Abstract
The goal of this mini review is to summarize the relevant contribution of some beneficial traits to the behavior of the species Pseudomonas chlororaphis, and using that information, to give a practical point of view using the model biocontrol strain P. chlororaphis PCL1606 (PcPCL1606). Among the group of plant-beneficial rhizobacteria, P. chlororaphis has emerged as a plant- and soil-related bacterium that is mainly known because of its biological control of phytopathogenic fungi. Many traits have been reported to be crucial during the multitrophic interaction involving the plant, the fungal pathogen and the soil environment. To explore the different biocontrol-related traits, the biocontrol rhizobacterium PcPCL1606 has been used as a model in recent studies. This bacterium is antagonistic to many phytopathogenic fungi and displays effective biocontrol against fungal phytopathogens. Antagonistic and biocontrol activities are directly related to the production of the compound 2-hexyl, 5-propyl resorcinol (HPR), despite the production of other antifungal compounds. Furthermore, PcPCL1606 has displayed additional traits regarding its fitness in soil and plant root environments such as soil survival, efficient plant root colonization, cell-to-cell interaction or promotion of plant growth.
Keywords: Pseudomonas chlororaphis, root colonization, biocontrol, avocado, antifungals
Introduction
Since the earliest studies, soil has been described as an infinite source of microorganisms with beneficial activities that promote plant health (Waksman and Woodruff, 1940). Inside the soil, the rhizosphere environment is considered the soil-plant root interphase where potentially beneficial rhizobacteria are established. The plant-beneficial microbial life can be actively recruited by the plant rhizosphere (Berendsen et al., 2018) and can finally result in the biological control of the disease (Babalola, 2010). These biocontrol rhizobacteria can use a wide range of mechanisms involved in the suppression of plant pathogens. A diverse range of bacterial genera, such as Bacillus, Pseudomonas, Serratia, Stenotrophomonas, and Streptomyces, has been commonly described asx beneficial rhizobacteria (Berg, 2009). Among them, representatives of the Pseudomonas genus have been commonly associated with the rhizosphere and soil habitats (Lugtenberg and Dekkers, 1999). This bacterial genus has also been widely studied due to its ability to produce antifungal compounds, compete for niche and/or nutrients on the rhizosphere, and elicit induced systemic resistance in plants (Haas and Défago, 2005). Currently, many strains belonging to the group of fluorescent Pseudomonas are known to enhance plant growth promotion and reduce the severity of various diseases (Ganeshan and Kumar, 2005; Mercado-Blanco and Bakker, 2007; Weller, 2007).
The Pseudomonas fluorescens complex is one of the most diverse bacterial groups within the Pseudomonas genus and comprises more than fifty validly named species and many unclassified isolates (Garrido-Sanz et al., 2017). Many strains of this complex have been isolated from plant-related environments, and several species can be considered beneficial since many are described as plant growth-promoting rhizobacteria and/or minimize the effects of phytopathogens (PGPR; Kang et al., 2006; Raaijmakers et al., 2009). The beneficial effects displayed by some bacteria result from the expression of multiple activities that act directly and indirectly inhibiting pathogen activities and promoting plant health (McSpadden, 2007). To date, a number of studies have characterized the environmental factors that affect the abundance of different pseudomonad populations below ground (Berg et al., 2002; Ownley et al., 2003; Mazzola et al., 2004; Bergsma-Vlami et al., 2005). Pseudomonas species most commonly reported to include plant beneficial rhizospheric strains are Pseudomonas aureofaciens, Pseudomonas brassicacearum, Pseudomonas chlororaphis, P. fluorescens, Pseudomonas Protegens, and Pseudomonas putida.
Beneficial Traits of Rhizospheric Pseudomonas chlororaphis Strains
Among the beneficial Pseudomonas spp., P. chlororaphis has evolved to be a common inhabitant of the root environment of many plants. Moreover, it has been extensively reported the role of specific traits that render this bacterium able to be used as an inoculant for biofertilization, phytostimulation, and biocontrol purposes (Bloemberg and Lugtenberg, 2001).
Subspecies of Pseudomonas chlororaphis
Pseudomonas chlororaphis, P. aureofaciens, and Pseudomonas aurantiaca were initially included in the Approved List of Bacterial Names (Skerman et al., 1980) and considered separate species in the first edition of Bergey’s Manual of Systematic Bacteriology by Palleroni (1984). However, the results obtained by Peix et al. (2007) of fatty acid analysis, phenotypic characterization, 16S rRNA gene sequencing, and DNA-DNA relatedness together with the results obtained by Hilario et al. (2004) on the phylogenetic analysis of several housekeeping genes, support the reclassification of P. aurantiaca as a later heterotypic synonym of P. chlororaphis. The results published by Peix et al. (2007) also reveal that strains of P. aurantiaca, P. aureofaciens, and P. chlororaphis form three clearly distinguishable groups within P. chlororaphis that merit the status of subspecies. Therefore, the current classification is P. chlororaphis subsp. chlororaphis subsp. nov., P. chlororaphis subsp. aureofaciens subsp nov., comb. nov. and P. chlororaphis subsp. aurantiaca subsp. nov., comb. nov. (Peix et al., 2007). Three years later, Burr et al. (2010) added a new subspecies to P. chlororaphis and placed it on a distinct branch within this species with the name P. chlororaphis subsp. piscium subsp. nov. The current reports of sequenced bacterial genomes of P. chlororaphis strains (Calderón et al., 2015; Deng et al., 2015; Town et al., 2016; Moreno-Avitia et al., 2017; Biessy et al., 2019) will help to refine the current classification of the P. chlororaphis group.
Main Traits Involved in Biocontrol by P. chlororaphis
These aerobic, Gram-negative bacteria are associated with soil and plant roots (González-Sánchez et al., 2010; Calderón et al., 2015; Vida et al., 2017a). Typically, this species possesses plant-colonizing and antagonistic activities against soil-borne plant pathogens. Products from secondary metabolism usually mediate antagonism, and can be regulated by the GacS-GacA two component regulatory system. GacS-GacA system governs a complex signal transduction pathway, involving regulatory RNAs and translational repression (Yan et al., 2018; Jahanshah et al., 2019). Simultaneously, Quorum Sensing (QS) is a regulatory systems which is involved in the general biology performance of P. chlororaphis, including biofilm formation, antifungal production or exoenzyme secretion. QS is a mechanism of intercellular signaling that makes the bacterial population to act co-ordinately, based in the secretion of diffusible signal molecules (mainly acyl homoserine lactones, or AHL; Venturi, 2006). The use of OMICs and functional studies have revealed a more complex scenario, where the presence of several QS systems can coexist inside the same bacterial cell (Morohoshi et al., 2017), but also the participation of secondary metabolites (such as the antifungals phenazines and/or the resorcinol-related compounds) in final QS regulation (Selin et al., 2010; Brameyer et al., 2015).
Recent reports using OMICs techniques, have allowed a more comprehensive understanding of the potential weaponry that P. chlororaphis group could uses to interact with the root plant. For example, presence of different antimicrobial and insecticidal compounds, cyclic peptides, siderophores, bacteriocins, molecules involved in beneficial plant-bacteria interactions, secretions systems, antibacterial proteins, etc., (Loper et al., 2012; Chen et al., 2015; Biessy et al., 2019). Below, the most relevant are summarized (Table 1).
Table 1.
Summary of main compounds produced by Pseudomonas chlororaphis subspecies with beneficial effects in plant pathogen control.
| Compound | Target/beneficial effect | Subspecies1 | Reference strain | References |
|---|---|---|---|---|
| Antibiotics | ||||
| Phenazine 1-carboxamide | Antifungal redox-active antibiotic | Pa, Pe, Pc, Pp | PCL1391 | Hernández et al., 2004 |
| Phenazine 1-carboxylic acid | Antifungal redox-active antibiotic | Pc, Pp | PCL1391 | Chin-A-Woeng et al., 1998 |
| 2-hydroxy phenazine 1-carboxylic acid | Fungistatic and bacteriostatic | Pa, Pe, Pc | GP72 | Liu et al., 2016 |
| Pyrrolnitrin | Antifungal compound | Pa, Pe, Pc, | PA23 | Nandi et al., 2015 |
| 2-hexyl, 5-propylresorcinol | Antifungal compound and signal molecule | Pa, Pe, Pc | PCL1606 | Cazorla et al., 2006 |
| 2,4 Diacetylphloroglucinol | Membrane damage, distribution of mitochondria electron transport chain and inhibition of V-ATPase activity. Antifungal | Pc | UFB2 | Deng et al., 2015 |
| Rhizoxin | Antifungal | Pc | MA 342 | Loper et al., 2008 |
| Insecticidal compounds | ||||
| Cyclic peptides | Insecticidal, surfactant and antagonistic activity | Pc | PCL1391 | Flury et al., 2017 |
| Fit toxin | Insecticidal activity | Pc, Pe, Pp | PCL1606 | Flury et al., 2016 |
| Siderophores | ||||
| Pyoverdine | Fe chelation and competition | Pa, Pe, Pc | D-TR133 | Barelmann et al., 2003 |
| Achromobactine | Fe chelation and competition | Pa, Pe, Pc | PCL1606 | Calderón et al., 2015 |
| Hemophore | Fe chelation | Pp | PCL1607 | Biessy et al., 2019 |
| Enzymes and hormones | ||||
| Chitinase | Chitin hydrolysis enzyme and antifungal | Pc, Pe, Pp | PCL1391 | Flury et al., 2016 |
| Protease | Protein hydrolysis enzyme and antifungal | Pa | M71 | Raio et al., 2017 |
| Phosphatase | Phosphorus solubilization enzyme | Pc | SZY6 | Ahemad, 2015 |
| ACC deaminase | Plant growth promotion | Pa, Pe, Pc, Pp | 6G5 | Glick, 2014 |
| PQQ | Plant growth promotion | Pa, Pe, Pc | B23 | Nishiyama et al., 1991 |
| IAA | Plant growth promotion | Pa, Pe, Pc | O6 | Kang et al., 2006 |
| Volatile | ||||
| 2,3 butanediol | Elicite plant resistance | Pa, Pe, Pc | O6 | Han et al., 2006 |
| Hydrogen cyanide | Metalloenzymes inhibitor and antifungal | Pa, Pc, Pe, Pp | PA23 | Nandi et al., 2015 |
| Hormones | ||||
| Indol acetic acid | Plant growth promotion | Pa, Pc, Pe, Pp | O6 | Kang et al., 2006 |
Reference strains published are included. 1Pa: P. chlororaphis subsp. aurantiaca; Pe: P. chlororaphis subsp. aureofaciens; Pc: P. chlororaphis subsp. chlororaphis; Pp: P. chlororaphis subsp. Piscium.
Phenazines are among the most copious secondary metabolites produced by fluorescent pseudomonads, and phenazine-producing microorganisms represent a ubiquitous group of antibiotic-producing bacteria in the environment (Chin-A-Woeng et al., 2000; Mavrodi et al., 2013). Phenazine compounds are redox-active nitrogen-containing heterocyclic molecules and its beneficial role on plant biology is not limited to antibiosis against phytopathogenic microbes (Pierson and Pierson, 2010; Biessy and Filion, 2018; Biessy et al., 2019). Additional effects have been shown for this compound such as triggering induced systemic resistance in plants, reducing the expression of key pathogenicity-related genes of the phytopathogen, or its involvement in the root persistence (Biessy and Filion, 2018). In relation to the bacterial interaction with the plant root, phenazines can be crucial for biofilm formation (Selin et al., 2010). An extensive colonization of the rhizosphere is a prerequisite in efficient disease suppression by preventing pathogen form access to the root (Lugtenberg and Kamilova, 2009). The involvement of phenazines on root colonization has been strengthened because some phenazine compounds could be terminal signaling factors in the QS network of some bacteria, and are directly involved in biofilm formation on biotic surfaces (Dietrich et al., 2006; Selin et al., 2012).
Pyrrolnitrin and the volatile compound hydrogen cyanide, are also among the additional antifungal compounds typically produced by P. chlororaphis strains. Pyrrolnitrin is considered a key compound for fungal biocontrol (Hill et al., 1994) and is becoming even more relevant than phenazines extending its action to eukaryotic organisms (Nandi et al., 2015; Huang et al., 2018). The same observation can be applied to the volatile compound hydrogen cyanide, which also has a broad spectrum of prokaryotic and eukaryotic targets (Nandi et al., 2017; Kang et al., 2018). The biological importance of this broad spectrum of both active compounds would be related to its typical environmental persistence, for example, allowing them to escape from predation (Nandi et al., 2017). Related to the insecticidal activity of this bacterial species, the most studied virulence factor against insects is the Fit toxin, which is similar to Mcf1 of the entomopathogenic bacterium Photorhabdus luminescens (Ruffner et al., 2015). Fit mutants of P. chlororaphis PCL1391 further showed reduced virulence, and the residual toxicity could be assigned to the wide range of other antimicrobial compounds produced by P. chlororaphis (previously listed) or cyclic lipopetides (Flury et al., 2017).
About Clps, these compounds can be involved in many biological functions, such as motility, biofilm formation, protection against predators and antagonism (De Souza et al., 2003; Raaijmakers et al., 2010). Clps produced by plants-beneficial bacteria were found to induce plant resistance and to contribute to plant protection against root pathogenic fungi (Olorunleke et al., 2015). But interestingly, Clps were demonstrated to be further insect pathogenicity factor in P. chlororaphis strains (Flury et al., 2016, 2017).
The production of exoenzymes has also been described to have a role in biocontrol activity (Haran et al., 1996). Enzymes such as chitinases, lipases or proteases have a broad distribution among the soil bacterial community and are probably related to general metabolism, but also inhibit the pathogen (degrading some cell structures) and stimulate plant growth by providing additional resources from the degradative activity (Vida et al., 2017a). Remarkably, P. chlororaphis strains can produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase (Nadeem et al., 2007), which is an enzyme produced by plant-associated bacteria that decrease the ethylene levels and protect the plant from its effect, which results in a general beneficial activity (Glick, 2014). In addition, the production of the biofertilizer hormone indole-3-acetic acid (IAA) has also been reported for P. chlororaphis strains (Dimkpa et al., 2012), and its production is important in microbe-microbe and microbe-plant signaling, and can also results in an promotion of plant growth (Kang et al., 2006).
Other compounds can also have an important role for P. chlororaphis, such as the production of siderophores, which can be considered as a general beneficial activity, at least, for all the soil-related Pseudomonas spp. (Zhang and Rainey, 2013). These molecules are secondary metabolites involved in iron quelation. The most known is pyoverdine, a water-soluble fluorescent pigment produced by fluorescent Pseudomonas species (Barelmann et al., 2003). However, the recent comparative genomic studies of P. chlororaphis genomes, revealed the putative presence of various secondary siderophores, such as achromobactine and hemophore (Biessy et al., 2019).
The Beneficial Rhizobacterium Pseudomonas chlororaphis PCL1606 (PcPCL1606) as a Model
In order to find potential bacterial biocontrol agents against the avocado white root rot caused by Rosellinia necatrix, a collection of bacterial isolates belonging to the genera Bacillus and Pseudomonas were isolated from avocado rhizosphere (Cazorla et al., 2006, 2007; Pliego et al., 2011). Interestingly, a number of P. chlororaphis were consistently isolated from avocado roots (Cazorla et al., 2006). The management of this crop could enhance this presence on avocado roots of P. chlororaphis isolates, since it has been reported that application of organic amendments can enhance the presence of specific groups of beneficial microbes, including antagonistic P. chlororaphis (Vida et al., 2016).
PcPCL1606 as a Biological Control Agent
Nearly all the P. chlororaphis isolated from avocado roots were antagonistic and produced a broad range of antimicrobials including phenazines. Among them, the strain PcPCL1606 do not produce phenazines; otherwise produce proteases, lipases and the antifungal metabolite 2-hexyl 5-propylresorcinol (HPR; Figure 1). Another unusual characteristic of this strain is the absence of plant growth promotion in the assayed plant models; however, siderophore production and phosphorous solubilization were detected (among other PGPR-related traits; Vida et al., 2017a). This strain displayed strong antagonism to many phytopathogenic fungi and showed biocontrol of crown and root rot of tomato, caused by Fusarium oxysporum f. sp. radicis-lycopersici and avocado white root, caused by R. necatrix (Cazorla et al., 2006; González-Sánchez et al., 2013). Effectiveness of biocontrol was directly related to the compound HPR (Cazorla et al., 2006; Calderón et al., 2013). HPR production was led by three biosynthetic genes located in a cluster (darA, darB, and darC) followed by two independent regulatory genes (darS and darR; Nowak-Thompson et al., 2003; Calderón et al., 2013). Further experiments revealed that HPR production was also under transcriptional regulation of the GacS-GacA two-component regulatory system, as previously described for other antifungal antibiotics (Haas and Keel, 2003), and also modulated by different growth parameters such as temperature, pH and the presence of salts in the medium (Calderón et al., 2014a).
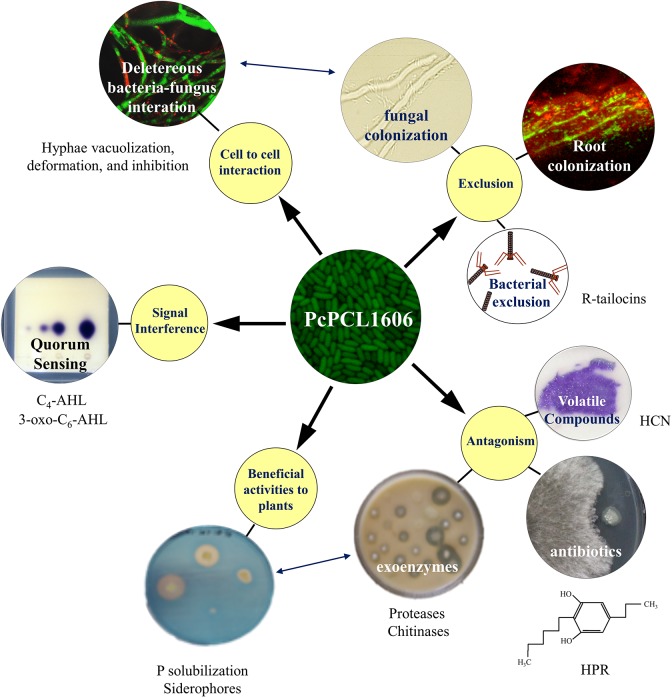
FIGURE 1.
Current knowledge of the strategies displayed during the multitrophic interactions with plants and fungal pathogens by the model rhizobacterial biocontrol strain PcPCL1606. The main strategies involved in the interaction (yellow circles) included cell-to-cell interaction, beneficial interaction with plants, exclusion, antibiotic production and signal interference. The different strategies can enhance a second one (double-ended blue arrows).
Main Features of PcPCL1606 Involved in Pathogen and Plant Interaction
PcPCL1606 showed strong antifungal activity (Figure 1), and HPR production was the main determinant in the antagonistic and biocontrol phenotypes (Calderón et al., 2013). In addition to HPR, other antifungals can be produced by PcPCL1606, such as pyrrolnitrin (PRN) or hydrogen cyanide (HCN), as well as several exoenzymes such as proteases, chitinases or phosphatases (Vida et al., 2017a). Nevertheless, HPR is more than a powerful compound against pathogenic fungi in the soil and could have additional roles. It has been reported that some alkylresorcinols (to which the compound HPR belongs) can behave as quorum sensing-like signal molecules in the genus Photorhabdus (Brameyer et al., 2015), and for this, could have a similar role in HPR-producing P. chlororaphis strains. Thus, additional HPR-dependent traits, which are different from antagonism, could have an essential role in the beneficial effects of PcPCL1606 on the plant, such as the root colonization or the biofilm formation (Calderón et al., 2014b, 2019).
Related to the possibility to physically exclude the pathogen from the plant root habitat (Figure 1), biological processes, such as biofilm formation or chemotaxis, are crucial for the PcPCL1606. PcPCL1606 is strongly attracted to the avocado root exudates by chemotactic processes (Polonio et al., 2017). As a result of this attraction, PcPCL1606 efficiently colonizes avocado roots (González-Sánchez et al., 2010) and can be found forming a biofilm on avocado root surfaces, located in the same area where R. necatrix can be found during the early stages of infection (Calderón et al., 2014b). Moreover, two bacteriocins (R-tailocins 1 and 2), recently described in PcPCL1606 would contribute to better competition against other rhizosphere-associated bacteria (Dorosky et al., 2017). However, PcPCL1606 bacterial cells also displayed a direct chemotaxis to fungal exudates and finally showed a direct contact with the fungal hyphae of R. necatrix. This cell-to-cell contact causes an increase in stress symptoms on the hyphae, among others, by the direct release of antifungal substances, which lead to an accelerated ageing process in the hyphae and hyphal death (Calderón et al., 2014b; Moore-Landecker, 1996). Moreover, the root colonization ability and biofilm formation of the wild-type strain was also related to HPR production, and the absence of HPR resulted in reduced root colonization levels and no biofilm formation by PcPCL1606 (Calderón et al., 2014b, 2019).
To obtain insight into the features of PcPCL1606, its complete genome sequencing was completed. Phylogenetic studies clustered this strain into the P. chlororaphis clade which is placed into the fluorescent Pseudomonas complex, however, as previously mentioned, PcPCL1606 it is not a typical P. chlororaphis strain (Biessy et al., 2019). Thus, phylogenetic analysis revealed clear differences with the genomes of other biocontrol P. chlororaphis, such as PcPCL1601 or PcPCL1607, also isolated from avocado root (Calderón et al., 2015; Vida et al., 2017b; Biessy et al., 2019). Analysis of PcPCL1606 genome confirmed a lack of phenazine biosynthetic genes, cyclic lipopeptides that are related to the surfactant and insecticidal properties, which are typical for P. chlororaphisx (Raaijmakers et al., 2006). However, PcPCL1606 exhibits a complete Fit toxin (fit) cluster (Calderón et al., 2015).
Futures Projects and Research
The future of P. chlororaphis as biocontrol agent is very promising. P. chlororaphis is ubiquitous in the environment, lacks known toxic or allergenic properties, and has a history of safe use in agriculture and in food and feed crops. P. chlororaphis is considered non-pathogenic to humans, wildlife or the environment according to the United States Environmental Protection Agency (EPA), and commercial products based on P. chlororaphis strains are already available. For example, Cedomon® (P. chlororaphis, BioAgri AB, Sweden), Spot-Less® (P. aureofaciens Tx-1, Turf Science Laboratories, Carlsbad, United States) or AtEze® (P. chlororaphis 63-28, Turf Science Laboratories, Carlsbad, United States) are based on P. chlororaphis strains, but many other products are already present in the market based on other Pseudomonas spp. These facts pointed out to a promising future for the use of biocontrol agents belonging to the specie P. chlororaphis.
Regarding the model bacterium PcPCL1606, studies revealed that PcPCL1606, as well as other P. chlororaphis isolates from avocado roots, displayed high persistence and reached a population density that was enough to reduce disease (González-Sánchez et al., 2013). Under commercial greenhouse conditions, applications of PcPCL1606 cells resulted in biocontrol against R. necatrix. Moreover, some other P. chlororaphis isolates from avocado roots, that have different beneficial traits (such as phenazine production or plant growth promotion), could also provide plant protection. These finding suggest that a promising approach to improve P. chlororaphis based biocontrol would be to develop consortia which combine strains with complementary traits resulting in more stable or even enhanced beneficial effects on plants.
Author Contributions
EA and FC designed the review content. EA, ST, CV, AV, and FC wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was supported by the Spanish Plan Nacional I + D + I. Grant AGL2017-83368-C2-1-R and partially supported by the European Union (FEDER). CV and ST were supported by a grant from FPI, Ministerio de Ciencia e Innovación, Spain.
References
- Ahemad M. (2015). Phosphate-solubilizing bacteria-assisted phytoremediation of metalliferous soils: a review. 3 Biotech 5 111–121. 10.1007/s13205-014-0206-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babalola O. O. (2010). Beneficial bacteria of agricultural importance. Biotechnol. Lett. 32 1559–1570. 10.1007/s10529-010-0347-0 [DOI] [PubMed] [Google Scholar]
- Barelmann I., Fernández D. U., Budzikiewicz H., Meyer J. M. (2003). The pyoverdine from Pseudomonas chlororaphis D-TR133 showing mutual acceptance with the pyoverdine of Pseudomonas fluorescens CHA0. Biometals 16 263–270. 10.1023/A:1020615830765 [DOI] [PubMed] [Google Scholar]
- Berendsen R. L., Vismans G., Yu K., Song Y., de Jonge R., Burgman W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12 1496–1507. 10.1038/s41396-018-0093-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84 11–18. 10.1128/AEM.68.7.3328-3338.2002 [DOI] [PubMed] [Google Scholar]
- Berg G., Roskot N., Steidle A., Eberl L., Zock A., Smalla K. (2002). Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microbiol. 68 3328–3338. 10.1128/AEM.68.7.3328-3338.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsma-Vlami M., Prins M. E., Raaijmakers J. M. (2005). Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol. Ecol. 52 59–69. 10.1016/j.femsec.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Biessy A., Filion M. (2018). Phenazines in plant-beneficial Pseudomonas spp.: biosynthesis, regulation, function and genomics. Environ. Microbiol. 20 3905–3917. 10.1111/1462-2920.14395 [DOI] [PubMed] [Google Scholar]
- Biessy A., Novinscak A., Blom J., Léger G., Thomashow L., Cazorla F. M., et al. (2019). Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ. Microbiol. 21 437–455. 10.1111/1462-2920.14476 [DOI] [PubMed] [Google Scholar]
- Bloemberg G. V., Lugtenberg B. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4 343–350. 10.1016/S1369-5266(00)00183-7 [DOI] [PubMed] [Google Scholar]
- Brameyer S., Kresovic D., Bode H. B., Heermann R. (2015). Dialkylresorcinols as bacterial signaling molecules. Proc. Natl. Acad. Sci. U.S.A. 112 572–577. 10.1073/pnas.1417685112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr S. E., Gobeli S., Kuhnert P., Goldschmidt-Clermont E., Frey J. (2010). Pseudomonas chlororaphis subsp. Piscium subsp. nov., isolated from freshwater fish. Int. J. Syst. Evol. Microbiol. 60 2753–2757. 10.1099/ijs.0.011692-0 [DOI] [PubMed] [Google Scholar]
- Calderón C. E., Carrión V. J., de Vicente A., Cazorla F. M. (2014a). darR and darS are regulatory genes that modulate 2-hexyl, 5-propyl resorcinol transcription in Pseudomonas chlororaphis PCL1606. Microbiology 160 2670–2680. 10.1099/mic0.082677-0 [DOI] [PubMed] [Google Scholar]
- Calderón C. E., de Vicente A., Cazorla F. M. (2014b). Role of 2-hexyl, 5-propyl resorcinol production by Pseudomonas chlororaphis PCL1606 in the multitrophic interactions in the avocado rhizosphere during the biocontrol process. FEMS Microbiol. Ecol. 89 20–31. 10.111/1574-6941.12319 [DOI] [PubMed] [Google Scholar]
- Calderón C. E., Pérez-García A., de Vicente A., Cazorla F. M. (2013). The dar genes of Pseudomonas chlororaphis PCL1606 are crucial for biocontrol activity via production of the antifungal compound 2-hexyl, 5-propyl resorcinol. Mol. Plant Microbe Interact. 26 554–565. 10.1094/MPMI-01-13-0012-R [DOI] [PubMed] [Google Scholar]
- Calderón C. E., Ramos C., de Vicente A., Cazorla F. M. (2015). Comparative genomic analysis of Pseudomonas chlororaphis PCL1606 reveals new insight into antifungal compounds involved in biocontrol. Mol. Plant Microbe Interact. 28 249–260. 10.1094/MPMI/10-14-0326-FI [DOI] [PubMed] [Google Scholar]
- Calderón C. E., Tienda S., Heredia-Ponce Z., Arrebola E., Cárcamo-Oyarce G., Eberl L., et al. (2019). The compound 2-hexyl, 5-propyl resorcinol has a key role in biofilm formation by the biocontrol rhizobacterium Pseudomonas chlororaphis PCL1606. Front. Microbiol. 10:396. 10.3389/fmicb.2019.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla F. M., Duckett S. D., Bergström E. T., Odijk R., Lugtenberg B. J. J., Thomas-Oates J. E., et al. (2006). Biocontrol of avocado dematophora root rot by antagonistic Pseudomonas fluorescens PCL1606 correlates with the production of 2-hexyl, 5-propyl resorcinol. Mol. Plant Microbe Interact. 19 418–428. 10.1094/MPMI-19-0418 [DOI] [PubMed] [Google Scholar]
- Cazorla F. M., Romero D., Pérez-García A., Lugtenberg B. J. J., de Vicente A., Bloemberg G. (2007). Isolation and characterization of antagonistic Bacillus subtillis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol. 103 1950–1959. 10.111/j.1365-2672.2007.03433.x [DOI] [PubMed] [Google Scholar]
- Chen Y., Shen X., Peng H., Hu H., Wang W., Zhang X. (2015). Comparative genomic analysis and phenazine production of Pseudomonas chlororaphis, a plant growth-promoting rhizobacterium. Gemon. Data 4 33–42. 10.1010/j.gdata.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-A-Woeng T. F. C., Bloemberg G. V., Mulders I. H., Dekkers L. C., Lugtenberg B. J. (2000). Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant Microbe Interact. 13 1340–1345. 10.1094/MPMI.2000.13.12.1340 [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng T. F. C., Bloemberg G. V., van der Bij A. J., van der Drift K. M. G. M., Schripsema J., Kroon B., et al. (1998). Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant-Microbe Interact. 11 1069–1077. 10.1094/MPMI.1998.11.11.1069 [DOI] [Google Scholar]
- De Souza J. T., De Boer M., De Waard P., Van Beek T. A., Raaijmakers J. M. (2003). Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 69 7161–7172. 10.1128/AEM.69.127161-7172.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Wang X., Baird S. M., Lu S. E. (2015). Complete genome of Pseudomonas chlororaphis strain UFB2, a soil bacterium with antibacterial activity against bacterial canker pathogen of tomato. Stand. Genomic Sci. 10:117. 10.1186/s40793-015-0106-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich L. E. P., Price-Whelan A., Petersen A., Whiteley M., Newman D. K. (2006). The phenacine pyocyanin is a terminal signaling factor in the quorum-sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61 1308–1321. 10.1111/j.1365-2958.2006.05306.x [DOI] [PubMed] [Google Scholar]
- Dimkpa C. O., Zeng J., McLean J. E., Britt D. W., Zhan J., Anderson A. J. (2012). Pathway in the plant-beneficial bacterium Pseudomonas chlororaphis O6 is inhibited by ZnO nanoparticles but enhanced by CuO nanoparticles. Appl. Environ. Microbiol. 78 1404–1410. 10.1128/AEM.07424-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorosky R. J., Yu J. M., Pierson L. S., III, Pierson E. A. (2017). Pseudomonas chlororaphis produces two distinct R-tailocins that contribute to bacterial competition in biofilm and on roots. Appl. Environ. Microbiol. 83:e706–17. 10.1128/AEM.00706-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury P., Aellen N., Ruffner B., Péchy-Tarr M., Fataar S., Metla Z., et al. (2016). Insect pathogenicity in plant-beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME 10 2527–2542. 10.1038/ismej.2016.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury P., Vesga P., Péchy-Tarr M., Aellen N., Dennert F., Hofer N., et al. (2017). Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 8:100. 10.3389/fmicb2017.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan G., Kumar A. M. (2005). Pseudomonas fluorescens a potential bacterial antagonist to control plant diseases. J. Plant Interact. 13 123–134. 10.1080/17429140600907043 [DOI] [Google Scholar]
- Garrido-Sanz D., Arrebola E., Martínez-Granero F., García-Méndez S., Muriel C., Blanco-Romero E., et al. (2017). Classification of isolates from the Pseudomonas fluorescens complex into phylogenomic groups based in group-specific markers. Front. Microbiol. 8:413. 10.3389/fmicb.2017.00413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169 30–39. 10.1016/j.micres.2013.09.009 [DOI] [PubMed] [Google Scholar]
- González-Sánchez M. A., de Vicente A., Pérez-García A., Pérez-Jiménez R., Romero D., Cazorla F. M. (2013). Evaluation of the effectiveness of biocontrol bacteria against avocado white root rot occurring under commercial greenhouse plant production conditions. Biol. Control 67 94–100. 10.1016/j.biocontrol.2013.08.009 [DOI] [Google Scholar]
- González-Sánchez M. A., Pérez-Jiménez R. M., Pliego C., Ramos C., de Vicente A., Cazorla F. M. (2010). Biocontrol bacteria selected by a direct plant protection strategy against avocado white root rot show antagonism as a prevalent trait. J. Appl. Microbiol. 109 65–78. 10.1111/j.1365-2672.2009.04628.x [DOI] [PubMed] [Google Scholar]
- Haas D., Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3 307–319. 10.1038/nrmicro1129 [DOI] [PubMed] [Google Scholar]
- Haas D., Keel C. (2003). Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41 117–153. 10.1146/annurev.phyto.41.052002.095656 [DOI] [PubMed] [Google Scholar]
- Han S. H., Lee S. J., Moon J. H., Park K. H., Yang K. Y., Cho B. H., et al. (2006). GacS-dependent production of 2R, 3R-butanediol by Pseudomonas chlororaphis O6 is a mayor determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. Tabaci in tobacco. Mol. Plant Microbe Interact. 19 924–930. 10.1094/MPMI-19-0924 [DOI] [PubMed] [Google Scholar]
- Haran S., Schickler H., Chet I. (1996). Molecular mechanisms of lytic enzymes involved in the biocontrol activity of Trichoderma harzianum. Microbiology 142 2321–2331. 10.1099/00221287-142-9-2321 [DOI] [Google Scholar]
- Hernández M. E., Kappler A., Newman D. K. (2004). Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70 921–928. 10.1128/AEM.70.2.921-928.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario E., Buckley T. R., Young J. M. (2004). Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. A. van Leeuw. J. Microb. 86 51–64. 10.1023/B.ANTO.0000024910.57117.16 [DOI] [PubMed] [Google Scholar]
- Hill D. S., Stein J. I., Torkewitz N. R., Morse A. M., Howell C. R., Pachlatko J. P., et al. (1994). Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 60 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Feng Z., Chi X., Sun X., Lu Y., Zhang B., et al. (2018). Pyrrolnitrin is more essential than phenazines for Pseudomonas chlororaphis G05 in its suppression of Fusarium graminearum. Microbiol. Res. 215 55–64. 10.1016/j.micres.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Jahanshah G., Yan Q., Gerhardt H., Pataj Z., Lämnerhofer M., Pianet I., et al. (2019). Dicovery of the cyclic lipopeptide gacamide A by genome mining and repair of the defective GacA regulator in Pseudomonas fluorescens Pf0-1. J. Nat. Prod. 82 301–308. 10.1021/acs.jnatprod.8b00747 [DOI] [PubMed] [Google Scholar]
- Kang B. R., Anderson A. J., Kim Y. C. (2018). Hydrogen cyanide produced by Pseudomonas chlororaphis O6 exhibits nematicidal activity against Meloidogyne hapla. Plant Pathol. J. 34 35–43. 10.5423/PPJ.OA.06.2017.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B. R., Yang K. Y., Cho B. H., Han T. H., Kim I. S., Lee M. C., et al. (2006). Production of indole-3-acetic acid in the plant-beneficial strain Pseudomonas chlororaphis O6 is negatively regulated by the global sensor kinase GacS. Curr. Microbiol. 52 473–476. 10.1007/s00284-005-0427-x [DOI] [PubMed] [Google Scholar]
- Liu K., Hu H., Wang W., Zhang X. (2016). Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-hydroxyphenazine. Microb. Cell Fact. 15:131. 10.1186/s12934-016-0529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper J. E., Hassan K. A., Mavrodi D. V., Davis E. W. I. I., Lim C. K., Shaffer B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insight into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. 10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper J. E., Henkels M. D., Shaffer B. T., Valeriote F. A., Gross H. (2008). Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl. Environ. Microbiol. 74 3085–3093. 10.1128/AEM.02848-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B. J. J., Dekkers L. C. (1999). What makes Pseudomonas bacteria rhizosphere competent? Environ. Microbiol. 1 9–13. 10.1046/j.1462-2920.1999.00005.x [DOI] [PubMed] [Google Scholar]
- Lugtenberg B. J. J., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Mavrodi D. V., Parejko J. A., Mavrodi O. V., Kwak Y. S., Weller D. M., Blankenfeldt W., et al. (2013). Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 15 675–686. 10.1111/j.1462-2920.2012.02846.x [DOI] [PubMed] [Google Scholar]
- Mazzola M., Funnell D. L., Raaijmakers J. M. (2004). Wheat cultivar-specific selection of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas species from resident soil populations. Microbiol. Ecol. 48 338–348. 10.1007/s00248-003-1067-y [DOI] [PubMed] [Google Scholar]
- McSpadden B. B. (2007). Diversity and ecology of biocontrol Pseudomonas spp. in agricultural systems. Phytopathology 97 221–226. 10.1094/PHYTO-97-2-0221 [DOI] [PubMed] [Google Scholar]
- Mercado-Blanco J., Bakker P. A. H. M. (2007). Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits from crop protection. Antonie Van Leeuw. 92 367–389. 10.1007/s10482-007-9167-1 [DOI] [PubMed] [Google Scholar]
- Moore-Landecker E. (1996). Fundamentals of the Fungi, 4th Edn. Saddle River, NJ: Prentice Hall. [Google Scholar]
- Moreno-Avitia F., Lozano L., Utrilla J., Bolívar F., Escalante A. (2017). Draft genome sequence of Pseudomonas chlororaphis ATCC 9446, a nonpathogenic bacterium with bioremediation and industrial potential. Genome Announc. 5:e474–17. 10.1128/genomeA.00474-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi T., Yamaguchi T., Xie X., Wang W. Z., Takeuchi K., Someya N. (2017). Complete genome sequence of Pseudomonas chlororaphis subsp. Auranthiaca reveals a triplicate quorum-sensing mechanism for regulation of phenazine production. Microbes Environ. 32 47–53. 10.1264/jsme2.ME16162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeem S. M., Zahir Z. A., Naveed M., Arshad M. (2007). Preliminary investigation on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can. J. Microbiol. 53 1141–1149. 10.1139/W07-081 [DOI] [PubMed] [Google Scholar]
- Nandi M., Selin C., Brassinga A. K. C., Belmonte M. F., Fernando W. G. D., Loewen P. C., et al. (2015). Pyrronitrin and hydrogen cyanide production by Pseudomonas chlororaphis PA23 exhibits nematicidal and repellent activity against Caenorhabditis elegans. PLoS One 10:e0123184. 10.1371/journaol.pone.0123184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi M., Selin C., Brawerman G., Fernando W. G. D., de Kievit T. (2017). Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol. Control 108 47–54. 10.1016/j.biocontrol.2017.02.008 [DOI] [Google Scholar]
- Nishiyama M., Horinouchi S., Kobayashi M., Nagasawa T., Yamada H., Beppu T. (1991). Cloning and characterization of genes responsible for metabolism of nitrile compounds from Pseudomonas chlororaphis B23. J. Bacteriol. 173 2465–2472. 10.1128/jb.173.8.2465-2472.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak-Thompson B., Philip E., Hammer D., Hill D. S., Staffords J., Torkewitz N., et al. (2003). 2, 5-diakylresorcinol biosynthesis in Pseudomonas aurantiaca: novel head-to-head condensation of two fatty acid-derived precursors. J. Bacteriol. 185 860–869. 10.1128/JB.185.3.860-869.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olorunleke F. E., Hua G. K., Kieu N. P., Ma Z., Höfte M. (2015). Interplay between orfamides, sessilins and phenazines in the control of rhizoctonia diseases by Pseudomonas sp. CMR12a. Environ. Microbiol. Rep. 7 774–781. 10.1111/1758-2229.12310 [DOI] [PubMed] [Google Scholar]
- Ownley B. H., Duffy B. K., Weller D. M. (2003). Identification and manipulation of soil properties to improve the biological control performance of phenazine-producing Pseudomonas fluorescens. Appl. Environ. Microbiol. 69 3333–3343. 10.1128/AEM.69.6.3333-3343.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleroni N. J. (1984). “Family I. Pseudomonadaceae Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 555,” in Bergey’s Manual of Systematic Bacteriology, 1st Edn, eds Sneath P. H. A., Mair N. S., Sharpe M. E., Holt J. G. (Baltimore, MD: Williams & Wilkins; ), 144–218. [Google Scholar]
- Peix A., Valverde A., Rivas R., Igual J. M., Ramírez-Bahena M. H., Mateos P. F., et al. (2007). Reclassification of Pseudomonas aurantiaca as a synonym of Pseudomonas chlororaphis and proposal of three subspecies, P. Chlororaphis subsp. Chlororaphis Subsp. nov., P. Chlororaphis subsp. Aureofaciens subsp. nov., comb. nov. and P. chlororaphis subsp. aurantiaca subsp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 57 1286–1290. 10.1099/ijs.0.64621-0 [DOI] [PubMed] [Google Scholar]
- Pierson L. S., III, Pierson E. A. (2010). Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 86 1659–1670. 10.1007/s00253-010-2509-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliego C., Ramos C., de Vicente A., Cazorla F. M. (2011). Screening for candidate bacterial biocontrol agents against soilborne fungal plant pathogens. Plant Soil 340 505–520. 10.1007/s11104-010-0615-8 [DOI] [Google Scholar]
- Polonio A., Vida C., de Vicente A., Cazorla F. M. (2017). Impact of motility and chemotaxis features of the rhizobacterium Pseudomonas chlororaphis PCL1606 on its biocontrol of avocado white root rot. Int. Microbiol. 20 95–104. 10.2436/20.1501.01.289 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., de Bruijn I., de Kock M. J. D. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant Microbe Interact. 19 699–710. 10.1094/MPMI-19-0699 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., De Bruijn I., Nybroe O., Ongena M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34 1037–1062. 10.1111/j.1574-6976.2010.00221.x [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., Paulitz T. C., Steinberg C., Alabouvette C., Moënne-Loccoz Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:20 10.1007/s11104-008-9568-6 [DOI] [Google Scholar]
- Raio A., Reveglia P., Puopolo G., Cimmino A., Danti R., Evidente A. (2017). Involvement of phenaazine-1-carboxylic acid in the interaction between Pseudomonas chlororaphis subsp. Aureofaciens strain M71 and Seiridium cardinal in vivo. Microbiol. Res. 199 49–56. 10.1016/j.micres.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Ruffner B., Péchy-Tarr M., Höfte M., Bloemberg G., Grunder J., Keel C.et al. (2015). Evolutionary patchwork of an insecticidal toxin shared between plant-associated pseudomonads and the insect pathogens Photorhabdus and Xenorhabdus. BMC Genomics 16:609. 10.1186/s12864-015-1763-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin C., Fernando D., de Kievit T. R. (2012). The Phzl/PhzR quorum-sensing system is required for pyrrolnitrin and phenazine production, and exhibits cross-regulation with RpoS in Pseudomonas chlororaphis PA23. Microbiology 158 896–907. 10.1099/mic.0.054254-0 [DOI] [PubMed] [Google Scholar]
- Selin C., Habibian R., Poritsanos N., Athukorala S. N., Fernando D., de Kievit T. R. (2010). Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol. Ecol. 71 73–83. 10.1111/j.1574-6941.2009.00792.x [DOI] [PubMed] [Google Scholar]
- Skerman V. B. D., McGoawna V., Sneath P. H. A. (1980). Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30 225–420. 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- Town J., Audy P., Boyetchko S. M., Dumonceaux T. J. (2016). Genome sequence of Pseudomonas chlororaphis strain 189. Genome Announc. 4:e581–16. 10.1128/genomeA.00581-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V. (2006). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30 274–291. 10.1111/j.1574-6976.2005.00012.x [DOI] [PubMed] [Google Scholar]
- Vida C., Bonilla N., de Vicente A., Cazorla F. M. (2016). Microbial profiling of a suppressiveness-induced agricultural soil amended with composted almond shells. Front. Microbiol. 7:4. 10.3389/fmicb.2016.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida C., Cazorla F. M., de Vicente A. (2017a). Characterization of biocontrol bacterial strains isolated from a suppressiveness-induced soil after amendment with composted almond shells. Res. Microbiol. 168 583–593. 10.1016/j.resmic.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Vida C., de Vicente A., Cazorla F. M. (2017b). Draft genome sequence of the rhizobacterium Pseudomonas chlororaphis PCL1606, displaying biocontrol against soilborne phytopatogens. Genome Announc. 5:e130–17. 10.1128/genomeA.00130-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman S. A., Woodruff B. H. (1940). The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 40 581–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller D. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97 250–256. 10.1094/PHYTO-97-2-0250 [DOI] [PubMed] [Google Scholar]
- Yan Q., Lopes L. D., Shaffer B. T., Kidarsa T. A., Vining O., Philmus B., et al. (2018). Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. mBio 9:e1845–17. 10.1128/mBio.01845-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. X., Rainey P. B. (2013). Exploring the sociobiology of pyoverdin-producing Pseudomonas. Evolution 67 3161–3174. 10.1111/evo.12183 [DOI] [PubMed] [Google Scholar]