Abstract
Substantial research has been devoted to elucidate the roles that extracellular vesicles (EVs) play in the regulation of both normal and pathological processes, and multiple studies have demonstrated their potential as a source of cancer biomarkers. However, several factors have slowed the development of liquid biopsy EV biomarkers for cancer diagnosis, including logistical and technical difficulties associated with reproducibly obtaining highly purified EVs suitable for diagnostic analysis. Significant effort has focused on addressing these problems, and multiple groups have now reported EV analysis methods using liquid biopsies that have the potential for clinical translation. However, there are still important issues that must be addressed if these discoveries and technical advances are to be used for clinical translation of EV cancer biomarkers from liquid biopsies. To address these issues, this review focuses on the potential application of EV biomarkers for diagnosis of major cancer types, discussing approaches for EV biomarker discovery and verification, EV clinical assay development, analytical and clinical validation, clinical trials, regulatory submission, and end user utilization for the intended clinical application. This review also discusses key difficulities related to these steps, and recommendations for how to best accomplish steps in order to translate EV-based biomarkers into clinical settings.
Introduction
Cancer is the second leading cause of death worldwide, being responsible for one in six deaths. New biomarkers are badly needed to improve cancer diagnosis and evaluation and thus improve patient outcomes. Extracellular vesicles (EVs) exhibit potential as such biomarkers, since these vesicles contain DNA, RNA and protein cargoes that reflect the status of their parent cells at the time of their formation. The term “EV” covers a broad array of vesicles secreted by cells, including exosomes, microvesicles, and apoptotic bodies. Most of the studies we discuss in this review focus on exosome populations, but due to the potential for contamination of such populations with small microvesicles we will use the term EV to refer to a population that may contain both exosomes and microvesicles in keeping with current practice. The unique composition, long in vivo half-life and physical durability of EVs support that EVs are qualified materials to serve as stable and sensitive biomarkers for various evolving malignancies. Thus, sophisticated EV-based diagnostic may one day refine the algorithms for cancer diagnosis and therapeutic responses (Fig 1). EV research is a rapidly evolving field, and researchers have proposed various diagnostic algorithms, incorporating multiple tumor-associated EV biomarkers, with rapid and sensitive quantifications.
Fig. 1.
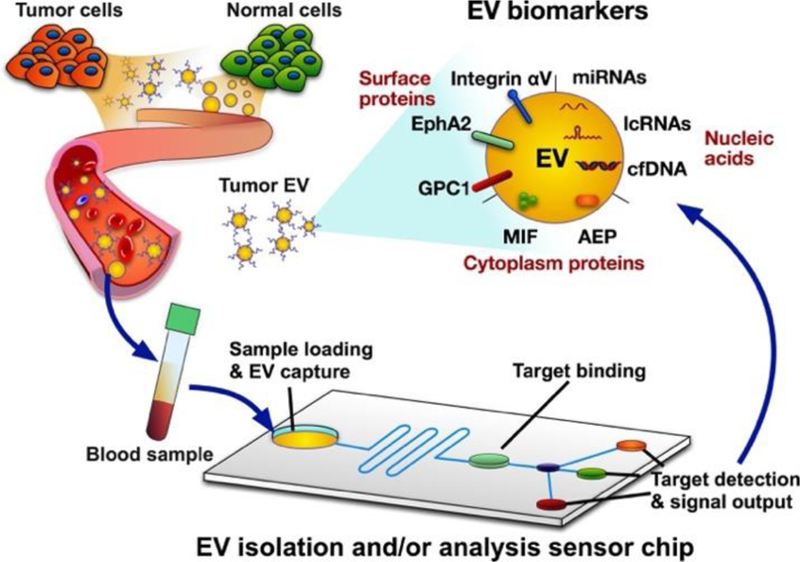
Schematic of extracellular vesicles (EVs) diagnostics from EV release to analysis. EVs secreted by diseased and healthy cells are secreted into most body fluids, including blood, and can be isolated by integrated EV capture and analysis platforms to detect and measure expression of disease-associated EVs for disease diagnosis and the analysis of treatment responses. Captured EVs are analyzed for disease-specific (e.g. pancreatic cancer-associated) EV biomarkers, including membrane proteins (glypican-1 (GPC1), Ephrin type-A receptor 2 (EphA2) and Integrin αV); cytosolic proteins (macrophage migration inhibitory factor (MIF) and asparaginyl endopeptidase (AEP)); microRNA (miRNA), long non-coding RNA (lncRNA), cell-free DNA (cfDNA). Microfluidic devices are often used for such devices, with most using one or more EV-specific antibodies (CD63, CD9 or CD81) for EV capture. Antibodies targeting disease-associated EVs can be used as probes to generate chromogenic, chemifluorescent, or electrochemical signals, among other readouts, or PCR-based approaches can be utilized to quantify disease-associated nucleic acid biomarkers.
Recent proteomic, metabolomic and genomic approaches applied to identify cancer-specific EV markers show promising results in exploratory studies using small patient populations. Emerging technologies for EV analysis, especially integrated platforms capable of EV isolation, enrichment, and analysis on one device, promise to advance the potential translation of EV liquid biopsy approaches for cancer diagnosis.
The development of integrated EV analysis platforms based on microfluidic, nano-plasmonic, electrokinetic, surface plasmon resonance, and electrochemistry technologies holds promise for the future production of high sensitivity and high-throughput assays for EV analysis. However, despite the growing number of research studies that have generated novel biomarkers, only a few new EV biomarkers/assays have progressed to actual applications in clinical settings. Technical, clinical, logistical, financial, and regulatory burdens have limited the rate of EV biomarker and platform translation. Understanding and overcoming these challenges is essential for continued progress in EV-based biomarker development.
Numerous reviews have addressed application of EVs as cancer biomarkers and recent progress in EV isolation and analysis methods 1–28 but few focus on the translation and clinical development of these EV biomarker assays. Extensive guidelines have been developed to provide a framework for discovery and validation of cancer markers 29–37, but there are substantial barriers to implementing advanced techniques in clinical settings, and significant effort needs to be spent to establish successful EV clinical assays with meaningful clinical benefits. Similar to other clinical assays, EV clinical assay development requires a sequence of essential phases carried out in a step-wise manner. These are: EV biomarker discovery and verification, EV clinical assay development, analytical and clinical validation, regulatory submission, and end user utilization for the intended clinical application (Fig. 2). This review will emphasize two fundamental processes critical for successful translation of an EV marker/assay into a clinical setting: following the standard process for clinical assay development and addressing the practical aspects of implementing EV assays in clinical settings. To address the first requirement, investigators should be familiar with procedures governing clinical assay development, and follow best practices for EV clinical assay development in compliance with current analytical, clinical, and manufacturing guidelines. The second focus requires consideration of practical aspects of implementing EV assays in clinical settings. Useful clinical EV assays should utilize an automated, user friendly, reliable, and inexpensive system for EV isolation, enrichment and analysis, ideally these processes would all be accomplished on an integrated platform.
Fig. 2.
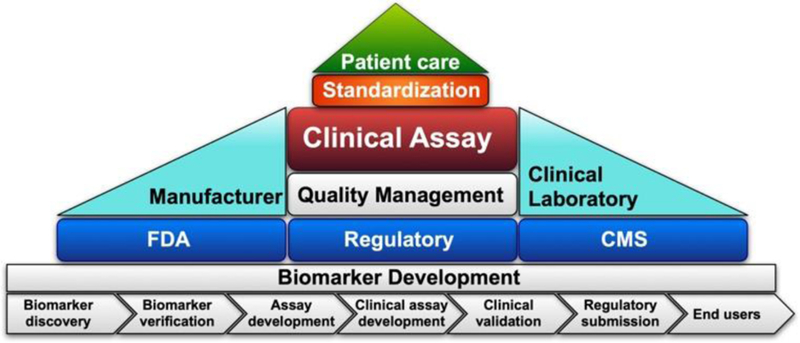
Essential aspects of clinical assay development and utilization
This review will also discuss the challenges and recommendations for translating potential EV biomarkers for clinical disease diagnostics. We will particularly emphasize critical steps required to translate emerging EV markers and EV analysis technologies into clinical practice. Due to this focus and the scope of activity in EV research, we direct the reader to recent review articles for summaries of other EV topics, such as EV biogenesis, secretion, and function; conventional methods for EV isolation and detection; and recent research on new EV applications as therapeutics 1–24.
EV biomarker discovery
EV biomarker discovery begins with the identification of EV targets that are specific to and associate with, a disease of interest. This discovery effort also aims to obtain a relative estimate of their clinical value, and prioritize these candidates for future evaluation. Traditionally, most cancer biomarker candidates have been identified through knowledge of the cancer’s pathophysiology, biochemistry and key processes. More recently, the use of untargeted analysis approaches (e.g., genomics, proteomics, and metabolomics) now enables the construction of comprehensive biomarker discovery pipelines. Many of the EV-based biomarker candidates for specific cancers discussed in this article were discovered through exploratory studies that employed mass spectrometry or second generation sequencing approaches for proteomic and genomic analyses, respectively. However, despite the power of these analytical approaches, one should exercise caution in the immediate clinical translation of these biomarkers. Due to the large number of reported EV biomarker candidates that have been proposed for various diseases and conditions, we will focus our discussion on EV biomarker discovery and validation for clinical application on EV factors associated with pancreatic cancer (PC), which is one of the most active areas for EV biomarker discovery.
Sample size:
Technological advances have propelled EV biomarker discovery for various cancers. However, as exemplified using PC biomarker discover studies as example (Table 2) most of these studies employ very small sample sets (10–20 samples or less in each group (e.g., cancer vs non-cancer). Only 8 out of 38 studies (for different clinical utilities) reviewed in Table 2 employed cohorts with ≥20 samples in each group. Studies that employ such small sample sizes at the discovery phase increase the risk of selecting false negative (failure to detect true biomarkers) and false positive (candidates that fail to replicate) candidates.
Table 2. Clinical application of EV markers for PC.
Abbreviations: AC: ampullary carcinoma; AEP: asparaginyl endopeptidase; AUC: area under curve; bPaTu: benign pancreatic tumor; BPD: benign pancreatic disease; BPT: benign pancreatic tumors; ELISA: enzyme-linked immunosorbent assay; ddPCR: droplet digital polymerase chain reaction; DFS: disease-free survival rates; ExoTENPO: Exosome Track-Etched Magnetic Nanopore; IPMN: intraductal papillary mucinous neoplasm; LOD: limit of detection; LSPR: localized surface plasmon resonance; MAF: mutant allele frequency; MIF: macrophage migration inhibitory factor; NED: no evidence of disease post-diagnosis; NC: normal control; nPES: nanoplasmon-enhanced scattering; OS: overall survival; PC: Pancreatic Cancer; PCPL: pancreatic cancer precursor lesions; PDAC: pancreatic ductal adenocarcinoma; Pt: chronic pancreatitis; POD: progression of disease post-diagnosis; PaCIC: pancreatic cancer initiating cells; SCA: serous cystadenoma; UC: ultracentrifugation.
| EV markers for pancreatic cancer | EV Isolation | EV marker analysis | Analytical performance | Clinical Utility | Sample size | Clinical performance /Statistical analysis | Blinded? Yes/No | Sample type/single site collection? | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Glypican-1 | UC | Flow cytometry | Not specified | Early detection | Discovery Cohort: NC: N=100, BPD: N=26; PCPL: N=5; PDAC: N=190, Validation Cohort:, NC: N=20; BPD: N=6; PDAC: N=56 | PC vs other groups AUC: 1 Sensitivity: 100% Specificity: 100% |
Yes | Serum/ discovery cohort yes; validation cohort yes | 38 |
| Metastatic disease burden | No metastases: N=18, Lymph node metastases: N=134, Distant metastases: N=32 | Distant metastases vs No metastases or Lymph node metastases statistically significant | Yes | Serum/not specified | |||||
| Monitor surgery responses | BPD: N=4, PCPL: N=4, PDAC: N=29 | Pre vs post resection (PDAC and PCPL statistically significant) | |||||||
| Prognosis | PDAC: N=29 | Improved OS and DFS if drop of exosomal GPC1+ >= median | |||||||
| Glypican-1 | UC | Tandem mass spectrometry | Accuracy Precision Sensitivity Matrix effect Spike recovery Sample Stability |
Diagnosis | NC: N= 6, Pt: N = 3, PDAC: N = 3 | Not statistically significant | Not specified | EDTA Plasma/ not specified | 39 |
| Monitor surgery responses | Matched pre- and post- surgical resection: N=3 | Not statistically significant | |||||||
| Glypican-1 | UC | ELISA | Not specified | Diagnosis | BPD: N=16, IPMN: N=7 Pt: N=6, SCA: N=3 PDAC: N=27 |
No significant difference AUC= 0.59, Sensitivity=74%specificity=44% for detecting PDAC. | Not specified | EDTA Plasma /Yes | 40 |
| Monitor surgery responses | Matched pre- and post- surgical resection: N=11 | Statistically significant | |||||||
| Glypican-1 and CD63 | None | AC electrokinetic microarray chip | Not specified | Diagnosis | NC: N=11, BPD: N=7, PDAC: N=20 | PDAC vs other groups, AUC: 0.79, Sensitivity: 81%, Specificity: 70% | Yes | Serum/Yes | 41 |
| Glypican-1 | Spin column-based method | Flow cytometry | Not specified | Diagnosis | NC: N=16, PC: N=28 | Statistically significant | Not specified | Plasma ((anticoagulant not specify)/Yes | 42 |
| Prognosis | PC: N=28 | Improved OS with great decrease of Glypican-1+ EV | |||||||
| EphA2-EV | Isolation free | nPES | Cutoff, Precision LOD, Linearity | Early detection | Discovery Cohort: NC: n = 10, Pt: n = 10, PC: n = 10 Validation Cohort: NC: n = 48, Pt: n = 48, PC: n = 49 (8 stage I, 29 stage II, and 12 stage III) | AUC: 0.93–0.96 Sensitivity: 86%−94% Specificity: 85% |
No | Plasma (anticoagulant not specify)/yes | 43 |
| Not specified | Monitor treatment responses | good/partial responses: N= 13 poor responses: N= 10 |
good/partial vs poor responses: statistically significant | No | Plasma (anticoagulant not specify) /yes | ||||
| EphA2 | UC | ELISA | LOD Linearity | Diagnosis | NC: n = 10, Pt: n = 10, PC: n = 10 | PC vs other groups Statistically significant |
No | Plasma (anticoagulant not specify) /yes | |
| Integrin αv | UC | ELISA | Not specified | Prognosis | NC: N=13, NED: N=14, POD to liver: N=13 | NED vs POD to liver, Statistically significant | Not specified | Plasma (anticoagulant not specify)or serum/No | 44 |
| MIF | UC | ELISA | Not specified | Prognosis | NC: N=15, NED= 10, POD N=12, POD to liver: N=18 | NED vs POD Statistically significant |
No | EDTA plasma/No | 45 |
| AEP | ExoQuick | Western blot | Not specified | Exploratory | PDAC: N=3,Pt: N=3 | Statistically significant | N/A | Serum/Yes | 46 |
| CD44v6, Tspan8, EpCAM, MET, CD104, miR‐1246, miR‐4644, miR‐3976 and miR‐4306 | UC | Flow cytometry, RT-qPCR | Cutoff | Diagnosis | Discovery cohort: NC: N=6, Non-PC cancer:
N=3, Pt: N=7, PC: N=37 bPaTu: N=5, Validation cohort: NC: N=12,Non-PC cancer: N=8, Pt: N=16, PC: N=75, bPaTu: N=17 |
Combining all markers PC vs non-PC sensitivity: 100% specificity: 80% |
Yes | Serum/No | 47 |
| miR-17–5p, miR-21 | UC | RT-qPCR | Not specified | Diagnosis | NC: N=8, Pt: N=6, AC: N=6, PC: N=22, BPT: N=7 | PC vs non-PC, AUC: 0.887 and 0.897, Sensitivity: 92.6% and 81.5%, Specificity: 72.7% and 95.5% | Not specified | Serum/Yes | 48 |
| miR-10b | UC | LSPR-based sensor | LOD, Method
comparison Linearity |
Exploratory | NC: N=3, Pt: N=3, PDAC: N=3 | Statistically significant | Yes | Trisodium citrate Plasma/Not specified | 49 |
| miR-1246 and miR-4644 | Total Exosome Isolation Reagent | RT-qPCR | Threshold cycle values Cutoff | Exploratory | PC: N=12 NC: N=13 |
AUC: 0.763–0.833 Sensitivity: 0.667–0.833 Specificity: 0.769–1 |
Not specified | Saliva/Yes | 50 |
| miR-196a and miR-1246 | Exoquick | RT-qPCR | Not specified | Early detection | NC: N=15; PC stage I or IIA: N=15 | AUC: 0.73–0.81 | Not specified | Plasma (anticoagulant not specify)/No | 51 |
| miR-23b-3p | UC | RT-qPCR | Not specified | Diagnosis | NC: N=20, Pt: N=18, PC: N=16 | PC vs NC and Pt, Statistically significant | Not specified | Serum/Yes | 52 |
| miR-10b, −21, −30c, −106b, −20a, −181a, −483, -let7a, and −122 | UC | RT-qPCR | Cutoff | Diagnosis | NC: N= 6, Pt: N = 11 PDAC: N = 29 |
PDAC vs Normal, AUC: 0.57–1, Sensitivity: 62%−100%, Specificity: 100% | Not specified | EDTA Plasma/Not specified |
39 |
| miR-301a-3p | Total exosomes isolation reagent | RT-qPCR | Not specified | Diagnosis | PC: N=50, NC: N=12 | Statistically significant | Not specified | Serum/No | 53 |
| Prognosis | PC: N=50, miR-301a-3p low: N=20; high: N=30 | higher miR-301a expression was associated with poor OS | |||||||
| miR-122–5p, miR-193b-3p | ExoQuick | RT-qPCR | Not specified | Diagnosis | PC: N=31 NC: N=37 |
AUC=0.849 | Not specified | EDTA plasma/yes | 54 |
| miR-451a | UC | RT-qPCR | Not specified | Early detection | PC: Stage I N=7, Stage II N=43, NC: N=20 | Statistically significant | Not specified | Plasma (anticoagulant not specify)/Yes | 55 |
| Prognosis | high-miR-451a expression group:
N=25; |
The high miR-451a group showed a
significantly worse OS (P = 0.001) and DFS Significantly shorter OS and DFS in the high-miR-451a group |
|||||||
| miR-191, miR-21 and miR-451a | ExoQuick | RT-qPCR | Not specified | Early detection | PC: N= 32, IPMN: N= 29, Control: N= 22 | AUC: 0.768–0.862 Sensitivity: 69.6%−86.4% Specificity: 79%*81% |
Not specified | Serum/Yes | 56 |
| miR-21 | cutoff | Prognosis | Not specified | Significantly shorter OS in the high-miR-21 group | |||||
| miR-21 and miR-451a | Not specified | Monitor treatment responses | Not specified | Statistically significant | |||||
| Sox2ot | ExoQuick | Microarray and qRT-PCR | Not specified | Prognosis | High Sox2ot expression group,
N=25; Low Sox2ot expression group, N=31 |
patients with high exosomal Sox2ot expression had lower overall survival rates | Not specified | Plasma (anticoagulant not specify)/Yes | 57 |
| Monitor surgery responses | Matched pre- and post- surgical resection: N=16 | Statistically significant | |||||||
| Mutant KRAS and TP53 DNA | UC | ddPCR | Threshold (intensity) limit for mutant alleles | Exploratory | PDAC: N=48 IPMN: N = 7, Pt: N = 9, Others:
N = 12, NC: N= 114 |
PDAC
KRASG12D:
39.6% TP53R273H : 4.2% |
Not specified | Serum/No | 58 |
| KRAS MAF | UC | ddPCR | Cutoff | Prognosis | Localized PDAC: N=13 | Longer DFS if exoKRAS MAF was <1% | Yes | Sodium Heparin plasma/not specify | 59 |
| KRAS MAF | UC vs UC+ immunocapture pulldown | ddPCR | Not specified | Exploratory | PDAC No-pulldown: N=136 Pulldown: N=37 |
Significantly higher KRAS MAF in the pulldown-cohort | Not specified | Acid Citrate Dextrose Plasma/Not specified | 60 |
| mRNA (ARG1, CK18, CD63, Erbb3, KRAS, GAPDH, H3F3A, ODC1) | EpCAM-based isolation on the ExoTENPO | RT-qPCR | Not specified | Exploratory | Training cohort, NC: N = 5, PC: N = 5, Validation cohort, NC: N = 12, PC: N = 12 | Sensitivity: 100% Specificity: 100% |
Yes for validation | EDTA plasma, Streck Cell-Free DNA BCT, or serum/Yes | 61 |
Sample quality:
EV biomarker discovery studies often begin with convenient samples that are obtained retrospectively from other studies or generic sample archives. Samples with incomplete information (e.g., draw time relative to diagnosis and treatment) and handling history (e.g., number of freeze–thaw cycles, or length of storage) can introduce additional challenges to achieving consistent analytical reproducibility. One common scenario that can produce this effect occurs when clinical samples are analyzed from different study sites that may have used non-uniform procedures to obtain, process, store and handle samples. For example, in the 38 studies reviewed in Table 2, there were 22 that utilized samples from a single site, 7 that employed samples from multiple sites, and 9 studies that did not specify if the samples utilized were obtained from single or multiple sites. EV biomarkers detected using such samples may thus be subject to unknown biases and experimental noise. EV-associated biomarkers derived from samples with missing demographic and clinical data (e.g., patient age and treatment) can also lead to diagnostic inaccuracy, and this problem can be further compounded in studies where diagnostic criteria and definitions are not well defined and consistent.
In robust studies, samples must be linked to appropriate data to confirm a subject’s clinical status, the subject’s demographic information (e.g., age, gender, and ethnicity), the study’s sampling methodology, and sample handling history. The “garbage-in, garbage-out” adage of computer science also directly applies to the analysis of biological specimens for EV biomarker discovery.
Sample type:
EVs have been identified in various types of body fluids for use as a liquid biopsy, including saliva, urine, plasma, serum and whole blood. Among the 38 studies for different clinical utilities reviewed here, the majority of studies used blood samples: 21 used plasma, whereas 11 used plasma without specifying the anticoagulant, 15 used serum, 2 used both plasma and serum, and one used saliva.
Sample consistency is a key factor during the discovery phase. For example, serum specimens experience EV contamination from activated and degranulated platelets. While quantification of EV-based biomarkers may not differ when analyzed in either serum or plasma specimen, it should be acknowledged that different EV-based biomarker thresholds should be defined in one specimen type should be applied only to that specimen type. Several recommendations and guidelines have been published recently regarding the selection of blood sample types for EV analysis 62–69. The key point is that specific analytical parameters (explained in the analytical validation section) and cut-off values (explained in the clinical validation section) should be determined for each specimen type.
Notably, there are several clinical trials in which EVs are evaluated as the primary or secondary outcome measure for the purpose of discovery of potential EV biomarkers using proteomics, sequencing, and PCR techniques. The prospective nature of sample collection will help overcome some of aforementioned sample related issues. Taking the PC clinical trials as examples: one study (NCT03334708) has been designed to recruit 750 participants to study the change in biomarkers to determine sensitivity and specificity of the assay to diagnose early stage PC. As a discovery study, initial biomarkers to be tested include proteins and proteases, functional DNA repair assays, EVs, stromal elements, circular RNAs and circulating tumor DNA (ctDNA). Another study (NCT02393703) has planned to recruit 70 participants to study the EV-mediated intercellular signalling in patients with PC. In this study, EV will be purified for downstream applications such as proteomics and RNA sequencing. The third study (NCT03250078) aims to explore the relationship between new-onset diabetes mellitus and a subsequent diagnosis of PC. As a secondary outcome measure, 800 serum from participants will be banked to isolate circulating EVs and ctDNA. Additional clinical trials will be described in the section: analytical and clinical validation of EV associated biomarkers-review of clinical trials.
EV Biomarker verification
Initially promising reports at the biomarker discovery phase are often not sufficient to develop a candidate biomarker into a valid clinical assay. Before moving to the validation phase, thorough verification of biomarker candidates is required and successful completion of this process is much more difficult than initial discovery. Verification has to be performed in several studies with large and well-described patient populations that are completely independent from those employed in the original discovery studies. Sample blinding is also strongly recommended when performing EV biomarker verification studies, as systematic review has observed pronounced bias due to the lack of patient blinding in clinical trials with patient-reported outcomes 70. Most of the discovery studies listed in Table 2 were not blinded.
The goal of candidate biomarker verification studies is to isolate true positive markers from a large pool of candidate factors, and to ensure that only the most promising biomarkers found in the discovery phase move on to the validation stage. As with drug development studies, the vast majority of leads identified in the discovery phase do not survive the subsequent verification step. This verification process may be the single greatest challenge in the EV clinical assay development process.
Table 2 lists the potential clinical applications for multiple candidate EV biomarkers of PC that were analyzed in separate early-stage exploratory studies, which were primarily proof-of-concept. Notably, at the early discovery stage certain EV biomarkers demonstrated promise for multiple clinical applications. For example, in a single exploratory study, GPC1-positive EVs exhibited superior clinical performance for early PC detection, and evaluation of metastatic disease burden, response to surgery, and disease prognosis 38. However, there are very few cancer biomarkers that are approved by the US FDA for multiple intended clinical usages. For example, the PC biomarker carbohydrate antigen 19–9 (CA19–9) has been approved only for monitoring and management of PC but not any other functions, due to lack of clinical evidence.
Verification studies require a clear definition of and justification for the intended clinical application, careful selection of the source populations and sample types, and an adequate number of high quality samples. For example, a biomarker for early PC diagnosis would require a patient population with confirmed early-stage PC and benign pancreatic disease (BPD), and appropriately matched normal controls (NC), while a biomarker for PC disease progression would require patients with BPD and various stage PC tumors. We will describe the verification process required for EV clinical applications in clinical trials in more detail in a later section (Analytical and Clinical Validation of EV Associated Biomarkers-Review of Clinical Trials) of this review.
Independent biomarker verification studies are essential during clinical development. Definitive clinical verification requires analysis of study cohorts that contain hundreds of subjects, preferably from multiple institutions, and that incorporate a broad range of cases and controls to mitigate environmental, genetic, and biological variation. The goal of these studies is to isolate true positive markers and ensure that only the most promising move on to validation, since the vast majority of leads do not survive verification. EV biomarker verification must be performed using multiple studies with large, well-described patient populations, independent from those analyzed in the original studies, before beginning the biomarker validation phase. There are some EV clinical trials that employ large cohorts (e.g., a few hundred or few thousand of participants), but the majority of clinical trial studies for EV biomarkers employ very small sample sizes (e.g., fewer than 20 participants), as exemplified in Table 4.
Table 4.
EV assay validation-review of clinical trials
| Study Title | Recruitment Status | Clinical Trial | Cancer type | Specimen Type | Target | Enrollment |
|---|---|---|---|---|---|---|
| Prostate Cancer | ||||||
| Clinical Validation of a Urinary Exosome Gene Signature in Men Presenting for Suspicion of Prostate Cancer | Completed | NCT02702856 | Prostate Cancer | Random, non-DRE, non catheter urine | Exosome RNA gene signature | 2000 |
| Clinical Evaluation of the ‘ExoDx Prostate IntelliScore’ in Men Presenting for Initial Biopsy; Additional Confirmation Study Including Impact on Decision-making and Health Economics | Recruiting | NCT03031418 | Prostate Cancer | Urine | ExoDx Prostate Intelliscore | 1000 |
| A Prospective, Randomized Blinded, Shared Decision Impact Trial of the ExoDx Prostate (IntelliScore), EPI Test, in Men Presenting for Initial Biopsy | Recruiting | NCT03235687 | Prostate Cancer | Urine | ExoDx Prostate Intelliscore | 1000 |
| Quantification and Purification of Circulating Prostasomes as Diagnostic Tool for Prostate Cancer Detection | Recruiting | NCT03694483 | Prostate Cancer | Plasma | Presence of prostasomes using the ExoPLA (Exosome in situ Proximity Ligation Assay) assay; prostasome miRNA sequencing. | 600 |
| Detection of ARv7 in the Plasma of Men With Advanced Metastatic Castrate Resistant Prostate Cancer (MCRP) | Recruiting | NCT03236688 | Metastatic Castrate Resistant Prostate Cancer | EDTA Plasma | ARv7 splice variant transcripts from exosomes | 30 |
| Pancreatic Cancer | ||||||
| Diagnostic Accuracy of Circulating Tumor Cells (CTCs) and Onco-exosome Quantification in the Diagnosis of Pancreatic Cancer - PANC-CTC | Completed | NCT03032913 | Pancreatic cancer | Blood | GPC1+ exosomes | 52 |
| Phase IB/II Trial of High Dose Ascorbic Acid (AA) + Nanoparticle Paclitaxel Protein Bound + Cisplatin + Gemcitabine (AA NABPLAGEM) in Patients Who Have No Prior Therapy for Their Metastatic Pancreatic Cancer | Recruiting | NCT03410030 | Pancreatic Cancer | Blood | GPC1+ exosomes | 36 |
| Development of Biomarkers for the Early Detection, Surveillance and Monitoring of Pancreatic Ductal Adenocarcinoma | Recruiting | NCT03334708 | Pancreatic Cancer | Blood | Not specified | 750 |
| Interrogation of Exosome-mediated Intercellular Signaling in Patients With Pancreatic Cancer | Recruiting | NCT02393703 | Pancreatic Cancer | Blood and tissue | Exosome proteomics and RNA sequencing | 70 |
| A Pancreatic Cancer Screening Study in High Risk Individuals Including Those With New-Onset Diabetes Mellitus | Recruiting | NCT03250078 | Pancreatic cancer | Serum | Biobank | 800 |
| Lung Cancer | ||||||
| Detection of Either the EML4-ALK Gene Rearrangements or the T790M EGFR Mutation in the Plasma of Advanced NSCLC Patients | Recruiting | NCT03236675 | NSCLC | EDTA Plasma | EML4-ALK Gene Rearrangements or the T790M EGFR Mutation in exosome | 60 |
| Detection of Circulating Biomarkers of Immunogenic Cell Death After Radiotherapy and Chemotherapy: An Exploratory Study | Recruiting | NCT02921854 | NSCLC | Blood | Not specified | 40 |
| Phase II, Multicenter, Single-arm, Open-label Study to Evaluate the Efficacy of Olmutinib(Olita®) in Patients With NSCLC Who Harboring T790M Mutation Confirmed Using DNA Extracted From Extracellular Vesicles in Bronchoalveolar Lavage Fluid | Active, not recruiting | NCT03228277 | NSCLC | Bronchoalveolar Lavage Fluid | T790M mutation in EV | 25 |
| Combined Diagnosis of CT and Exosome in Early Lung Cancer | Not yet recruiting | NCT03542253 | Early Lung Cancer | cancer tissue and para cancerous tissue | Exosomal micor-A | 80 |
| Clinical Research for the Consistency Analysis of PD-L1 in Cancer Tissue and Plasma Exosome | Not yet recruiting | NCT02890849 | NSCLC | Plasma | PD-L1 mRNA in plasma exosomes (pExo) | 60 |
| Clinical Research for the Consistency Analysis of PD-L1 in Lung Cancer Tissue and Plasma Exosome Before and After Radiotherapy | Not yet recruiting | NCT02869685 | NSCLC | Plasma | PD-L1 mRNA in plasma exosomes (pExo) | 60 |
| Breast Cancer | ||||||
| A Randomized, Open-label Phase III Trial to Evaluate the Efficacy and Safety of Pertuzumab Retreatment in Previously Pertuzumab, Trastuzuamb and Chemotherapy Treated Her2-Positive Metastatic Advanced Breast Cancer | Recruiting | NCT02514681 | HER2-positive Locally Advanced or Metastatic Breast Cancer | Blood | micro RNAs expression in EV | 370 |
| A Phase 2 Clinical Trial of the Combination of Pembrolizumab and Selective Androgen Receptor Modulator (SARM) GTX-024 in Patients With Metastatic Androgen Receptor (AR) Positive Triple Negative Breast Cancer (TNBC) | Recruiting | NCT02971761 | Breast Cancer | Blood | Temporal profile of tumor-derived exosomes | 29 |
| A Phase II Window of Opportunity Trial of Ipilimumab and Nivolumab in Metastatic Recurrent HER2- Inflammatory Breast Cancer (IBC) The Win Trial | Recruiting | NCT02892734 | Breast Cancer | Blood | ctDNA and Immune signature in exosome | 29 |
| A Pilot Study of Tumor-Derived Exosomes as Diagnostic and Prognostic Markers in Breast Cancer Patients Receiving Neoadjuvant Chemotherapy | Withdrawn | NCT01344109 | Breast cancer | Serum | protein surface markers and RNA profiles in tumor derived exosomes | 0 |
| Other Cancers | ||||||
| Pilot Study With the Aim to Quantify a Stress Protein in the Blood and in the Urine for Early Diagnosis of Malgnant Solid Tumors | Recruiting | NCT02662621 | Cancer | Blood and Urine | HSP-70 exosome | 80 |
| Pimo Study: Extracellular Vesicle-based Liquid Biopsy to Detect Hypoxia in Tumours | Recruiting | NCT03262311 | Cancer | Blood | Pimonidazole staining in EV | 20 |
| Phase I/II Clinical Trial of Daratumumab and Donor Lymphocyte Infusion in Patients With Relapsed Acute Myeloid Leukemia Post-Allogeneic Hematopoietic Stem Cell Transplant | Not yet recruiting | NCT03537599 | Acute Myeloid Leukemia | bone marrow | Exosome number and content (protein, mRNA), and miRNAs | 30 |
| Evaluation of MicroRNA Expression in Blood and Cytology Specimens as a Novel Method for Detecting Barrett’s Esophagus | Unknown | NCT02464930 | Barrett’s
Esophagus Gastroesophageal Reflux Esophageal Adenocarcinoma |
Serum and Bile | miRNA in exosome from serum and bile: miRNAs −192–5p, −215–5p and −194–5p | 220 |
| Early Biomarkers of Tumor Response in High Dose Hypofractionated Radiotherapy Word Package 3 : Immune Response | Active, not recruiting | NCT02439008 | Carcinoma, Hepatocellular Colorectal Neoplasms Melanoma Kidney Neoplasms |
Blood | Quantification of secreted exosomes | |
| Exosomes-derived ncRNAs As Biomarkers In Cholangiocarcinoma Patients | Recruiting | NCT03102268 | Cholangiocarcinoma | plasma | Exosome ncRNAs | 80 |
| A Phase 2, Single Arm, European Multi-center Trial Evaluating the Efficacy of Afatinib as First-line or Later-line Treatment in Advanced Chordoma | Recruiting | NCT03083678 | Chordoma | Blood | Circulating exosomes identification by PCR | 40 |
| Development of Novel Imaging and Laboratory Biomarkers to Monitor the Liver Pre-metastatic Niche and Guide Treatment of Colon Cancer: A Pilot Study | Recruiting | NCT03432806 | Colon Cancer | Blood | Exosomal protein | 80 |
| Circulating Exosomes As Potential Prognostic And Predictive Biomarkers In Advanced Gastric Cancer Patients: A Prospective Observational Study (“EXO-PPP Study”) | Unknown | NCT01779583 | Gastric Cancer | Serum | Plasma level and kinetics of gastric cancer derived exosomes | 80 |
| An Open-label Phase Ib, Study, to Determine Safety of Oral AL3810 in Patients With Locally Advanced or Metastatic Gastric, Hepatocellular or Nasopharyngeal Carcinoma | Not yet recruiting | NCT03260179 | Gastric, Hepatocellular or Nasopharyngeal Carcinoma | Plasma | Growth factor such as INFγ and cMyc in exosome | 60 |
| Newly diagnosed HGG: surgically eligible patients AdV-tk into wall of resection cavity; 1–3 days post-surgery Valacyclovir d1–14; Day 8 RT for 6 wks; day 15 TMZ 75mg/m2 daily; Nivo 240mgIV every 2 weeks | Active, not recruiting | NCT03576612 | Glioma, Malignant | Serum | Immune characterization of surface and content protein in EV | 36 |
| Non-coding RNA in the Exosome of the Epithelia Ovarian Cancer | Recruiting | NCT03738319 | High Grade Serous Ovarian Cancer | Blood | Exosomal miRNA/lncRNA by next gen sequencing | 160 |
| A Pilot Study of Circulating Exosome RNA as Diagnostic and Prognostic Markers in Lung Metastases of Primary High-Grade Osteosarcoma | Recruiting | NCT03108677 | Lung Metastases Osteosarcoma |
Blood | Levels and mutations of circulating exosome RNA | 40 |
| Pilot Study of Exosomes Before and After BRAF Inhibitor Therapy in Patients With Advanced Unresectable or Metastatic BRAF Mutation-positive Melanoma | Unknown | NCT02310451 | Metastatic Melanoma | Blood | Not specified | 15 |
| An Observational, Single-Institution Pilot/Feasibility Study of Exosome Testing as a Screening Modality for Human Papillomavirus-Positive Oropharyngeal Squamous Cell Carcinoma | Recruiting | NCT02147418 | Oropharyngeal Cancer | Primary cell culture and Oropharyngeal Rinse | Exosome Protein Signature | 30 |
| A Study of Circulating Exosome Proteomics In Gallbladder Carcinoma Patients | Recruiting | NCT03581435 | Proteinosis Gallbladder Carcinoma |
Blood | Exosome proteomics | 50 |
| A Prospective Study of Predicting Prognosis and Recurrence of Thyroid Cancer Via New Biomarkers, Urinary Exosomal Thyroglobulin and Galectin-3 | Recruiting | NCT03488134 | Thyroid Cancer | Urine | Galectin-3, Calprotectin A8, Calprotectin A9, TKT, Annexin II, Afamin, Keratin 8, Keratin 9, Angiopoietin-1 and TIMP in exosome | 150 |
| Anaplastic Thyroid Cancer and Follicular Thyroid Cancer-derived Exosomal Analysis Via Treatment of Lovastatin and Vildagliptin and Pilot Prognostic Study Via Urine Exosomal Biological Markers in Thyroid Cancer Patients | Active, not recruiting | NCT02862470 | Thyroid Cancer | Urine | Not specified | 22 |
EV clinical assay development
An EV biomarker candidate that survives the verification step moves to the next phase, in which a clinical assay is developed and subjected to analytical validation. Ideally, the assay used in the EV biomarker discovery and validation phases should be the one intended for routine clinical use, but this rarely happens in reality.
Commercial clinical assays are not yet available for most EV biomarkers and a research assay may therefore be utilized as an alternative assay in an early discovery phase study. In the context of developing EV biomarkers, conventional methods for EV isolation (e.g., ultracentrifugation (UC)) and characterization (e.g., ELISA, flow cytometry, and reverse transcriptase polymerase chain reaction (RT-qPCR)) have been utilized for EV or EV content analysis in research settings and discovery phases.
Most of the studies outlined in Table 2 used UC for EV isolation, but UC is not practical for high-throughput assays or clinical settings due to its long run time, high instrumentation consumables cost, technical expertise requirement, and undesirable performance characteristics, including low purity and high variability. However, there is currently no gold standard method for the isolation and analysis of EVs. A recent survey of 1742 published EV experiments identified more than 190 distinct isolation methods and 1038 different protocols used to isolate EVs from biofluids 67. A plurality of these experiments (45%) used UC, but employed different parameters. Among the studies reviewed in this work in Table 2, 62% employed UC although the parameters used differed among these studies. For actual EV analysis, most miRNA studies utilized RT-PCR, but several different methods were employed for EV protein analysis in these studies, including nanoplasmon-enhanced scattering (nPES), ELISA, an alternating current (AC) electrokinetic microarray, flow cytometry, mass spectrometry, a localized surface plasmon resonance (LSPR)-based sensor, and Western blot analysis. Different methodologies have also been used to analyze the same EV target in different studies, which may also contribute to the inconsistent clinical performance of these EV markers. For example, ELISA, AC electrokinetic microarray, flow cytometry, and mass spectrometry were employed to measure EV expression of Glypican-1 in different studies 38–42. These studies showed very different clinical performance, with AUCs for PC diagnosis ranging from 0.59–1 (Table 2). In addition to their use of different methodologies, these studies also employed distinct study designs, patient populations, and sample sizes, all of which can all affect assay results.
Most of the studies listed in Table 2 performed only rudimentary discovery or verification studies using very small patient cohorts, and 60% of these studies did not perform or report any analytical performance parameters. Of those that did report such data, 23% (6) reported a cut-off threshold, 12% (3) reported limit of detection (LOD) and/or linearity information, and 7% (2) reported precision data. Further, very little technical detail was provided for many of these studies, which may contribute to the growing concern that a large fraction of published discovery studies lack reproducibility. We highly recommend that researchers perform analytical analysis on EV data intended for biomarker discovery and validation studies, and provide product and manufacturer information, and at least basic analytical performance data for any assay used to quantify the biomarker of interest, including accuracy, precision, linearity and analytical sensitivity (e.g., limit of detection (LOD)).
The above mentioned research assays are may not be suitable for routine clinical use due to their multiple manual operation steps, long analytical times and undesirable performance characteristics, which include low recovery, sensitivity, specificity, precision and reproducibility. In order to translate such research data to general clinical testing, effort must be taken to develop EV assays that are suitable for routine and high-volume use, with acceptable performance characteristics, in clinical laboratory settings.
A standard EV clinical assay development process therefore normally requires multiple time-consuming and technically demanding steps, including purification of the EV subtype of interest for analytical studies, generation of monoclonal antibodies specific for the target EV biomarkers, development and optimization of a rapid and reliable assay and procedure for the analysis of these biomarkers, and an assessment of its analytical performance. In order to translate research findings to clinical tests, it is first necessary to improve EV isolation methodology to deliver consistent results. Preferably a clinical assay would employ an integrated platform with acceptable performance characteristics for reproducible and high-throughput isolation and quantification of target EV populations in routine clinical laboratory use. Methods that may be suitable for these types of analyses are described in the following section, keeping in mind that they are still in the technology development phase and have not yet been assessed by thorough validation studies.
Novel integrated systems for EV isolation and analysis
The clinical utility of EVs as cancer biomarkers when utilizing conventional methods for EV isolation and analysis is very limited since these approaches are technically complex, labor-intensive, time-consuming, and require highly trained technical personnel and relatively expensive and specialized analytical instruments. For EV analysis methods to be useful in clinical settings, they should generally exhibit several common features. Specifically, they should use relatively small sample volumes; require minimal sample processing; and be rapid, sensitive, specific, high-throughput, inexpensive and amenable to automation. Ideally, these methods should use direct patient samples, without pre-treatment steps, or employ simple and reproducible EV isolation steps. Several methods have been developed in the past few years to isolate, enrich, and analyze EVs by incorporating different technologies into integrated platforms for EV analysis. These approaches represent a promising step towards translating EV-biomarkers into clinical settings.
For example, multiple groups have employed microfluidic approaches to isolate and analyze EVs, as discussed below, using platforms that require small sample volumes and exhibit rapid assay completion times 16, 25, 27, 61, 71–79. These approaches tend to utilize a limited number of EV enrichment methods, including EV capture by specific antibodies, by microfiltration, or by electric fields. Quantification of specific EV populations or biomarkers is likewise determined using a relatively limited array of methods, including the detection signals produced by plasmon resonance, or the chromogenic conversion or electrochemical reduction of a substrate.
Proof-of-concept studies with prototypes of these platforms have analyzed total EV abundance, EV subpopulations, EV RNA targets, and EV intravesicular and membrane proteins. These integrated EV analysis platforms offer many technical advantages that hold great promise for enhancing the clinical translation of new EV biomarkers. It should be emphasized, however, that most of these platforms are in very early stages of development, and that rigorous analytical and clinical validation of these platforms are still needed to evaluate their true potential for clinical translation. We will discuss some of these approaches and specific prototypes in the following sections.
Sensor chip assays:
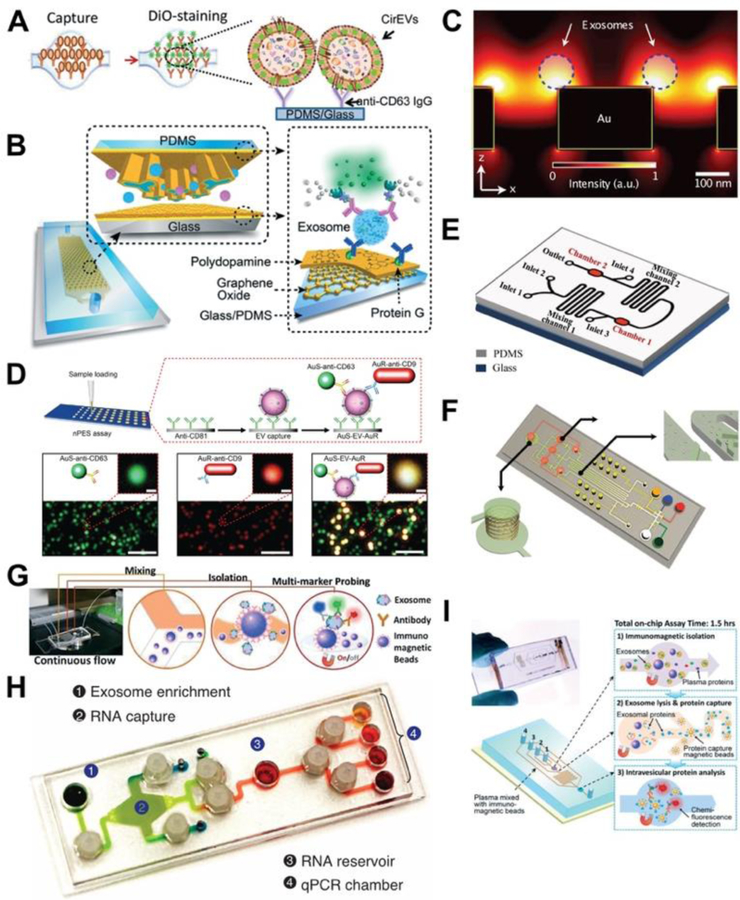
Several groups have developed platforms to isolate and/or analyze EVs upon binding to an antibody-conjugated sensor chip. For example, Kanwar et al developed a simple “ExoChip” microfluidic device for the on-chip isolation, quantification and characterization of EVs that utilizes high surface area to volume ratios and chaotic mixing properties for rapid and efficient EV capture by an anti-CD63 antibody, and subsequent EV labeling and quantification (Fig. 3A), or in situ lysis to isolate protein or RNA for subsequent analyses. A small clinical validation experiment using this platform analyzed total EV levels in serum from 5 PC patients and 5 healthy controls, and found a >2-fold EV increase in the PC vs. healthy control samples.
Fig. 3.
Examples of microfluidic devices applications for EV analysis. (A) ExoChip EV platform73. (B) Nano-interfaced microfluidic exosome (nano-IMEX) assay78. (C) Nano-plasmonic exosome (nPLEX) assay80. (D) Nanoplasmonic-enhanced scattering (nPES) assay43. (E) Fluorescent immunoassay71. (F) µNMR assay. (G) ExoSearch assay79. (H) Immuno-magnetic exosome RNA (iMER) assay75. (I) Immunomagnetic ELISA81. Panels A and G were adapted with permission from The Royal Society of Chemistry. Panels. B, E, G, H, and I were adapted under the Creative Common license http://creativecommons.org/licenses/by/4.0/.
Panels C, D and F were adapted with permission from Springer Nature
Similarly, Zhang et al developed a nano-interfaced microfluidic exosome (nano-IMEX) platform where EVs were captured on the chip surface with an anti-CD81 antibody, and specific detection antibodies were used for an on-chip ELISA in which EV abundance was measured by detection antibody-mediated substrate conversion by an inverted epiflouresence microscope (Fig. 3B) 78, with a reported detection limit of 50 EV/μL and a 4-log dynamic range. In a proof-of-concept analysis, higher EV levels were detected in plasma samples from 7 ovarian cancer patients than 5 healthy controls, which decreased after treatment.
Employing a different approach, Im et al. designed a nano-plasmonic exosome (nPLEX) assay platform where antibody-mediated EV binding on an array of periodic nanoholes produced a transmission surface plasmon resonance effect where changes in the intensity and wavelength of transmitted light could quantitate EV binding or the binding of specific proteins in EV lysates (Fig. 3C) 80. The reported LOD of ~3,000 EVs was ~100× that of a chemiluminescent ELISA comparator, and labeling captured EVs with nanoparticle-conjugated antibody could increase signal intensity 300%, depending upon the nanoparticle size and conformation. Results from a small clinical study (20 cancer cases and 10 non-cancerous controls) indicated that this platform could distinguish patients with ovarian cancer from cirrhosis patients (non-cancer controls).
Our group independently developed a nano-plasmon enhanced scattering (nPES) assay to quantify total and disease-derived EVs from unpurified biological samples (Fig. 3D) 43. EVs that bind both an antibody-conjugated gold nanosphere (AuS) and nanorod (AuR) produce a plasmon that increases the intensity and shift the wavelength of their scattered light. We employed this assay to evaluate plasma samples of PC, chronic pancreatitis and NC subjects in a trial cohort (10/group) and a validation cohort (48–49/ group) and found that nPES signal from EVs labeled with antibodies to EV-selective (CD9) and cancer-selective (EphA2) markers distinguished PC patients from patients with chronic pancreatitis or healthy subjects, outperforming results obtained using a standard EV ELISA; corresponded with tumor burden, stage and early response to neoadjuvant therapy. This study also examined the discriminatory ability (AUC 0.93–0.96), sensitivity (86–94%) and sensitivity (85%) of this assay to discriminate PC patients with stage I-II tumors or any stage tumor from healthy controls or patients with chronic pancreatitis.
Immunomagnetic EV capture assays:
Several groups have now employed magnetic particles (MPs) in EV assay platforms. For example, Fang et al. developed a microfluidic chip that permits EVs captured on magnetic particles to be sequentially hybridized with primary and secondary antibodies, and then analyzed for fluorescent signal 71 (Fig. 3E). This group analyzed EV expression of two cancer-associated proteins (EpCAM and HER2) in plasma samples. The EpCAM-positive EV level was significantly increased in 6 breast cancer patients compared to that found in 3 healthy controls. EV HER2 levels in 19 breast cancer patients corresponded with its tissue expression.
Shao et al. developed a micro-nuclear magnetic resonance (µNMR) platform to quantify the 1H NMR decay signal associated with EVs rendered superparamagetic by the binding of antibody-conjugated MPs specific to EV target proteins (Fig. 3F) 82. NMR signal was found to be proportional to EV number (R2 >98%), strongly correlated (R2 >99%) with fluorescent ELISA results, and revealed a detection threshold of ~104 EVs. NMR analysis of four glioblastoma markers (EGFR, EGFRvIII, PDPN and IDH1 R132H) distinguished glioblastoma-derived EVs from other EVs. Results from a mouse model revealed that a composite score of glioma-derived EV abundance and the biomarker panel paralleled tumor progression and decreased with treatment. Analysis in glioma patients before and after treatment also found that this composite EV score could differentiate responders from non-responders. However, this assay requires UC-purified EV samples that are not suitable for use in clinical assays, and it is not clear if this approach can be readily adapted to use minimally processed liquid biopsy samples.
He’s group subsequently developed a continuous flow microfluidic device (ExoSearch) for quantitative capture and release of plasma EVs over a large range of sample volumes (Fig. 3G) 79. In this approach, plasma samples and antibody-conjugated MPs are injected in separate channels and, after on-chip mixing, MP-bound EVs are retained by a magnetic field and then hybridized with a mixture of fluorescently labeled antibodies for multiplex analyses of EV composition. EV capture efficiency was inversely related to flow rate, ranging from 97% at 50nL/min to 42% at 10µL/min. Study assays were performed at a 1µL/min, with a 72% EV capture efficiency and a LOD of 7.5 × 105 particles/mL at a signal-to-noise ratio of 3. It was estimated that this platform capture EVs from 2 mL of plasma in 3 hours at 10µL/min (42% efficiency), favorably comparing with standard UC approaches that require 3 times as long with a 25% EV recovery rate. Multiplex analysis of the EV expression of the cancer-associated markers CA-125, EpCAM and CD24 in serum from 15 patients with ovarian cancer and 5 healthy controls found that all three markers were elevated on cancer patient EVs, and observed AUC values of 1 for CA-125 and EpCAM and 0.91 for CD24. The speed, sensitivity, scalability, multiplex capability, cost and potential for automation are important features that may enhance the clinical translation of this method.
Shao et al. developed an immuno-magnetic exosome RNA (iMER) microfluidic chip to analyze EV mRNA levels in circulating EV populations 75. This chip contains four functional regions: an EV enrichment area, an RNA isolation site, a reservoir for the reverse transcription of eluted RNA targets, and parallel chambers for on-chip qPCR analysis (Fig. 2H). Serum or cell culture EVs incubated with MPs conjugated with an anti-EV antibody (CD63 or EGFR) were captured 93% efficiency upon on-chip magnetic enrichment, and on-chip EV lysis and RNA isolation generated EV RNA size distributions similar to those produced by a commercial RNA isolation column, although the chip yielded 50% more RNA for a given sample volume. The iMER qPCR results also strongly correlated (R2=0.986) with conventional qPCR. A small clinical study found that EphA2 and EGFR, but not PDPN, were significantly elevated in a small cohort of GBM patients versus control subjects. The accuracy for GBM diagnosis was 84% (EphA2) and 78% (EGFR) for single marker iMER and increased to 90% when using three markers. An analysis of pre- and post-treatment GBM patient serum samples also detected a significant inverse association between the EV level of two mRNAs associated with the repair of chemotherapy-induced DNA damage and favorable treatment response.
He et al. developed a microfluidic platform that used a cascading microchannel circuit to sequentially isolate, enrich and lyse human plasma EVs to capture EV biomarker targets, and analyze them in an chemifluorescence sandwich immunoassay (Fig. 3I) 72. Plasma samples are pre-mixed with antibody-labeled MPs, which are captured and lysed on-chip, after which EV lysates are mixed in a serpentine channel with antibody-labeled MPs recognizing specific EV proteins. These MP are retained in a second chamber by a magnetic field and incubated with detection antibodies and detection substrate. This approach was used to quantitate total and phosphorylated IGF-1R expression in EpCAM+ EVs, and found that mean IGF-1R, but not p-IGF-1R expression, was increased in EpCAM+ EVs from plasma samples of 5 non-small cell lung cancer (NSCLC) patients with early-stage disease (stage II) versus their 6 NCs. IGF-1R and p-IGF-1R were quantifiable over a 4 log dynamic range with a detection limit of 0.28 and 0.38 pg/mL−1, respectively, at a signal noise of 3, which was approximately 100× more sensitive than a commercial ELISA.
Immunomagnetic assays with electrochemical detection approaches:
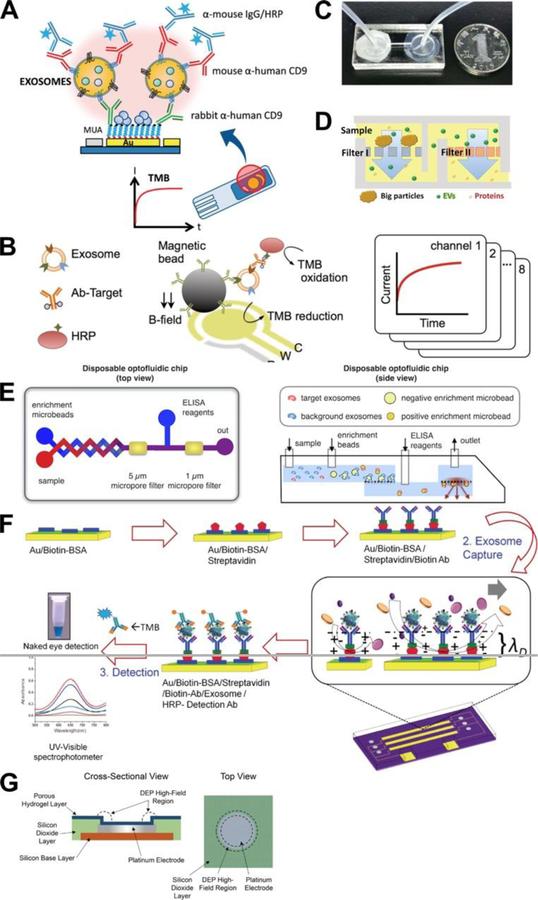
Several groups have also utilized electrochemical detection approaches to analyze EVs. For example, Doldan et al developed an amperometric biosensor (Fig. 4A) 81. In this method, EV are bound by a capture antibody linked to the gold electrodes of a biosensor chip and then incubated with an EV detection antibody and HRP-labeled secondary antibody and the HRP substrate TMB, after which EVs are detected by the measuring the electrochemical reduction of HRP-oxidized TMB. The linearity range of this assay found to be 102 to 106 EVs/µL, with a calculated LOD of 200 EVs/µL. EV specificity was quite good, since a concentration standard of 106 MVs/µL produced a signal that was only slightly greater than the EV LOD signal, and a 103 EVs/µL standard demonstrated similar signal in the presence or absence of a 1000× excess of MVs. No clinical samples were analyzed in this system, but it was demonstrated that EVs could be detected in 1000× dilutions of EV-depleted fetal bovine serum with an average 90% recovery in EV spike-in experiments, suggesting that this approach could be used to quantitate EVs present in diluted serum samples.
Fig. 4.
Examples of microfluidc devices applications for EV analysis (continued). (A) Immunomagnetic assays with electrochemical detection approaches81. (B) Integrated magnetic–electrochemical exosome (iMEX) assay83. (C) Microfiltration for direct EV capture74. (D) Microfiltration for direct EV capture - Exodisc assay77. (E) Microfiltration of EV affinity beads for indirect EV capture84. (F) Alternating current electrohydrodynamic (ac-EHD) assay method76. (G) Alternating current electrokinetic (ACE) assay method52. (H) Surface plasmon resonance (SPR) detection85. Panels A, B, D, F and G were adapted with permission from the American Chemical Society. Panels C and E were adapted under Creative Common license.
Jeong et al developed a similar integrated magnetic–electrochemical exosome (iMEX) analysis platform that employed MPs for EV enrichment prior to on-chip analysis (Fig. 4B) 83. In this method, MPs conjugated with an EV capture antibody were incubated with undiluted plasma samples, and then isolated and incubated with a HRP-labeled antibody specific to an EV biomarker, loading on the iMEX chip, and captured on its sensor electrode by a magnetic field. EV levels were measured by the signal from the electrochemical reduction of the HRP-oxidized TMB. This assay was faster (1 hr vs. 5 hr), and required less sample (10 µL vs. 100 µL), than ELISA and demonstrated a dynamic measurement range spanning 4 orders of magnitude, with a LOD of 3 × 104 EVs while ELISA revealed a LOD of 3 × 107 EVs. A small clinical study found that EpCAM and CD24 levels on plasma EVs were higher in ovarian cancer patients than normal control subjects, and that pre- vs. post-therapy EV levels of both proteins decreased in patients that responded to therapy.
EV analyses employing microfiltration-based EV capture approaches:
Multiple groups have used microfiltration-based approaches to isolate EVs for subsequent analyses. Liang et al. developed a microfluidic device comprised of two serially connected 10 mm diameter chambers bounded by polycarbonate membranes with 200 nm and 30 nm to retain EVs in the second chamber of the chip (Fig. 4C) 74, demonstrating EV isolation efficiency 74% of the comparable UC yield. Captured EVs were analyzed by an on-chip ELISA which was read by a smart phone camera and transferred to a laptop for data analysis. In a small clinical study, mean EV levels were elevated in the urine of 16 patients with bladder cancer versus 8 healthy controls, with a ROC demonstrating AUC of 0.96 and 81% sensitivity and 90% specificity. Urine samples were centrifuged and sterile-filtered prior to analysis, and 8 mL was injected into the chip at a flow rate of 40 µL/min (>3.5 hours) while the on-chip ELISA required an additional 3 hours.
Woo et al. developed an automated “exodisc” microfluidic system that permits rapid and efficient centrifugal isolation of EVs from biological samples, including cell culture supernatants and urine, and subsequent in situ ELISA analysis of captured EV samples (Fig. 4D) 77. Cell debris and large particles are removed by a centrifugation step prior to sample loading, large particles are retained by the 600 nm filter, and EVs are retained by the 20 nm filter. This platform demonstrated a >95% recovery rate for EVs from cell culture supernatants, markedly more than UC (3.9×) or the commercial Exospin procedure (2.8×), with higher RNA (>100×) and protein (13×) concentrations for analyzed candidates. From an operational standpoint, the Exodisc EV isolation method also required relatively modest g-force and processing time (500g, 30 minutes) compared to standard UC (150,000g, 6 hours) or Exospin (16000g, 4 hours) EV isolation procedures, and exhibited better performance than UC and Exospin methods in urine from 5 bladder cancer patients and 5 healthy controls.
Using an alternative fractionation approach, Ko et al developed a disposable optofluidic chip where serial filtration of EV-capture beads with different diameters is utilized to negatively select undesired EVs and enrich target EVs before an on-chip ELISA that is read by the LED and optics of a standard smartphone (Fig. 4E) 84. Negative selection beads (7 µm) displaying antibodies to CD45 and CD61 capture leukocyte-derived EVs, which are removed by filtration, after which positive selection beads (2.2 µm) conjugated with antibodies to CD81 capture the remaining EVs and are incubated with HRP-linked antibodies specific to a target biomarker, and the LED and camera of a smartphone is used to detect chemiluminescent assay signal. The disposable optofluidic chip is designed to slot into a housing that fits a smartphone and contains two optical filters to restrict the incident light and the captured fluorescent signal. A custom smartphone app is used to control the LED, measure the emitted light and analyze the resulting data. A proof-of-principle clinical study found that injury samples demonstrated an increased rate and amplitude of target-specific fluorescent signal development. However, this system demonstrated an LOD of 107 EVs, and a significant signal flattening as it approached this value.
EV analyses employing electric fields for EV capture:
Multiple groups now have now generated assay platforms that utilize alternating current to driven EVs across a sensor chip. Vaidyanathan et al. developed an alternating current electrohydrodynamic (ac-EHD) microfluidic device for multiplex exosome capture and detection (Fig. 4F) 76. In this approach, chip microelectrodes are conjugated with specific EV capture antibodies and purified EVs suspended in PBS are driven through the device by an applied ac-EHD field, employing a 30 min on and 15 min off cycle for 2 hours. EV detection antibodies labeled with FITC or HRP are then drawn through the device by either ac-EHD force or applied pressure, and EV-bound FITC signal is captured with a fluorescence microscope. Samples hybridized with FITC-conjugated detection antibodies are then hybridized with anti-FITC antibodies conjugated with HRP, and signal from HRP-labeled primary and secondary antibodies is analyzed by incubating these assays with TMB and recovering this substrate for off-chip analysis. The linear detection range was found to be 2.76 × 103 to 4.15 × 104 EV/μL in a pilot study, which was up to 3× more sensitive than assays using pressure-driven hydrodynamic flow (LOD 8.30 103 EV/μL). Serum sample from single individuals with HER2+ and HER2− breast cancer analyzed in a proof-of-concept study, also revealed a significant difference in target EV level (2 × 104 vs 3.7 × 103 EV/μL). The narrow dynamic range of this assay may limit its translational potential for assays requiring quantitation, but may be suitable for assays that require only sensitive detection for a yes-or-no diagnosis.
Similarly, Ibsen et al developed an alternating current electrokinetic (ACE) microarray device in which EVs and other nanoscale particles from undiluted plasma samples could be driven to accumulate at the edges of microelectrodes in response to dielectrophorectic high-field regions that form when these chips are exposed to an alternating current (Fig 4G) 52. A 10 min application of AC current to the chip causes dielectrophoretic separation and isolation of EVs and other particulates at the chip electrodes. EVs, but not plasma proteins, are retained by the electric field during a wash step and can be stained in situ with fluoresce6nt antibodies to quantitate total or specific EV subpopulation levels or with an RNA-specific dye to quantitate EV RNA abundance. Bound EVs can also be released from the device by using a series of low-frequency electric pulses for off-chip analyses or lysed in situ to generate high quality EV RNA samples. This process requires only a small amount (30–50 µL) of plasma, serum or whole blood for EV analysis, and can be rapidly performed to analyze or isolate EVs present in these samples. Lewis et al. subsequently utilized this approach to isolate EVs from blood samples of patients with pancreatic or colon cancer to analyze specific EV protein biomarkers for association with these cancers 41. This study found that in situ staining of EV GPC1 expression distinguished PC patients from NCs, but did not differentiate PC and BPD patients, likely due to the small sample size and the inclusion of arguably premalignant subjects in the BPD group. GPC1 also did not show a trend to differentiate lymph node-negative and -positive PC patients, or early and late stage PC cases. Conversely, analysis of a small set of samples from patients with colon cancer and NC subjects found that high level EV GPC1 expression differentiated metastatic from non-metastatic cases or NC subjects, but did not distinguish non-metastatic colon cancer patients from NC subjects.
Current Limitations:
Many of the above proof-of-concept studies offer advantages that could facilitate the clinical translation of EV biomarker assays for disease diagnosis and prognosis, including the potential for more rapid, higher purity and more reproducible EV purifications that require less operator time and less specialized and expensive equipment. However, all of these studies performed only partial analytical validation of the method and most used small sample size for both analytical and clinical validation. Complete analytical validation studies need to be performed to more fully characterize operating parameters of these platforms for the analysis of total EV populations and their performance with specific EV biomarkers, while further clinical studies with large sample size and robust statistical designs, need to be conducted to determine their clinical application in the appropriate populations.
Fig 5 indicates that most integrated systems for EV isolation and analysis have used small numbers of patient samples (mean of 30, 95%CI: 7–54). By contrast, the patient recruitment numbers for the EV clinical trial studies described in the next section are much larger (mean of 230, 95%CI: 90–370). This is a key issue, since when a laboratory develops a novel device, it may exhibit adequate performance handling small numbers of samples. However, when translating such devices to the much larger scale required for clinical trials, and ultimately clinical applications, these devices need to consider scaling issues, including fabrication, assembly, and operation issues associated with utilizing the device to consistently analyze hundreds of samples. Research conducted on EV detection methods has made significant progress with new innovations allowing rapid small analysis of clinical samples for preclinical tests. However, several issues contribute to the bottleneck in the translation of such tests from small scale research laboratory tests to large scale tests suitable for validation in clinical trials for eventual clinical applications. One such issue is the difficulty of scaling up the fabrication of such assay platforms, which is often not a concern during the conception of a novel device but which can present significant challenges when the time comes to mass produce the assay platform. Production capacity may vary widely depending on the type of device and its requirement for specific manufacturing processes. Another non-trivial issue is the relative scarcity of manufacturing facilities available to perform such fabrication at the intermediate scale required for clinical trials. This is a particular concern for proposed EV assay platforms that require manufacturing facilities suitable for the production of microfluidic platforms or devices with nanoscale components. Given the specialized expertise and equipment and the relatively high costs required to produce such devices, particularly at limited scale, this appears likely to remain a significant barrier to EV assay development. Similar to efforts to standardize EV purification and characterization procedures, solving this issue may require the development of a consortium focused on mid-scale production of devices with such specialize properties, although it does not appear that any such initiatives are now under consideration. Precision and reproducibility of device fabrication and associated quality controls may also become a significant issue when scaling the production of a novel device, as device fabrication parameters may become an issue when the device fabrication process is streamlined or automated to allow increase production. Careful consideration should therefore be given to the design of any new device intended for clinical application to ensure that its fabrication can be readily scaled-up during the transition from research laboratory testing studies to clinical trials and its ultimate use in clinical settings. Such manufacturing needs should be considered at the early stages of the initial development process to avoid potentially costly project delays required to troubleshoot fabrication issues and the potential need to redesign the device or fabrication process to overcome them. In addition to fabrication issues outlined above, additional effort may also be required for method optimization to enhance the reproducibility of the assay procedure and streamline the hands-on time required for its performance.
Fig 5.
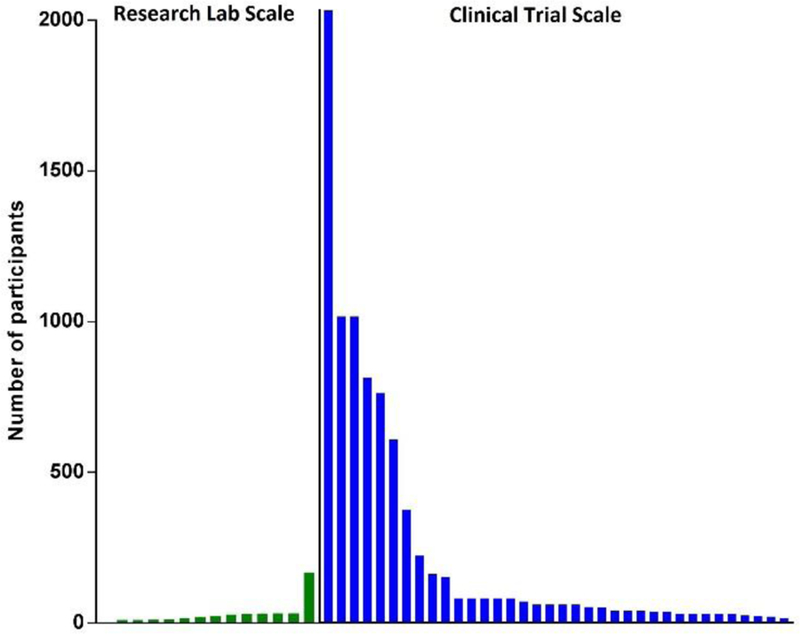
Comparison of the different scale of patient sample sizes employed in the research laboratory studies described in the section on novel integrated systems for EV isolation and analysis (green bars) and the clinical trial studies described in the section on EV assay validation-clinical validation (blue bars).
There is also an urgent need for more robust and reproducible methods for the isolation of a pure vesicular population, since the quality of the EVs used for method development and validation is critical for reproducibility studies. The lack of EV standards is a significant issue for such studies, since there is currently no central source of such standards or a consensus on the best method available to isolate high-purity EV populations for required analytical validation studies. This absence requires that the process used to isolate such EV standards and the purity and reproducibility of the standards generated in these studies be carefully documented during all analyses. This is complicated by the potential for overlap between exosome and microvesicle physical properties and biomarkers.
In the future method development studies, the characterization and study design should be included to ensure the EV quality and validity, even in the face of consensus standards, and thereby improve method reproducibility. We will further discuss the requirements and recommendations of analytical validation of the EV platforms in the following section.
EV assay validation-overview
EV assays intended for clinical use must undergo both analytical and clinical validation. Analytical validation and clinical validation are two distinct processes with separate evaluation characteristics. In the analytical validation process, procedures for the clinical assay are established and the assay’s analytical performance characteristics are optimized through a series of well-defined experiments. The clinical validation process establishes the validity and utility of the assay through testing many thousands of patient samples to determine its clinical performance in context of intended use. It has to be emphasized that the diagnostic performance (e.g. diagnostic sensitivity and specificity) should not be confused with analytical performance (e.g. analytical sensitivity and specificity). An analytically optimized assay with adequate analytical performance characteristics will not necessarily demonstrate satisfactory performance during its subsequent clinical validation.
Many national and international consensus standards have been developed to address aspects of performance characteristics relevant to clinical assays. For example, standards and guidelines have been published by the Clinical and Laboratory Standards Institute (CLSI), the International Organization for Standardization (ISO), and the European Committee for Standardization (CEN). The Food and Drug Administration (FDA) has recognized more than 90 of these consensus standards in in vitro diagnostic (IVD) premarket submission (Medical Device Databases, http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/Databases/default.htm) and published guidelines (http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfggp/search.cfm) for a variety of products.
EV assay validation-analytical validation
Analytical validation addresses the following fundamental assay parameters: accuracy, precision, analytical sensitivity, specificity, reportable range, and reference intervals. Analytical performance of an assay must be thoroughly evaluated for the intended use or application before validating its clinical utility. Most EV assays that have been reported as potential EV cancer biomarker assay platforms report only a few of these parameters, if any, and few of them describe validation protocols. Comprehensive description of experiments required to determine the necessary analytical performance characteristics can be found in Clinical and Laboratory Standards Institute (CLSI) guidelines (Table 3), which should be followed to validate EV analysis systems intended for potential clinical use.
Table 3. CLSI guidelines for validation performance characteristic.
Source of guidelines: https://clsi.org/
| validation performance characteristic | CLSI guideline |
|---|---|
| Accuracy |
EP09c: Measurement Procedure
Comparison and Bias Estimation Using Patient
Samples EP15-A3: User Verification of Precision and Estimation of Bias |
| Precision |
EP05-A3: Evaluation of Precision
of Quantitative Measurement Procedures EP15-A3: User Verification of Precision and Estimation of Bias |
| Detection capability | EP17-A2: Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures |
| Analytical specificity | EP7-A3: Interference Testing in Clinical Chemistry |
| Reportable range | EP6-A: Evaluation of the Linearity of Quantitative Measurement Procedures: A Statistical Approach |
| Reference intervals | EP28-A3C: Defining, Establishing, and Verifying Reference Intervals in the Clinical Laboratory |
| Diagnostic accuracy | EP24-A2: Assessment of the Diagnostic Accuracy of Laboratory Tests Using Receiver Operating Characteristic Curves, 2nd Edition |
Accuracy:
Accuracy describes how closely the measured concentration and the true concentration of a sample agree. Several approaches are available for accuracy determination, such as comparing the results measured by an assay under validation with those determined by a reference method or by measuring certified reference materials with values assigned by a reference method (CLSI EP09c). However, if there are neither reference methods nor definitive reference materials available for the EV biomarkers being analyzed, an acceptable alternative approach is to perform a recovery study (CLSI EP15-A3). In a recovery study, known amounts of the analyte are added to samples at different concentrations, and the measured concentrations are corrected to correlate with the true amounts.
In the current context, not all EV assays perform equally well in terms of accuracy, as evident by a huge variability in recovery rates obtained by different purification and detection methods. Beside well-recognized differences owing to differences in EV isolation techniques, two other factors significantly contribute to poor method agreement: 1) the use of antibodies with different epitope specificities for the same biomarker target, and 2) the lack of a suitable reference EV material due to heterogeneity of EVs, which are presents in multiple forms in circulation and may vary in response to various physiologic stimuli.
Precision:
Precision describes how closely two independent test results agree. Precision should be assessed by replication experiments that measure multiple aliquots of the same sample, or same pool of samples, in independent analyses performed over a defined period of time, usually several days. It is highly recommended that one measure multiple concentrations (low, intermediate, and high) of the EV biomarkers of interest since precision is concentration dependent. Measured imprecision at each concentration analyzed is expressed as a standard deviation (SD) or a CV. CLSI documents EP05-A3 and EP15-A3 describe the details of these procedures.
Detectable capability (analytical sensitivity):
According to CLSI guidelines, detection capability is an umbrella term for a set of performance characteristics, including limit of blank (LOB), LOD, and limit of quantitation (LOQ). For EV biomarkers, the LOB is the highest measurement result likely to be observed for a blank sample known to be free of the EV biomarker of interest, while the LOD is the lowest concentrations of the EV biomarker that can be consistently detected. The LOQ is the smallest amount of the EV biomarker that can be quantitatively determined with acceptable precision. Procedures used to determine these values are described in CLSI document EP17-A2.
Specificity:
The analytical specificity of an assay is concerned with questions such as “does the assay measure the analyte that it’s supposed to measure without cross-reacting with non-targeted substances?” and “are there any factors that interfere with the measurement of this biomarker?” Analytical specificity thus differs from clinical specificity, which refers to the ability of a test to give a negative result for subjects who do not have the disease or condition for which they are being tested. In designing experiments to determine the analytical specificity of an EV assay, the most straightforward approach is to evaluate the behavior of the assay in the presence and absence of potential cross-reacting or interfering substances (CLSI document EP7-A2), which might include vesicles similar to the specific EV target of interests, or compounds that can directly inhibit the assay reaction.
Reportable range:
The reportable range of an assay is the span of test result values for which the laboratory can verify the accuracy of the measurement response. To establish the reportable range for an assay, a linearity experiment is performed to establish the range of values over which there is a constant relationship between observed and expected values (the linear range). This range can be found by making series of known dilutions of a highly elevated patient specimen or standard with a known high quantity of the EV biomarker of interest (CLSI document EP6-A).
Reference interval:
The reference interval of a biomarker refers to the range of values typically found in individuals who do not have the disease or condition that is being assayed for by the test in question. Due to the potential for significant biomarker variability in even healthy populations, the CSLI guidelines recommend that one test 120 samples obtained from healthy subjects to establish this reference interval (CLSI document EP28-A3C). The healthy population examined in the study should be similar to the population that is the intended target of the assay to reduce the potential for influence by cryptic confounding factors. For most biomarkers that are present in apparently healthy individuals, the reference interval is usually defined as encompassing the central 95% of ranked values in a population bounded at the lower reference limit by the 2.5th percentile and at the upper reference limit by the 97.5th percentile. The reference interval is different from the clinical decision limit, which is a cut-off value derived from statistical methods (e.g., receiver operating characteristic (ROC) analysis) applied to both the affected and healthy populations. Cut-off determinations will be discussed in the clinical validation section below.
EV assay validation-clinical validation
Clinical validation is the process through which an assay is determined to be clinically meaningful. Characterizing the diagnostic performance of novel clinical assays is an extensive process. Clinical validation starts with a sound and careful study design that is critical to minimize bias that can arise from systemic errors in laboratory measurements, unrepresentative sampling, or uneven distribution of confounding variables. Evaluation of clinical assays is ideally conducted following standard principles such as those outlined in Standards for Reporting of Diagnostic Accuracy (STARD) and CLSI guidelines.
A series of clinical validation studies are necessary to determine the diagnostic accuracy (clinical sensitivity and specificity) and diagnostic predictability (positive and negative predictive values) through testing many thousands of samples in context of the intended clinical use. An extremely important consideration in the clinical validation process is whether the assay exhibits adequate performance characteristic for the intended clinical use.
One critical component of a clinical validation study is to determine the decision (cut-off) level for the assay, the threshold to be used in making a medical decision. This differs from the reference interval, and represents a clinical decision limit derived from statistical methods (e.g., ROC analysis). Specific examples will be discussed in the following section.
EV assay validation-review of clinical trials
As of December 2018, there were about 40 trials testing EV-associated biomarkers for various cancers. Table 4 lists the study title, recruitment status, cancer type, sample type, EV biomarker target (if specified), and actual or estimated enrollment numbers for these clinical trials found at https://clinicaltrials.gov/. This section will focus on EV-associated biomarkers that are being evaluated in Table 4 studies as the primary outcome measure for prostate cancer (PCA), PC, lung cancer and breast cancer, the four most-studied cancers in these clinical trials. Previously published studies and data related to these EV biomarkers will also be reviewed. There are also several trials in which EVs are evaluated as the secondary outcome measure using proteomics, sequencing, and PCR techniques to discover potential EV biomarkers (as described earlier in this review).
While these EV-based biomarkers were evaluated in the clinical trial setting, only a few of these studies disclosed detailed methods on EV recovery and EV sample evaluation. Due to the limited information provided, it is not possible to completely evaluate the analytical performance of the methods used to analyze these EV biomarkers. Since many of the proposed EV analytes are also present in the non-EV fraction of a specimen, it is particularly important that investigators realize and strive to address the potential risk of contamination during studies intended to analyze the specificity of an EV-based biomarker.
No EV associated biomarkers or EV assays have yet been approved by the US FDA. A few EV assays are performed as laboratory developed tests (LDTs) in clinical laboratories with Clinical Laboratory Improvement Amendments of 1988 (CLIA) certifications for these tests. We will review the current commercial status of these clinically available EV tests. Detailed description of the regulatory requirements will be discussed in the section titled regulatory requirements and end user utilization.
PCA:
PCA is the most common solid malignancy and second leading cause of cancer death in men worldwide. Several urine and plasma EV biomarkers have been reported and examined for PCA diagnosis and treatment-selection.
ExoDx Prostate IntelliScore (EPI) based on PCA3 and ERG
One of the key diagnostic challenges for PCA is the inability to accurately differentiate high-risk PCA from low-risk (indolent) PCA or benign disease. Attempting to address this hurdle, three large clinical trials (NCT02702856, NCT03031418, NCT03235687), have been registered to study the ExoDx Prostate IntelliScore (EPI), which is based on EV RNA expression, in urine samples from men who present with an elevated prostate-specific antigen (PSA) score at their initial prostate biopsy. EPI normalizes the results of Prostate Cancer antigen 3 (PCA3) and v-ets erythroblastosis virus E26 oncogene homolog (ERG), which are commonly identified in PCA tissue, with SAM pointed domain-containing Ets transcription factor (SPDEF) expressed in normal prostatic epithelial cells 86. The intended use of EPI is to discriminate clinically significant PCA from indolent PCA or benign disease for men aged ≥50 yr with PSA levels 2–10 ng/ml in the initial biopsy setting. In the aforementioned JAMA Onclology study 87, in a cohort of 519 patients, EPI revealed a higher area under the receiver operating characteristics curve (AUC) score than the standard of care (SOC) based on PSA, age, race, family history (0.73 vs. 0.63, respectively) for the ability to discriminate between PSA cases with Gleason score (GS) values ≥ GS7 and those with GS6 values or benign disease.
The EPI assay utilizes a proprietary spin column process to isolate EVs from urine and extract RNA for downstream RT-qPCR analysis. In the clinical trials and published manuscripts, this test was performed in a CLIA-certified clinical laboratory. Sample exclusion for this test was based on any of several pre-analytical and analytical-related factors 87, which were: 1) failure of the internal control (bacteriophage Qβ spike-in); 2) SPDEF detected at >30 copies per reaction; or 3) any sample outside the first-catch urine volumes of 25–49 ml. In the prospective, multi-site trial study published in JAMA Oncology 87, 6% (training cohort) and 9% (validation cohort) of samples were excluded due to internal control failure and 15–17% of samples were excluded due to urine volumes > 49 mL. In the follow-up trial 88, 5% samples were excluded due to gene expression and/or internal control levels outside assay acceptance limits. A two-phase adaptive clinical utility trial (NCT03031418) was designed to further evaluate the clinical performance of the EPI test in the intended use population (cohort 1, estimated enrollment: 500 patients) and to evaluate how the results of the EPI test influenced the decision process when the biopsy decision was uncertain (cohort 2, estimated enrollment: 500 patients). The results of the cohort 1 study were published recently 88, reporting AUC values of 0.70 and 0.62 for EPI and SOC, respectively, in a cohort of 503 patients. These results are comparable with previously published results87.
To reduce the false positive rate for unnecessary biopsy, a clinically useful test that follows PSA screening should exhibit significantly improved clinical specificity compared to other available methods. EPI is a non-invasive test that uses non-digital rectal examination (DRE) urine in the initial biopsy setting. Its clinical utility is to potentially avoid an initial biopsy when the EPI value is below the diagnostic cut-off point. Importantly, cut-off points were carefully evaluated and defined in both the above studies. The sensitivity and specify of the EPI test was 92–93% and 26–34%, respectively, when using a cut-off point of 15.6, and the sensitivity decreased to 87–97% and the specificity increased to 37–40% when this cut-off was raised to 20. In cohort 2 of the clinical utility trial (NCT03031418), the investigators reached a consensus recommending use of the 15.6 cut-off point to evaluate how the results of the EPI test influenced the decision process for determining whether or not to perform a prostate biopsy, although these results have not been published to date. Furthermore, the authors suggested that EPI may be combined with other parameters such as clinical preference and comorbidities for personalized risk assignment.
The EPI test is now available in a CLIA-certified clinical laboratory, which is a major milestone for the translation of an EV-based assay into clinical practice, but medical practices based on this assay result should carry some reservations. First, the assay is performed as a LDT in a CLIA-certified laboratory, but details of its analytical validation data, such as its linearity, analytical sensitivity, analytical specificity, and precision, have not been reported. However, since the assay is commercialized, reimbursed by insurance, and regulated by the CLIA, this validation data should be available upon the inquiry. Second, even though the initial proof-of-concept results are promising, the reported specificity and sensitivity of the EPI test for clinically significant PCA cases are still suboptimal for ruling in or ruling out the need for a prostate biopsy. There are also commercially available plasma or urine based assays that show comparable or better performance, such as the Prostate Health Index (PHI, Beckman Coulter, Brea, CA), 4Kscore (Opko, Miami, FL), PCA3 (Gen-Probe, San Diego, CA), and SelectDx (MDx Health, Herstal, Belgium). Further studies are warrant to evaluate the added value of this test when combined or compared with existing PCA risk markers, including imaging results, prostate volume, free PSA, HOXC6, TDRD1, DLX1 and/or some kallikreins 89. Finally, the EPI test has also not been mentioned or recommended in clinical guidelines for incorporation into daily practice.
Androgen receptor splice variant 7 (AR-V7)
AR splice variants are truncated receptor isoforms lacking the C-terminal ligand-binding domain (LBD) but which retain the N-terminal transcriptional elements. The deletion of the AR LBD, a key regulatory region, results in loss of the anti-androgen binding site and constitutive activation of AR signaling, independent of ligand. Mounting studies 90–93 have suggested that detection of AR-V7 in circulating tumor cells (CTCs) could represent a hormone therapy treatment-selection marker in metastatic castration-resistant PCA (mCRPC). CTC tests for AR-V7 are clinically available in CLIA-certified academic 94–96 and commercial 97 clinical laboratories. Several clinical trials have been designed to evaluate the clinical utility of CTC-based AR-V7 as a biomarker to guide treatment in mCRPC (e.g., NCT02269982, NCT03700099, NCT03601143, NCT02601014, and NCT03050866).
In a recent study to examine the potential clinical utility of EV-associated AR-V7, Marzia Del Re et al. 98 described a digital droplet PCR (ddPCR) method to detect AR-V7 mRNA in EVs isolated from small plasma samples (1–2mL). In this study, exoEasy spin columns (Qiagen, Valencia, CA) were used for EV isolation, RNA was extracted from bound EVs using a QIAzol phenol/guanidine-based lysis solution, and ddPCR to analyze AR-V7 was performed using the One-Step RT-ddPCR kit and endogenous full-length AR was used as an internal control. The CV for this assay ranged from 3.1% to 36% as target abundance ranged from 1.5 to 448 copies/mL. The authors measured EV-associated AR-V7 expression in 26 patients who received abiraterone and 10 who received enzalutamide. Among this population, 14 patients were AR-V7-positive. Median progression-free survival (PFS) was 20 months in AR-V7-negative patients, who did not reach the mortality threshold for median overall survival (OS) after 40 months of follow-up. These values significantly differed from the median 3 month PFS and 8 month OS observed in AR-V7-positive patients (p < 0.001). This OS data is similar to that in a previous study using a CTC-based method to detect AR-V7 91. However, there was a larger difference in median PFS between AR-V7-negative and -positive patients using the EV method and the CTC method between these studies. Larger studies are warranted to determine if the EV method has higher sensitivity to detect AR-V7-positive status in the same patient population.
One clinical trial (NCT03236688) is recruiting 30 patients to demonstrate the ability to detect AR-V7 transcripts from EVs in plasma drawn from mCRPC patients before and after treatment with selective Androgen pathway inhibitors (abiraterone and enzalutamide), using the PSA response rate in AR-V7 positive patients as the primary outcome measure. This study is sponsored by EV Diagnostics, Inc., which has presented a poster on its preliminary method development 99 that employs a proprietary spin column methodology to extract total EV RNA from 0.5–4 mL of plasma for subsequent detection of full-length AR and AR-V7 by qPCR. The LOD of this method was reported to be 4 copies/reaction, and the AR-V7 status was determined in 6 PCA patients using this assay.
PC:
PC is the third leading cause of cancer-related death in the US, with a 5-year survival rate of only 9% 100. Biomarkers for early detection, prognosis, and treatment monitoring are crucial for PC patient management due to the aggressiveness of this disease and its frequent late diagnosis. Various EV-associated biomarkers have been reported for early detection (screening), diagnosis, treatment monitoring, metastasis burden, prediction, and prognosis in small cancer patient populations.
Despite the accumulating number of reports that have shown proof-of-concept results for EV-associated PC biomarkers (Table 2), Glypican-1 (GPC1) has thus far been the only EV candidate that has entered clinical trials for evaluating its performance as a clinical biomarker.
GPC1+ EV studies and clinical trials
One highly-cited EV biomarker study identified GPC1 as an EV-associated marker for pancreatic ductal adenocarcinoma (PDAC), reporting that circulating level of GPC1+ EVs demonstrated excellent clinical performance for early detection of PDAC 38. In this study, serum EVs were isolated by UC and bound to EV capture beads, which were then hybridized with a fluorescently labeled anti-GCP1 antibody and analyzed by flow cytometry to determine the frequency of circulating GPC1+ EVs. Reasonable numbers of patients were included for both the discovery (190 PDAC) and validation (56 PDAC) cohorts in a blind clinical validation study, which found that circulating GPC1+ EV abundance exhibited 100% sensitivity and specificity for early detection of PDAC. Circulating GPC1+ EV level was also found to be a promising indicator of disease prognosis and metastatic disease burden, and for monitoring surgery responses. Further studies are required to confirm these results, particularly since the diagnostic performance of this EV biomarker significantly outperforms the performance of other cancer biomarkers. To date, subsequent studies have yielded mixed results about the diagnostic potential of GPC1+ EVs, although most such studies have analyzed GPC1 expression levels in total EV samples isolated from plasma or serum samples rather than the relative abundance of GPC1+ EVs among the total circulating EV population.
For example, one subsequent study employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantitatively measure GPC1 protein concentration in UC-isolated plasma EVs, since the authors observed that highly specific antibodies to GPC1 are not readily available 39. This study observed that EV GPC1 levels did not significantly differ among patients with PDAC or chronic pancreatitis (CP) and NCs, or between pre- and post-surgical resection samples of the same PDAC patients (N=3/group). However, another study that employed ELISA to measure GPC1 levels in UC-isolated plasma EVs found there was a significant difference between matched pre- and post- surgical resection plasma samples from 11 patients, although no significant differences in EV GPC1 levels were detected between a group of 27 patients with PDAC and 16 patients with different forms of BPD 40. Correspondingly, the AUC value for discriminating PDAC from BPD in this study was only 0.59, with a sensitivity of 74% and specificity of 44%.
Two recent studies more closely resemble the original study methodology. One study employed an ACE chip assay to directly analyze GPC1 on the surface of circulating EVs 41. This study found that mean GPC1 expression of total EVs from PDAC patients was significantly higher than that of NCs, but not patients with various BPD diagnoses. A bivariate model incorporating EV surface expression of GPC1 and the EV-associated surface protein CD63 exhibited an AUC of 0.99, such that a probability cut-off point of 0.7 yielded 94% sensitivity and 91% specificity to discriminate samples from PDAC patients and NCs. A similar analysis performed with PDAC and BPD patient samples, yielded an AUC of 0.79 with 81% sensitivity and 70% specificity to distinguish these groups. However, the clinical validation component of this study was performed with a small patient population (20 PDAC, 7 BPD and 11 NC), limiting confidence in these results. A more recent study that employs methodology that more closely resembles that of the original GPC1 EV study found that GPC1+ EV levels were significantly increased in purified plasma EVs from patients with advanced PC versus those from age- and gender-matched NCs 42. This study also found that plasma GPC1+ EV levels in matched patient samples significantly decreased following radiotherapy, and a greater decrease in GPC1+ EV level following therapy was associated with improved median OS rate.
All four of these studies focused on GPC1 in PC, but with different study designs, sample sizes, and methodologies. The original study 38 employed flow cytometry to measure the number of GPC1 positive EV particles, while the subsequent studies employed LC-MS/MS, ELISA, and the ACE chip to measure EV GPC1 protein concentrations 39–41. One flow cytometry study used a similar analytical approach but employed a different patient population, analyzing the association of GPC1+ EV with advanced rather than early PC disease. This lack of a standard analytical approach makes it impossible to directly compare the results of these studies, and illustrates a fundamental issue in the development of EV biomarkers: the lack of useful replication studies.
One clinical trial study (NCT03032913) has completed its recruitment to evaluate and compare diagnostic accuracy of CTCs and GPC1+ EV quantification for PC diagnosis, with a target enrollment of 20 PDAC and 20 non-cancer patients. This study will compare several methods (not specified) for tumor cell recovery and the best method will be used to detect and enumerate CTCs. GPC1+ EVs from patient plasma samples will also be quantified by flow cytometry. This study has not yet published any results. Another on-going study (NCT03410030) was designed to test if a treatment regimen using a combination of protein bound paclitaxel, gemcitabine, cisplatin and high dose ascorbic acid is safe and effective in untreated metastatic PC, using an estimated 36 recruited study subjects. GPC1+ EVs were proposed as one of the potential blood biomarkers to be evaluated in the blood samples of these trial participants. However, the samples sizes in both these studies is rather small, and neither study has specified the details of its methodology for EV purification or quantification.
Other PC EV biomarker clinical trials
Additional clinical trials related to EV biomarkers for PC are exploratory and do not describe specific EV markers. For example, a study (NCT03334708) that will recruit 750 participants, with the goal of developing a minimally invasive test for early diagnosis of PC and treatment monitoring, will analyze various blood-based biomarkers, including proteins and proteases, functional DNA repair assays, EVs, stromal elements, circular RNAs and circulating tumor DNA (ctDNA). Another study (NCT02393703) titled “Interrogation of EV-mediated Intercellular Signaling in Patients With PC” will recruit 70 participates, and purify and analyze exosomes for downstream applications such as proteomics and RNA sequencing, and investigate whether EV activity has a connection to disease recurrence and patient outcomes.
Lung Cancer:
Lung cancer remains the leading cause of cancer deaths worldwide. 80–85% of lung cancer cases are non-small cell lung cancer (NSCLC). Several reviews 101–105 have summarized lung cancer-related EV biomarkers reported for diagnosis, prognosis, prediction and treatment selection, including various miRNAs, RNA, DNA and proteins.
Targeted therapy and immune checkpoint inhibitor (ICI) therapy have made great progress in improved the effective treatment of lung cancer. Diagnostic assays are being specifically developed to identify the right lung cancer patient population for specific therapy. To date, clinical trials have been designed to analyze specific genetic abnormalities and programmed cell death-1 protein ligand (PDL1) expression in EVs to guide targeted therapy and immunotherapy.
T790M epidermal growth factor receptor (EGFR)
Numerous targeted therapies have recently been developed for lung cancer, and management strategies for lung cancer patients have improved and gradually shifted to personalized therapies that are based on the detection of specific mutations. The first such important oncogenic target identified for NSCLC was the presence of EGFR gene mutations. The reported prevalence of such EGFR mutations varies, ranging from about 40% in Asian patients with NSCLC to 11–17% in Caucasian NSCLC patients 106. The development of the first and second-generation EGFR tyrosine kinase inhibitors (EGFR-TKIs) such as erlotinib, gefitinib and afatinib has greatly improved the treatment of NSCLC patients with kinase domain EGFR mutations, although about 40–60% of these patients acquire drug resistance after treatment 107. The T790M missense point mutation in EGFR exon 20 is the most common mechanism of drug resistance to the first and second-generation EGFR-TKIs. NSCLC patients with the T790M mutation respond well to the third-generation TKIs, therefore detecting the T790M mutation during first and second-generation TKI treatment will aid in guiding treatment selection to a third generation EGFR-TKI.
Initial and repeated invasive tissue biopsy for molecular testing is not always possible and for many cases such tissue biopsies are challenging due to comorbidities, insufficient tumor tissue, safety concerns, and cost. Liquid biopsy using ctDNA has emerged as a noninvasive strategy 108 and many clinical laboratories and companies now offer molecular diagnostics for detection of specific mutations using ctDNA. However, these tests reveal wide variations in their reported sensitivities (21%−100%) and specificities (60–100%) 108. Results from a meta-analysis 109 have reported sensitivity of 67% and specificity of 80%. The low sensitivity of ctDNA assays may relate to the low amount of ctDNA available for analysis that may yield target copy numbers that are too low to reproducibly reach the LODs of technologies employed in these analyses, including NGS, qPCR, and ddPCR.
EVs are a significant source of cell-free circulating nucleic acids. Recently, methods have been developed to co-isolate the major sources of circulating nucleic acid (ctDNA and EV RNA and DNA; together called exoNA) from the plasma of NSCLC patients, using a proprietary spin-column extraction technology (Exosome Diagnostics, Inc.) 110, 111. This exoNA is then reverse-transcribed and the mixture of ctDNA and EV cDNA and DNA is analyzed using NGS or qPCR, as decried below.
In the TIGER-X (NCT01526928) study 110, 84 patients were enrolled from a population of 548 patients. This study is a phase 1/2 trial of rociletinib in NSCLC patients who are known to have the T790M EGFR mutation and to have previously failed treatment with a thymidine kinase inhibitor such as erlotinib, gefitinib or afatinib. A MiSeq System (Illumina CA) was utilized to analyze the exoNA (EXO1000). In contrast, the ctDNA extraction and analysis was performed in different laboratories by BEAMing (Sysmex Inostics GmbH, Germany) and the cobas® EGFR mutation test v2 (Roche Molecular Systems, Inc.), the later has been approved by the US FDA. The status of the clinical laboratory certification in this study was not specified. In subgroup A (n = 56), which is representative of the TIGER-X study population, the sensitivities of exoRN (EXO1000) and ctDNA (BEAMing) for T790M were 90% and 84%, respectively. The sensitivities of exoRN (EXO1000) and ctDNA (BEAMing) for EGFR L858R or del19 were 98% and 82%, respectively. The difference in sensitivity between co-isolating exoNA and ctDNA (BEAMing) alone is larger for EGFR L858R or del19 compared to T790M. In a subgroup (N=50) enriched for low-copy plasma ctDNA samples, the sensitivity was higher using exoNA (EXO1000) (81% compared with 58% for ctDNA (BEAMing)). Interestingly, in a subset of 22 cases, the sensitivities of exoRN and ctDNA (cobas) for T790M were 72% and 67%, respectively. These detection rates are much lower than those in the subgroup A but comparable to the previously reported rates in the meta-analysis. The difference between exoRN (EXO1000) and ctDNA (cobas) is small, if unremarkable. As mentioned above, there were big variations of reported sensitivities using different ctDNA platforms in different studies. Unfortunately, the manuscript does not provide detailed information of analytical performance of the assays used in this study. The observed difference in sensitivity could be due to various factors such as nucleic acid extraction methods as suggested by the authors or different analytical performance of the assays used for mutation analysis. The landscape of the EV diagnostic tests is complicated by the fact that each test has its own EV extraction system and mutations are anlayed using different platforms. To demonstrated the superiority of exoNA, the same analysis platform, eg. exoRN (EXO1000) or ctDNA (EXO1000) should be used to make an apple to apple comparison.
A larger study 111 from the same group included biopsy-confirmed 102 T790M-positive and 108 T790M-negative samples, where the T790M mutation status was determined using an analytically validated qPCR assay in a CLIA-certified clinical laboratory, and analytical validation was performed to establish precision, LOD, and analytical specificity. The T790M mutation frequency in exoNA achieved sensitivity of 92% sensitivity and 89% specificity in this study, and showed higher sensitivity and specificity to the meta-analysis of ctDNA alone. However, as aforementioned, a direct comparison using the same qPCR between exoNA and ctDAN is warrant.
Currently, one clinical trial (NCT03236675) is recruiting 60 participants to demonstrate the feasibility of detecting either EML4-ALK gene rearrangements or the T790M EGFR mutation in plasma EVs isolated from patients with advanced NSCLC. This trial refers to an “institutionally accepted assay” but does not provide more detail. Another clinical trial (NCT03228277) is a phase 2 study to assess the efficacy of olmutinib in 25 NSCLC patients with the T790M mutation, where mutation status will be confirmed using DNA extracted from EVs present in bronchoalveolar lavage fluid.
PD-L1
PD-L1 is a classical immune surface protein and a ligand of the programmed death receptor 1 (PD-1) protein, a cell-surface receptor present on certain immune cells, including T, B and NK cells. Many cancer cells also express PD-L1 and the PD-1/PD-L1 interaction between tumor and immune cells can inhibit the anti-tumor function of immune cells and enable these tumors to evade immune recognition. ICI therapies targeting the PD-1/PD-L1 pathway have the potential to advance the treatment of cancer. However, reliable methods are required to identify cancer patients who are likely to respond ICI therapy. According to a recent systematic literature review 112, most of the published validation data for PD-L1 tests relates to immunohistochemistry tests in the context of lung cancer. There is also significant variability and lack of standardization in types of samples (biopsy or surgical-resection), types of antibodies, types of cells tested, test cut-offs, and scoring algorithms. The discussion of PD-L1 immunohistochemistry assays is beyond of the scope of this review and can be found in other review articles 104, 112, 113.
Whether PD-L1 expression is enhanced in cancer cell-derived EVs, whether it plays a role in immune surveillance, and whether it can function as a biomarker for patient treatment stratification and tumor progression has been unclear until recently when these points were addressed in three EV studies examining EV-derived PDL1 in cell culture, a mouse model and cancer patients, including patients with head and neck cancer (HNSCC), melanoma, or NSCLC.
Using cell culture and a mouse model, Yang et al. 114 reported the presence of PD-L1 protein in EVs in the cell culture media of breast cancer cells and mouse mammary tumor cells. This study found that EV-derived PD-L1 could bind PD-1 to inhibit T-cell mediated cytotoxicity, that EVs could transport PD-L1 from PD-L1-positive to PD-L1-negative breast cancer cells, and that treatment of mouse tumor models with the EV secretion inhibitor GW4869 enhanced the efficiency of an anti-PD-L1 antibody mediated immunotherapy.
Theodoraki and colleagues identified PD-L1 expression in plasma EVs of HNSCC patients 115. EV was isolated by mini size-exclusion chromatography from plasma of 40 HNSCC patients. Purified EVs were captured on beads using anti-CD63 antibodies, stained for PD-1 and PD-L1 and analyzed by flow cytometry. The authors demonstrated that PD-L1 levels on EVs, but not PD1 on EVs or soluble PD-L1, corrected with HNSCC patients’ disease progression.
Marzia Del Re et al. 116 demonstrated plasma EV PD-L1 mRNA level changes during the ICI therapy and is significantly associated with response to treatment in melanoma (n=18) and NSCLC (n=8). The EV isolation and RNA extraction was as previously described methods 98. ddPCR was employed to analyze PD-L1 mRNA. Human ß-actin ddPCR assay was used as internal control. EV PD-L1 mRNA levels in plasma significantly decreased in patients responding to treatment and increased in patients with disease progression at 2 months compared with baseline. There was no significant changes in patients with stable disease.
Two clinical trials (NCT02890849 and NCT02869685) have been registered but not recruiting yet focusing on EV PDL1 in NSCLC. The studies expect that the detection of PD-L1 mRNA in plasma EVs should be simple, rapid, non-invasive. The first trial (NCT02890849) is designed to explore the consistency of PD-L1 protein expression level in tissues and mRNA levels in plasma EV of 60 NSCLC patients. The second trial (NCT02890849) will measure the match rate of tissue PD-L1 protein expression and plasma EV PD-L1 mRNA level before and after radiotherapy of 60 NSCLC patients.
Breast Cancer:
Several recent reviews 117–119 have summarized EV biomarkers associated with breast cancer, including HER2, CD47, DEL-1, GSTP1, TRPC5, TRPC5, NANOG, NEUROD1, HTR7, KISS1R, HOXC, KDR, CD49d, CXCR4 CD44 and various miRNAs (e.g., miR-1246, miR-21, miR-340–5p, miR-17–5p, miR-130a-3p, miR-93–5p). Four clinical trials (NCT02514681, NCT02971761, NCT02892734, and NCT01344109) are set to study EV biomarkers in blood, plasma, or serum. However, these studies are designed for treatment evaluation or as pilot studies for biomarker discovery with no specific EV targets mentioned. One study (NCT02514681) will recruit 370 patients to participate in a phase III trial of pertuzumab retreatment in previously pertuzumab, trastuzuamb and chemotherapy treated Her2-positive metastatic advanced breast cancer. One of the secondary outcome measures is to find prognostic and predictive biomarker markers for patients receiving anti-HER2 treatment. The study design indicates that miRNA expression in plasma EVs is one of the potential biomarkers slated for evaluation, but does not specify which miRNAs are to be analyzed as biomarkers. Two small phase II studies (NCT02971761 and NCT02892734) will recruit < 30 breast cancer patients each to evaluate differences in treatment efficacy and safety. One of the tertiary objectives of NCT02971761 is to measure the temporal profile of tumor-derived EVs, while NCT02892734 will evaluate ctDNA and EV-associated immune signatures.
Regulatory requirements and end user utilization
After the analytical and clinical evidence for the performance and utility of a clinical assay has been established, the validated assay must comply with a set of rules and regulations before it can move into commercialization. Commercially distributed clinical laboratory assays intended for diagnostic use in the US are considered to be in vitro diagnostics (IVDs) and their development and marketing are regulated by the US FDA. The specific type of regulatory requirements applied for such tests is determined by the specific device classification. IVDs are classified into three regulatory classes (Class I, II, or III) based on the risk they pose to patients (low-, moderate-, and high-risk, respectively), with the stringency of regulatory requirements increasing from Class I to Class III. Many low-risk devices are deemed exempt from pre-market review and can be legally marketed upon registration alone, but most moderate- and high-risk devices must receive FDA authorization, after complying with a series of regulatory requirements, before they can be legally commercialized in the US market. No EV biomarker assay has been approved by the US FDA, and the regulatory approval processes can be very complicated and challenging for this type of assay.
Under the FDA guidelines, LDTs are the subset of IVDs intended for clinical use that are designed, manufactured, and used in a single laboratory, as opposed to other IVDs that are made by a conventional manufacturer and used by many laboratories. Many LDTs serve increasingly critical roles for patient care and some involve innovative and/or advanced assay approaches, including mass spectrometry and second generation sequencing. LDTs as medical devices are subject to the United States Federal Food, Drug, and Cosmetic Act (FD&C Act), although the FDA has generally exercised enforcement discretion toward LDTs.
In the US, LDTs are subject to regulation by the Centers for Medicare & Medicaid Services (CMS) under the CLIA, which established quality standards for laboratories conducting testing on human specimens for the purpose of providing information for the diagnosis, prevention, treatment of disease, or on the impairment or assessment of health. Laboratories performing these types of tests, including LDTs, must be certified by CMS-approved accreditation organizations. There are now examples of EV cancer assays which are performed (e.g., EPI by Exosome Diagnostic, Inc.) or are under development as LDTs in CLIA-certified clinical laboratories. However, even though the FDA generally does not enforce the applicable provisions under the FD&C Act and FDA regulations, clinical laboratories which develop and perform LDTs are still required by CLIA and state-specific regulations to conduct rigorous analytical and clinical validation.
EV Assay standardization
The goal of testing standardization is to generate accurate test results regardless of the laboratory, the analytical system, the location, or the time of testing. Equivalent laboratory test values are crucial to allow application of consistent standards of patient care and disease management. This is extremely challenging for EV assays, due to the lack of a gold standard method and standard EV materials for method calibration. Efforts have been made to standardize critical parameters of EV assays, such as blood collection, sample type, and sample processing and storage, etc. 63–65, 68, 120, 121. For EV quantification, however, there is still no consensus on how to define EV dosage, whether it should be number of vesicles, protein amount, or a vesicle number to protein ratio 15. There is still a critical need to standardize EV isolation and quantification methods. It would also be extremely valuable to have generic biological standards for EVs, or EV subtypes, to calibrate different assay platforms. Although it should be recognized that EV composition and/or characterization could differ by source and cancer types.
Conclusions
As illustrated in this review, there is considerable activity focused on the development of EV assays for clinical applications. However, we emphasize that each phase listed in Fig. 2 is essential for the ultimate development of a successful EV assay. The critical question is which candidate EV biomarker from a large pool of biomarker pipelines justifies the significant investment of time and money required to complete the entire assay development process. We reiterate that rigorous study design and validation is essential, since a successful EV biomarker must address essential criteria in each of the development phases.
Table 1.
Glossary
| Analytical validation | Assessment of performance characteristics of an assay, including accuracy, precision, specificity, limits of detection and quantitation, linearity and range, ruggedness, and robustness |
| Analytical accuracy | Closeness of agreement between a measured quantity value and a true quantity value of a measurand |
| Analytical sensitivity | Lowest analyte concentration reliably determined as nonzero with a minimum reliably detectable level |
| Analytical specificity | Ability of a measurement procedure to measure solely the measurand |
| Biomarker | A characteristic or compound that is objectively measured and evaluated as an indicator of normal biologic or pathogenic processes or pharmacologic responses to a therapeutic intervention |
| Clinical validation | The process through which one shows that test results are clinically meaningful, ie, finding whether the test is able to detect or predict the disorder or condition of interest in targeted patient groups |
| Clinical accuracy (Diagnostic accuracy): | The ability of a diagnostic test to discriminate between diseased and non-diseased subjects, or between two or more clinical states |
| Clinical sensitivity (Diagnostic sensitivity) | The proportion of patients with well-defined clinical disorders whose test values are positive or exceed a defined decision limit (eg, a positive result and identification of the patients who have a disease) |
| Clinical specificity (Diagnostic specificity) | The proportion of subjects who do not have a specified clinical disorder whose test results are negative or within the defined decision limit |
| Decision Level (cut-off level) | A test value or statistic that marks the upper (or lower) boundary between diagnostic categories, ie, between negative (acceptable or unaffected) results and positive (unacceptable or affected) results |
| Diagnostic test | A measurement or examination of a diagnostic specimen for the purpose of diagnosis, prevention, or treatment of any disease or the assessment of health or impairment of health of an individual patient |
| Extracellular vesicles# | Extracellular vesicles are a heterogeneous group of cell-derived membranous structures comprising exosomes and microvesicles, which originate from the endosomal system or which are shed from the plasma membrane, respectively. |
| Intended use | The clinical use for which the measurement procedure was originally designed |
| In vitro diagnostics (IVD) | Tests done on samples such as blood or tissue that have been taken from the human body. In vitro diagnostics can detect diseases or other conditions, and can be used to monitor a person’s overall health to help cure, treat, or prevent diseases. |
| Laboratory developed test (LDT) | A type of in vitro diagnostic test that is designed, manufactured and used within a single laboratory. |
| Limit of detection (LOD) | lowest amount of analyte (measurand) in a sample that can be detected with (stated) probability, although perhaps not quantified as an exact value |
| Precision | Closeness of agreement between indications or measured quantity values obtained by replicate measurements on the same or similar objects under specified conditions |
| Quality control (QC) | Operational techniques and activities that are used to fulfil requirements for quality |
| Reference interval (reportable range) | Interval between, and including, the lower reference value limit and the upper reference value limit of the biological reference population |
| Reference method | A methodology that has exact and clear descriptions of the necessary conditions and procedures that provide sufficiently accurate and precise laboratory data for it to be used to assess the validity of other laboratory methods |
| Reportable range | The range of test values over which the relationship between the instrument, kit, or system’s measurement response is shown to be valid |
| Validation | Confirmation, through the provision of objective evidence, that the requirements for a specific intended use or application have been fulfilled. Validation is the establishment and/or confirmation, through extensive testing, of the analytical and/or clinical performance characteristics of the measurement procedure |
Acknowledgements
The work was primarily supported by research funding provided by NIH (U01CA214254, R01HD090927, R01AI122932, R01AI113725 and R21Al126361-01), Arizona Biomedical Research Commission (ABRC) young investigator award and ASU-Mayo seed funds.
Footnotes
Footnotes relating to the title and/or authors should appear here.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
References
- 1.Bu H, He D, He X and Wang K, Chembiochem : a European journal of chemical biology, 2018, DOI: 10.1002/cbic.201800470. [DOI] [Google Scholar]
- 2.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW and Simpson RJ, Nature reviews. Clinical oncology, 2018, 15, 617–638. [DOI] [PubMed] [Google Scholar]
- 3.Ruhen O and Meehan K, Proteomics, 2018, DOI: 10.1002/pmic.201800155, e1800155. [DOI] [PubMed] [Google Scholar]
- 4.Jabalee J, Towle R and Garnis C, Cells, 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalishwaralal K, Kwon WY and Park KS, Biotechnology journal, 2019, 14, e1800430. [DOI] [PubMed] [Google Scholar]
- 6.Marrugo-Ramirez J, Mir M and Samitier J, International journal of molecular sciences, 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, Xu YM, Huang LF and Wang XZ, TheScientificWorldJournal, 2015, 2015, 657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarzenbach H, Expert review of molecular diagnostics, 2015, 15, 1159–1169. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita T, Yip KW, Spence T and Liu FF, Journal of human genetics, 2017, 62, 67–74. [DOI] [PubMed] [Google Scholar]
- 10.Dreyer F and Baur A, Methods in molecular biology, 2016, 1448, 201–216. [DOI] [PubMed] [Google Scholar]
- 11.Greening DW and Simpson RJ, Expert review of proteomics, 2018, 15, 887–910. [DOI] [PubMed] [Google Scholar]
- 12.Hessvik NP and Llorente A, Cellular and molecular life sciences : CMLS, 2018, 75, 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalluri R, The Journal of clinical investigation, 2016, 126, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruivo CF, Adem B, Silva M and Melo SA, Cancer research, 2017, 77, 6480–6488. [DOI] [PubMed] [Google Scholar]
- 15.Xu R, Greening DW, Zhu HJ, Takahashi N and Simpson RJ, J Clin Invest, 2016, 126, 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW and Kersaudy-Kerhoas M, Lab on a chip, 2015, 15, 2388–2394. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Li S, Li L, Li M, Guo C, Yao J and Mi S, Genomics, proteomics & bioinformatics, 2015, 13, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szatanek R, Baran J, Siedlar M and Baj-Krzyworzeka M, International journal of molecular medicine, 2015, 36, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV and Govorun VM, Scientific reports, 2015, 5, 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucchetti D, Fattorossi A and Sgambato A, Biotechnology journal, 2019, 14, e1700716. [DOI] [PubMed] [Google Scholar]
- 21.Brenner AW, Su GH and Momen-Heravi F, Methods in molecular biology, 2019, 1882, 229–237. [DOI] [PubMed] [Google Scholar]
- 22.Juan T and Furthauer M, Semin Cell Dev Biol, 2018, 74, 66–77. [DOI] [PubMed] [Google Scholar]
- 23.Konoshenko MY, Lekchnov EA, Vlassov AV and Laktionov PP, Biomed Res Int, 2018, 2018, 8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vader P, Mol EA, Pasterkamp G and Schiffelers RM, Adv Drug Deliv Rev, 2016, 106, 148–156. [DOI] [PubMed] [Google Scholar]
- 25.Contreras-Naranjo JC, Wu HJ and Ugaz VM, Lab on a chip, 2017, 17, 3558–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thind A and Wilson C, Journal of extracellular vesicles, 2016, 5, 31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He M and Zeng Y, Journal of laboratory automation, 2016, 21, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Niel G, D’Angelo G and Raposo G, Nature reviews. Molecular cell biology, 2018, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- 29.Pepe MS, Feng ZD, Janes H, Bossuyt PM and Potter JD, J Natl Cancer I, 2008, 100, 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepe MS, Li CI and Feng ZD, Cancer Epidem Biomar, 2015, 24, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duffy MJ, Sturgeon CM, Soletormos G, Barak V, Molina R, Hayes DF, Diamandis EP and Bossuyt PM, Clinical chemistry, 2015, 61, 809–820. [DOI] [PubMed] [Google Scholar]
- 32.Goossens N, Nakagawa S, Sun X and Hoshida Y, Translational cancer research, 2015, 4, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ransohoff DF, Nature reviews. Cancer, 2004, 4, 309–314. [DOI] [PubMed] [Google Scholar]
- 34.Diamandis EP, Hoffman BR and Sturgeon CM, Clinical chemistry, 2008, 54, 1935–1939. [DOI] [PubMed] [Google Scholar]
- 35.Sturgeon CM, Duffy MJ, Hofmann BR, Lamerz R, Fritsche HA, Gaarenstroom K, Bonfrer J, Ecke TH, Grossman HB, Hayes P, Hoffmann RT, Lerner SP, Lohe F, Louhimo J, Sawczuk I, Taketa K, Diamandis EP and National B Academy of Clinical, Clinical chemistry, 2010, 56, e1–48. [DOI] [PubMed] [Google Scholar]
- 36.Sturgeon CM, Duffy MJ, Stenman UH, Lilja H, Brunner N, Chan DW, Babaian R, Bast RC Jr., Dowell B, Esteva FJ, Haglund C, Harbeck N, Hayes DF, Holten-Andersen M, Klee GG, Lamerz R, Looijenga LH, Molina R, Nielsen HJ, Rittenhouse H, Semjonow A, Shih Ie M, Sibley P, Soletormos G, Stephan C, Sokoll L, Hoffman BR, Diamandis EP and National B Academy of Clinical, Clinical chemistry, 2008, 54, e11–79. [DOI] [PubMed] [Google Scholar]
- 37.Sturgeon CM, Hoffman BR, Chan DW, Ch’ng SL, Hammond E, Hayes DF, Liotta LA, Petricoin EF, Schmitt M, Semmes OJ, Soletormos G, van der Merwe E, Diamandis EP and National B Academy of Clinical, Clinical chemistry, 2008, 54, e1–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D and Kalluri R, Nature, 2015, 523, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai X, Wang M, McElyea SD, Sherman S, House M and Korc M, Cancer Lett, 2017, 393, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frampton AE, Prado MM, Lopez-Jimenez E, Fajardo-Puerta AB, Jawad ZAR, Lawton P, Giovannetti E, Habib NA, Castellano L, Stebbing J, Krell J and Jiao LR, Oncotarget, 2018, 9, 19006–19013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis JM, Vyas AD, Qiu Y, Messer KS, White R and Heller MJ, ACS Nano, 2018, 12, 3311–3320. [DOI] [PubMed] [Google Scholar]
- 42.Qian JY, Tan YL, Zhang Y, Yang YF and Li XQ, Oncol Lett, 2018, 16, 1253–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z and Hu Y, Nat Biomed Eng, 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J and Lyden D, Nature, 2015, 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, Garcia-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J and Lyden D, Nat Cell Biol, 2015, 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Q, Yuan WB, Sun X, Zhang MJ, Cen F, Zhou SY, Wu WB, Xu YC, Tong LH and Ma ZH, Int J Oncol, 2018, DOI: 10.3892/ijo.2018.4318. [DOI] [PubMed] [Google Scholar]
- 47.Madhavan B, Yue S, Galli U, Rana S, Gross W, Muller M, Giese NA, Kalthoff H, Becker T, Buchler MW and Zoller M, International journal of cancer, 2015, 136, 2616–2627. [DOI] [PubMed] [Google Scholar]
- 48.Que R, Ding G, Chen J and Cao L, World journal of surgical oncology, 2013, 11, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi GK, Deitz-McElyea S, Liyanage T, Lawrence K, Mali S, Sardar R and Korc M, ACS Nano, 2015, 9, 11075–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T, Miyai H, Mizuno H, Kato H, Tsutsumi K, Uchida D, Takaki A, Okada H and Morita M, Oncology reports, 2016, 36, 2375–2381. [DOI] [PubMed] [Google Scholar]
- 51.Xu YF, Hannafon BN, Zhao YD, Postier RG and Ding WQ, Oncotarget, 2017, 8, 77028–77040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ibsen SD, Wright J, Lewis JM, Kim S, Ko SY, Ong J, Manouchehri S, Vyas A, Akers J, Chen CC, Carter BS, Esener SC and Heller MJ, ACS Nano, 2017, 11, 6641–6651. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, Liu B, Su L and Qiu Z, Cancer research, 2018, 78, 4586–4598. [DOI] [PubMed] [Google Scholar]
- 54.Zhou X, Lu Z, Wang T, Huang Z, Zhu W and Miao Y, Gene, 2018, 673, 181–193. [DOI] [PubMed] [Google Scholar]
- 55.Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F and Sano K, Journal of hepato-biliary-pancreatic sciences, 2018, 25, 155–161. [DOI] [PubMed] [Google Scholar]
- 56.Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, Sakatani A, Tanaka K, Nomura Y, Ueno N, Kashima S, Moriichi K, Mizukami Y, Kohgo Y and Okumura T, BMC cancer, 2018, 18, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, Chen K, Zhang Y, Liu H, Gan L, Bi H, Zhen P, Zhu J and Li X, Oncogene, 2018, DOI: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]
- 58.Yang S, Che SP, Kurywchak P, Tavormina JL, Gansmo LB, Correa de Sampaio P, Tachezy M, Bockhorn M, Gebauer F, Haltom AR, Melo SA, LeBleu VS and Kalluri R, Cancer Biol Ther, 2017, 18, 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, Davis G, Kumar T, Katz M, Overman MJ, Foretova L, Fabianova E, Holcatova I, Janout V, Meric-Bernstam F, Gascoyne P, Wistuba I, Varadhachary G, Brennan P, Hanash S, Li D, Maitra A and Alvarez H, Ann Oncol, 2017, 28, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, Gascoyne P, Mulu FC, Stephens BM, Huang J, Wang H, Momin AA, Jacamo RO, Katz M, Wolff R, Javle M, Varadhachary G, Wistuba II, Hanash S, Maitra A and Alvarez H, Ann Oncol, 2018, 29, 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T, Aiello NM, McKenzie L, O’Hara M, Redlinger C, Romeo J, Carpenter EL, Stanger BZ and Issadore D, ACS Nano, 2017, 11, 11182–11193. [DOI] [PubMed] [Google Scholar]
- 62.Sodar BW, Kovacs A, Visnovitz T, Pallinger E, Vekey K, Pocsfalvi G, Turiak L and Buzas EI, Expert Rev Proteomics, 2017, 14, 1073–1090. [DOI] [PubMed] [Google Scholar]
- 63.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH and Hochberg F, J Extracell Vesicles, 2013, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lacroix R, Judicone C, Mooberry M, Boucekine M, Key NS, Dignat-George F and The ISSCW, J Thromb Haemost, 2013, DOI: 10.1111/jth.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW and Thery C, J Extracell Vesicles, 2014, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, Buck AH, de Candia P, Chow FW, Das S, Driedonks TA, Fernandez-Messina L, Haderk F, Hill AF, Jones JC, Van Keuren-Jensen KR, Lai CP, Lasser C, Liegro ID, Lunavat TR, Lorenowicz MJ, Maas SL, Mager I, Mittelbrunn M, Momma S, Mukherjee K, Nawaz M, Pegtel DM, Pfaffl MW, Schiffelers RM, Tahara H, Thery C, Tosar JP, Wauben MH, Witwer KW and Nolte-’t Hoen EN, J Extracell Vesicles, 2017, 6, 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Consortium E-T, Van Deun J, Mestdagh P, Agostinis P, Akay O, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, Bertier L, Berx G, Boere J, Boukouris S, Bremer M, Buschmann D, Byrd JB, Casert C, Cheng L, Cmoch A, Daveloose D, De Smedt E, Demirsoy S, Depoorter V, Dhondt B, Driedonks TA, Dudek A, Elsharawy A, Floris I, Foers AD, Gartner K, Garg AD, Geeurickx E, Gettemans J, Ghazavi F, Giebel B, Kormelink TG, Hancock G, Helsmoortel H, Hill AF, Hyenne V, Kalra H, Kim D, Kowal J, Kraemer S, Leidinger P, Leonelli C, Liang Y, Lippens L, Liu S, Lo Cicero A, Martin S, Mathivanan S, Mathiyalagan P, Matusek T, Milani G, Monguio-Tortajada M, Mus LM, Muth DC, Nemeth A, Nolte-’t Hoen EN, O’Driscoll L, Palmulli R, Pfaffl MW, Primdal-Bengtson B, Romano E, Rousseau Q, Sahoo S, Sampaio N, Samuel M, Scicluna B, Soen B, Steels A, Swinnen JV, Takatalo M, Thaminy S, Thery C, Tulkens J, Van Audenhove I, van der Grein S, Van Goethem A, van Herwijnen MJ, Van Niel G, Van Roy N, Van Vliet AR, Vandamme N, Vanhauwaert S, Vergauwen G, Verweij F, Wallaert A, Wauben M, Witwer KW, Zonneveld MI, De Wever O, Vandesompele J and Hendrix A, Nat Methods, 2017, 14, 228–232. [DOI] [PubMed] [Google Scholar]
- 68.Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, Emanueli C, Gasecka A, Hendrix A, Hill AF, Lacroix R, Lee Y, van Leeuwen TG, Mackman N, Mager I, Nolan JP, van der Pol E, Pegtel DM, Sahoo S, Siljander PRM, Sturk G, de Wever O and Nieuwland R, Circ Res, 2017, 120, 1632–1648. [DOI] [PubMed] [Google Scholar]
- 69.Ridger VC, Boulanger CM, Angelillo-Scherrer A, Badimon L, Blanc-Brude O, Bochaton-Piallat ML, Boilard E, Buzas EI, Caporali A, Dignat-George F, Evans PC, Lacroix R, Lutgens E, Ketelhuth DFJ, Nieuwland R, Toti F, Tunon J, Weber C and Hoefer IE, Thromb Haemost, 2017, 117, 1296–1316. [DOI] [PubMed] [Google Scholar]
- 70.Hrobjartsson A, Emanuelsson F, Skou Thomsen AS, Hilden J and Brorson S, Int J Epidemiol, 2014, 43, 1272–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang S, Tian H, Li X, Jin D, Li X, Kong J, Yang C, Yang X, Lu Y, Luo Y, Lin B, Niu W and Liu T, PLoS One, 2017, 12, e0175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He M, Crow J, Roth M, Zeng Y and Godwin AK, Lab Chip, 2014, 14, 3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanwar SS, Dunlay CJ, Simeone DM and Nagrath S, Lab Chip, 2014, 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P, Yu T, Inci F, Kuo WP, Li LJ, Demirci U and Wang S, Sci Rep, 2017, 7, 46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H and Weissleder R, Nat Commun, 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJ and Trau M, Anal Chem, 2014, 86, 11125–11132. [DOI] [PubMed] [Google Scholar]
- 77.Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, Choi HI, Kim YK and Cho YK, ACS Nano, 2017, 11, 1360–1370. [DOI] [PubMed] [Google Scholar]
- 78.Zhang P, He M and Zeng Y, Lab Chip, 2016, 16, 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao Z, Yang Y, Zeng Y and He M, Lab Chip, 2016, 16, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R and Lee H, Nat Biotechnol, 2014, 32, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doldan X, Fagundez P, Cayota A, Laiz J and Tosar JP, Anal Chem, 2016, 88, 10466–10473. [DOI] [PubMed] [Google Scholar]
- 82.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R and Lee H, Nature Medicine, 2012, 18, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeong S, Park J, Pathania D, Castro CM, Weissleder R and Lee H, ACS Nano, 2016, 10, 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ko J, Hemphill MA, Gabrieli D, Wu L, Yelleswarapu V, Lawrence G, Pennycooke W, Singh A, Meaney DF and Issadore D, Sci Rep, 2016, 6, 31215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sina AA, Vaidyanathan R, Dey S, Carrascosa LG, Shiddiky MJ and Trau M, Sci Rep, 2016, 6, 30460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Donovan MJ, Noerholm M, Bentink S, Belzer S, Skog J, O’Neill V, Cochran JS and Brown GA, Prostate cancer and prostatic diseases, 2015, 18, 370–375. [DOI] [PubMed] [Google Scholar]
- 87.McKiernan J, Donovan MJ, O’Neill V, Bentink S, Noerholm M, Belzer S, Skog J, Kattan MW, Partin A, Andriole G, Brown G, Wei JT, Thompson IM Jr. and Carroll P, JAMA oncology, 2016, 2, 882–889. [DOI] [PubMed] [Google Scholar]
- 88.McKiernan J, Donovan MJ, Margolis E, Partin A, Carter B, Brown G, Torkler P, Noerholm M, Skog J, Shore N, Andriole G, Thompson I and Carroll P, European urology, 2018, 74, 731–738. [DOI] [PubMed] [Google Scholar]
- 89.Nordstrom T, Lindberg J and Eklund M, European urology, 2018, 74, 739–740. [DOI] [PubMed] [Google Scholar]
- 90.Steinestel J, Luedeke M, Arndt A, Schnoeller TJ, Lennerz JK, Wurm C, Maier C, Cronauer MV, Steinestel K and Schrader AJ, Oncotarget, 2015, 6, 12035–12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA and Luo J, The New England journal of medicine, 2014, 371, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, Johnson A, Jendrisak A, Bambury R, Danila D, McLaughlin B, Wahl J, Greene SB, Heller G, Marrinucci D, Fleisher M and Dittamore R, JAMA oncology, 2016, 2, 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Zhu Y, Silberstein JL, Taylor MN, Maughan BL, Denmeade SR, Pienta KJ, Paller CJ, Carducci MA, Eisenberger MA and Luo J, Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2017, 35, 2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lokhandwala PM, Riel SL, Haley L, Lu C, Chen Y, Silberstein J, Zhu Y, Zheng G, Lin MT, Gocke CD, Partin AW, Antonarakis ES, Luo J and Eshleman JR, The Journal of molecular diagnostics : JMD, 2017, 19, 115–125. [DOI] [PubMed] [Google Scholar]
- 95.Markowski MC, Silberstein JL, Eshleman JR, Eisenberger MA, Luo J and Antonarakis ES, JCO precision oncology, 2017, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y, Liu F, Yuan Y, Jin C, Chang C, Zhu Y, Zhang X, Tian C, He F and Wang J, J Proteome Res, 2017, 16, 170–178. [DOI] [PubMed] [Google Scholar]
- 97.Valimaki E, Miettinen JJ, Lietzen N, Matikainen S and Nyman TA, Mol Cell Proteomics, 2013, 12, 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Del Re M, Biasco E, Crucitta S, Derosa L, Rofi E, Orlandini C, Miccoli M, Galli L, Falcone A, Jenster GW, van Schaik RH and Danesi R, European urology, 2017, 71, 680–687. [DOI] [PubMed] [Google Scholar]
- 99.Solinas G, Marchesi F, Garlanda C, Mantovani A and Allavena P, Cancer Metastasis Rev, 2010, 29, 243–248. [DOI] [PubMed] [Google Scholar]
- 100.Siegel RL, Miller KD and Jemal A, CA Cancer J Clin, 2016, 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 101.Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng J and Fan S, Journal of Experimental & Clinical Cancer Research, 2018, 37, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cui S, Cheng Z, Qin W and Jiang L, Lung cancer, 2018, 116, 46–54. [DOI] [PubMed] [Google Scholar]
- 103.Reclusa P, Taverna S, Pucci M, Durendez E, Calabuig S, Manca P, Serrano MJ, Sober L, Pauwels P, Russo A and Rolfo C, Journal of thoracic disease, 2017, 9, S1373–S1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Q, Xu Z, Zheng L, Zhang L, You Q and Sun J, American journal of cancer research, 2018, 8, 1689–1696. [PMC free article] [PubMed] [Google Scholar]
- 105.Huang C, Liu S, Tong X and Fan H, Cancer chemotherapy and pharmacology, 2018, 82, 171–183. [DOI] [PubMed] [Google Scholar]
- 106.Garinet S, Laurent-Puig P, Blons H and Oudart JB, Journal of clinical medicine, 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tanaka K, Nosaki K, Otsubo K, Azuma K, Sakata S, Ouchi H, Morinaga R, Wataya H, Fujii A, Nakagaki N, Tsuruta N, Takeshita M, Iwama E, Harada T, Nakanishi Y and Okamoto I, Oncotarget, 2017, 8, 68123–68130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vendrell JA, Mau-Them FT, Beganton B, Godreuil S, Coopman P and Solassol J, International journal of molecular sciences, 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Passiglia F, Rizzo S, Di Maio M, Galvano A, Badalamenti G, Listì A, Gulotta L, Castiglia M, Fulfaro F, Bazan V and Russo A, Scientific reports, 2018, 8, 13379–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, Brinkmann K, Emenegger J, Grimm DG, Castellanos-Rizaldos E, Goldman JW, Sequist LV, Soria JC, Camidge DR, Gadgeel SM, Wakelee HA, Raponi M, Noerholm M and Skog J, Ann Oncol, 2018, 29, 2143. [DOI] [PubMed] [Google Scholar]
- 111.Castellanos-Rizaldos E, Grimm DG, Tadigotla V, Hurley J, Healy J, Neal PL, Sher M, Venkatesan R, Karlovich C, Raponi M, Krug A, Noerholm M, Tannous J, Tannous BA, Raez LE and Skog JK, Clinical cancer research : an official journal of the American Association for Cancer Research, 2018, 24, 2944–2950. [DOI] [PubMed] [Google Scholar]
- 112.Udall M, Rizzo M, Kenny J, Doherty J, Dahm S, Robbins P and Faulkner E, Diagnostic pathology, 2018, 13, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen X and Zhao B, Bmj, 2018, 362, k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, Cha JH, Hou J, Hsu JL, Sun L and Hung MC, Cell research, 2018, 28, 862–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE and Whiteside TL, Clinical cancer research : an official journal of the American Association for Cancer Research, 2018, 24, 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Del Re M, Marconcini R, Pasquini G, Rofi E, Vivaldi C, Bloise F, Restante G, Arrigoni E, Caparello C, Bianco MG, Crucitta S, Petrini I, Vasile E, Falcone A and Danesi R, British journal of cancer, 2018, 118, 820–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang M, Ji S, Shao G, Zhang J, Zhao K, Wang Z and Wu A, Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico, 2018, 20, 906–911. [DOI] [PubMed] [Google Scholar]
- 118.Jia Y, Chen Y, Wang Q, Jayasinghe U, Luo X, Wei Q, Wang J, Xiong H, Chen C, Xu B, Hu W, Wang L, Zhao W and Zhou J, Oncotarget, 2017, 8, 41717–41733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Halvaei S, Daryani S, Eslami SZ, Samadi T, Jafarbeik-Iravani N, Bakhshayesh TO, Majidzadeh AK and Esmaeili R, Molecular therapy. Nucleic acids, 2018, 10, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hill AF, Pegtel DM, Lambertz U, Leonardi T, O’Driscoll L, Pluchino S, Ter-Ovanesyan D and Nolte-’t Hoen EN, J Extracell Vesicles, 2013, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reiner AT, Witwer KW, van Balkom BWM, de Beer J, Brodie C, Corteling RL, Gabrielsson S, Gimona M, Ibrahim AG, de Kleijn D, Lai CP, Lotvall J, Del Portillo HA, Reischl IG, Riazifar M, Salomon C, Tahara H, Toh WS, Wauben MHM, Yang VK, Yang Y, Yeo RWY, Yin H, Giebel B, Rohde E and Lim SK, Stem Cells Transl Med, 2017, 6, 1730–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]