Abstract
The molecular determinants of substrate specificity and selectivity in the proprotein convertase (PC) family of proteases are poorly understood. Here we demonstrate that the natural serpin family inhibitor, serpin B8, is a specific and selective inhibitor of furin relative to the other PCs of the constitutive secretion pathway, PC4, PC5, PACE4, and PC7 (PCs 4–7), and identify reactive-site (P6-P5’ residues) and exosite elements of the serpin that contribute to this specificity/selectivity through studies of chimeras of serpin B8 and α1PDX, an engineered serpin inhibitor of furin. Kinetic studies revealed that the specificity and selectivity of the serpin chimeras for inhibiting PCs was determined by P6-P5 and P3-P2 residue-dependent recognition of the P4Arg-X-X-P1Arg PC consensus sequence and exosite-dependent recognition of the reactive loop P2’ residue of the chimeras by the PCs. Both productive and non-productive binding of the chimeras to PCs 4–7 but not to furin contributed to a decreased specificity for inhibiting PCs 4–7 and an increased selectivity for inhibiting furin. MDS simulations suggested that non-productive binding of the chimeras to the PCs was correlated with a greater conformational variability of the catalytic sites of PCs 4–7 relative to furin. Our findings suggest a new approach for designing selective inhibitors of PCs using α1PDX as a scaffold, as evidenced by our ability to engineer highly specific and selective inhibitors of furin and PCs 4–7.
Introduction
Proprotein convertases (PCs) are ubiquitous calcium dependent serine proteases of the subtilisin fold. In mammals, PCs are complex multi-domain proteins that carry out the proteolytic posttranslational modification of many secreted peptides and proteins, and regulate central cellular processes like growth and proliferation (1, 2). The PCs of the Kexin-like subtype, furin, PC4, PC5, PACE4 and PC7, localize to the trans-Golgi network and endosomes of the constitutive protein secretion pathway and cleave precursors of a large diversity of proteins at polybasic sites consisting of the general P4Arg-X-X-P1Arg PC substrate specificity motif. Many important viral and bacterial pathogens exploit these PCs to promote and regulate their own growth. For this reason, specific PC inhibitors are sought as potential therapeutic agents (3).
Furin reactivity is regulated by changes of pH and calcium concentrations to impact enzymatic activity and autocatalytic activation (4, 5). Protein crystallography and molecular dynamic simulations (MDS) revealed that the furin catalytic site is in equilibrium between active and inactive conformations (6, 7). Given the similarities at the catalytic site among PCs (8), it is predictable that furin shares with PC4, PC5, PACE4 (PC6) and PC7 (PCs 4–7) similar mechanisms to regulate reactivity.
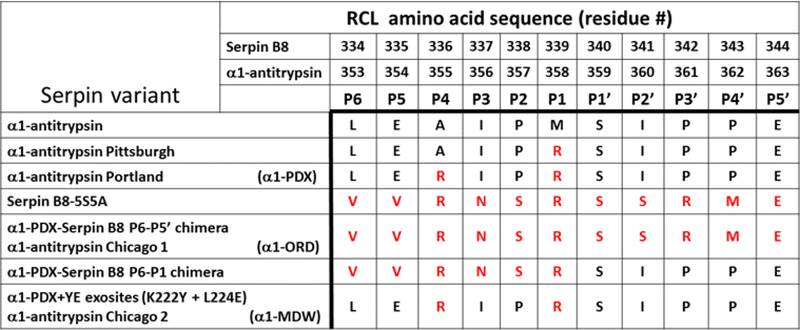
The assignment of natural substrates to individual PCs has been speculative due to the great deal of cross-reactivity among these proteases, and substrate preferences have been assumed to depend mainly on differences of expression and cell type distribution among PCs. Efforts to identify amino acid residue preferences at the substrate cleavage site by individual PCs have been attempted with the use of peptide libraries with limited success (9). We developed a more robust approach in which the serpin-type protease inhibitor, α1-antitrypsin, was used as a model PC substrate to engineer changes in its reactive center loop (RCL) site of cleavage (10). The validity of this approach is supported by the fact that serpin B8 is the only mammalian natural furin inhibitor known so far, and PCs from a variety of organisms are regulated by serpins (10–14). Here, we found that serpin B8 is a selective furin inhibitor and used the serpin α1-Antitrypsin as a scaffold to graft serpin B8 RCL and exosite amino acid residues to elucidate the basis for this selectivity. α1-Antitrypsin is known to inhibit furin efficiently when arginine residues are engineered at its RCL P4 and P1 positions (α1PDX) (15, 16). This approach is supported by our previous study showing that RCL and exosite determinants of serpin B8 reactivity, when substituted into their homologous regions in α1-antitrypsin, regulate reactivity with furin (10). The same approach was here extended to the other PCs of the constitutive secretion pathway to identify the serpin B8 determinants responsible for the specific and selective inhibition of furin compared to PCs 4–7. Knowledge of these determinants enabled us to engineer α1-PDX derivatives that were highly specific and selective inhibitors of furin and PCs 4–7.
Materials and Methods
Production of PCs.
Recombinant proprotein convertases were produced in truncated form as described for furin (10). They included the first 579 residues of furin (UniProtKB P09958), 584 of PC4 (UniProtKB Q6UW60), 605 of PC5 (UniProtKB Q92824), 638 of PACE4 (UniProtKB P29122) and 621 of PC7 (UniProtKB Q16549), up to the end of their P-domains. The gene constructs were synthesized (Integrated DNA Technologies) with a 10×His tag extension at the C-terminus. The proteins were expressed for 24 h in 1L Hi5 or sf9 insect cells using the baculovirus expression system. The PCs were purified from secreted proteins to homogeneity using Nickel-affinity and size exclusion chromatography as shown previously for furin (10). Protein yields were between 100 μg and 1 mg of purified protein. PCs 4–7 were expressed less abundantly than furin, especially PC7.
Production and Engineering of α1-Antitrypsin Mutants.
The serpin α1-antitrypsin (UniProtKB P01009) was expressed in bacteria, refolded from inclusion bodies, and purified by ion exchange chromatography as described (10). Mutagenesis of the serpin was done by PCR using specifically designed oligonucleotides (Integrated DNA Technologies) and Pfu Ultra II HF DNA polymerases (Agilent Technologies).
Production of Serpin B8.
Recombinant serpin B8 (UniProtKB P50452) was produced in insect cells using the baculovirus system as described before (10). Five of its cysteine residues were mutated into serine and five into alanine (serpin B8–5S5A) in order to improve stability.
Protease Activity Assay.
Protease activity assays and determinations of Michaelis-Menten enzyme kinetic parameters (Km and kcat) with the fluorogenic substrates, Boc-Arg-Val-Arg-Arg-7-amido-methylcoumarin and pyr-Arg-Thr-Lys-Arg-amido-methylcoumarin (Bachem) were performed in 100 mM Hepes pH 7.5, 1 mM CaCl2, 1 mM β-mercaptoethanol, 0.5 % Triton X-100, and 0.1 % polyethylene glycol 8000 at 25 °C employing the same methodology as described previously for furin (10). The same conditions were used with all PCs in order to compare their activities.
Protease Active-site Titrations.
Concentration of active enzymes and turnover number values were determined from titrations of fixed enzyme concentrations with the inhibitor dec-Arg-Val-Lys-Arg-chloromethyl ketone (Bachem) to reach end points of zero activity at pH 5.5, as described (10). The stoichiometric inhibition of PCs can only be achieved at acidic conditions. At the commonly used neutral pH of the enzyme activity assay, the competing hydrolysis of the inhibitor results in curved instead of straight line, stoichiometric titrations, due to depletion of the inhibitor. Specific activity values of 6.4, 1.9, 1.8, 1.3 and 1.5 ΔF/min/nM PC were measured for furin, PC4, PC5, PACE4 and PC7, respectively, with 10 μM pyr-Arg-Thr-Lys-Arg-amido-methylcoumarin at protease activity assay conditions and used to calculate the concentration of the enzyme from linear changes in fluorescence.
Protease Inhibition Assays.
The second order association rate constant (ka) values for the reaction of PCs with serpin B8 and α1-antitrypsin variants were determined from the kinetics of protease inhibition by the serpins as described previously (10). All reactions were performed in protease activity assay conditions and at pseudo-first order reaction conditions in which the inhibitor concentrations exceeded that of the enzyme at least 10-fold. Measurements of fast reactions (ka in the range 105 to 107 M−1s−1) were obtained from the continuous exponential loss of enzyme activity observed in protease assay conditions that consumed minimal amounts of the 10 μM fluorogenic substrate pyr-Arg-Thr-Lys-Arg-amido-methylcoumarin present in the assay. The observed first order rate constants (kobs) were obtained by fitting the time-dependent increase in observed fluorescence (ΔFobs) in reaction progress curves to the exponential rise equation : ΔF = ΔFmax × (1−exp(−kobs × t)) where ΔFmax is the maximum fluorescence change at the endpoint. Apparent association rate constant values were derived from slopes of the linear dependency of the kobs values against inhibitor concentrations that were corrected for the competitive effect of the substrate by dividing by the factor, 1+([S]o/Km). For slower reactions (ka values under 105 M−1s−1), longer time assays of discontinuous protease inhibition were performed and residual activities determined from aliquots removed from the inhibition reactions. Plots of time dependent residual protease activity (Aobs) were fitted to an exponential decay equation, Aobs = Ao × exp(−kobs × t) to obtain kobs values. Apparent association rate constant values were derived from slopes of the linear dependency of the kobs values against inhibitor concentrations. For those reactions in which the initial Michaelis encounter complex was formed rapidly and the subsequent chemical step was comparatively slow, association rate constant (ka) values were determined from kcat/Kd ratios (equal to kcat/Km) where kcat = kobs/(fractional occupation of the Michaelis complex). Apparent association rate constant values were corrected by factoring the stoichiometry of protease inhibition (SI) to yield the true association rate constant (ka) value. The stoichiometry for protease inhibition was determined under protease assay conditions as the serpin to protease molar ratio that yielded zero enzyme activity and was obtained from the linear decrease of enzyme activity as a function of increasing inhibitor concentrations.
PC-serpin Binding Affinity.
The reaction of some of the α1-antitrypsin variants with PCs yielded fast enzyme activity drops that were dependent on the concentration of the inhibitor in a manner that reflected equilibrium formation of the Michaelis complex. The rapid loss of reactivity was followed by a slower loss of the remaining activity that reflected acylation of the Michaelis complex to form the conformationally trapped covalent serpin-enzyme complex. Equilibrium binding dissociation constants for the Michaelis complex (Kd) were determined from nonlinear least squares computer fits of plots of the dependency of residual PC activity, calculated from the initial linear segment of residual activity, on inhibitor concentrations to the tight binding quadratic equation by Morrison, [E]/[E]o = 1 – (([E]o + [I]o + Kd) - (([E]o + [I]o + Kd)^2 – 4 × [E]o × [I]o)^0.5 / (2 × [E]o) where [E] and [E]o are the residual active and total PC concentrations, respectively, and [I]o is the total inhibitor concentration (21). Inhibitor concentrations were corrected for the competitive effect of the substrate by dividing by the factor, 1+([S]o/Km).
Molecular Dynamic Simulations.
Principal component analyses on two furin crystal structures and models of the catalytic domain of PCs 4–7 were carried out as described (7). MD simulations were performed using the GROMACS (v5.0.3) molecular simulation package as described (7). Models of the catalytic domain of PC4, PC5, PACE4 and PC7 were obtained using the SWISS-MODEL tool for protein homology modeling, and based on the x-ray crystal structures of the ligand-free form (PDB code 5JXG) and the ligand-bound form (PDB code 5JXH) of human furin as templates.
Results
Furin Hydrolyzes Two Small Peptide Substrates at a Faster Chemical Conversion Rate than PC4, PC5, PACE4 and PC7.
Recombinant furin, PC4, PC5, PACE4 and PC7, consisting of only their catalytic and P-domains, were produced in a fully active and highly purified form (10). To determine the relative specificity and selectivity of these PCs for two of the most commonly used PC substrates, we measured the Michaelis-Menten kinetic parameters for the PC-catalyzed hydrolysis of the substrates. (Table 1). This comparative study showed that furin is 2 to 20-fold more reactive than PCs 4–7 primarily due to the faster chemical conversion step (kcat).
Table 1.
Michaelis-Menten Kinetic Constant Value & for the Hydrolysis of Two Tetrapeptide Fluorogenic Substrates by PCs
| Boc-Arg-Va1-Arg-Arg-AM C | pyr-Arg-Thr-Lys-Arg-AMC | |||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km(M−1s−1) | Km (μM) | kcat (s−1) | kcat/Km(M−1s−1) | |
| FURIN | 28 ± 1 | 0.27 ± 0.01 | 9.2 ± 1.0 × 103 | 4.7 ± 0.5 | 0.16 ± 0.00 | 3.4 ± 0.7 × 104 |
| PC4 | 51 ± 4 | 0.040 ± 0 001 | 7.9 ± 1.0 × 102 | 2.6 ± 0.2 | 0.041 ± 0.00 | 1.6 ± 0.1 × l04 |
| PC5 | 40 ± 2 | 0.031 ± 0.001 | 7.6 ± 0.5 × 102 | 2.0 ± 0.1 | 0.038 ± 0.001 | 1.9 ± 0.1 × l04 |
| PACE4 | 45 ± 3 | 0.021 ± 0.001 | 4.6 ± 0.4 × 102 | 2.6 ± 0.2 | 0.028 ± 0.001 | 1.1 ± 0.1 × l04 |
| PC7 | 27 ± 3 | 0.018 ± 0.001 | 6.8 ± 1.0 × 102 | 2.2 ± 0.3 | 0.032 ± 0 002 | 1.5 ± 0.3 × 104 |
Michaelis-Menten constant (Km and kcat) values were determined from initial velocities of product formation from the catalyzed hydrolysis of two fluorogenic peptide substrates. Reactions were carried out in protease activity assay conditions as described in the Materials and Methods section. Errors represent S.E from linear regression fits of data. Fixed concentrations of active-site titrated PCs were used in reactions with substrate concentrations ranging from 0.2 to 5 times the Km values. Parameters defining the dependence of initial velocities on substrate concentrations were adjusted to the Michaelis-Menten.
Serpin B8 is a Specific and Selective Furin Inhibitor.
We have previously determined that serpin B8 inhibits furin at a physiologically relevant rate (10). If serpin B8 is a specific and selective natural furin inhibitor, then it should react much more rapidly with furin than with PCs 4–7. In order to test this hypothesis, the rate of inhibition of PC4, PC5, PACE4 and PC7 by serpin B8 was measured using our stabilized recombinant form of serpin B8 that we previously described (10), in which five external and internal cysteine residues were mutated to serine or alanine, respectively (Table 2). Serpin B8 reacted about 20-fold slower with PCs 4–7 (ka 1 × 104 M−1s−1) than with furin (ka 2 × 105 M−1s−1). Serpin B8 is thus significantly more selective for inhibiting furin over PCs 4–7.
Table 2.
Rate Constant Values for the Reaction of PCs with Serpin B8 and αPDX
| Serpin B8 | α1PDX | |||
|---|---|---|---|---|
| ka(M−1s−1) | SI | ka(M−1s−1) | SI | |
| FURIN | 2 3 ± 0.2 × 105 | 2.2 ± 0.1 | 1.l ± 0.1 × 106 | 1.1 ± 0.1 |
| PC4 | 14 ± 0.2 × 104 | 1.2 ± 0.1 | 1.6 ± 03 × 106 | 1.2 ± 0.2 |
| PC5 | 1.1 ± 0.2 × 104 | 0.9 ± 0.1 | 2.0 ± 0.1 × 106 | 1.3 ± 0.1 |
| PACE4 | 1.1 ± 0.3 × 104 | 0.9 ± 0.1 | 1.2 ± 0.3 × 106 | 1.0 ± 0.2 |
| PC7 | 1.2 ± 0.2 × 104 | 1.0 ± 0.1 | 2.3 ± 0.8 × 106 | 1.4 ± 0.2 |
Second order association rate constant (ka) values for the reaction of PCs with serpin B8 and α1PDX, and their stoichiometry of inhibition (SI) were measured in protease activity assay conditions as described in the Materials and Methods section. The SI measures the partition between the inhibitory and substrate pathways of the serpin mechanism of protease inhibition. A value of 1 corresponds to an efficient inhibition (figure S1). Errors represent S.E from linear regression fits of data. The ka and SI values for furin were taken from reference (10).
The serpin B8 P6-P5’ RCL Segment Confers Greater Furin Selectivity in α1PDX than in Serpin B8.
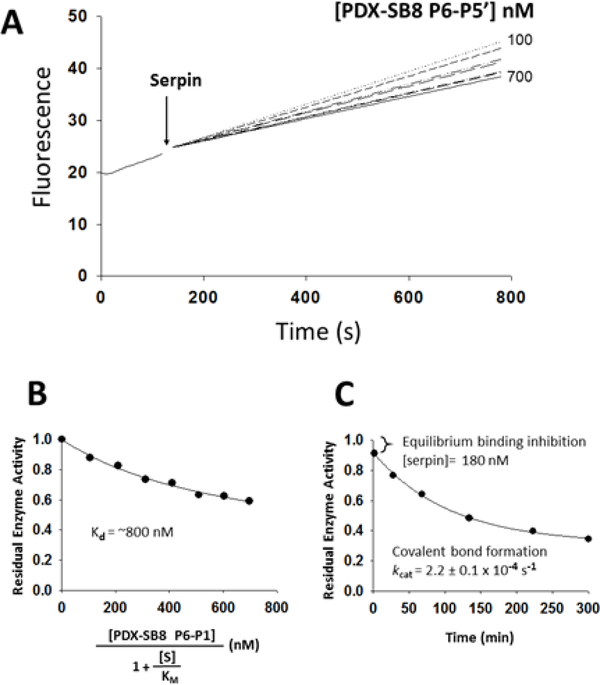
We previously determined that the primed and non-primed regions of the serpin B8 RCL, when grafted onto the RCL of the serpin α1PDX, play critical roles in influencing reactivity with furin (10). The same analysis was applied here to investigate the role that these two serpin B8 RCL regions play in determining the differential reactivity between furin and PCs 4–7. Serpins share the same general structure and a suicide substrate mechanism of inhibition of proteases that results in the formation of a conformationally trapped serpin-protease covalent complex (Fig. S1) (17). α1PDX is a derivative of the serpin α1-antitrypsin that was developed as a specific furin inhibitor by grafting the P4Arg-X-X-P1Arg PC recognition sequence onto the RCL (18). We found that α1PDX is equally reactive with furin, PC4, PC5, PACE4 and PC7, all the PCs being inhibited efficiently by α1PDX at similar rates (106 M−1s−1) and with stoichiometries of inhibition of 1:1 (Table 2). A previous report (16) found differences in α1PDX reactivity with some of these PCs based on an erroneous characterization of serpin reactivity by Ki values instead of ka (19). The serpin B8 RCL P6-P5’ segment (Fig. 1), when placed at its homologous location in the RCL of α1PDX resulted in a chimera that reacted with furin 10-fold slower (105 M−1s−1) than α1PDX, similar to serpin B8 (Table 3). However, the P6-P5’ chimera reacted with PCs 4–7 much slower than serpin B8 and in a biphasic manner distinct from the monophasic reaction with furin (Fig. 2A). The P6-P5’ chimera thus caused an immediate inhibition of a fraction of the protease activity followed by a slower exponential inhibition of the remaining activity (Fig. 2C). This behavior suggested that an initial rapid equilibrium binding of the serpin and protease to form a Michaelis complex was kinetically separated from the slower acylation of the Michaelis complex to form the conformationally trapped serpin-protease covalent complex. Indeed, the serpin concentration dependence of the initial rapid inhibition phase was consistent with Kd values of about 1 μΜ for formation of the Michaelis complex (Fig. 2B, Table 4), whereas the subsequent exponential reaction phase indicated first-order rates of acylation of the Michaelis complex (kobs ~ 10−4 s−1) that were about 100-fold slower than kcat values measured for the hydrolysis of the tetrapeptide fluorogenic substrates by PCs 4–7 (Table 5). Because of the kinetic separation, the contributions to overall reactivity (ka = kcat/Km) by the binding (Km) and the chemical (kcat) steps can be calculated separately. The Km equals the Kd for the rapid equilibrium formation of the Michaelis complex, and the kcat can be calculated by dividing kobs for formation of the covalent complex by the fractional saturation of the Michaelis complex (Fig. 2C, Table 5). Values of ka calculated from kcat/Kd ratios (102 M−1s−1) were several 1,000-fold lower than ka values for reactions of α1PDX with PCs 4–7 (Table 3). The serpin B8 P6-P5’ segment when grafted onto α1PDX thus amplified the differences in reactivity observed for serpin B8 with furin and PCs 4–7.
Figure 1.
Amino acid residue sequence at the RCL of α1-antitrypsin derivatives and serpin B8. The amino acid residue sequences from the RCL of α1-antitrypsin variants and of serpin B8 were aligned based on the location of the protease cleavage site between the residues P1 and P1’. A natural mutant of α1-antitrypsin, the Pittsburgh variant, has an arginine substitution at the P1 residue that changes the serpin specificity to inhibit trypsin-type proteases (20). The α1-antitrypsin Portland variant (α1PDX) was engineered with arginine residues at the P4 and P1 positions to inhibit furin (15, 16). The serpin B8–5S5A variant is a stabilized form of serpin B8 in which cysteine residues were mutated, five to serine and five to alanine, including the one at the P1’ position (10). The α1PDX-serpin B8 Ρ6-Ρ5’ chimera is now designated the Chicago 1 derivative (α1ORD) due to its high furin selectivity. The α1PDX+YE exosites variant, is now designated the Chicago 2 derivative (α1MDW) due to its high selectivity for PC4, PC5, PACE4 and PC7 over furin.
Table 3.
Rate Constant Values for the Reaction of PCs with α1PDX-serpin B8 RCL Chimeras
| α1PDX-Serpin B8 RCL Chimera | Second order association rate, ka(M−1s−1) | ||||
|---|---|---|---|---|---|
| FURIN | PC4 | PC5 | PACE4 | PC7 | |
| α1PDX | 1.1 ± 0.1 × 106 | 1.6 ± 0.3 × 106 | 2.0 ± 0.1 × 106 | 1.2 ± 0.3 × 106 | 2.3 ± 0.8 × 106 |
| P6-P5’* | 1.1 ± 0.1 × 105 | 7.1 ± 1.0 × 102 | 0.0 ± 0.2 × 102 | 2.9 ± 0.l × 102 | 7.7 ± 2.2 × 102 |
| P6-P1* | 2.5 ± 0.3 × l06 | <103 | <103 | 7.5 ± 0.8 × 103 | l.5 ± 0.2 × lO3 |
| P6-P5* | 2.2 ± 0.2 × 106 | 7.9 ± 1.8 × 105 | 2.0 ± 0.3 × 106 | 4.7 ± 0.8 × 105 | 3.3 ± 1.2 × 106 |
| P3-P2 | 1.3 ± 0.1 × 105 | >2 × 103 | >2 × 103 | >9 × 102 | >4 × 102 |
| P1’-P5’ | 5.5 ± 0.8 × 104 | 3.1 ± 0.9 × 104 | 4.9 ± 1.3 × 104 | l.5 ± 0.3 × 104 | 4.8 ± 2.0 × 104 |
| P2’ | 6.4 ± 0.8 × 104 | 2.5 ± 0.6 × 104 | 3.3 ± 0.6 × 104 | 1.0 ± 0.2 × 104 | 2.4 ± 0.3 × 104 |
| P3’ | 4.7 ± 0.2 ×l06 | 3.8 ± 0.5 ×l06 | 1.8 ± 0.4 ×l06 | 8.6 ± 1.3 ×l05 | 4.8 ± 2.4 ×l06 |
| P4’ | 7.9 ± 0.5 ×l05 | 3.9 ± 0.6 ×l05 | 5.8 ± 0.6 ×l05 | 1.9 ± 0.5 ×l05 | 4.0 ± 0.1 ×l05 |
Amino acid sequences in the RCL of α1PDX were substituted with the homologous sequences from the RCL of serpin B8. Second order association rate constant (ka) values for the reaction of PCs with α1PDX-serpin B8 chimeras were determined as described in the Materials and Methods section. Errors represent S.E from linear regression fits of data. The ka values for the reaction of furin with α1PDX and the P6-P5’, P6-P1 and Ρ1’-Ρ5’ chimeras were taken from reference (10).
> Unstable covalent complexes prevented the SI to be determined so the rate constant values are larger than here reported.
ka values for PCs 4–7 were determined from kcat/Kd ratios as described in the Materials and Methods section. The reactions with furin were monophasic, therefore the effect of the Kd and kcat on reactivity were not separated.
Figure 2.
Reaction of the α1PDX-serpin B8 P6-P5’ chimera with PACE4. (A) Shown are plots of PACE4 residual activity at increasing concentrations of the α1PDX-serpin B8 P6-P5’ chimera. Progressive reactions of 2 nM PACE4 activity with 10 μΜ of the fluorogenic substrate pyr-Arg-Thr-Lys-Arg-amido-methylcoumarin under protease activity assay conditions were run as described in the Materials and Methods section. (B) Shown is the fitted plot of the dependence of residual PACE4 activity on increasing concentrations of the serpin chimera that were corrected for the fluorogenic substrate concentration [S]. The plot was fitted to the quadratic tight binding equation by Morrison (21). Similar results were obtained with PC4, PC5 and PC7, and Kd values are listed in Table 4. (C) Shown is a plot of residual PACE4 activity for the inhibition of 50 nM PACE4 by 1.4 μΜ serpin chimera. The initial drop in activity reflects the fast equilibrium binding for the formation of the Michaelis complex that precedes slow covalent complex formation.
Table 4.
Dissociation Constant Values for the Binding of PCs to α1PDX-serpin B8 RCL chimeras
| α1PDX-Serpin B8 chimera | Equilibrium binding dissociation constant, Kd (nM) | ||||
|---|---|---|---|---|---|
| FURIN | PC4 | PC5 | PACE4 | PC7 | |
| P6-P5’ | -------- | ~1400 | ~1000 | ~800 | ~1500 |
| P6-P1 | -------- | 13 ± 1 | 7.8 ± 0.8 | 9.1 ± 0.7 | 15 ± 0.9 |
| P6-P5 | -------- | 0.52 ± 0.01 | 0.62 ± 0.01 | 1.0 ± 0.1 | 0.41 ± 0.06 |
Table 5.
Rate Constant Values for the Catalytic Chemical Step in the Reaction of PCs with α1PDX-serpin B8 RCL Chimeras and Tetrapeptide Substrates
| Chemical reaction step, kcat (s−1) | |||||
|---|---|---|---|---|---|
| FURIN | PC4 | PC5 | PACE4 | PC7 | |
| Peptide substrates | 0.27 – 0.16 | 0.040 – 0.041 | 0.031 – 0.038 | 0.021 – 0.023 | 0.018 −0.032 |
| P6-P5’ | -------- | 0.00029 | 0.00029 | 0.00022 | 0.00034 |
| P6-P1 | -------- | Non-measurable | Non-measurable | 0.000068 | 0.000023 |
| P6-P5 | -------- | 0.00011 | 0.00035 | 0.00024 | 0.00029 |
Comparison of kcat values for the catalyzed hydrolysis of two tetrapepnde fhiorogenic substrates by PCs (taken from Table 1), against the kcat values obtained from the reaction of PCs with three αPDX-serpin B8 RCL chimeras. These reactions, which are described in Figure 2, Figure 3 and Figure 4, operate based on initial rapid formation of high affinity Michaelis complexes followed by slower chemical step reactivity leading to covalent complex formation. The kcat values for the chemical step were obtained from kobs = kcat × fractional occupation. The reactions of furin with serpins were monophasic, therefore the value of kcat could not be determined.
The Serpin B8 P6-P1 RCL Segment Determines PC Selectivity.
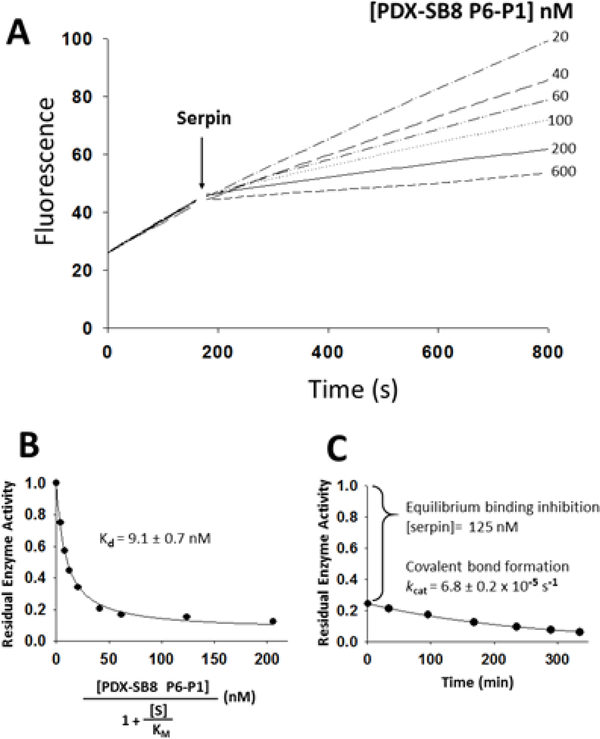
In order to define the origin of the large difference in reactivity between furin and PCs 4–7 observed with the P6-P5’ chimera, the serpin B8 P6-P1 segment was alone grafted onto the RCL of α1PDX. In contrast to the P6-P5’ segment, the P6-P1 segment resulted in a two-fold increase in reactivity with furin (2 × 106 M−1s−1) (Table 3). However, the P6-P1 chimera reacted with PCs 4–7 similar to the P6-P5’ chimera, i.e., with a greatly reduced overall reactivity relative to α1PDX and biphasic kinetics of inhibition (Fig. 3A). Surprisingly, the chimera formed Michaelis complexes in an initial rapid equilibrium binding step that exhibited about 100-fold stronger affinities than those of the P6-P5’ chimera (Kd values of 7–15 nM) (Fig. 3B, Table 4). Moreover, the subsequent slower exponential phase reflecting conversion of the non-covalent Michaelis complex to the covalent inhibitory complex (Fig. 3C), exhibited kcat values minimally 10-fold slower than those observed with the P6-P5’ chimera (10−5 s−1 or less) (Table 5). This slow reactivity was confirmed by electrophoresis analysis of the serpin chimera-PC reaction which showed minimal reaction of the serpin to form a covalent complex or to be cleaved as a substrate over the timeframe in which covalent complexes formed with the P6-P5’ chimera. Calculated ka values for PACE4 and PC7, for which kcat values were measurable, were determined from the kcat/Kd ratios (103 M_1s_1) to be 100 to 1000-fold lower than those for the reaction of α1PDX with PCs 4–7 (Table 3).
Figure 3.
Reaction of the α1PDX-serpin B8 P6-P1 chimera with PACE4. (A) Shown are plots of PACE4 residual activity at increasing concentrations of the α1PDX-serpin B8 P6-P1 chimera. Progressive reactions of 5 nM PACE4 were run as described in legend to figure 2. (B) Shown is the fitted plot of the dependence of residual PACE4 activity on increasing concentrations of the serpin chimera as described in legend to figure 2. Similar results were obtained with PC4, PC5 and PC7, and Kd values are listed in Table 4. (C) Shown is a plot of residual PACE4 activity for the time course of inhibition of 50 nM PACE4 by 1.0 μΜ of the chimera. The large initial drop in activity reflects the fast equilibrium binding for the formation of the Michaelis complex that precedes slow covalent complex formation.
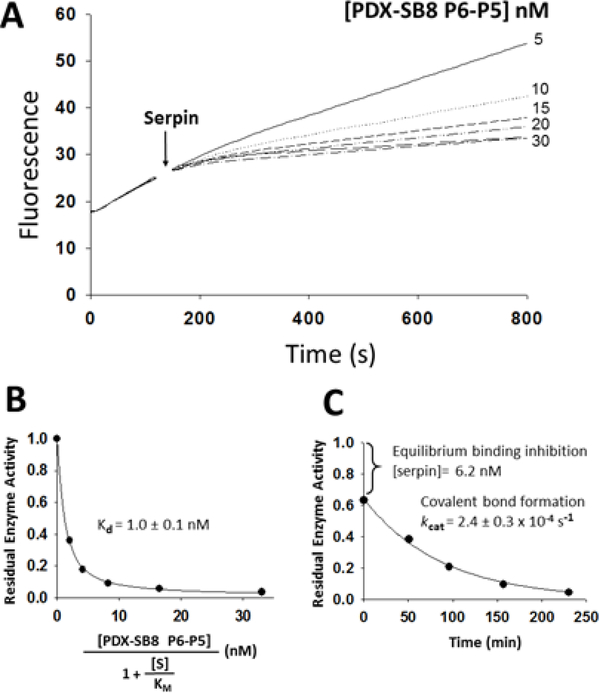
Further reducing the size of the grafted serpin B8 RCL to only the P6-P5 segment produced a chimera with PC reactivity that resembled that of the P6-P1 chimera. This chimera thus increased reactivity with furin 2-fold and reduced reactivity with PCs 4–7 relative to α1PDX (Fig. 4A). Moreover, the kinetics of PC inhibition was monophasic for furin and biphasic for PCs 4–7. However, unlike the immediate equilibrium formation of the Michaelis complex observed for the P6-P1 chimera reaction, the P6-P5 chimera showed a slower approach to the Michaelis complex binding equilibrium (Fig. 4A). Notably, Michaelis complex affinities for the P6-P5 chimera interaction with PC 4–7 were 10–30-fold greater than those for the P6-P1 chimera (Kd values of 0.4–1 nM) (Fig. 4B, Table 4). Moreover, the conversion of the Michaelis complex to the covalent complex in the subsequent slow exponential phase indicated that kcat values were 3–12-fold faster for the P6-P5 chimera than the P6-P1 chimera reactions (1–3 × 10−4 s−1) (Fig. 4C, Table 5). As a result, ka values calculated from kcat/Kd ratios were larger for the P6-P5 chimera than the P6-P1 chimera reactions (105 to 106 M−1s−1) and approached those for the reactions of α1PDX with PCs 4–7 (Table 3). Like the P6-P5’ and P6-P1 chimeras, the serpin B8 P6-P5 substitution in α1PDX thus resulted in the formation of stable high-affinity Michaelis complexes with PCs 4–7 that were poorly reactive in forming a conformationally trapped covalent complex.
Figure 4.
Reaction of the α1PDX-serpin B8 P6-P5 chimera with PACE4. (A) Shown are plots of PACE4 residual activity at increasing concentrations of the α1PDX-serpin B8 P6-P5 chimera. Progressive reactions of 3 nM PACE4 were run as described in legend to figure 2. (B) Shown is the fitted plot of the dependence of residual PACE4 activity on increasing concentrations of the serpin chimera as described in legend to figure 2. Similar results were obtained with PC4, PC5 and PC7, and Kd values are listed in Table 4. (C) Shown is a plot of residual PACE4 activity for the time course of inhibition of 2 nM PACE4 by 50 nM of the chimera. The large initial drop in activity reflects the fast equilibrium binding for the formation of the Michaelis complex that precedes slow covalent complex formation.
The grafting of the serpin B8 RCL P3-P2 segment led to losses of α1PDX reactivity of about 10-fold with furin (105 M−1s−1) and about 1000-fold with PCs 4–7 (~103 M−1s−1) (Table 3). Contrasting the reaction of the P6-P5 chimera, the reaction of the P3-P2 chimera with PCs 4–7 showed an initial equilibrium binding of low affinity that was not measurable over the inhibitor concentrations used. The serpin B8 P3-P2 segment thus decreased reactivity of α1PDX with all PCs, but especially with PCs 4–7.
Together, these results suggest that the RCL P6-P1 segment contributes to the enhanced substrate selectivity between furin and PCs 4–7 observed with the P6-P5’chimera.
The P2’ Residue Determines PC Reaction Specificity.
Contrasting the effect of the P6-P1 segment, the serpin B8 RCL P1’-P5’ segment decreased α1PDX reactivity with furin, 20-fold, and PCs 4–7, 40–80 fold (104 M−1s−1) (Table 3). No kinetic separation of the formation of the Michaelis and covalent complexes was observed for this chimera. Single residue substitutions within the P2’-P4’ region identified the P2’ substitution as the most detrimental to reactivity as it reduced α1PDX reactivity 17-fold with furin and 60–120 fold with PCs 4–7 (l04 M−1s−1), similar to the effect produced by substituting the full P1’-P5’ region (Table 3). The effect of the P3’ and P4’ serpin B8 residue substitutions on α1PDX reactivity with all PCs was comparatively modest (105 to l06 M−1s−1). The lack of importance of the P1’ residue on PC substrate specificity and selectivity was shown by a serine to aspartic acid mutation that had less than 10-fold detrimental effect on α1PDX reactivity with all PCs (Table 6).
Table 6.
Second Order Association Rate Constant Values for the Reaction of PCs with α1PDX RCL Non-seroin B8 Substitutions
| α1PDX-Serpin B8 RCL Chimera | Second order association rate, ka(M−1s−1) | ||||
|---|---|---|---|---|---|
| FURIN | PC4 | PC5 | PACE4 | PC7 | |
| P4A (Pittsburgh) | 3.2 ± 0.1 × 101 | 6.7 ± 0.1 × 101 | ND | 6.7 ± 0 2 × 101 | ND |
| P3E | 1.5 ± 0.2 × 104 | 4.6 ± 0.3 × 102 | 4.6 ± 0.7 × 102 | 5.6 ± 0.3 × 102 | 2.2 ± 0.5 × 102 |
| P2G | 1.5 ± 0.3 × 106 | > 4 × 103 | 1.2 ± 0.3 × 105 | 5.3 ± 0.4 × 104 | > 7 × 103 |
| Pl’D | 2.3 ± 0.6 × 105 | 4.5 ± 1.0 × 105 | 3.4 ± 0.5 × 105 | 9.5 ± 3 1 × 105 | 2.S, ±0.4 × 105 |
Amino acid sequences in the RCL of α1PDX was mutated at single amino acid positions to non-serpin B8 residues. Second order association rate constant (ka) values for the reaction of PCs with α1PDX mutants were determined as described in Table 3. Errors represent S.E from linear regression fits of data.
> Unstable covalent complexes prevented the SI to be determined so the rate constant values are larger than here reported.
ND Not determined
Exosites Selectively Regulate Reactivity with PCs.
We have previously shown that the homologous grafting of the serpin B8 strand 3C exosite residues onto α1PDX or the α1PDX-serpin B8 P6-P5’ chimera leads to losses of reactivity with furin of 5-fold (l05 M−1s−1) and 70-fold (l03 M−1s−1), respectively (10). In stark contrast, the serpin B8 exosite residues increased α1PDX reactivity with PCs 4–7 by 3 to 9-fold (l06 to l07 M−1s−1), but reduced reactivity of the α1PDX-serpin B8 P6-P5’ chimera with PCs 4–7 by 8 to 30-fold (101 M−1s−1) (Table 7).
Table 7.
Effect of the YE Exosite Residues on PC Reactivity with α1PDX and the α1PDX-serpin B8 RCL P6-P5’ Chimera
| α1PDX-Serpin B8 Chimera | Second order association rate, ka(M−1s−1) | ||||
|---|---|---|---|---|---|
| FURIN | PC4 | PC5 | PACE4 | PC7 | |
| P6-P5’ | 1.1 ± 0.1 × 105 | 7.1 ± 1.0 × 102 | 6.0 ± 0.2 × 102 | 2.9 ± 0.1 × 102 | 7.7 ± 2.2 × 102 |
| P6-PS’+YE exosites | 1.5 ± 0.2 × 103 | 9.1 ± 0.1 × 101 | 6.9 ± 0.4 × 101 | 8.1 ± 0.2 × 101 | 2.2 ± 0.1 × 101 |
| α1PDX | 1.1 ± 0.1 × 106 | 1.6 ± 0.3 × 106 | 2.0 ± 0.1 × 106 | 1.2 ± 0.3 × 106 | 2.3 ± 0.8 × 106 |
| α1PDX+YE exosites | 2.1 ± 0.3 × 105 | 1.4 ± 0.4 × 107 | 8.6 ± 2.9 × 106 | 4.4 ± 1.0 × 106 | 6.7 ± 2.0 × 106 |
The tyrosine (Y) and glutamic acid (E) exosite residues were grafted at their homologous location in strand 3 of sheet C of α1PDX and the P6-P5’ chimera. Serpin B8 has the conserved residue phenylalanine at the position of the tyrosine residue. Second order association rate constant (ka) values for the reaction of PCs with the chimeras α1PDX+YE exosites and the α1PDX-serpin B8 RCL P6-P5’+YE exosites were determined as described in Table 3. Errors represent S.E from linear regression fits of data. The ka values for the reaction of furin with both chimeras were taken from reference (10). The ka values for the reaction of α1PDX and the P6-P5’ chimera with all PCs were taken from tables 2 and 3 and included for comparison.
The Catalytic Site Conformations of Furin and PCs 4–7 Differ.
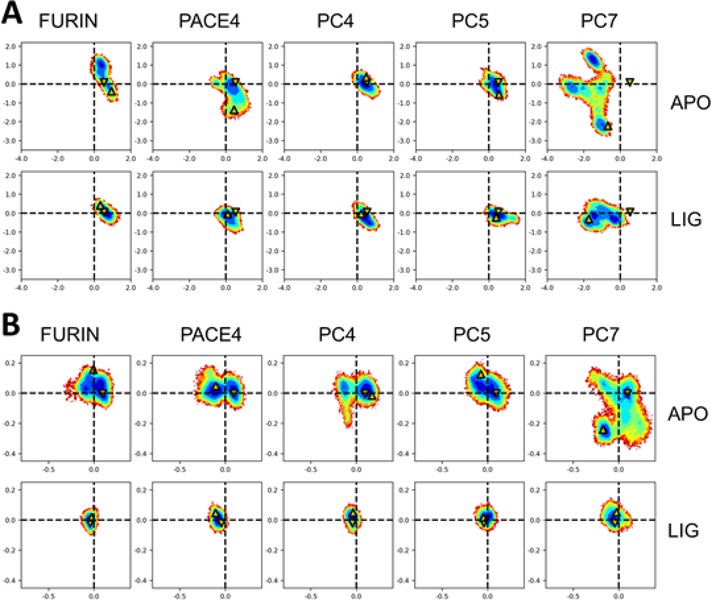
Protein crystallography and MDS studies have shown that the catalytic site of furin adopts active and inactive conformations and have suggested that substrates preferentially bind to the active conformation through conformational selection or induced-fit binding mechanisms (7, 8). The kinetic studies described above show that unlike furin, the catalytic site of PCs 4–7 may adopt inactive conformations that preferentially bind substrate non-productively. To investigate the basis for such differences, we compared the catalytic site conformations of furin and PCs 4–7 by MDS. Homology models of the whole catalytic domain for PCs 4–7 were built based on the x-ray crystal structures of the ligand-free and ligand-bound forms of furin. Although the amino acid sequence is 50–65% conserved between furin and PCs 4–7, the structures are expected to be highly conserved (8). MDS were performed on ten simulated systems of five proteins and two different conditions: 1) ligand-free form associated with active and inactive configurations, and 2) ligand-bound form associated only with active configurations. A principal component analysis (PCA), constructed on a common structural core of 313 Cα atoms in the catalytic domain, was performed employing MDS trajectories for a global evaluation of the structures (Fig. 5A). The analysis showed well resolved and distinct active and inactive furin conformations in agreement with previous studies (7). PCs 4–7 showed the most pronounced differences in PACE4 and, especially, in PC7 for the ligand-free forms. The ligand-bound forms were more consistent with furin, except for PC7. A second PCA was performed on 5 Cα atoms located in the catalytic site. Again, furin produced two clearly distinguishable sets of conformations corresponding to active and inactive states (Fig. 5B). In agreement, all PCs 4–7 also produced clearly differentiated active and inactive conformations. Only the ligand-free conformations of PCs 4–7 showed more variability compared to furin, especially that of PC7.
Figure 5.
Molecular dynamic simulation analysis of the structure of PCs. The configuration of amino acid residues in the catalytic domain of furin, PC4, PC5, PACE4 and PC7 was simulated employing furin crystal structures and molecular models of the catalytic domain of PCs 4–7 based on the x-ray crystal structure of the ligand-free and ligand-bound furin, as described in the Materials and Methods section. Two different starting structure conditions were employed, i) ligand-free form, which is associated with inactive configurations, and ii) ligand-bound form, which is associated with active configurations. Shown are molecular dynamic trajectories from the simulated systems projected on the first two principal components (nm). These projections are presented as heat maps where blue represents highly populated states. The maps derived from (i) are labeled as APO, and the ones from (ii) as LIG. The arrows pointing down correspond to the starting frame, which is the furin crystal structure. The arrows pointing upward correspond to the last frame of the analysis. (A) Comparative MDS analysis of the PC catalytic domain. PCA performed over 313 Cα atoms of amino acid residues. (B) Comparative MDS analysis of the PC catalytic site. PCA performed over 5 Cα atoms of catalytic site amino acid residues (Aspl53, Hisl94, Gly255, Asn295 and Ser368).
Discussion
Despite the large body of information regarding how PCs regulate the growth, proliferation and differentiation of cells in numerous physiological and pathological processes, a molecular understanding of how PCs perform their specific regulatory functions has remained elusive (22). Because PCs are co-expressed and overlap at their cellular location, significant cross-reactivity seems unavoidable, and the significance of the redundant targeting of substrates by PCs is unclear. However, examples of PCs with opposite regulatory effects imply that PCs should be able to target their substrates selectively (23). A recurrent theme is that of furin and PCs 4–7 playing opposite roles by targeting the same protein but in a selective manner involving different recognition sites (24, 25). The basis for this PC selectivity has not been well characterized. In this study, serpin B8 was demonstrated to react with furin selectively based on a 20-fold faster reactivity with furin than with PC4, PC5, PACE4 and PC7 (Table 2). In addition, it was shown that furin catalyzes the hydrolysis of two tetrapeptide substrates at a faster catalytic rate, especially with the substrate Boc-Arg-Val-Arg-Arg-AMC (20-fold), than PCs 4–7 (Table 1). These observations suggested that furin and PCs 4–7 differ in substrate specificity and selectivity. Following the same rationale that we used to study the reactive-site and exosite elements of serpin B8 that determine reactivity with furin (10), we used α1PDX as a model substrate to determine the basis for the differences in serpin B8 reactivity between furin and PCs 4–7 by the homologous transferring of RCL and exosite amino acid residues from serpin B8 to α1PDX.
Notably, the serpin B8 P6-P5’ RCL segment induced a difference in reactivity between furin and PCs 4–7 significantly larger within α1PDX (140 to 400-fold) than within serpin B8 (20-fold) (Tables 2 & 3). This disparity argues that the conformation that the serpin B8 RCL P6-P5’ segment adopts in the α1PDX context accentuates the difference of reactivity between furin and PCs 4–7. Unexpectedly, the α1PDX-serpin B8 P6-P5’ chimera reacted with furin with monophasic kinetics, but reacted with PCs 4–7 with biphasic kinetics. The serpin thus bound rapidly to PCs 4–7 to form a Michaelis complex in the first kinetic phase and this complex then slowly underwent acylation to form the trapped covalent serpin-enzyme complex in the second phase. The marked reduction in the rate of the second catalytic step therefore accounted for the biphasic kinetics and large reduction in overall reactivity of the chimera with PCs 4–7. The chimera thus by binding to the more variable catalytic site of PCs 4–7 than furin, as shown by the MDS experiments (Fig. 5), may be favoring non-productive conformations of the catalytic site.
The grafting of smaller segments of the RCL showed the P6-P1 segment, and in particular the P6-P5 residues, were responsible for the marked difference in reactivity between furin and PCs 4–7. By contrast, the serpin B8 P3-P2 residues did not produce biphasic reaction kinetics with PCs 4–7, but did result in marked reductions in reactivity with both furin (10-fold) and PCs 4–7 (1000-fold). The effects of other mutations of the P3 and P2 residues in α1PDX are also consistent with such mutations affecting reactivity more with PCs 4–7 than with furin (Table 6).
The major effects of the P6-P5 and P3-P2 residues, which flank the P4 Arg residue, on the reactivity of the P1-P6 chimera with PCs 4–7 suggests that the conformation of the P6-P1 segment plays a key role in regulating the productive insertion of the P4 arginine side chain into the S4 pocket of the enzyme catalytic site. Consistent with this idea, the level of PC reactivity of the P6-P1 chimera (l01 to l02 M−1s−1) is comparable to that of α1-antitrypsin Pittsburgh (l01 M−1s−1) (Table 6), a natural mutant of α1-antitrypsin (20), which has an arginine for methionine substitution at the P1 position, but lacks the P4 arginine (Fig. 1).
The P1’-P5’ chimera led to significant losses of reactivity with all PCs (Table 3). The finding that the P2’ chimera largely accounted for the losses in reactivity indicated that the P2’ residue was principally responsible for the detrimental effect of the serpin B8 primed region on α1PDX reactivity. Like α1PDX, the P1’-P5’ and P2’ α1PDX chimeras showed little selectivity in their reaction with furin and PCs 4–7. This region thus contributes to general PC specificity but not to selectivity.
The contribution of the primed region of the RCL to PC reactivity was also shown to be influenced by exosite interactions. In our previous study on furin reactivity, we showed that of the four furin active-site loops, only the 298–300 loop is involved in exosite regulation of furin reactivity (10). We also demonstrated that the 298–300 furin loop, of which Arg298 forms part of the S2’ pocket in the catalytic site (8), mediates the interactions of the exosite residues in strand 3C of the serpin with the primed region of the RCL (Fig. S2). In this study, we further advanced this notion by establishing that the exosite residues selectively affected PC reactivity depending on the RCL primed region amino acid sequence. While the serpin B8 exosite residues were detrimental to reactivity with all the PCs in the presence of the serpin B8 primed region, they negatively affected furin reactivity but promoted reactivity with PCs 4–7 in the context of the α1PDX primed region (Table 7). These observations strongly suggest that the exosite residues modulate the P2’ residue insertion into the S2’ pocket and can regulate substrate selectivity between furin and PCs 4–7. Thus, the P1’-P5’ segment serves as a determinant of general PC substrate specificity that can be modulated by exosites (10, 25).
Our demonstration of differences in substrate specificity and selectivity between furin and PCs 4–7 significantly advances understanding of the molecular determinants of specificity and selectivity in the PC family of proteases. Our findings thus clearly establish the ability of PCs to differentiate target substrates based on the recognition of different preferred substrate cleavage sequences. Moreover, our findings underscore the importance of the structural context of such recognition sequences and the ability of exosites to influence this context. Finally, they establish that substrate recognition can be favored or disfavored by differential abilities of furin and PCs 4–7 to bind their substrates productively versus non-productively, due to differences in catalytic site conformational states of PCs. Interestingly, such differences may be exploited to engineer selective inhibitors of PCs 4–7 as shown by the nanomolar Michaelis complex affinities observed for P1-P6 and P6-P5 chimera interactions with these PCs (Table 4).
Serpins are the only known natural PC inhibitors (10–14). Protein-based inhibitors have the advantage over small molecule inhibitors in that they can be expressed by the targeted cell and directed to specific subcellular locations (18, 27). We have demonstrated that serpin B8 is a specific and selective furin inhibitor, and shown that introducing serpin B8 reactivity determinants into the engineered general PC inhibitor, α1PDX, results in added selectivity for furin or for PCs 4–7 (Fig. 1). The P6-P5’ chimera (now named α1ORD) achieves a 140 to 400-fold higher reactivity with furin than with PCs 4–7 (Table 3). Although this chimera forms inhibitory Michaelis complexes with PCs 4–7, the binding affinity is weak (Kd 1 μΜ) and minimal inhibition is expected at concentrations below 1 μΜ. The α1PDX+YE derivative (now named α1MDW) is selective for PCs 4–7 with 20 to 60-fold higher reactivity with these PCs than with furin (Table 7). These inhibitors, and their future improved derivatives, should become useful tools in the search to understand the regulatory role of PCs, and to discover new therapeutic drugs.
Supplementary Material
Acknowledgements
We thank Dirección General de Cómputo y de Tecnologías de Información y Comunicación at Universidad Nacional Autónoma de México for granting the use of the supercomputer Miztli (LANCAD-UNAM-dGTIC-320), and Dr. Steve Olson, from the University of Illinois at Chicago, for helpful discussions and critical comments on the manuscript.
This work was supported by the National Institutes of Health (R37 HL 39888), and from Dirección General de Asuntos del Personal Académico at Universidad Nacional Autónoma de México (PAPIIT-IA202917).
Footnotes
Supporting information: Descriptions of the serpin mechanism of protease inhibition and of the furin-serpin B8 Michaelis complex.
References
- 1.Seidah NG, and Prat A (2012) The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov 11: 367–383 [DOI] [PubMed] [Google Scholar]
- 2.Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R, and Prat A (2008) The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol 40: 1111–1125 [DOI] [PubMed] [Google Scholar]
- 3.Shiryaev SA, Remacle AG, Ratnikov BI, Nelson NA, Savinov AY, Wei G, Bottini M, Rega MF, Parent A, Desjardins R, Fugere M, Day R, Sabet M, Pellecchia M, Liddington RC, Smith JW, Mustelin T, Guiney DG, Lebl M, and Strongin AY (2007) Targeting host cell furin proprotein convertases as a therapeutic strategy against bacterial toxins and viral pathogens. J. Biol. Chem 282: 20847–20853 [DOI] [PubMed] [Google Scholar]
- 4.Feliciangeli SF, Thomas L, Scott GK, Subbian E, Hung CH, Molloy SS, Jean F, Shinde U, and Thomas G (2006) Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J. Biol. Chem 281: 16108–16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson DM, Elferich J, Ramakrishnan P, Thomas G, and Shinde U (2013) The mechanism by which a propeptide-encoded pH sensor regulates spatiotemporal activation of furin. J. Biol. Chem 288: 19154–19165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omotuyi IO (2015) Ebola virus envelope grycoprotein derived peptide in human furin-bound state: computational studies. J. Biomol. Struct. & Dyn 33: 461–470 [DOI] [PubMed] [Google Scholar]
- 7.Dahms SO, Arciniega M, Steinmetzer T, Huber R, and Than ME (2016) Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc. Natl. Acad. Sci. USA 113: 11196–11201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrich S, Lindberg I, Bode W, and Than ME (2005) Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J. Mol. Biol 345: 211–227 [DOI] [PubMed] [Google Scholar]
- 9.Remacle AG, Shiryaev SA, Oh ES, Cieplak P, Srinivasan A, Wei G, Liddington RC, Ratnikov BI, Parent A, Desjardins R, Day R, Smith JW, Lebl M, and Strongin AY (2008) Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J. Biol. Chem 283: 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izaguirre G, Qi L, Lima M, and Olson ST (2013) Identification of serpin determinants of specificity and selectivity for furin inhibition through studies of α1PDX (α1-protease inhibitor Portland)-serpin B8 and furin active-site loop chimeras. J. Biol. Chem 288: 21802–21814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leblond J, Laprise MH, Gaudreau S, Grondin F, Kisiel W, and Dubois CM (2006) The serpin proteinase inhibitor 8: an endogenous furin inhibitor released from human platelets. Thromb. Haemost 95: 243–252 [DOI] [PubMed] [Google Scholar]
- 12.Osterwalder T, Kuhnen A, Leiserson WM, Kim YS, and Keshishian H (2004) Drosophila serpin 4 functions as a neuroserpin-like inhibitor of subtilisin-like proprotein convertases. J. Neurosci 24: 5482–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richer MJ, Keays CA, Waterhouse J, Minhas J, Hashimoto C, and Jean F (2004) The Spn4 gene of Drosophila encodes a potent furin-directed secretory pathway serpin. Proc. Natl. Acad. Sci. USA 101: 10560–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentele C, Krüger O, Tödtmann U, Oley M, and Ragg H (2006) A proprotein convertase-inhibiting serpin with an endoplasmic reticulum targeting signal from Branchiostoma lanceolatum, a close relative of vertebrates. Biochem. J 395: 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson ED, Thomas L, Hayflick JS, and Thomas G (1993) Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed α1-antitrypsin variant. J. Biol. Chem 268: 24887–24891 [PubMed] [Google Scholar]
- 16.Jean F, Stella K, Thomas L, Liu G, Xiang Y, Reason AJ, and Thomas G (1998) α1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: Application as an antipathogenic agent. Proc. Natl. Acad. Sci. USA 95: 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, and Whisstock JC (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem 276: 33293–33296 [DOI] [PubMed] [Google Scholar]
- 18.Benjannet S, Savaria D, Laslop A, Munzer JS, Chrétien M, Marcinkiewicz M, and Seidah NG (1997) Alpha1-antitrypsin Portland inhibits processing of precursors mediated by proprotein convertases primarily within the constitutive secretory pathway. J. Biol. Chem 272: 26210–26218 [DOI] [PubMed] [Google Scholar]
- 19.Gettins PG (2002) Serpin structure, mechanism, and function. Chem. Rev 102: 4751–4804 [DOI] [PubMed] [Google Scholar]
- 20.Vidaud D, Emmerich J, Alhenc-Gelas M, Yvart J, Fiessinger JN, and Aiach M (1992) Met 358 to Arg mutation of alpha 1-antitrypsin associated with protein C deficiency in a patient with mild bleeding tendency. J. Clin. Inves.t 89: 1537–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison JF (1969) Kinetics of the reversible inhibition of enzyme-catalyzed reactions by tight binding inhibitors. Biochim. Biophys. Acta 185: 269–286 [DOI] [PubMed] [Google Scholar]
- 22.Scamuffa N, Calvo F, Chrétien Μ, Seidah NG, and Khatib ΑΜ (2006) Prorpotein convertases: lessons from knockouts. FASEB J. 12: 1954–1963 [DOI] [PubMed] [Google Scholar]
- 23.Seidah NG, Sadr MS, Chrétien Μ, and Mbikay M (2013) The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J. Biol. Chem 288: 21473–21481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapierre M, Siegfried G, Scamuffa N, Bontemps Y, Calvo F, Seidah NG, and Khatib AM (2007) Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells’ malignant phenotypes: role of tissue inhibitors of metalloproteinase-1. Cancer Res. 19: 9030–9004 [DOI] [PubMed] [Google Scholar]
- 25.Maret D, Sadr MS, Sadr ES, Colman DR, Del Maestro RF, and Seidah NG (2012) Opposite roles of furin and PC5A in N-cadherin processing. Neoplasia 14: 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komiyama T, and Fuller RS (2000) Engineered eglin c variants inhibit yeast and human proprotein processing proteases, Kex2 and furin. Biochemistry 39, 15156–15165 [DOI] [PubMed] [Google Scholar]
- 27.Richer MJ, Juliano L, Hashimoto C, and Jean F (2004) Serpin mechanism of hepatitis C virus nonstructural 3 (NS3) protease inhibition: induced fit as a mechanism for narrow specificity. J. Biol. Chem 279: 10222–10227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.