Abstract
The ability to regulate gene expression in a cell-specific and temporally restricted manner provides a powerful means to test gene function, bypass the action of lethal genes, label subsets of cells for developmental studies, monitor subcellular structures, and target tissues for selective ablation or physiological analyses. The galactose-inducible system of yeast, mediated by the transcriptional activator Gal4 and its consensus UAS binding site, has proven to be a highly successful and versatile system for controlling transcriptional activation in Drosophila. It has also been used effectively, albeit in a more limited manner, in the mouse. While zebrafish has lagged behind other model systems in the widespread application of Gal4 transgenic approaches to modulate gene activity during development, recent technological advances are permitting rapid progress. Here we review Gal4-regulated genetic tools and discuss how they have been used in zebrafish as well as their potential drawbacks. We describe some exciting new directions, in large part afforded by the Tol2 transposition system, that are generating valuable new Gal4/UAS reagents for zebrafish research.
Introduction to the Gal4/UAS Transcriptional Activation System
The yeast Gal4 regulatory protein activates transcription of genes required for utilization of galactose by binding to short, defined DNA sequences upstream of target genes, the so-called upstream activating sequence or UAS.1 Gal4 is a modular 881 amino acid protein that has discrete DNA binding, dimerization, and activator domains, with the latter recruiting transcriptional machinery to adjacent promoters (refer to Traven et al.2). Gal4 binds to DNA as a dimer3 and can bind cooperatively in the presence of multiple, tandem UAS sites for further enhancement of gene expression.4,5 In the absence of galactose, Gal4 is inactive due to the repressor protein Gal 80, which binds to the Gal4 activation domain and thereby inhibits its interaction with the transcriptional machinery.6,7
On account of the modular nature of the Gal4 functional domains, the transcription factor can be modified for a variety of purposes. A truncated protein or “mini-Gal4” was constructed that consists of the Gal4 N-terminal DNA binding domain (contained within residues 1–147)3 joined by a seven amino acid linker to residues 840–881 that comprise the acidic C-terminal activation domain.8 This compact version of the transcription factor still contains the activity of full-length Gal4 and is sufficient to drive transcription in both yeast and Drosophila.9,10 Another valuable Gal4 reagent involves fusion of only the DNA binding domain to the highly acidic activating region of the herpes simplex virus VP16 protein, producing a significantly more potent hybrid transcriptional activator (Gal4-VP16).11 Gal4-VP16 has been widely adopted to express exogenous and endogenous genes of interest at high levels in a number of plant and animal systems.12–14 An additional elegant refinement is the split Gal4 expression system developed for Drosophila.15 In this technique, transcription is restricted to the intersection of expression from two different promoters, with one regulating the Gal4 DNA binding domain fused with a synthetic leucine zipper and the other regulating a complementary leucine zipper fused to the activation domain of Gal4 or VP16. When both components are coexpressed with spatial and temporal overlap, they heterodimerize, reconstituting transcriptional activity and expression of UAS-regulated genes.16
The Versatile Gal4/UAS Tool Box of Drosophila
Ever since the first description of the Gal4/UAS interaction in yeast, the potential of this binary system to manipulate gene expression in vivo has been well appreciated. Importantly, the mechanism for transcriptional activation is conserved throughout eukaryotes, which has enabled variations on the Gal4/UAS approach to be applied in a wide range of organisms and cell culture paradigms.
Following the first demonstration that yeast Gal4-dependent transcriptional activation functions effectively in Drosophila,17 a major advance was its implementation in large-scale enhancer or gene trap screens which was made possible by the controlled mobilization of transposable elements. Enhancer trapping for Drosophila traditionally involved incorporation of a β-galactosidase (lacZ) reporter into P-element vectors containing a minimal promoter element, so that lacZ expression required insertion nearby an endogenous enhancer.18 Taking advantage of P-element transposition, enhancer trap vectors were later designed to integrate the gal4 gene randomly into the fly genome so that adjacent, endogenous regulatory regions would control its transcription.19,20 Spatially restricted patterns of Gal4 expression were revealed through the activation of UAS-regulated “reporter” genes such as lacZ or green fluorescent protein (GFP). Once generated, a transgenic enhancer trap line expressing Gal4 in a particular cell type or tissue-specific manner could be crossed to any UAS line and used as a general resource to drive gene expression in the same spatially restricted manner. Close to 10,000 transgenic “driver” lines that confer specific patterns of Gal4 activity are currently available to Drosophila researchers (http://flymap.lab.nig.ac.jp/getdb.html; http://flystocks.bio.indiana.edu), and other large-scale efforts are underway to create many more.
Drosophila transgenic lines bearing UAS insertions have also been generated using transposition technology. A misexpression screen relied on random P-element–mediated integration of a 14 copy (14 × ) UAS construct that promoted transcription when juxtaposed to an endogenous gene.21 In this method, ectopic activation of gene expression by virtue of an adjacent UAS insertion is limited to cells of interest by employing particular Gal4 driver lines. Researchers using this “modular misexpression” approach mapped nearly 3000 independent insertions of the multicopy UAS element and characterized the phenotypes produced by ectopic expression of each targeted “effector” gene with tissue-specific Gal4 drivers.22 As with Gal4 transgenic lines, the panel of UAS lines driving misexpression of endogenous genes has served as a general resource for Drosophila researchers. A further ingenious application of the modular misexpression approach was the identification of genes that when overexpressed, can suppress or enhance a mutant phenotype, thereby revealing new components of a genetic pathway.22
Modifications of the Gal4/UAS system in Drosophila have become increasingly sophisticated, and have been applied to study all stages of development, as well as adult physiology and behavior. Methods that rely on Gal4/UAS tools in the fly include in vivo visualization of subcellular structure, time-lapse imaging, mapping of axonal projections, single-cell lineage tracing, targeted cell ablation, disruption of synaptic transmission in neuronal subclasses, and partial rescue of mutant phenotypes. Notable technical advances are the plethora of inducible methods that allow conditional regulation of Gal4 activity, and the use of Gal4/UAS in conjunction with other well-established systems such as FLP/FRT, which induces cellular clones by mitotic recombination and enables visualization of single cells within a tissue.23–25 As noted above, the split Gal4 technique also brings a new level of regulatory finesse to transcriptional activation in the fly, but relies on the availability of promoters that have partially overlapping domains of expression.16
Three other general strategies have been used to achieve greater temporal control over Gal4 transcriptional activation in Drosophila while maintaining spatial or tissue specificity: (1) conditional regulation of Gal4 activity by the Gal4 inhibitor Gal80, (2) the introduction of temperature dependency, and (3) the use of chemically inducible Gal4 chimeras.
As described above, the yeast Gal80 protein inhibits Gal4 by binding to its 30 amino acid carboxyl terminal activation domain and blocking transcriptional activation of UAS-regulated genes.7,26 Temporal and regional gene expression targeting (TARGET) technology was developed in Drosophila to enable temporal dissection of the pleiotropic action of genes involved both in neural development and in adult brain functions such as learning and memory.27 In this approach, Gal4-dependent gene expression is developmentally restricted through the use of a temperature-sensitive (TS) mutation of Gal80 adopted from yeast. At permissive temperatures (20°C), Gal80TS binds to Gal4, whereas at restrictive temperatures (30°C), it no longer binds and Gal4-mediated transcriptional activation is recovered.
An alternative to Gal80TS is the use of the wild-type (WT) yeast Gal80 gene under control of a heat shock promoter.25 Using such temperature-dependent methods, gene activity can be restored in correct spatial patterns and in temporally defined periods in a mutant, or alternatively, abolished in WT. In addition to temporal regulation, Gal80 permits a greater refinement of spatially restricted transcription. For example, Gal80 can be used to inhibit Gal4 in only a subset of Gal4-expressing cells using a different promoter that drives expression in a more restricted or partially overlapping pattern.24,25
Two methods have been reported to confer temperature sensitivity directly on the yeast Gal4 transcription factor that are effective in Drosophila. The first is the recent isolation of TS mutations in the DNA binding domain of the Gal4 protein.10 From the analysis of the crystal structure of Gal4 bound to DNA,28 four amino acid residues that contact critical nucleotide repeats within the UAS were identified and randomly mutated by PCR. Each Gal4 variant was tested in a yeast viability assay that relied on Gal4 induction of an essential UAS-regulated gene. Several Gal4TS mutants were found to restore viability at low temperatures (21°C), but not when shifted to higher ones (30°C or 37°C). Remarkably, the two Gal4TS mutants that were the most susceptible to temperature regulation in yeast also showed a strong temperature-dependent response in transgenic flies.10
The second method used for temperature inducibility involves introducing a short protein sequence from yeast within the Gal4 coding sequence, which is removed posttranslationally in an autocatalytic process akin to protein splicing. The inserted amino acid sequence, termed an intein, serves as an “intronic” protein that when removed also ligates the N-and C-terminal polypeptides of the Gal4 protein to regenerate Gal4 activity. To make this system regulatable for the fly, Zeidler et al. (2004) used TS versions of an intein that only self-splice out of Gal4 polypeptide precursors at permissive temperatures.29 At restrictive temperatures, the intein behaves as an insertion mutation, abolishing Gal4 activity. A Gal80-inteinTS, which potentially can block the activity of any Gal4 driver line in a temperature-dependent manner, was also produced. While the use of inteins as disruptors of Gal4 or Gal80 function was convincingly shown,29 this method is somewhat more complex, is not reversible, and, as yet, has not been widely adopted in Drosophila research.
A number of ligand-based technologies have been successfully applied to regulate Gal4 activity in Drosophila, including the tetracycline responsive system of Escherichia coli30,31 and mammalian steroid hormone responsive transcription factors.25,32–34 A notable example is the “GeneSwitch Gal4” system, in which a GAL4-progesterone receptor fusion protein is activated by the synthetic steroid mifepristone (Ru-486).35 The many variations on these general approaches and their particular pros and cons have been reviewed extensively.24,25,36,37 The inducible embellishments to the Gal4/UAS system, which allow gene expression to be regulated at defined developmental stages and in defined cell types in Drosophila, have opened up a whole new range of experimental opportunities. It is now possible to monitor the phenotypic consequences of modulating gene dosage in the same individual over time, to explore later functions of early lethal genes (such as signaling pathways important both in gastrulation and organogenesis), and to dissect precisely when genes are required for a given function, to name but a few. By design, several of these methods, including approaches capitalizing on Cre recombinase and the LexA binary system,38 can be used in conjunction with the preexisting collection of Gal4 driver and UAS effector transgenic lines to activate gene expression with extraordinary control.
Application of Gal4/UAS Regulation to the Mouse
In contrast to the substantial legacy of Gal4/UAS resources in Drosophila, Gal4-mediated regulation of gene activity has been relatively limited in the mouse. Although other binary systems for targeted transgene expression, including tTA/tetO, Cre/loxP, and Flp/FRT, have subsequently achieved more widespread use (refer to Lewandoski39), the Gal4/UAS system was actually one of the first driver/responder systems to be developed for the mouse.40 In this study, a mouse mammary tumor virus (MMTV) long terminal repeat that directed expression of a modified Gal4 to mammary epithelium caused pronounced mammary hyperplasia upon activating transcription of an int-2 target gene through a 4 × UAS repeat fused to a minimal elastase promoter. More recently, stable transgenic lines expressing Gal4 or Gal4-VP16 under the regulation of neural-, prostate-, chondrocyte-, or myeloid-specific promoters were shown to mediate UAS-regulated gene expression in the appropriate tissue-restricted manner.41–44 Further refinements of the Gal4/UAS technology in mice, including steroid hormone-inducible variants of Gal4, have also been used to modulate UAS-regulated transgene expression.45,46 In addition, fusions of the Gal4 DNA binding domain to the ligand binding domain of the thyroid hormone,47 retinoid,48,49 or NR4A250 nuclear receptors enabled mapping of active signaling domains in developing mouse embryos, with either UAS:lacZ or UAS:GFP reporters.
Although there are reports of productive use of Gal4/UAS technology in the mouse, several studies have suggested that Gal4 and/or Gal4-VP16 expression in certain tissues might be associated with toxicity or developmental abnormalities, even in the absence of UAS responder alleles. For instance, when Gal4 was expressed in cardiac myocytes under regulation of the Mhc promoter, it resulted in progressive cardiomyopathy associated with biventricular heart failure and eventual demise.51 Similar tissue-specific abnormalities were reported in mice expressing a Pax6:Gal4-VP16 transgene, in which intracorneal positioning of the lens and microophthalmia were noted.52 In both cases, the reported phenotypes did not appear to result from insertional disruption of endogenous genes, as effects were observed in lines generated from multiple founders. Instead, the incidence and severity of observed phenotypes correlated with levels of Gal4 or Gal4-VP16 expression.
Adoption of the Gal4/UAS System in Zebrafish
In contrast to the fly, the application of Gal4/UAS technology to zebrafish has moved rather slowly. Scheer and Campos-Ortega (1999) pioneered the use of Gal4 in zebrafish and produced the first stable transgenic lines.53 Although they could demonstrate Gal4-dependent transcriptional activation of an independent 5 × UAS-regulated reporter, the levels of gene expression were disappointingly low. Addressing this concern, the Gal4/UAS system was further modified for zebrafish by Köster and Fraser (2001) with both driver and reporter modules introduced into a single-plasmid vector.54 The vector included a Gal4-VP16 fusion protein and GFP under the control of a minimal promoter and the 14 × UAS repeat produced for ectopic misexpression in the fly.21 Highly robust GFP labeling was observed in transient assays following injection into 1–2 cell–stage embryos; however, stable transgenic lines were not recovered and some negative effects on embryonic development were observed.54 Nevertheless, Gal4-VP16 and 14 × UAS constructs originally designed for overexpression schemes in the fly have become standard tools for activating reporters or genes of interest in zebrafish. Several laboratories have generated stable Gal4, mini-Gal4, or Gal4-VP16 driver lines using identified promoters and successfully demonstrated transcriptional activation of reporters such as GFP. Expression was achieved at early embryonic stages with Gal4-VP16 driven from the goosecoid promoter,55 and in a number of differentiated tissues using a variety of cell-type specific promoters.56–60
Other efforts for increasing Gal4 activity in zebrafish and in medaka have involved expressing the gal4 gene under the control of the inducible heat shock promoter hsp70.13,61 At permissive temperatures, overexpression of UAS-regulated genes of interest was accomplished throughout the zebrafish embryo using hsp70:Gal4 transgenic fish.56,57,62 Although this approach yields high levels of temperature-dependent gene expression, it precludes restricting Gal4 activity in a precise spatial manner. Focal induction of heat shock promoter activity by warming selected cells or regions of the embryo with a targeted laser beam63 might overcome this limitation.
Additional methods for temporal regulation of Gal4 activity in zebrafish have combined the transcription factor with hormone-inducible systems. When the ligand binding domain of the glucocorticoid receptor is fused to Gal4 (Gal4-GR), the chimeric protein remains inactive in the cytosol. Following application of the glucocorticoid agonist dexamethasone, it translocates to the nucleus and activates transcription.64 This system is routinely used in Xenopus to regulate transcription factors,65,66 but has had only limited application in transient assays of zebrafish embryos.67 One potential drawback is that perturbation of endogenous glucocorticoid signaling during normal development may preclude the usefulness of Gal4-GR regulation in stable transgenic lines (refer to Esengil and Chen68).
An inducible system for mammalian cells used the arthropod hormone ecdysone69 with the premise that the lack of ecdysone receptor (EcR) orthologs in vertebrates would prevent spurious activity. The EcR-based inducible system was improved upon further for use in plants70 and, recently, was successfully employed for induction of gene expression in zebrafish.71 This strategy involves a chimeric transactivator comprised of Gal4-VP16 fused with the ligand binding domain of the silkworm EcR (GV-EcR). Minimizing the VP16 domain to reduce basal activity and replacing amino acids in the EcR to increase sensitivity to ecdysone agonists optimized the transactivator further. An impressive 1000-fold induction in luciferase activity was recorded when zebrafish embryos were injected with the modified GV-EcR and a UAS:luciferase reporter and treated with an EcR agonist. Further, the system was shown to be reversible by simply removing the larval fish from the agonist.71 While convincing data for agonist-dependent and tissue-specific induction of UAS:GFP were provided using promoters from cardiac or skeletal myosin genes to drive the modified GV-EcR hybrid gene, to date, this method has only been tested in one stable transgenic line.68 The ability to regulate gene expression in a conditional and reversible manner is an important innovation for zebrafish that definitely warrants the production and validation of many more tissue-specific transgenic lines.
Identification of Gal4 Driver Lines by Enhancer/Gene Trapping in Zebrafish
In spite of a growing number of examples in which tissue-specific promoters have been used to direct Gal4 or Gal4-VP16 expression in transgenic zebrafish, this approach has been somewhat limited by the fact that characterization of zebrafish regulatory elements lags behind other species, in part due to challenges with assembly of the genome (refer to Ekker et al.72). As an alternative to discrete promoter/enhancer elements from known genes, several studies have taken advantage of either retroviral or transposon technology for efficient integration of gene-and enhancer-trap vectors into the zebrafish genome. Similar to the fly, novel regulatory elements have been identified on the basis of expression patterns of reporter genes. For example, enhancer detection screens were undertaken in zebrafish that built on experience from large-scale retroviral insertional mutagenesis efforts using sequences derived from the Maloney mouse leukemia virus.73–76 This work revealed a host of regulatory elements demonstrating cell- and tissue-restricted patterns.77,78
Gene- and enhancer-trapping efforts have been further facilitated by the expanding repertoire of transposable elements that are active in the zebrafish genome, including Tol2,79–81 Sleeping Beauty,82,83 and PiggyBAC.84 Both Tol2 and Sleeping Beauty have been utilized to distribute gene and/or enhancer trap vectors randomly throughout the zebrafish genome.79,85,86 Tol2 transposition, in particular, has dramatically increased the frequency of chromosomal integration of exogenous DNA, allowed the recovery of single-copy transgenic insertions and improved germ-line transmission of transgenes.87
Several groups have capitalized on Tol2 transposition to generate expanded collections of Gal4 driver lines.88–90 Our group initially reported the use of Tol2 elements to distribute a self-reporting Gal4-VP16; 14 × UAS:GFP gene/enhancer trap vector throughout the zebrafish genome, with the goal of isolating a diverse group of transgenic Gal4-VP16 driver lines. In a pilot screen, 15 stable lines were recovered that displayed spatially and temporally restricted patterns of GFP expression encompassing a wide variety of tissues, including notochord, brain, spinal cord, retina, muscle, pineal gland, and pancreas. In addition, Gal4-VP16-dependent GFP expression also marked previously uncharacterized cell populations, including a migratory population of ventral cells that ultimately came to reside in the liver capsule.88 The majority of the selected lines were demonstrated to transactivate additional UAS-regulated reporters, confirming that they were functional drivers. Eight Tol2 insertions segregating with GFP expression were mapped to specific chromosomal loci using linker-mediated PCR, and one was associated with a late-onset lethal phenotype.
Two other studies made use of Tol2 transposition with an emphasis on expanding the repertoire of neuron-specific driver lines. Scott et al. (2007) used Tol2 vectors to distribute a Gal-VP16 enhancer trap throughout the zebrafish genome, and assessed the ability of the resulting inserts to activate in trans a UAS:Kaede reporter89 (described in following section on UAS-regulated reporters). Many of the 22 stably recovered lines displayed highly restricted patterns of Gal4-VP16–dependent Kaede expression in discrete forebrain, midbrain, and hindbrain nuclei and small subsets of neurons in both larvae and adults, allowing for new insights on the anatomy, location, and connectivity of individual neurons.
Instead of using Gal4-VP16, for their Tol2 trapping vector Asakawa et al. (2008) coupled the Gal4 DNA binding domain to two smaller acidic domains derived from VP16 (Gal4FF).90 They also produced a number of valuable reporter and effector transgenic lines under the control of a 5 × UAS, including a novel fluorescently tagged effector bearing the tetanus toxin light chain that was previously shown to inhibit synaptic transmission in other organisms.91,92 From screening of a remarkable 121 Gal4FF-independent lines in the presence of this neuronal activity blocker, nine driver lines were found to elicit an abnormal touch response, in what is the first demonstration of a Gal4-based behavioral screen for a vertebrate system. These recent reports highlight the power of the Gal4/UAS approach to reveal neuronal circuitry and function in a brain significantly more complicated than that of a fruit fly.
Generation of UAS-Regulated Reporter and Effector Transgenic Lines
The clarity of the zebrafish embryo and larva makes it an ideal organism for tracking specific cell types in real time using fluorescent reporters. In addition to the more routinely used fluorescent proteins (i.e., GFP, YFP, CFP, dsRed, or mCherry), genes encoding newer variants derived largely from corals, and possessing special photoswitching properties, have also been placed under UAS control in transgenic zebrafish. Two notable examples are Kaede89,93 and Dronpa.94 The former has an emission profile similar to GFP, but undergoes a permanent green to red conversion following exposure to UV light, while the latter can exist stably in two interchangeable states, in either brightly fluorescent or essentially nonfluorescent forms. Photoconversion of Kaede has permitted the visualization of neuronal subpopulations and their processes as well as extended fate mapping of unidentified cell populations88,89,93 and Dronpa has revealed detailed neuronal morphology at single-cell resolution94 in the zebrafish nervous system.
Another valuable application of UAS-regulated reporters is their use as visual markers of subcellular domains or specific organelles. For example, selective labeling of the nucleus in a particular cell type facilitates counting cell numbers, observing proliferation, and tracking cell position over time, providing a valuable reagent in studies of cell migration, morphogenesis, tissue growth, or regeneration. We have begun to assemble a collection of zebrafish UAS lines capable of marking specific subcellular domains in the context of appropriate Gal4 driver lines. For nuclear labeling, we created fused proteins of GFP with various chromatin-associated proteins. To label cell membranes and projections, we modified mCherry by incorporating a C-terminal CAAX domain for prenylation-dependent membrane targeting. To visualize mitochondria, a mitochondrial localization domain from the zebrafish cox5a gene was fused to GFP. Using Tol2-based vectors, we have generated stable lines in which these subcellular markers are expressed under the control of UAS-regulatory elements (Fig. 1). Such transgenic tools, as well as related reagents being generated with the Gateway cloning system,95 provide a powerful way to label and image subcellular structures during developmental processes and in adult tissues. Another exciting prospect is the adaptation of recently described cell cycle–specific, fluorescently tagged proteins for zebrafish transgenesis, which will allow direct monitoring of cell cycle dynamics.96
FIG. 1.
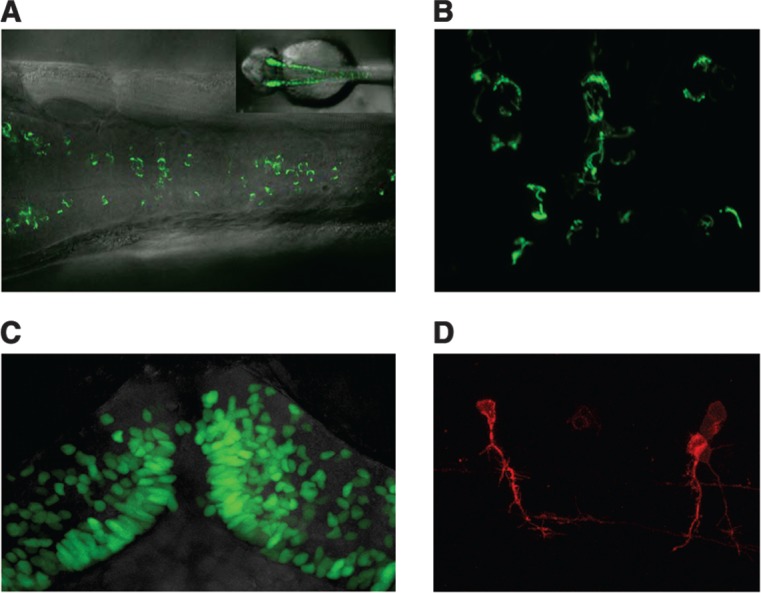
UAS-regulated transgenic tools for subcellular labeling. All embryos were generated by crossing transgenic UAS reporter lines with a stable BAC transgenic line expressing Gal4-VP16 under the control of ptf1a regulatory elements.128 (A, B) Mitochondrial labeling from a 14 × UAS:cox5a-eGFP fusion in the hindbrain (shown at low power in inset in A). (C) Nuclear localization of 14 × UAS:hmgb1-GFP in hindbrain neurons. (D) Membrane-targeted 14 × UAS:mCherry-CAAX expression outlines the morphology of spinal cord interneurons. (A) and (B) are dorsal views with embryos oriented with anterior to the left, (C) is a dorsal view with anterior to the top, and (D) is a lateral view. Constructs and transgenic lines are available upon request.
In addition to utilizing Gal4/UAS technology to image individual cell types and their subcellular organelles, we have also exploited this system to develop methods for ablation of specific zebrafish cell types.88 To create a transgenic line with the dual function of promoting targeted cell ablation, while fluorescently labeling those cells destined for death, we fused the E. coli nfsB gene to mCherry and placed this fusion under UAS transcriptional control to produce the 14 × UAS:nfsB-mCherry transgene.97 The nfsB gene encodes nitroreductase B (NTR), which can convert prodrugs such as metronidazole (MET) into toxic cellular metabolites.98 The NTR fusion protein simultaneously renders cells visible due to mCherry fluorescence and susceptible to prodrug treatment, triggering autonomous cell death. Stable 14 × UAS:nfsB-mCherry transgenics have been crossed onto a wide variety of Gal4 driver lines, and utilized for targeted ablation of cells in a variety of tissues, including notochord, floor plate, and pancreas.88,97
Potential Problems with the Gal4/UAS System in Zebrafish
One reason for the delay in the widespread adoption of the Gal4/UAS system to transgenic zebrafish could be the unregulated method that, up until recently, was the standard practice for creating transgenic animals. Transgenic zebrafish were typically produced following stable integration of plasmid DNA injected into 1–2 cell–stage embryos. The resultant insertions were usually high in copy number and consisted of complex concatemeric arrays.99 It was argued that elevated levels of expression of the Gal4-VP16 activator due to high copy number might also be toxic to early developmental stages in zebrafish.54 Although transposon-mediated integration is expected to alleviate this problem, high-level expression derived from many independent insertions of Gal4 transgenes could remain an issue. Moreover, transposons are obviously mutagenic when integrated within or nearby coding regions.88,100–102 Recovery of multiple independent transgenic founders and the isolation and mapping of unique insertional events should address such concerns.
A more pressing issue that impacts on the production of effective, stable transgenic lines is that they frequently show variegated or diminished expression over time.99 This has been observed from constructs under the control of ubiquitous promoters that nevertheless generate tissue- or cell-specific differences in expression, or in transgenic lines that initially produce high levels of expression but are prone to transcriptional silencing in subsequent generations.103 Initially, it was proposed that the use of promoters from heterologous species in zebrafish might lead to imperfect binding and impaired regulation by endogenous transcription factors, and result in reduced or no transgene expression.99,104 However, variable expression from transgenes has also been reported using zebrafish promoters,105,106 and this phenomenon has been described in other organisms as well (refer to Dorer and Henikoff107). Variegated or mosaic expression is frequently observed when exogenous DNA integrates in close proximity to regions of heterochromatin, such as at centromeres or telomeres.108,109 Transcriptional silencing has been attributed to position effects for a number of transgenes in zebrafish; however, the specific sites of integration were not identified.99,103,110,111 Position effects resulting from integration near heterochromatin have been ruled out as unlikely because of the consistent finding of mosaicism from many independent insertions of the same construct.99 One study did suggest that position effects have an important influence on transgene expression in zebrafish because border elements or insulator sequences isolated from other species seemed to protect insertions from transcriptional silencing.112 A further complication, noted above, is that insertions are usually present in complicated tandem arrays when transgenesis is accomplished by injection of plasmid DNA constructs. Repetitive DNA sequence can itself serve as a trigger for silencing (refer to Dorer113). A rigorous reexamination of the basis for transcriptional silencing of zebrafish transgenes is warranted now that techniques are in place for producing single-copy transgenic insertions by transposition.
Although the Gal4/UAS system is used widely in Drosophila, the issue of mosaic expression is acknowledged, but largely overlooked. Even in the earliest studies it was realized that, as in zebrafish, ubiquitous promoters do not produce equivalent Gal4 activity in all tissues of the fly.17 Variegated expression is observed for many Drosophila Gal4/UAS lines,19 and some lines have been shown to reduce expression further, or suppress variegation completely and show uniformly high expression, when paired with mutations in proteins that influence chromatin structure.114 In examples of Gal4/UAS transgene silencing in other model systems, such as in plants, reduced expression has been correlated with DNA methylation. Progressive loss of expression of a UAS::β-glucuronidase transgene was observed in transgenic tobacco plant lines, despite the presence of active Gal4.115 Unlike the Drosophila genome, the tobacco genome has significant DNA methylation of cytosine and guanine nucleotides separated by a phosphate group (CpG).116,117 In tobacco lines undergoing transgene silencing, UAS elements were indeed found to be methylated, but could be reactivated when plants were grown on the methyl-transferase inhibitor 5-azacytidine.115 Based on the Gal4 crystal structure, methylation of the UAS should prevent Gal4 binding and, accordingly, Gal4 does not bind to methylated DNA in in vitro assays.115 Moreover, CpG methylation was recently found to inhibit Gal4-mediated activation of a UAS-regulated luciferase gene in a human cell culture system.118
Several studies support a link between transgene silencing and DNA methylation in the zebrafish as well,111,119 and the 14 × UAS is a CpG-rich repeated structure that is a prime target for methylation. DNA methylation does play a role in the silencing of UAS-regulated gene expression in transgenic zebrafish, but weakly or nonexpressing UAS-regulated transgenes can be transcriptionally reactivated in the presence of excess Gal4, suggesting that silencing is a reversible process that potentially can be manipulated experimentally (M. Goll, unpublished observations). In addition, although many stable transgenic lines show evidence of progressive transcriptional repression, high expressing larvae can be preferentially selected each generation and used for continued propagation of the line. While mosaic in nature, transient UAS assay systems that rely on injections of driver and responder plasmids or UAS-regulated genes in the context of transgenic Gal4 driver lines can also be useful experimental approaches to bypass the problem of transcriptional repression in stable UAS lines.
Future Prospects for the Gal4/UAS System in Zebrafish
Clearly, an essential requirement for broad applicability and reliability of the Gal4 system in transgenic zebrafish is the sequence optimization and streamlining of the multicopy UAS to reduce its appeal as a target for methylation. Although this may prove challenging because CpG dinucleotides appear necessary for Gal4 binding,28 efforts to test UAS sequence and copy number variants are currently underway and the outcome will likely influence the construction of future UAS-regulated reagents. Other practical directions to increase the utility of the Gal4 system for zebrafish are determining the fidelity of Gal4TS alleles within a temperature range compatible with viability at different developmental stages, and the effectiveness of Gal80 at Gal4 repression in vivo. More work is also needed to validate inducible systems, including extensive examination of hormone-regulatable Gal4 drivers in stable transgenic lines.
Progress in the application of Gal4/UAS technology has occurred in parallel with other significant advances in methods for visualizing and regulating transgene expression in zebra-fish. These include the application of “self-cleaving” viral 2A peptides for the simultaneous and equimolar expression of independent protein products from a single open reading frame,120 and Cre/loxP methodology for conditional transgene activation and inactivation.121,122 A host of additional reverse genetic methodologies await evaluation in zebrafish, including use of phiC31 integrase for targeting transgenes to predesignated chromosomal positions,123 drug-regulated destabilization domains for dynamic regulation of transgenic proteins,124 and Cre/loxP modifications for recombinase-mediated cassette exchange.125,126 In exciting recent work that heralds advances to come, single neurons were marked with exquisite precision in the zebrafish brain by Cremediated excision in combination with Gal4/UAS regulation.59,127 By intersecting state-of-the-art techniques with the ever-expanding battery of zebrafish Gal4/UAS reagents, the ambitious goal of expressing any gene of interest in select zebrafish cell types and within discrete temporal windows is rapidly being achieved.
Acknowledgments
We acknowledge Jose Campos-Ortega and Scott Fraser and their coworkers for their pioneering work on the Gal4/UAS system for zebrafish. We thank Allan Spradling for his helpful advice. M.E.H. was supported by NIH Grant HD42215, M.G.G. by a Damon Runyon Postdoctoral Research Fellowship DRG1945-07, M.P. by the Juvenile Diabetes Research Foundation, and S.D.L. by NIH Grants DK56211 and DK61215 and a grant from the Lustgarten Foundation for Pancreatic Cancer Research.
Note Added in Proof
Asakawa and Kawakami just published online a useful complementary review on the Gal4/UAS system in zebrafish.
Asakawa K, Kawakami K. Target gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ, May 13, 2008 [Epub ahead of print].
References
- 1. Guarente L, Yocum RR, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA 1982;79:7410–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep 2006;7:496–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol 1989;209:423–432 [DOI] [PubMed] [Google Scholar]
- 4. Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 1985;40:767–774 [DOI] [PubMed] [Google Scholar]
- 5. Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature 1987;330:670–672 [DOI] [PubMed] [Google Scholar]
- 6. Johnston SA, Salmeron JM, Jr, Dincher SS. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell 1987;50:143–146 [DOI] [PubMed] [Google Scholar]
- 7. Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 1987; 50:137–142 [DOI] [PubMed] [Google Scholar]
- 8. Wu Y, Reece RJ, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J 1996;15:3951–3963 [PMC free article] [PubMed] [Google Scholar]
- 9. Chakshusmathi G, Mondal K, Lakshmi GS, Singh G, Roy A, Ch RB, et al. Design of temperature-sensitive mutants solely from amino acid sequence. Proc Natl Acad Sci USA 2004;101:7925–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mondal K, Dastidar AG, Singh G, Madhusudhanan S, Gonde SL, VijayRanghavan K, et al. Design and isolation of temperature-sensitive mutants of Gal4 in yeast and Drosophila. J Mol Biol 2007;370:939–950 [DOI] [PubMed] [Google Scholar]
- 11. Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature 1988;335:563–564 [DOI] [PubMed] [Google Scholar]
- 12. Xu L, Schaffner W, Rungger D. Transcriptional activation by recombinant GAL4-VP16 in the Xenopus oocyte. Nucleic Acids Res 1993;21:2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grabher C, Wittbrodt J. Efficient activation of gene expression using a heat-shock inducible Gal4/Vp16-UAS system in medaka. BMC Biotechnol 2004;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Engineer CB, Fitzsimmons KC, Schmuke JJ, Dotson SB, Kranz RG. Development and evaluation of a Gal4-mediated LUC/GFP/GUS enhancer trap system in Arabidopsis. BMC Plant Biol 2005;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 2006;52:425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luan H, White BH. Combinatorial methods for refined neuronal gene targeting. Curr Opin Neurobiol 2007;17:572–580 [DOI] [PubMed] [Google Scholar]
- 17. Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature 1988; 332:853–856 [DOI] [PubMed] [Google Scholar]
- 18. O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA 1987;84:9123–9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993;118:401–415 [DOI] [PubMed] [Google Scholar]
- 20. Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, et al. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn 1997; 209:310–322 [DOI] [PubMed] [Google Scholar]
- 21. Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA 1996;93:12418–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rorth P, Szaboc K, Bailey A, Laverty T, Rehm J, Rubin GM, et al. Systematic gain-of-function genetics in Drosophila. Development 1998;125:1049–1057 [DOI] [PubMed] [Google Scholar]
- 23. Lee T, Luo L. Mosaic analysis with a repressible cell marker ( MARCM) for Drosophila neural development. Trends Neurosci 2001;24:251–254 [DOI] [PubMed] [Google Scholar]
- 24. Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 2002;34:1–15 [DOI] [PubMed] [Google Scholar]
- 25. McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004;2004:pl6. [DOI] [PubMed] [Google Scholar]
- 26. Thoden JB, Sellick CA, Reece RJ, Holden HM. Understanding a transcriptional paradigm at the molecular level. The structure of yeast Gal80p. J Biol Chem 2007;282:1534–1538 [DOI] [PubMed] [Google Scholar]
- 27. McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 2003;302:1765–1768 [DOI] [PubMed] [Google Scholar]
- 28. Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 1992;356:408–414 [DOI] [PubMed] [Google Scholar]
- 29. Zeidler MP, Tan C, Bellaiche Y, Cherry S, Hader S, Gayko U, Perrimon N. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat Biotechnol 2004;22:871–876 [DOI] [PubMed] [Google Scholar]
- 30. Bello B, Resendez-Perez D, Gehring WJ. Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system. Development 1998;125:2193–2202 [DOI] [PubMed] [Google Scholar]
- 31. Stebbins MJ, Yin JC. Adaptable doxycycline-regulated gene expression systems for Drosophila. Gene 2001;270:103–111 [DOI] [PubMed] [Google Scholar]
- 32. Han DD, Stein D, Stevens LM. Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development 2000; 127:573–583 [DOI] [PubMed] [Google Scholar]
- 33. Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA 2001;98:12596–12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA 2001;98:12602–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nicholson L, Singh GK, Osterwalder T, Roman GW, Davis RL, Keshishian H. Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics 2008;178:215–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burcin MM, O'Malley BW, Tsai SY. A regulatory system for target gene expression. Front Biosci 1998; 3:c1–c7 [DOI] [PubMed] [Google Scholar]
- 37. Phelps CB, Brand AH. Ectopic gene expression in Drosophila using GAL4 system. Methods 1998; 14:367–379 [DOI] [PubMed] [Google Scholar]
- 38. Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci 2006;9:703–709 [DOI] [PubMed] [Google Scholar]
- 39. Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet 2001;2:743–755 [DOI] [PubMed] [Google Scholar]
- 40. Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci USA 1991;88:698–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rowitch DH, St. Jacques B, Lee SM, Flax JD, Snyder EY, McMahon AP. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J Neurosci 1999;19:8954–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iyer M, Salazar FB, Lewis X, Zhang L, Carey M, Wu L, et al. Non-invasive imaging of a transgenic mouse model using a prostate-specific two-step transcriptional amplification strategy. Transgenic Res 2005;14:47–55 [DOI] [PubMed] [Google Scholar]
- 43. Kobayashi T, Lyons KM, McMahon AP, Kronenberg HM. BMP signaling stimulates cellular differentiation at multiple steps during cartilage development. Proc Natl Acad Sci USA 2005;102:18023–18027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ovchinnikov DA, van Zuylen WJ, DeBats CE, Alexander KA, Kellie S, Hume DA. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. J Leukoc Biol 2008;83:430–433 [DOI] [PubMed] [Google Scholar]
- 45. Bo J, Yu W, Zhang YM, Demayo FJ, Wei L. Cardiac-specific and ligand-inducible target gene expression in transgenic mice. J Mol Cell Cardiol 2005;38:685–691 [DOI] [PubMed] [Google Scholar]
- 46. Matsumoto T, Kiguchi K, Jiang J, Carbajal S, Ruffino L, Beltran L, et al. Development of transgenic mice that inducibly express an active form of c-Src in the epidermis. Mol Carcinog 2004;40:189–200 [DOI] [PubMed] [Google Scholar]
- 47. Quignodon L, Legrand C, Allioli N, Guadano-Ferraz A, Bernal J, Samarut J, et al. Thyroid hormone signaling is highly heterogeneous during pre- and postnatal brain development. J Mol Endocrinol 2004;33:467–476 [DOI] [PubMed] [Google Scholar]
- 48. Solomin L, Johansson CB, Zetterstrom RH, Bissonnette RP, Heyman RA, Olson L, et al. Retinoid-X receptor signalling in the developing spinal cord. Nature 1998;395:398–402 [DOI] [PubMed] [Google Scholar]
- 49. Mata de Urquiza A, Solomin L, Perlmann T. Feedback-inducible nuclear-receptor-driven reporter gene expression in transgenic mice. Proc Natl Acad Sci USA 1999;96:13270–13275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wallen-Mackenzie A, Mata de Urquiza A, Petersson S, Rodriguez FJ, Friling S, Wagner J, et al. Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev 2003;17:3036–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Habets PE, Clout DE, Lekanne Deprez RH, van Roon MA, Moorman AF, Christoffels VM. Cardiac expression of Gal4 causes cardiomyopathy in a dose-dependent manner. J Muscle Res Cell Motil 2003;24:205–209 [DOI] [PubMed] [Google Scholar]
- 52. Govindarajan V, Harrison WR, Xiao N, Liang D, Overbeek PA. Intracorneal positioning of the lens in Pax6-GAL4/VP16 transgenic mice. Mol Vis 2005;11:876–886 [PubMed] [Google Scholar]
- 53. Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev 1999;80:153–158 [DOI] [PubMed] [Google Scholar]
- 54. Köster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol 2001;233:329–346 [DOI] [PubMed] [Google Scholar]
- 55. Inbal A, Topczewski J, Solnica-Krezel L. Targeted gene expression in the zebrafish prechordal plate. Genesis 2006;44:584–588 [DOI] [PubMed] [Google Scholar]
- 56. Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development 2001;128: 1099–1107 [DOI] [PubMed] [Google Scholar]
- 57. Hans S, Scheer N, Riedl I, Weizsacker E, Blader P, Campos-Ortega JA. her3, a zebrafish member of the hairy-E(spl) family, is repressed by Notch signalling. Development 2004;131:2957–2969 [DOI] [PubMed] [Google Scholar]
- 58. Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and -independent pathways. Neuron 2007;55:231–245 [DOI] [PubMed] [Google Scholar]
- 59. Sato T, Hamaoka T, Aizawa H, Hosoya T, Okamoto H. Genetic single-cell mosaic analysis implicates ephrinB2 reverse signaling in projections from the posterior tectum to the hindbrain in zebrafish. J Neurosci 2007;27:5271–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zecchin E, Filippi A, Biemar F, Tiso N, Pauls S, Ellertsdottir E, et al. Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish. Dev Biol 2007;301:192–204 [DOI] [PubMed] [Google Scholar]
- 61. Scheer N, Riedl I, Warren JT, Kuwada JY, Campos-Ortega JA. A quantitative analysis of the kinetics of Gal4 activator and effector gene expression in the zebrafish. Mech Dev 2002;112:9–14 [DOI] [PubMed] [Google Scholar]
- 62. Jeong JY, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, Guo S. Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development 2007;134:127–136 [DOI] [PubMed] [Google Scholar]
- 63. Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development 2000;127:1953–1960 [DOI] [PubMed] [Google Scholar]
- 64. Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell 1988;54:1073–1080 [DOI] [PubMed] [Google Scholar]
- 65. Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol 1995;171:267–272 [DOI] [PubMed] [Google Scholar]
- 66. Tada M, O'Reilly MA, Smith JC. Analysis of competence and of Brachyury autoinduction by use of hormone-inducible Xbra. Development 1997;124:2225–2234 [DOI] [PubMed] [Google Scholar]
- 67. de Graaf M, Zivkovic D, Joore J. Hormone-inducible expression of secreted factors in zebrafish embryos. Dev Growth Differ 1998;40:577–582 [DOI] [PubMed] [Google Scholar]
- 68. Esengil H, Chen JK. Gene regulation technologies in zebrafish. Mol Biosyst 2008;4:300–308 [DOI] [PubMed] [Google Scholar]
- 69. No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA 1996;93:3346–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Padidam M, Gore M, Lu DL, Smirnova O. Chemical-inducible, ecdysone receptor-based gene expression system for plants. Transgenic Res 2003;12:101–109 [DOI] [PubMed] [Google Scholar]
- 71. Esengil H, Chang V, Mich JK, Chen JK. Small-molecule regulation of zebrafish gene expression. Nat Chem Biol 2007;3:154–155 [DOI] [PubMed] [Google Scholar]
- 72. Ekker SC, Stemple DL, Clark M, Chien CB, Rasooly RS, Javois LC. Zebrafish genome project: bringing new biology to the vertebrate genome field. Zebrafish 2007;4:239–251 [DOI] [PubMed] [Google Scholar]
- 73. Amsterdam A. Insertional mutagenesis in zebrafish: genes for development, genes for disease. Brief Funct Genomic Proteomic 2006;5:19–23 [DOI] [PubMed] [Google Scholar]
- 74. Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA 2004;101:12792–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature 1996;383:829–832 [DOI] [PubMed] [Google Scholar]
- 76. Wang D, Jao LE, Zheng N, Dolan K, Ivey J, Zonies S, et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci USA 2007;104:12428–12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kikuta H, Fredman D, Rinkwitz S, Lenhard B, Becker TS. Retroviral enhancer detection insertions in zebrafish combined with comparative genomics reveal genomic regulatory blocks—a fundamental feature of vertebrate genomes. Genome Biol 2007;8 Suppl 1:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Laplante M, Kikuta H, Konig M, Becker TS. Enhancer detection in the zebrafish using pseudotyped murine retroviruses. Methods 2006;39:189–198 [DOI] [PubMed] [Google Scholar]
- 79. Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn 2005;234:244–254 [DOI] [PubMed] [Google Scholar]
- 80. Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 2007;8 Suppl 1:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene 1998;225:17–22 [DOI] [PubMed] [Google Scholar]
- 82. Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol 2003;263:191–202 [DOI] [PubMed] [Google Scholar]
- 83. Hermanson S, Davidson AE, Sivasubbu S, Balciunas D, Ekker SC. Sleeping Beauty transposon for efficient gene delivery. Methods Cell Biol 2004;77:349–362 [DOI] [PubMed] [Google Scholar]
- 84. Lobo NF, Fraser TS, Adams JA, Fraser MJ., Jr Interplasmid transposition demonstrates piggyBac mobility in vertebrate species. Genetica 2006;128:347–357 [DOI] [PubMed] [Google Scholar]
- 85. Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 2004;5:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn 2004;231:449–459 [DOI] [PubMed] [Google Scholar]
- 87. Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol 2004;77:201–222 [DOI] [PubMed] [Google Scholar]
- 88. Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol 2007;304:811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods 2007;4:323–326 [DOI] [PubMed] [Google Scholar]
- 90. Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA 2008;105:1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 1995;14:341–351 [DOI] [PubMed] [Google Scholar]
- 92. Yamamoto M, Wada N, Kitabatake Y, Watanabe D, Anzai M, Yokoyama M, et al. Reversible suppression of glutamatergic neurotransmission of cerebellar granule cells in vivo by genetically manipulated expression of tetanus neurotoxin light chain. J Neurosci 2003;23:6759–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc 2006;1:960–967 [DOI] [PubMed] [Google Scholar]
- 94. Aramaki S, Hatta K. Visualizing neurons one-by-one in vivo: optical dissection and reconstruction of neural networks with reversible fluorescent proteins. Dev Dyn 2006;235:2192–2199 [DOI] [PubMed] [Google Scholar]
- 95. Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007;236:3088–3099 [DOI] [PubMed] [Google Scholar]
- 96. Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008;132:487–498 [DOI] [PubMed] [Google Scholar]
- 97. Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitror-eductase. Mech Dev 2007;124:218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Drabek D, Guy J, Craig R, Grosveld F. The expression of bacterial nitroreductase in transgenic mice results in specific cell killing by the prodrug CB1954. Gene Ther 1997;4:93–100 [DOI] [PubMed] [Google Scholar]
- 99. Stuart GW, Vielkind JR, McMurray JV, Westerfield M. Stable lines of transgenic zebrafish exhibit reproducible patterns of transgene expression. Development 1990;109:577–584 [DOI] [PubMed] [Google Scholar]
- 100. Sivasubbu S, Balciunas D, Amsterdam A, Ekker SC. Insertional mutagenesis strategies in zebrafish. Genome Biol 2007;8 Suppl 1:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nagayoshi S, Hayashi E, Abe G, Osato N, Asakawa K, Urasaki A, et al. Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: tcf7 and synembryn-like. Development 2008;135: 159–169 [DOI] [PubMed] [Google Scholar]
- 102. Kotani T, Kawakami K. Misty somites, a maternal effect gene identified by transposon-mediated insertional mutagenesis in zebrafish that is essential for the somite boundary maintenance. Dev Biol 2008;316:383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Amsterdam A, Lin S, Hopkins N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev Biol 1995;171:123–129 [DOI] [PubMed] [Google Scholar]
- 104. Culp P, Nusslein-Volhard C, Hopkins N. High-frequency germ-line transmission of plasmid DNA sequences injected into fertilized zebrafish eggs. Proc Natl Acad Sci USA 1991;88:7953–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol 1997;192:289–299 [DOI] [PubMed] [Google Scholar]
- 106. Hackett P, Alvarez MC. The molecular genetics of transgenic fish. Recent Adv Mar Biotech 2000;4:77–145 [Google Scholar]
- 107. Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 1994;77:993–1002 [DOI] [PubMed] [Google Scholar]
- 108. Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 1995;9:1263–1277 [DOI] [PubMed] [Google Scholar]
- 109. Cryderman DE, Cuaycong MH, Elgin SC, Wallrath LL. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 1998;107:277–285 [DOI] [PubMed] [Google Scholar]
- 110. Gibbs PD, Peek A, Thorgaard G. An in vivo screen for the luciferase transgene in zebrafish. Mol Mar Biol Biotechnol 1994;3:307–316 [PubMed] [Google Scholar]
- 111. Thummel R, Bai S, Sarras MP, Jr, Song P, McDermott J, Brewer J, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn 2006;235:336–346 [DOI] [PubMed] [Google Scholar]
- 112. Caldovic L, Agalliu D, Hackett PB. Position-independent expression of transgenes in zebrafish. Transgenic Res 1999;8:321–334 [DOI] [PubMed] [Google Scholar]
- 113. Dorer DR. Do transgene arrays form heterochromatin in vertebrates? Transgenic Res 1997;6:3–10 [DOI] [PubMed] [Google Scholar]
- 114. Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev 2002;16:2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Galweiler L, Conlan RS, Mader P, Palme K, Moore I. Technical advance: the DNA-binding activity of gal4 is inhibited by methylation of the gal4 binding site in plant chromatin. Plant J 2000;23:143–157 [DOI] [PubMed] [Google Scholar]
- 116. Kovarik A, van Houdt H, Holy A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Lett 2000;467:47–51 [DOI] [PubMed] [Google Scholar]
- 117. Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature 2000;408: 538–540 [DOI] [PubMed] [Google Scholar]
- 118. Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. Chimeric DNA methyl-transferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res 2007;35:100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Martin CC, McGowan R. Genotype-specific modifiers of transgene methylation and expression in the zebrafish, Danio rerio. Genet Res 1995;65:21–28 [DOI] [PubMed] [Google Scholar]
- 120. Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 2007;45:625–629 [DOI] [PubMed] [Google Scholar]
- 121. Dong J, Stuart GW. Transgene manipulation in zebrafish by using recombinases. Methods Cell Biol 2004;77:363–379 [DOI] [PubMed] [Google Scholar]
- 122. Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci USA 2007;104:9410–9415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat Biotechnol 2003;21:321–324 [DOI] [PubMed] [Google Scholar]
- 124. Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods 2007;4:1007–1009 [DOI] [PubMed] [Google Scholar]
- 125. Liu WY, Wang Y, Qin Y, Wang YP, Zhu ZY. Site-directed gene integration in transgenic zebrafish mediated by cre recombinase using a combination of mutant lox sites. Mar Biotechnol (NY) 2007;9:420–428 [DOI] [PubMed] [Google Scholar]
- 126. Jones JR, Shelton KD, Magnuson MA. Strategies for the use of site-specific recombinases in genome engineering. Methods Mol Med 2005;103:245–257 [DOI] [PubMed] [Google Scholar]
- 127. Okamoto H, Sato T, Aizawa H. Transgenic technology for visualization and manipulation of the neural circuits controlling behavior in zebrafish. Dev Growth Differ 2008;50:S167–S175 [DOI] [PubMed] [Google Scholar]
- 128. Park SW DJ, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology 2008; in press [DOI] [PMC free article] [PubMed] [Google Scholar]


