heterozygous, AA = alleles from susceptible parent; Exp. = expected ratio; RG = root galling, EM = egg masses per root system; Vu = cowpea chromosome naming (Lonardi et al. 2017); Isolate = Nematode isolate; Avr = avirulent M. incognita Beltran, M.j = M. javanica; balso fit a 3:1 ratio.
Inheritance of resistance and allelism test
Segregation for the FN-2-9-04 resistance to root-galling and reproduction by M. javanica and root-galling by avirulent M. incognita isolates was determined using both phenotypic (root-galling and egg-masses) and genotypic data. In addition, phenotypic data of F, F and F populations, and SNP marker genotypes of F populations at mapped QTL regions were processed for goodness-of-fit analysis to determine the genetic model underlying resistance to RKN in FN-2-9-04. Analysis of goodness-of-fit of segregation ratio between resistant-susceptible lines in the F was performed through marker-trait association analysis using marker genotypes within mapped QTL regions (see Table 2) and phenotypic response of F and F populations. Each F line was scored for presence of parental alleles at each locus within the mapped QTL, and scores 2, 1 and 0 were assigned to homozygous favorable allele (BB = resistant parent), heterozygous (AB) and homozygous non-favorable allele (AA = susceptible parent), respectively. The genotype of each F line, within the QTL region, was determined as the mean score across all marker loci, and it was associated with its RG or EM phenotypic response determined at the F and F generations. The data for frequency distribution of genotypes (BB, AB and AA) (Table 3) were processed for goodness-of-fit analysis, and the chi-square values were determined following Yates correction for continuity (Little and Hills 1978). The numbers of genetic determinants associated with resistance were estimated using the Castle (1921) estimator of gene number, , where n is the estimated number of genes influencing the trait, and are the mean phenotypic values of the parents of the population and is the genetic variance of the trait. To estimate the number of genes governing response to root-galling and egg-mass production, the influencing these traits was derived as the genetic variance in the mapped QTL regions, flanked by known SNP markers.
Table 2. Chromosome locations of root-knot nematode (RKN) resistance determinants in cowpea accession FN-2-9-04, mapped using F and F populations of the cross CB46-Null x FN-2-9-04 and the F population of the cross CB46 x FN-2-9-04.
| Pop a | Trait | RKN | Vu | Position | Flanking markers | -logp | PVE (%) | A | D/A |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 34.4 | 2..04038-2..26991 | 5.4 | 33.0 | −1.3 | 0.5 | |||
| F | RG | Avr-M.i | 4 | 24.7-27.6 | 2..44685-2..10583 | 20 | 73.3 | −2.0 | 0.5 |
| F | RG | M.j | 1 | 27.7-42.0 | 2..47796-1.0027 | 20 | 95.1 | −2.3 | 0.3 |
| F | RG | M.j | 1 | 30.3-38.7 | 2..32677-2..19840 | 20 | 47.3 | −2.8 | 0.4 |
| RG | M.j | 1 | 19.2-72.9 | 2..53036-2..18359 | 20 | 65.9 | 2.7 | 0.8 | |
| F | EM | M.j | 1 | 31.5-36.9 | 2..21671-2..07103 | 10.9 | 34.1 | −17.0 | 0.5 |
| EM | M.j | 1 | 47.1-52.1 | 2..21671-2..12209 | 8.8 | 24.7 | −16.4 | 0.4 |
Pop = mapping population; the F were phenotyped in the field whereas the F were phenotyped in greenhouse and growth chamber (seedling-growth pouches) screens; RG = root-galling; EM = egg-masses per root system; Avr-M.i = avirulent M. incognita isolate Beltran; M.j = M. javanica;
mapping population CB46 x FN-2-9-04 phenotyped for RG and EM; Vu = cowpea chromosome pseudomolecule numbering (Lonardi et al. 2017); -logp = level of significance of the detected QTL (P < 0.05); PVE = percent of total phenotypic variation explained; A = additive effect of favorable alleles from the resistant parent (negative values indicate the extent of average reduction in RG or EM production due to the presence of favorable alleles); D = dominance effect due to substitution of favorable allele; and D/A = degree of dominance.
Broad-sense heritability () of resistance was estimated using two methods, midparent-offspring regression analysis (Fernandez and Miller 1985; Falconer and Mackay 1996) and the phenotypic variation among F lines and among F families accounted for by at the QTL regions associated with resistance. The proportion of phenotypic variance, , in root-galling or egg-masses attributed to genetic factors, , was estimated using SNP marker genotype scores () and SNP marker effects () at the mapped QTL regions plus the observed root-galling or egg-masses phenotypes using the algorithm: . In this algorithm (adapted from Xu 2013), the product is the associated with the variation in root-galling or egg-masses phenotypes in tested F and 2 F populations. To estimate the narrow-sense heritability (), the genetic variance () was partitioned into additive and dominance variances, and the component was used to compute the of the trait. Root-galling data of seven F populations (populations in Table 1 plus their subsets) and parental genotypes were used to perform midparent-offspring regression analysis, and four mapping populations (two F and two F, Exps. 1, 4, 8 and 9, Table 1) were used to derive genetic variances () within the QTL regions, influencing the response to galling and egg-mass production. Allelic relationships between the Rk locus present in cv. CB46 (Roberts et al. 1995; Hall and Frate1996; Roberts et al. 1996; Roberts et al. 1997; Ehlers et al. 2009; Huynh et al. 2016) and the genetic determinants of resistance in FN-2-9-04 were determined using the four F population sets of CB46 x FN-2-9-04 phenotyped with M. incognita isolate Project 77 and M. javanica infestation (Table 1).
Genotyping and QTL mapping
Leaf samples were collected from parents and each of 119 and 137 F lines of populations CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04, respectively (Exp. 1, 5, Table 1) 30 days after transplanting and dried in plastic ziploc bags containing silica gel packs. Genomic DNA was extracted from dried leaves using Plant DNeasy (Qiagen protocol) and quantified using Quant-iTTM dsDNA Assay Kit and fluorescence measured using a microplate reader. In addition, each F plant of population CB46-Null x FN-2-9-04 was selfed to generate F seeds for field phenotyping (Table 1). The 119 F lines are part of the 163 lines tested for egg-mass production (Exp. 1) and transplanted for root-galling assay (Exp. 4, Table 1).
Each DNA sample was assayed for single nucleotide polymorphism (SNP) using the Cowpea iSelect Consortium Array containing 51128 SNPs (Muñoz-Amatriaín et al. 2017). The SNP data were filtered for quality as follows: (i) elimination of SNPs with > 20% missing data; (ii) elimination of monomorphic SNPs; (iii) elimination of SNPs with minor allele frequency (MAF) < 0.4 and < 0.3 for populations CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04, respectively; iv) and elimination of duplicated lines. No loci were detected with non-parental alleles.
Linkage-maps of the CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04 F populations were constructed with MSTmap (Wu et al. 2015), and linkage groups were determined at LOD threshold = 10 and marker placement followed the Kosambi mapping function. The options “no mapping size threshold” and “no mapping distance threshold” were fixed at 2 units and 10 cM, respectively. In addition, the no mapping distance threshold option was set at 15 cM and the detection of genotyping errors was not solicited. The linkage groups of the final genetic map were numbered and ordered following the cowpea consensus genetic map order (Muñoz-Amatriaín et al. 2017) and the cowpea pseudomolecules (Lonardi et al. 2017 in preparation; https://phytozome.jgi.doe.gov/). Also, the cowpea reference genome was used to determine the physical positions of the SNPs and the QTL associated with the traits (Lonardi et al. 2017 in preparation; https://phytozome.jgi.doe.gov/).
QTL mapping was performed using five phenotypic data sets comprising two F populations of crosses CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04, and two F populations of cross CB46-Null x FN-2-9-04 (Exps. 1, 4, 5, 8 and 9, Table 1). QTL analysis was performed following the mixed-model for QTL mapping described by Xu (2013) using RStudio v1.1.442, and significant QTL were declared using Bonferroni adjusted threshold value -log (P-value) at P < 0.05. Reported QTL regions associated with resistance were based on the SNP markers with the most significant threshold values. In the mixed-model for QTL analysis (Xu 2013), the analysis comprises three input data sets; phenotypic and genotypic data sets, and a kinship data set matrix generated using genotypic data. The phenotypic response is associated with polygenic and marker effects; and these components are considered as random and fixed effects, respectively. The polygenic effect (total genetic variance) influencing the phenotype is further partitioned in to additive, dominance and epistatic effects. The proportion of phenotypic response explained comprises the genotypic and marker effects (see details in the previous section). All phenotypic data sets comprised raw phenotypic data.
Data Availability
All F and F populations and root-knot nematode isolates are available upon request. Phenotypic and genotypic data are included in data (D) files 1 - 9. These data files, including their description, and supplementary tables and figures are available at Figshare: https://doi.org/10.25387/g3.7324211.
Results
Linkage and QTL mapping
The linkage map of the F population CB46-Null x FN-2-9-04 (n = 119) contained 17208 polymorphic SNP markers distributed on 11 chromosomes and spanned 985.89 cM (Supplementary file S1A). Of the total SNPs, 90.79% (15624 SNPs) were mapped on the cowpea consensus genetic map (Muñoz-Amatriaín et al. 2017), while 9.21% (1585 SNPs) were unique to this population, and this portion corresponds to 2.5% of SNPs not mapped to the cowpea pseudomolecules. The linkage map comprised 1392 bins distributed at an average density of 1 bin per 0.71 cM. The linkage map of the F population CB46 x FN-2-9-04 (n = 137 lines) contained a total of 17903 polymorphic SNPs and spanned 1158.68 cM (Supplementary file S1B). Of these SNPs, 97.6% (17465 SNPs) mapped to the cowpea consensus genetic map, while 9.4% (1675 SNPs) are not part of the cowpea consensus genetic map, and this portion makes 2.4% of the total SNPs not mapped on the cowpea pseudomolecules (Lonardi et al. 2017 in preparation; https://phytozome.jgi.doe.gov/).
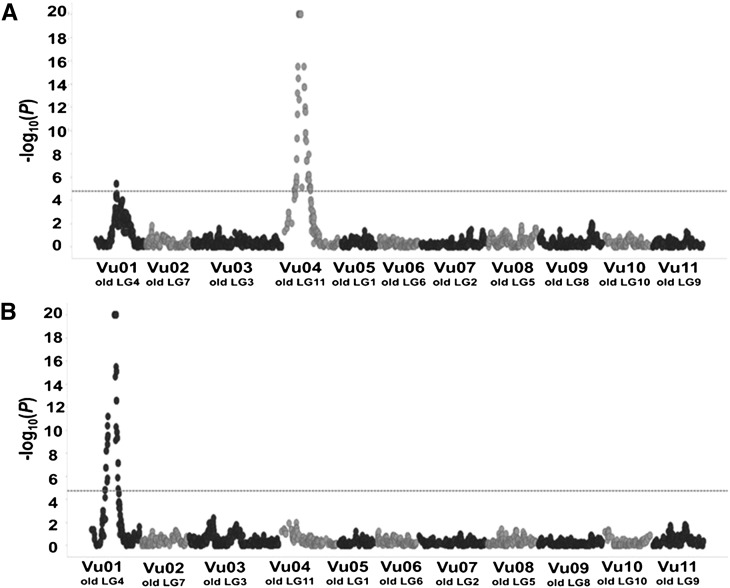
QTL analysis revealed two major QTL associated with resistance to root-galling (RG) and egg-mass (EM) production in FN-2-9-04 (Table 2; Figures 1 and 2); these QTL were mapped on chromosomes Vu01 and Vu04 of the CB46-Null x FN-2-9-04 population and chromosome Vu01 of the CB46 x FN-2-9-04 population. The QTL region on Vu01 consistently mapped almost within the same genomic location using F and F populations phenotyped under greenhouse, seedling-growth pouch and field conditions using two RKN isolates (Table 2; Supplementary file S1C). Two QTL controlling resistance to RG by avirulent M. incognita Beltran were detected and mapped on Vu01 and Vu04 (P < 0.05, threshold value -log(p) = 4.8) (Figure 1A) of the CB46-Null x FN-2-9-04 F population. The resistance QTL on Vu01 mapped to position 34.4 cM which spanned 0.1 Mb (28855569 - 28960128 bp) on the cowpea pseudomolecules (Supplementary file S1C) between flanking markers 2..04038 and 2..04039; it accounted for 33% of the total phenotypic variation (Vp) of the RG resistance response and had a likelihood of occurrence expressed by -log10(p) = 5.4 (Table 2). Input data files used for this QTL discovery are D1 and D2.
Figure 1.
Genomic localization of QTL associated with resistance to root-galling (RG) by: A, avirulent M. incognita and B, aggressive M. javanica. The QTL were detected in the CB46-Null x FN-2-9-04 F population phenotyped for RG under field infestation. Horizontal dashed line represents the Bonferroni threshold of significance at P < 0.05 [-log(p) = 4.8]. Old LG represents former cowpea linkage group numbering and Vu indicates the new cowpea linkage group numbering based on the cowpea pseudomolecules (Lonardi et al. 2017).
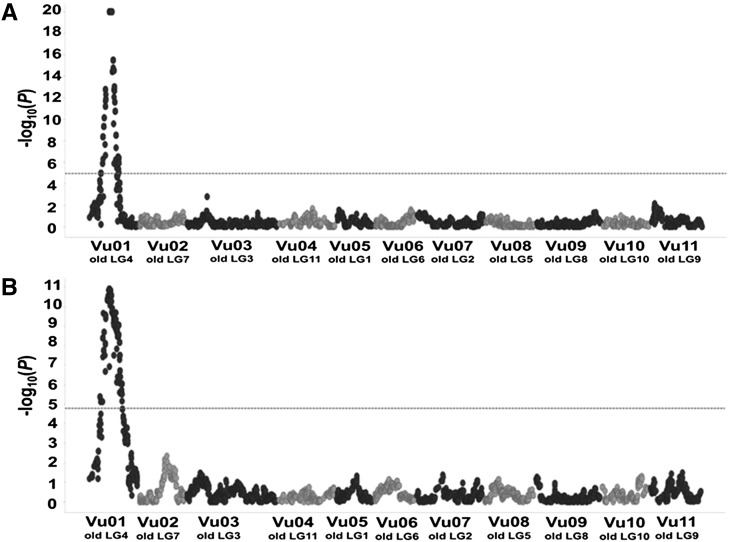
Figure 2.
Genomic localization of QTL associated with resistance to A, root-galling (RG) and B, egg-mass production (EM) by aggressive M. javanica. The QTL were detected in the F population CB46-Null x FN-2-9-04 phenotyped for RG in the greenhouse and for EM in seedling growth-pouch inoculations, respectively. Horizontal dashed-line represents the Bonferroni threshold of significance at P > 0.05 [-log(p) (A and B = 4.9 and 4.8, respectively). Old LG represents former cowpea linkage group numbering and Vu indicates the new cowpea linkage group numbering based on the cowpea pseudomolecules (Lonardi et al. 2017).
The resistance QTL on Vu01 (Figure 1A) detected under plant infection by avirulent M. incognita, exhibited additive and dominance effects of -1.3 and -0.6, respectively, and the degree of dominance, measured as a ratio between dominance and additive effects (D/A), indicated that the resistance in this QTL has partial dominant effect (D/A = 0.5) (Table 2). A second resistance QTL associated with response to the avirulent M. incognita was detected on Vu04 (Figure 1A, Table 2) at chromosome position 24.7 - 27.6 cM of the CB46-Null x FN-2-9-04 F population and spanned 2.9 cM which corresponds to approximately 1 Mb (3141521 – 4138458 bp) on the cowpea pseudomolecules (Supplementary file S1C), and it was flanked by SNP markers 2..44685 and 2..10583 (Table 2). This QTL explained 73.3% of the total Vp of the resistance response, and it had an infinite likelihood of occurrence which was represented by -log10(p) = 20 (Table 2). In addition, the additive (A = -2) and dominance (D = -1) effects of the QTL on Vu04 were slightly higher than those of the QTL on Vu01, but both QTL showed the same degree of dominance (D/A = 0.5).
On Vu01, an additional genomic region controlling resistance to M. javanica RG (Figures 1B; 2A) and EM production (Figure 2B) was consistently mapped on the same chromosomal region of the CB46-Null x FN-2-9-04 F2 and F populations using RG and EM phenotypic data from field, greenhouse and seedling-growth pouch experiments (Table 2). The M. javanica root-galling resistance QTL mapped to positions 30.3 - 38.7 cM and 27.7 - 42.0 cM on Vu01 using F (greenhouse experiment - input data files D1 and D5) and F (Field experiment - input data files D1 and D3) populations from the CB46-Null x FN-2-9-04 cross, respectively. These genomic regions spanned 8.4 and 14.3 cM, which correspond to 4.4 (26617356 - 31070755 bp) and 6.2 Mb (25784028 - 31953708 bp) on the cowpea pseudomolecules (Supplementary file S1C) and were flanked by SNP markers 2..32677 - 2..19840 and 2..47796 - 1..0027, respectively (Table 2). In both F and F populations, the RG resistance QTL was detected with infinite likelihood represented by -log10(p) = 20 (Figures 1B, 2A, Table 2). The percent of total phenotypic variation in RG explained by the QTL effect in the F (PVE = 95.1%) was higher than in the F (PVE = 47.2%), while the contributions of the additive and dominance effects in the total phenotypic variation in the F and F were similar (Table 2). Also, the degree of dominance in both generations were comparable, D/A = 0.4 and 0.3, respectively, indicating resistance with partial dominance.
The QTL on Vu01 associated with resistance to M. javanica reproduction (EM - input data files D1 and D4) mapped to position 31.5-36.9 cM of the CB46-Null x FN-2-9-04 F population (Figure 2B; Table 2). This QTL spanned 5.5 cM which corresponds to 2.7Mb (27254299 - 29984745 bp) on the cowpea pseudomolecules (Supplementary file S1C), and it was flanked by SNP markers 2..21671 and 2..07103. This QTL accounted for 34.1% of the total phenotypic variation in EM production with additive and dominance effects of 17.1 and 7.8, respectively; the gene action measured within the same QTL region indicated resistance with partial dominance (D/A = 0.5). Although this QTL was detected with high likelihood, -log10(p) = 10.9 (critical threshold = 4.8) (Figure 2B), it was lower than that observed for the RG QTL (Table 2).
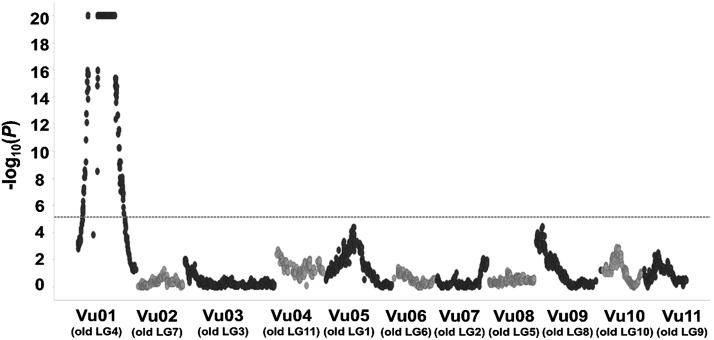
QTL mapping using the F population of CB46 x FN-2-9-04 (input data files D6 and D7) validated that the genomic region on Vu01 is associated with resistance to M. javanica RG (Figure 3; Table 2). This Vu01 genomic region was mapped to position 19.2-72.9 cM in the CB46 x FN-2-9-04 F2 population, and it spanned 53.7 cM which corresponds to 13.5 Mb (20889089 - 34401992 bp) on the cowpea pseudomolecules with flanking SNP markers 2..53036 - 2..18359 (Table 2; Supplementary file S1C). The QTL on Vu01 explained 65.9% of the total phenotypic variation in M. javanica root-galling, and the contribution of the additive and dominance effects were 2.7 and 2.1, respectively. The estimated gene action within this region indicated resistance with partial dominance (D/A = 0.8) (Table 2). This QTL was detected with high likelihood, -log10(p) = 20 (critical threshold = 5.1) (Figure 3). In addition, a genomic region associated with resistance to M. javanica EM production was mapped on Vu01 of the CB46 x FN-2-9-04 F (input data files D8 and D9) at position 46.7 – 53.5 cM, and it spanned 6.8 cM corresponding to 3.2 Mb (27254299 - 30434421 bp) on the cowpea pseudomolecules flanked by SNP markers 2..21671 – 2..12209. This QTL explained 24.7% of the total phenotypic variation in M. javanica EM production. (Table 2; Supplementary file S1C).
Figure 3.
Genomic localization of QTL associated with resistance to root-galling induced by aggressive M. javanica. The QTL was detected in the CB46 x FN-2-9-04 F2 population phenotyped for RG in the greenhouse. Horizontal dashed-line represents the Bonferroni threshold of significance at P < 0.05 [-log(p) = 5.1. Old LG represents former cowpea linkage group numbering and Vu indicates the new cowpea linkage group numbering based on the cowpea pseudomolecules (Lonardi et al. 2017).
Inheritance of resistance in FN-2-9-04
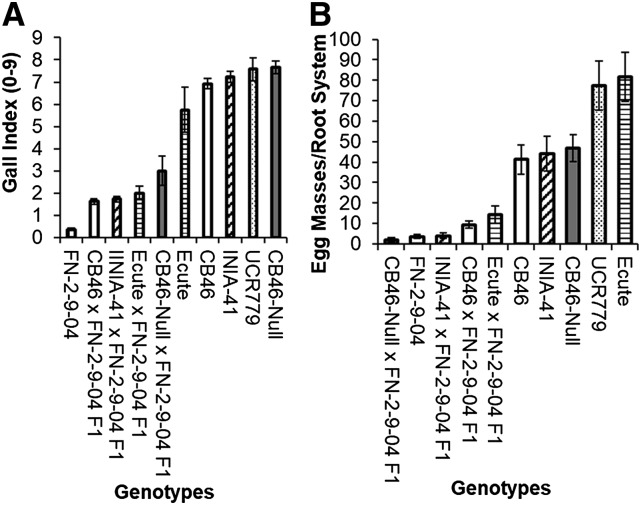
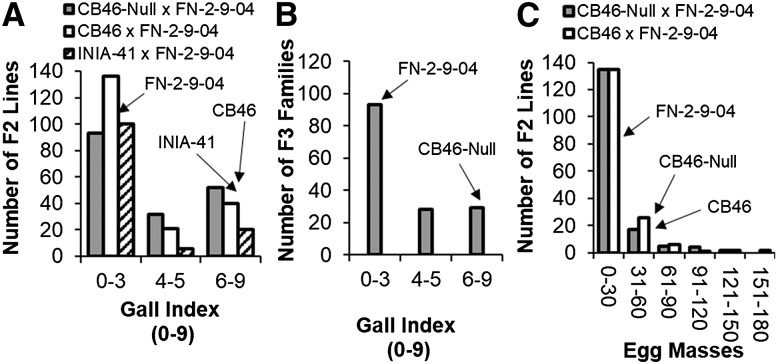
Figures 4A and 4B show the response of four F populations and their parental genotypes to root-galling (RG) and egg-mass (EM) production, respectively by M. javanica. All recurrent parents (Ecute, CB46, INIA-41 and CB46-Null) exhibited susceptible phenotypes for RG and EM, and their mean RG scores and EM scores ranged from 5.8 to 7.7 and 41 to 82, respectively, whereas the resistant parent, FN-2-9-04 had mean RG and EM scores of 0.4 and 4, respectively.
Figure 4.
Mean response of F populations and their parents to: A, root-galling and B, egg-mass production by M. javanica in greenhouse-pot and seedling growth-pouch inoculations, respectively. Bars represent +/− SE.
All F populations were resistant to M. javanica (Figure 4), with mean RG and EM scores below the mid-parent RG and EM score (GI = 6.9 and EM = 53). The CB46-Null x FN-2-9-04 F had the highest mean RG (GI = 3) of the four F populations. The observed differences in RG and EM between the resistant and susceptible parents were significant (P < 0.05), but the RG phenotype of the resistant parent was only different from F populations CB46-Null x FN-2-9-04 and Ecute x FN-2-9-04. The EM phenotypes of the resistant parent and F were not different. Significant differences among the genotypes were detected at GI = 1.3 and EM = 31.4 (Figure 4A and 4B).
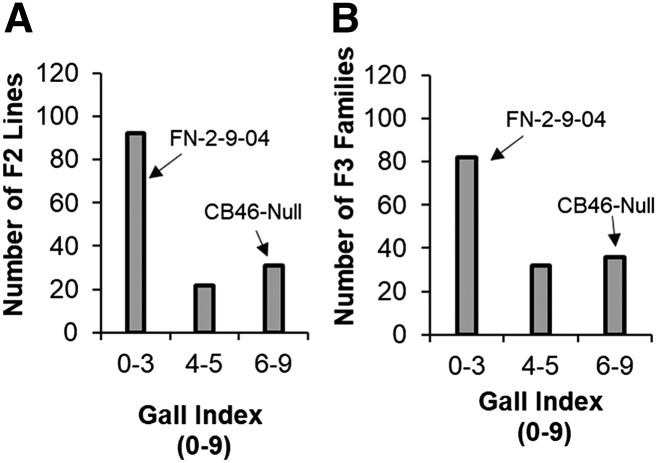
The segregation of F (Figure 5A) and F (Figure 5B) populations for M. javanica RG response appeared to follow a bimodal distribution, skewed toward lower RG phenotype. Also, a bimodal segregation pattern was observed for M. javanica EM production in the CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04 F populations (Figure 5C). In these same experiments, the average RG observed for parents CB46-Null, CB46, INIA-41 and FN-2-9-09 in greenhouse pots was 7.7, 6.9, 7.2 and 0.4, respectively. In the field experiment (Figure 5B), RG of 6.7 and 0.1 were observed for parents CB46-Null and FN-2-9-09, respectively, while egg-mass counts per root system equal to 46.7, 45 and 1.8 were observed for parents CB46-Null, CB46 and FN-2-9-09, respectively (seedling-growth pouches).
Figure 5.
Distribution of root-galling responses in A, F populations (greenhouse), B, F population CB46-Null x FN-2-9-04 (field), and C, egg-mass production in F populations CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04 (seedling growth-pouch) under M. javanica infestation.
A similar pattern of root-galling distribution was observed in F (Figure 6A) and F (Figure 6B) populations of CB46-Null x FN-2-9-04 under field infestation by avirulent M. incognita Beltran. This segregation pattern was consistent across all phenotyping environments (greenhouse, field and seedling growth-pouches) and traits (RG and EM). Egg-mass phenotypes ranged from 0 – 180 (Figure 5C), and RG across environments and generations ranged from 0 – 9 (Figures 5 and 6). The resistant parent FN-2-9-04 had consistently lower (P < 0.05) RG compared to all susceptible parents. The average M. incognita root-galling indices for parents CB46-Null and FN-2-9-04 in the field experiment were 6.4 and 0, respectively.
Figure 6.
Distribution of root-galling response in the F (A) and F (B) populations of CB46-Null x FN-2-9-04 under field infestation with avirulent M. incognita isolate Beltran.
The broad-sense heritability () of resistance to M. javanica root-galling estimated through regression of 7 field phenotyped F populations to the mean performance of their parents (CB46-Null, CB46, FN-2-9-04 and INIA-41,) was high (b = 0.76 ± 0.07, P = 0.00004) (Figure 7), while estimates of for the same trait computed using the genetic variance (Vg*) directly derived from the QTL region located on Vu01 were moderate (0.47) and high (0.95) for greenhouse and field phenotyped F and F populations, respectively. For these populations, the estimates of narrow-sense heritability () of RG were 0.33 and 0.71, respectively. Egg mass production (EM) response in the F had low (0.34) (Table 2) and (0.23). The estimated and for resistance to avirulent M. incognita RG were 0.33 and 0.23 on Vu01 and 0.73 and 0.49 on Vu04, respectively.
Figure 7.
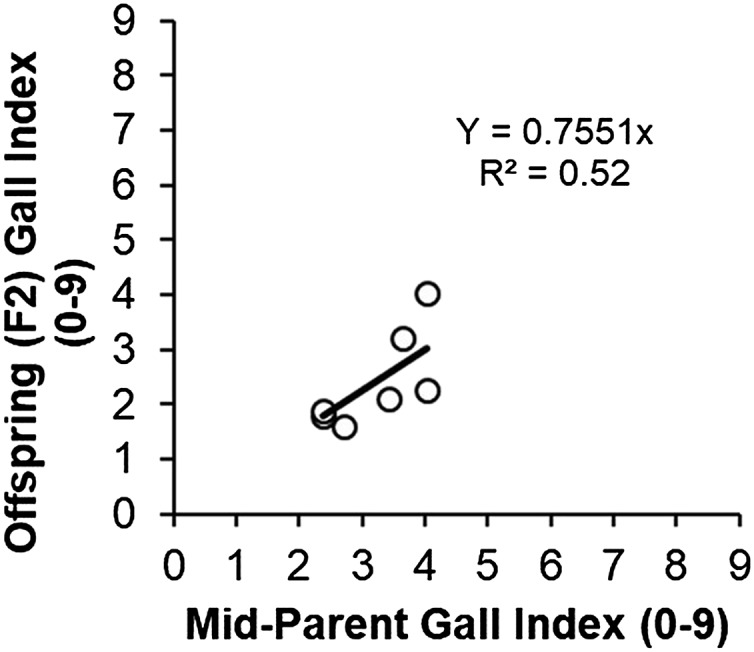
Midparent – offspring regression for F population means regressed on the midparent root-galling values.
Because the M. javanica RG and EM resistance QTL were co-located (Figures 1B, 2A and 2B), analysis of correlation between RG and EM responses was performed using RG and EM data of F populations CB46 x FN-2-9-04 and CB46-Null x FN-2-9-04. These traits were highly correlated in both populations, CB46 x FN-2-9-04 and CB46-Null x FN-2-9-04 (r = 0.78, P = 0.008 and r = 0.62, P = 0.06, respectively), although the correlation in the F population CB46-Null x FN-2-9-04 was not significant (P = 0.06) (Figure 8). The relationship between RG and EM in populations CB46 x FN-2-9-04 and CB46-Null x FN-2-9-04 was explained at 60.3% and 38.1%, respectively, based on the estimated coefficient of determination.
Figure 8.
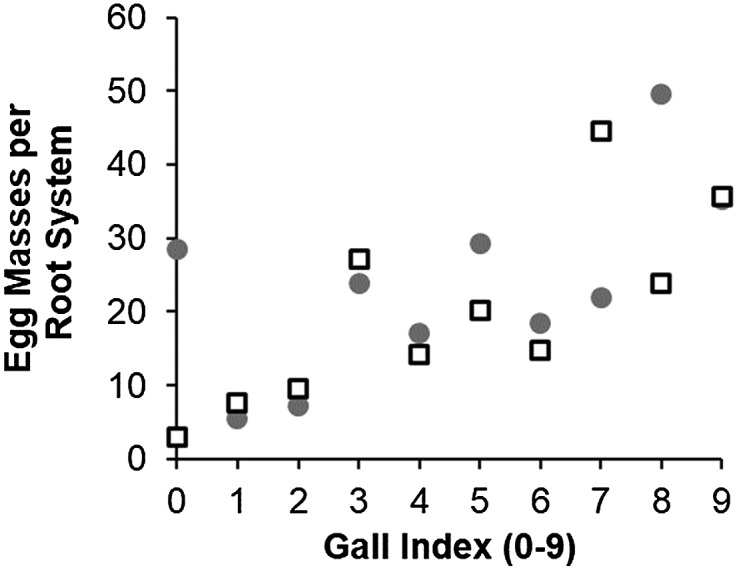
Correlation between M. javanica root-galling (greenhouse) and egg-mass production (seedling growth-pouch) in F populations Circle marker = CB46-Null x FN-2-9-04 (r = 0.62) and square marker = CB46 x FN-2-9-04 (r = 0.78).
The 119 and 137 F lines of populations CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04, respectively, assayed for 51128 SNP markers segregated for resistance-susceptibility to RG and EM within each mapped QTL, and it fit closely a ratio of 13:3 for phenotypic traits (Table 3). Also, a 3:1 ratio was significant, suggesting that the resistance at both QTL regions is mainly governed by one dominant gene or a combination of genes acting under dominant-recessive interaction. The fit to a 13:3 ratio could also indicate genetic distortion for a single dominant gene.
To validate the genetic models of segregation for resistance-susceptibility to avirulent M. incognita and M. javanica, gene enumerations were estimated at the mapped QTL regions associated with resistance to RG (Vu01 and Vu04) and EM production (Vu01) following the Castle (1921) algorithm. The estimates indicated that the resistance to avirulent M. incognita RG is under control primarily by 2 and 5 genes residing in QTL regions mapped on Vu04 and Vu01, respectively; whereas, the responses to M. javanica RG and EM production mapped on Vu01 are governed mainly by 2 genes each (Supplementary file S3).
Because two QTL, on Vu01 and Vu04, were associated with resistance to avirulent M. incognita RG, analysis of QTL allele combinations were performed to understand the interaction of both QTL. Through SNP marker-trait association, the genotype (AA, AB and BB) of each of the 119 F lines was determined at the QTL regions on Vu01 and Vu04 associated with resistance to avirulent M. incognita RG, and each genotype was associated with the average RG phenotypic response of the corresponding F. Based on this association, nine QTL combinations (Vu01/Vu04) (Figure 9) were derived by combining all possible haplotypes on Vu01 and Vu04 contributed from resistant (FN-2-9-04 – favorable allele donor) and susceptible (CB46-Null – non-favorable allele donor) parents.
Figure 9.
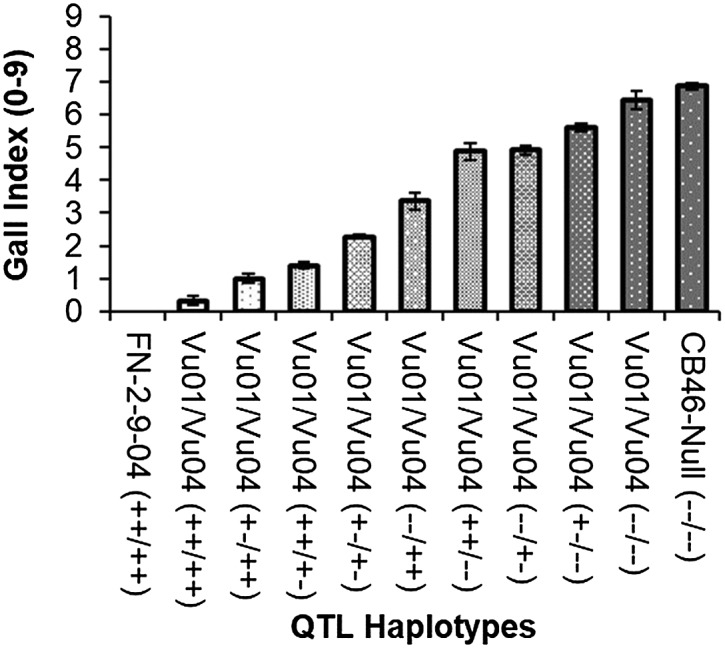
Avirulent M. incognita root-galling values for QTL allele combinations for the resistance traits in accession FN-2-9-04 mapped to Vu01 and Vu04 of the cowpea consensus genetic map. The zygosity status within each QTL is indicated by ++, +- and --, representing homozygous favorable, heterozygous and homozygous un-favorable, respectively, in each QTL. Bars are standard errors.
Analysis of variance showed significant effect (P 0.05) of combining QTL on avirulent M. incognita RG response; significant mean differences in RG phenotypes between genotypes carrying combined QTL were detected at gall index (GI) = 0.88. The resistant parent FN-2-9-04 [Vu01/Vu04(++/++)] did not show any root-galling (Figure 9), and its response was different (P < 0.05) from all genotypes carrying QTL haplotypes with favorable allele dosage different from this parent. Any of the genotypes carrying at least a single favorable allele on at least one of the chromosome regions had less galling than the susceptible parent CB46-Null [Vu01/Vu04(–/–)]. Absence of a single favorable allele in either chromosome predisposed the plants to root-galling, and substantial allele effect was observed for Vu04 [Vu01/Vu04(++/+-)] (Figure 9). At both loci the favorable alleles must be in the homozygous condition for fully effective M. incognita RG resistance.
Resistance relationship Between CB46 and FN-2-9-04
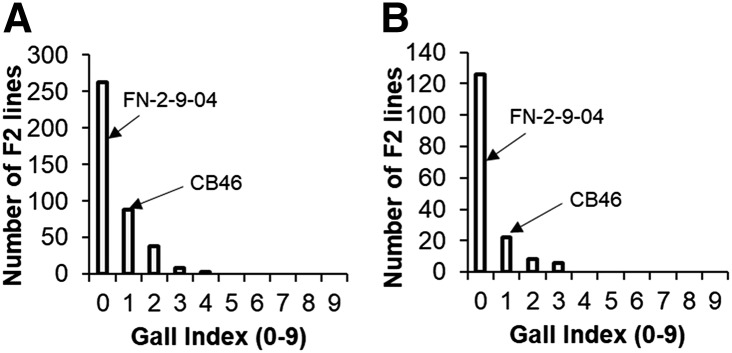
The relationship between the root-galling and nematode reproduction resistance in accession FN-2-9-04 and resistance conferred by the Rk gene in CB46 (Huynh et al. 2016) was determined through allelism tests using F populations of CB46 x FN-2-9-04. In addition, analysis of similarity was performed between FN-2-09-04, CB46 and breeding line CB46-Null within the mapped QTL regions to identify putative haplotypes associated with resistance in FN-2-9-04. In 2015 (Table 1), 400 and 162 F plants plus parents were phenotyped for avirulent M. incognita root-galling under field infestation at CVARS and KARE, respectively. At both sites (Figure 10), all F plants were resistant with no obvious segregation for root-galling response between plants, indicating that FN-2-9-04 carries a resistance locus allelic to or equivalent to the Rk gene found in CB46. The average root-gall indices for CB46 and FN-2-9-04 were 0.7 and 0.2, respectively.
Figure 10.
Distribution of root-galling response in the F population CB46 x FN-2-9-04 under field infestation by avirulent M. incognita (A): Coachella Valley Agricultural Research Station and (B): Kearney Agricultural Research and Extension Center, respectively.
To validate the allelic relationship between resistance determinants conferring resistance to RKN in CB46 and FN-2-9-04, F population subsets of CB46 x FN-2-9-04 were also phenotyped for resistance to M. javanica RG and EM, since these parents exhibited significant differences in M. javanica RG and EM production responses (Figure 4). Using 197 and 172 F lines for RG and EM phenotyping, respectively (Table 1), segregation occurred for M. javanica RG and EM in these F populations as shown in Figures 5A and 5C. Analysis of similarity between FN-2-09-04, CB46 and CB46-Null within the Vu04 genomic region associated with avirulent M. incognita RG resistance (Table 2; Figure 1A) revealed a putative haplotype associated with the resistance (Supplementary file S4). The location of the Rk locus on Vu04 identified in CB46 (Huynh et al. 2016) overlapped with the resistance region on the same chromosome in FN-2-9-04 within 2.9 cM of the CB46-Null x FN-2-9-04 F2 population and within 1.59 cM on the cowpea consensus genetic map (Muñoz-Amatriaín et al. 2017), corresponding to approximately 1 Mb on the cowpea pseudomolecules. Within this region, based on SNP marker haplotypes, FN-2-9-04 is 39% identical to CB46 and completely different from CB46-Null (identity = 0%) which is 60% identical to CB46. Conversely, in the region on Vu01 where an additional resistance QTL was detected in FN-2-09-04 (Table 2; Figures 1B, 2A, 2B), this resistant parent shares no SNP haplotype similarity with either CB46 or CB46-Null (identity = 0%), whereas CB46 and CB46-Null are 100% identical.
Discussion
Characterization of the resistance to avirulent M. incognita and aggressive M. javanica present in cowpea accession FN-2-9-04 from Mozambique revealed that the resistance is determined by two major QTL which were mapped on chromosomes Vu01 (old LG4) and Vu04 (old LG11) in the CB46-Null x FN-2-9-04 populations and on Vu01 in the CB46 x FN-2-9-04 population.
The QTL mapped on Vu04 overlaps with the previously mapped genomic region which harbors the Rk resistance locus (Huynh et al. 2016), suggesting that the Rk locus is also present in FN-2-9-04. In our previous RKN resistance QTL mapping of QRk-vu4.1 (old QRk-vu11.1) (Huynh et al. 2016), this region associated with the Rk resistance spanned about 8.35 cM compared to 2.9 cM in this study. This difference in mapping resolution is attributed in part to the current availability of the high-density SNP genotyping platform and high-density cowpea consensus genetic map (Muñoz-Amatriaín et al. 2017). If the genomic region harboring the Rk locus is a multi-allelic or multi-gene locus, the overlap between QRk-ʋu4.1 and the QTL mapped in this study on Vu04 indicates that the resistance alleles are within 2.9 cM interval of the CB46-Null x FN-2-9-04 population corresponding to approximately 1 Mb on the cowpea pseudomolecules. This locus provides effective resistance against avirulent M. incognita populations. The resistance to avirulent M. incognita present on Vu01 in FN-2-9-04 is confined to 0.1 Mb of the cowpea pseudomolecules, and its relatively low contribution to the total phenotypic variation in root-galling response (33%) compared to the resistance in Vu04 (73.3%) supports that the resistance in Vu04 is the main resistance for this nematode although both are required in the FN-2-9-04 background for fully effective resistance. The estimated values of contribution of each resistance QTL to the total phenotypic variance (Vu01 + Vu04; 33% + 73.3%) give a reliable indication of activity of each resistance QTL to the observed root-galling phenotypic response, with the excess in estimation attributed to error.
The resistance to M. javanica in FN-2-9-04 consistently mapped to Vu01 using root-galling and egg-mass production phenotypic data from F and F populations phenotyped under distinct environmental conditions (greenhouse, growth chamber and field). The QTL associated with resistance to M. javanica egg-mass production was co-located with the QTL controlling root-galling response, and based on the physical positions, on the cowpea pseudomolecules, of the mapped resistance QTL, the resistance to M. javanica root-galling and egg-mass production are confined within 6.2 Mb. The resistance QTL on Vu01 is distinct from the Rk locus (QRk-ʋu4.1, Huynh et al. 2016) which was mapped on Vu04, also it is distinct from the recently mapped RKN resistance locus on Vu11 (Old LG9) which also confers resistance to M. javanica (Santos et al. 2018). Therefore, it represents a novel RKN resistance QTL in cowpea designated here as QRk-vu1.1.
The response of four F populations to root-galling and egg-mass production relative to the resistant parent, and the skewed segregation of these nematode-induced phenotypes in the F2 and F populations indicated that these responses are under control by major genes with partial dominance effects, as also indicated by the estimated degrees of dominance (D/A). Resistance to RKN under control by major genes with partial dominance effect has been reported in several studies (Ali et al. 2014; Huynh et al. 2016).
Analysis of segregation for resistance against M. javanica and avirulent M. incognita through marker-trait association better fit a 13:3 ratio expected for a genetic control under a single dominant gene plus a recessive gene on both Vu01 and Vu04, also suggesting that the major genes controlling resistance are putatively aided by minor/recessive genes, and collectively in a dominant-recessive interaction to confer substantially stronger, broad-based resistance than that conferred by the Rk gene alone. A similar genetic phenomenon of major gene and minor/recessive gene interaction was described in cowpea cultivar CB27, where gene Rk acts together with a recessive gene to enhance and broaden root-knot nematode resistance (Ehlers et al. 2000a). The data also fit a 3:1 ratio expected for a single major gene, and the better fit to the 13:3 of the SNP haplotypes could represent genetic distortion within each locus. However, using the Castle (1921) algorithm for gene enumeration, the estimates also supported that two genes on Vu01 and two genes on Vu04, may be responsible for the resistance against M. javanica and avirulent M. incognita, respectively, but the estimates of genes involved in resistance against avirulent M. incognita on Vu01 did not support the observed segregation for resistance. The extent of genetic distortion in these regions or multi-allelic effects require further study.
Estimates of heritability of resistance in FN-2-9-04 to avirulent M. incognita and aggressive M. javanica in the F generation using greenhouse phenotypic data were lower than those estimated in the F generation using phenotypic data from field experiments. This can be accounted for by the segregation in both populations and because greenhouse phenotyping is less variable compared to field testing. The estimates of narrow-sense heritability of resistance to root-galling induced by both RKN species were in the range 0.23 – 0.71, indicating that the resistance in FN-2-9-04 can be transferred successfully into elite cowpea cultivars to broaden the genetic base of root-knot resistance which currently relies on the Rk gene. The resistance response to M. javanica reproduction had lower heritability estimates (H = 0.25 and 0.34; h = 0.17 and 0.24) compared to those for M. javanica induced root-galling (H = 0.47 - 0.95; h = 0.33 - 0.71), which could be due to egg-mass production data being generally more variable compared to root-galling data. High correlation between root-galling and nematode reproduction responses, and the co-location of resistance QTL associated with both phenotypes suggests that both traits may be governed by the same genes determining resistance. Similarly, significant correlation between root-galling and reproduction phenotypes in cowpea recombinant inbred populations was reported by Huynh et al. (2016) for the Rk locus on Vu04. In contrast, in lima bean (Phaseolus lunatus L.) the responses to root-galling and nematode reproduction were reported to be under control by independent genetic factors (Roberts et al. 2008). Since genetic factors explained 38.1 and 60.3% of the association between root-galling and egg-mass production in this study, these data suggest that although the genomic regions governing both traits are co-located, these traits may be under distinct regulatory mechanisms, or that the resistance to both traits may reside within a multi-allelic locus or tandemly arranged loci.
The heritability of resistance to avirulent M. incognita root-galling comprised two components, one on Vu01 (H = 0.33; h = 0.23,) and the other on Vu04 (H = 0.73; h = 0.49) indicating that the major locus for this resistance in FN-2-9-04 is housed on Vu04, and it is aided by the additional locus on Vu01 with low resistance heritability. Also, the differential activity between the resistance loci on Vu01 and Vu04 points to specificity of resistance to avirulent M. incognita and M. javanica. Huynh et al. (2016) reported that, although the QTL harboring the Rk locus had a significant effect on controlling both avirulent M. incognita and M. javanica, its resistance activity was lower against M. javanica. Marker-trait association analysis in the current study indicated that resistances on both Vu01 and Vu04 are required for effective resistance under avirulent M. incognita infestation.
The allelism test between CB46 and FN-2-9-04 revealed a lack of resistance segregation in the CB46 x FN-2-9-04 F population under avirulent M. incognita infestation, indicating that both parents carry the same major gene Rk locus previously mapped by Huynh et al. (2016) on Vu04 (old LG11) of the cowpea consensus genetic map (Muñoz-Amatriaín et al. 2017), also supporting that the resistance mapped in this study on Vu04 corresponds to the Rk locus. Rk was the first identified RKN resistance locus in cowpea, and it has been bred into many commercial cowpea cultivars (Fery and Dukes 1980; Helms et al. 1991; Ehlers et al. 2009). In contrast, the segregation found in F population CB46 x FN-2-9-04 for M. javanica root-galling and reproduction responses, and the mapping of resistance QTL for root-galling and egg-mass production confirmed that the heightened and broad-based resistance response in FN-2-9-04 relative to CB46 is conferred by novel resistance determinants located on Vu01.
Flanking markers associated with the mapped genomic regions on Vu01 and Vu04 can be used to assist the introgression of the resistance into elite cowpea cultivars. In particular, the novel resistance detected on Vu01 confers the most effective M. javanica resistance known to date in cowpea. The resistance on Vu01 appears to be more specifically effective against aggressive M. javanica, while both the Vu01 and Vu04 QTL have activity against avirulent M. incognita, but with the QTL on Vu04 playing the major role in resistance. This was also demonstrated by QTL pyramiding of resistance on Vu01 and Vu04. Thus, both resistance QTL on Vu01 and Vu04 are responsible for the strong and broad-based resistance observed in FN-2-9-04, which is more effective than the narrow-based resistance provided by the Rk gene alone. The mechanism of resistance displayed by this novel broad-based resistance is yet to be determined.
The genetic linkage maps of the F populations CB46-Null x FN-2-9-04 and CB46 x FN-2-9-04 are additional valuable genetic resources, especially because they are the first cowpea linkage maps constructed using a genotype from the cowpea gene-pool II from southeastern Africa (Huynh et al. 2013), and because 9.2% of the 17209 SNP markers on the CB46-Null x FN-2-9-04 map were unique to this population and were not mapped on the most recent version of the cowpea consensus genetic map (Muñoz-Amatriaín et al. 2017).
Acknowledgements
Funding for this work was provided by the USAID (Cooperative Agreements EDH-A-00-07-00005-00 and AID-OAA-A-13-00070) and California Agricultural Experiment Station (project CA-R-NEM-6964-H). We thank Dr. Rogerio M. Chiulele (Eduardo Mondlane University, Mozambique) for providing cowpea germplasm which led to the discovery of the root-knot resistant accession FN-2-9-04. We thank the staff at the UC Agricultural Research Stations, where field experiments were carried out, for their technical support.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7324211.
Communicating editor: J. Ma
Literature Cited
- Ali A., Matthews W. C., Cavagnaro P. F., Iorizzo M., Roberts P. A., et al. , 2014. Inheritance and mapping of Mj-2, a new source of root-knot nematode (Meloidogyne javanica) resistance in carrot. J. Hered. 105: 288–291. 10.1093/jhered/est090 [DOI] [PubMed] [Google Scholar]
- Amosu J. O., Franckowiak J. D., 1974. Inheritance of resistance to root-knot nematode in cowpea. Plant Dis. Rep. 58: 361–363. [Google Scholar]
- Atamian H. S., Roberts P. A., Kaloshian I., 2012. High and low throughput screens with root-knot nematodes Meloidogyne spp. Journal of Visualized Experiments. 61: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. F., Loveys B. R., 1975. The incorporation of photosynthates by Meloidogyne javanica. J. Nematol. 7: 111–113. [PMC free article] [PubMed] [Google Scholar]
- Bridge J., Page S. L. J., 1980. Estimation of root-knot nematode infestation levels on roots using a rating chart. Trop. Pest Manage. 26: 296–298. 10.1080/09670878009414416 [DOI] [Google Scholar]
- Castagnone-Sereno P., 2002. Genetic variability in parthenogenetic root-knot nematodes, Meloidogyne spp. and their ability to overcome plant resistance genes. Nematology 4: 605–608. 10.1163/15685410260438872 [DOI] [Google Scholar]
- Castle W. E., 1921. An improved method for estimating the number of genetic factors concerned in cases of blending inheritance. Science 54: 223 10.1126/science.54.1393.223 [DOI] [PubMed] [Google Scholar]
- Ehlers J., Hall A., 1997. Cowpea. Field Crops Res. 53: 187–204. 10.1016/S0378-4290(97)00031-2 [DOI] [Google Scholar]
- Ehlers J. D., Hall A. E., Patel P. N., Roberts P. A., Matthews W. C., 2000a Registration of ‘California Blackeye 27’ cowpea. Crop Sci. 40: 854–855. [Google Scholar]
- Ehlers J. D., Matthews W. C., Hall A. E., Roberts P. A., 2000b Inheritance of a broad-based form of root-knot nematode resistance in cowpea. Crop Sci. 40: 611–618. 10.2135/cropsci2000.403611x [DOI] [Google Scholar]
- Ehlers, J. D., W. C. Matthews, A. E. Hall, and P. A. Roberts, 2002 Breeding and evaluation of cowpeas with high levels of broad-based resistance to root-knot nematodes. In Fatokum, C., S. Tarawali, B. Singh, P. Kormawa, M. Tamo, editors. Challenges and opportunities for enhancing sustainable cowpea production. Proceedings for the World cowpea conference III held at the International Institute of Tropical Agriculture (IITA); 2000, Sept 4–8. Ibadan, Nigeria. [Google Scholar]
- Ehlers J. D., Sanden B. L., Frate C., Hall A. E., Roberts P. A., 2009. Registration of ‘California Blackeye 50’ cowpea. J. Plant Regist. 3: 236–240. 10.3198/jpr2009.01.0039crc [DOI] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to quantitative genetics, Ed. 4th Longman, Essex, UK. [Google Scholar]
- Fernandez G. C. J., Miller J. C., Jr, 1985. Estimation of heritability by parent-offspring regression. Theor. Appl. Genet. 70: 650–654. 10.1007/BF00252291 [DOI] [PubMed] [Google Scholar]
- Fery R. L., Dukes P. D., 1980. Inheritance of root-knot resistance in the cowpea (Vigna unguiculata (L.) Walp.). J. Am. Soc. Hortic. Sci. 105: 671–674. [Google Scholar]
- Fery R. L., Dukes P. D., Thies J. A., 1994. Characterization of new sources of resistance in cowpea to the southern root-knot nematode. Hortic. Sci. 29: 678–679. [Google Scholar]
- Hall A. E., Frate C. A., 1996. Blackeye Bean Production in California. UC DANR, Davis, CA. [Google Scholar]
- Helms D., Panella L., Buddenhagen I. W., Tucker C. L., Gepts P. L., 1991. Registration of California blackeye 46 cowpea. Crop Sci. 31: 1703. [Google Scholar]
- Hoagland D. R., Arnon D. I., 1950. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular 347, University of California, Berkely, CA. [Google Scholar]
- Huynh B. L., Close T. J., Roberts P. A., Hu Z., Wanamaker S., et al. , 2013. Gene pools and the genetic architecture of domesticated cowpea. The Plant Genome. 6: 1–8. [Google Scholar]
- Huynh B. L., Matthews W. C., Ehlers J. D., Lucas M. R., Santos J. R. P., et al. , 2016. A major QTL corresponding to the Rk locus for resistance to root-knot nematodes in cowpea (Vigna unguiculata L. Walp.). Theor. Appl. Genet. 129: 87–95. 10.1007/s00122-015-2611-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little T. M., Hills F. J., 1978. Agricultural experimentation – design and analysis. California, Wiley, CA. [Google Scholar]
- Lonardi, S., T. Zhu, M. Muñoz-Amatriaín, Q. Liang, S. Wanamaker et al., 2017 Assembly of eleven pseudomolecules representing the cowpea genome sequence. Plant and Animal Genome XXV P0688 https://phytozome.jgi.doe.gov/pz/portal.html!info?alias=Org-Vunguiculata-er.
- Luc M., Bridge J., Sikora R. A., 2005. Reflections on Nematology in Subtropical and Tropical Agriculture, pp. 1–10 in Plant Parasitic Nematodes in Subtropical and Tropical Agriculture, edited by Luc M., Bridge J., Sikora R. A. CABI Bioscience, Egham, UK: 10.1079/9780851997278.0001 [DOI] [Google Scholar]
- Lucas M. R., Diop N.-N., Wanamaker S., Ehlers J. D., Roberts P. A., et al. , 2011. Cowpea-soybean synteny clarified through an improved genetic map. Plant Genome 4: 218–225. 10.3835/plantgenome2011.06.0019 [DOI] [Google Scholar]
- McClure M. A., 1977. Meloidogyne incognita: A metabolic sink. J. Nematol. 9: 68–90. [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M., Mirebrahim H., Xu P., Wanamaker S. I., Luo M. C., et al. , 2017. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 89: 1042–1054. 10.1111/tpj.13404 [DOI] [PubMed] [Google Scholar]
- National Research Council , 2006. Cowpea, pp. 105–116 in Lost Crops of Africa: Volume II: Vegetables. The National Academies Press, Washington, DC. [Google Scholar]
- Ndeve A. D., Matthews W. C., Santos J. R. P., Huynh B. L., Roberts P. A., 2018. Broad-based root-knot nematode resistance identified in cowpea gene-pool two. J. Nematol. 50: 545–558. 10.21307/jofnem-2018-046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo M. D., Roberts P. A., 2005. Isofemale line analysis of Meloidogyne incognita virulence to cowpea resistance gene Rk. J. Nematol. 37: 448–456. [PMC free article] [PubMed] [Google Scholar]
- Petrillo M. D., Matthews W. C., Roberts P. A., 2006. Dynamics of Meloidogyne incognita virulence to resistance genes Rk and Rk2 in cowpea. J. Nematol. 38: 90–96. [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., 1992. Current status of the availability, development, and use of host plant resistance to nematodes. J. Nematol. 24: 213–227. [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., Frate C. A., Matthews W. C., Osterli P. P., 1995. Interaction of virulent Meloidogyne incognita and Fusarium wilt on resistant cowpea genotypes. Phytopathology 85: 1288–1295. 10.1094/Phyto-85-1288 [DOI] [Google Scholar]
- Roberts P. A., Matthews W. C., Ehlers J. D., 1996. New resistance to virulent root-knot nematodes linked to the Rk locus of cowpea. Crop Sci. 36: 889–894. 10.2135/cropsci1996.0011183X0036000400012x [DOI] [Google Scholar]
- Roberts P. A., Ehlers J. D., Hall A. E., Matthews W. C., 1997. Characterization of new resistance to root-knot nematodes in cowpea, pp. 207–214 in Advances in Cowpea Research, edited by Singh B. B., Mohan Raj D. R., Dashiel K. E., Jackai L. E. N. IITA, JIRCAS, Ibadan, Nigeria. [Google Scholar]
- Roberts P. A., Matthews W. C., Ehlers J. D., 2005. Root-knot nematode resistant cowpea cover crops in tomato production systems. Agron. J. 97: 1626–1635. 10.2134/agronj2004.0290 [DOI] [Google Scholar]
- Roberts P. A., Matthews W. C., Ehlers J. D., Helms D., 2008. Genetic determinants of differential resistance to root-knot nematode reproduction and galling in lima bean. Crop Sci. 48: 553–561. 10.2135/cropsci2007.07.0384 [DOI] [Google Scholar]
- Roberts P. A., Huynh B. L., Matthews W. C., Frate C. A., 2013. University of California Dry Bean Research, Progress Report. California Dry Bean Advisory Board, Dinuba, CA. [Google Scholar]
- Santos J. R. P., Ndeve A. D., Huynh B. L., Matthews W. C., Roberts P. A., 2018. QTL mapping and transcriptome analysis of cowpea reveal candidate genes for root-knot nematode resistance. PLoS One 13: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser J. N., 1980. Root-Knot Nematodes: a global menace to crop production. Plant Dis. 64: 36–41. 10.1094/PD-64-36 [DOI] [Google Scholar]
- Sikora R. A., Greco N., Silva J. F. V., 2005. Nematode Parasites of Food Legumes, pp. 259–318 in Plant parasitic nematodes in subtropical and tropical agriculture, edited by Luc M., Bridge J., Sikora R. A. CABI Bioscience, Egham, UK: 10.1079/9780851997278.0259 [DOI] [Google Scholar]
- Singh D. B., Reddy P. P., 1986. Inheritance of resistance to root-knot nematode in cowpea. Indian Journal of Nematology 16: 284–285. [Google Scholar]
- Swanson T. A., Van Gundy S. D., 1984. Cowpea resistance to root-knot caused by Meloidogyne incognita and M. javanica. Plant Dis. 68: 961–964. 10.1094/PD-68-961 [DOI] [Google Scholar]
- Thomason I. J., McKinney H. E., 1960. Reaction of cowpeas, Vigna sinensis to root-knot nematodes, Meloidogyne spp. Plant Dis. Rep. 44: 51 (Abstract). [Google Scholar]
- Taylor A. L., Sasser J. N., 1978. Biology, identification and control of root-knot nematodes (Meloidogyne species), Raleigh, North Carolina, North Carolina State University and USAID, NC. [Google Scholar]
- Williamson V. M., Hussey R. S., 1996. Nematode pathogenesis and resistance in plants. Plant Cell 8: 1735–1745. 10.1105/tpc.8.10.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., P. Bhat, T. J. Close, and S. Lonardi, 2015. MSTmap. University of California Riverside. http://www.mstmap.org/
- Xu S., 2013. Mapping quantitative trait loci by controlling polygenic background effects. Genetics 195: 1209–1222. 10.1534/genetics.113.157032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All F and F populations and root-knot nematode isolates are available upon request. Phenotypic and genotypic data are included in data (D) files 1 - 9. These data files, including their description, and supplementary tables and figures are available at Figshare: https://doi.org/10.25387/g3.7324211.