Abstract
HIV infection is a major problem in Indonesia. The number of people living with HIV has been increasing from year to year, especially among injecting drug users (IDUs). Since there were only limited data about molecular epidemiology profiles of HIV/AIDS in Indonesia, a cross-sectional study involving 208 HIV-1-seropositive individuals was conducted in 2007 in Jakarta. The majority of participants were 16–30 years of age (64.9%) and 74.5% were male. The most frequent risk factor was injecting drug use (IDU) (45.7%) followed by heterosexual transmission (34.1%). Phylogenetic analysis of gag (p17 and p6) and env C2V3 regions showed 200 (96.2%) of 208 DNA samples were CRF01_AE and only 3 (1.4%) were subtype B. Five samples (2.4%) indicated discordant subtypes between the three aforementioned regions: three of them showed unique CRF01_AE/B recombination patterns in 2.3-kbp nucleotide sequences (from p17 to part of RT), including one sample showing similarity to CRF33_01B, reported previously in Malaysia. This study shows the current predominant subtype is CRF01_AE in every risk group, with a decreasing number of pure subtype B, and the first identification of CRF01_AE/B recombinant forms among HIV-1-seropositive Indonesians.
Introduction
Since the first case was reported in 1986, HIV/AIDS has become one of the most important health problems in Indonesia. The number of people living with HIV is increasing from year to year, and since 1999, there has been a dramatic increase in HIV/AIDS cases.1 The Ministry of Health, Republic of Indonesia, as of March 2008, reported that cumulative HIV/AIDS cases reached 17,998 (11,868 AIDS and 6130 HIV). The highest number of HIV/AIDS cases was found in Jakarta, followed by West Java and Papua.2 The Joint United Nations Programme on AIDS (UNAIDS), however, estimated the number of people living with HIV in Indonesia was 270,000 with a prevalence of 0.2% in 2007, and warned the HIV epidemic in Indonesia was one of the fastest growing in Asia.3
The dramatic increase of HIV/AIDS patients in Indonesia was mostly caused by the increasing number of injecting drug users (IDUs). Among high-risk behaviors, IDU accounts for 49.2% of all cases, followed by heterosexual transmission (42.8%). This condition has been observed in most big cities and areas with high HIV prevalence, with the exception of several areas such as Papua, where 99.7% of HIV/AIDS cases were due to heterosexual transmission.2 However, the situation was different in the early HIV/AIDS period, when the known prevalence was low and most patients were males who had sex with males ( MSM) and the heterosexual group. The unique geographic characteristics of Indonesia also contributed to HIV spreading variably, depending on the culture, habits, and customs of the local people in each area.1,2
HIV-1 group M includes 11 different subtypes, and there are 43 circulating recombinant forms (CRFs) reported so far. In Asia, three major dominant subtypes, C, CRF01_AE, and B, are distributed relatively independently in different areas of the region: subtype C is the most common subtype in India, CRF01_AE is dominant in Southeast Asian countries, and subtype B predominates in Japan, Taiwan, and the Philippines.4 The segregated distribution was also found in different risk factor groups. At the beginning of the HIV pandemic in Thailand, CRF01_AE was distributed mostly among people at risk of sexual exposure, and subtype B′ (Thai B variant) was distributed mainly among IDUs.5,6 However, in current cases, despite the increasing number of IDUs, CRF01_AE remains predominant while the number of circulating pure subtype B cases decreased.7–9
As a retrovirus, HIV-1 mutates readily during the reverse transcription process, leading to a diversification of its strains.10 Intersubtype recombination contributes to HIV-1 sequence diversity, which is generated by cocirculation of two or more different subtypes and/or CRFs in high-risk populations.8,11,12 Cocirculation of subtype B and CRF01_AE in Southeast Asia has led to the emergence of new CRFs such as CRF15_01B and CRF34_01B in Thailand and CRF33_01B in Malaysia.13–15 This apparent evolution continued as evidence indicated the presence of other multiple recombinants related to the previously declared new CRFs.16–18
In Indonesia, previous studies focusing on subtype distribution in HIV-1 patients involved only a small number of subjects, and, therefore, cannot represent the current status of HIV/AIDS in Indonesia.19–20 The present study was conducted to investigate the current situation of HIV/AIDS with respect to its molecular, epidemiological, and clinical aspects, and to gain a better understanding of the possibility of HIV-1 intersubtype recombinant generation in Jakarta, Indonesia.
Materials and Methods
Subjects and specimens
HIV-seropositive patients were selected with random sampling from January to September 2007, and 214 subjects were enrolled in two public hospitals and one private hospital located in Central and North Jakarta. Every patient involved in this study signed an informed consent form for approval to join the study. Ethical clearance was released by the University of Indonesia Ethical Committee for Medical Research. Clinical data were obtained by interviewing each subject and collecting data from medical records. Blood samples were collected during enrollment; plasma and peripheral blood mononuclear cells (PBMCs) were separated by gradient centrifugation using Mono Poly Resolving Medium (Dainippon Pharmaceutical Company, Tokyo, Japan) and stored at −80°C until studied.
PCR amplification and DNA sequencing
DNA was extracted from PBMCs using DNeasy Blood and Tissue Kit (Qiagen Science, Maryland). For genetic subtyping, the gag p17 and env C2V3 regions were amplified by nested polymerase chain reaction (PCR) using Takara ExTaq Hot Start Version (Takara Bio Inc, Otsu, Japan) with the following primer sets: for the gag p17 region the first round primers are 172A/JA152 (sense) and 173B/JA155 (antisense) and the second round primers are 174A/JA153 (sense) and 175B/JA154 (antisense).12,21 For the env C2V3 region, the first round primers are 106A (sense, 5′-CATACATTATTGTGCCCCG GCTGG-3′) and 17B (antisense, 3′-AGAAAAATTCCCCTCTACAATTAA-5′) and the second round primers are 14A (sense, 5′-AATGTCAGCTCAGTACAATGCACAC-3′) and SD (antisense, 3′-AAATTCCCCTCCACAATTAAA-5′). For the p6 region analysis, the first round primers were P7Pr-F1 (sense, 5′-GAAGAAATGATGACAGCATGYCAGG-3′) and Pro-6 (antisense, 3′-ACTTTTGGGCCATCCATTCC-5′) and second round primers were P7Pr-F2 (sense, 5′-GCCATAAA GCAAGRGTTTTGGCTGA-3′) and JA20 (antisense, 3′-CCTGGCTTTAATTTTACTGG-5′), which amplify the gag p7 to protease gene. The second PCR products were purified with Autoseq G-50 (GE Healthcare UK Ltd, Amersham Place, Little Chalfont, England) and then directly sequenced with BigDye Terminator v3.1 Cycle Sequencing Kit and ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
To examine the samples showing different subtypes between the three regions analyzed in this study, 2.3-kbp nucleotide sequences from p17 to part of the RT region were analyzed. Nested PCR was performed using the Expand Long Template PCR System (Roche Diagnostic GmbH, Penzberg, Germany) with the following primers: for first round primers 172A/JA152 and RT2-29 (antisense, 3′-GGCTCTAAGATTTTTGTC-5′) and for second round primers 174A/JA153 and RT2-28 (antisense, 3′-TGGAATATTGCTGGTGATCC-5′). PCR products were TA-cloned using TOPO XL PCR Cloning Kit (Invitrogen, Carlsbad, CA). Positive clones were selected and directly sequenced, using primers designed to sequence overlapping regions.
Sequence and recombinant analysis
Nucleotide sequence alignment and phylogenetic analysis were performed using MEGA version 4.22 Phylogenetic trees were constructed by the neighbor-joining (NJ) method based on the Kimura two-parameter model with 500 bootstrap replicates. Bootscanning analyses for 2.3-kbp fragments were performed using SimPlot version 3.5, and bootstrap values were plotted for a window with a 200 bp.23
Statistical analysis
Statistical analysis for epidemiological and clinical data was performed using Statistical Package for Social Sciences (SPSS) version 16.0 (SPSS Inc, Chicago, IL).
Results
Epidemiological and clinical profile
Of 214 participants enrolled in this study, 208 DNA samples were successfully amplified and sequenced. The epidemiological and clinical characteristics of 208 patients are summarized in Table 1. Subjects were mainly male (74.5%), and the most common risk factor was IDU (45.7%), followed by 34.1% for heterosexual transmission. The age distribution ranges from 2 to 60 years old (mean 29.35 ± 8.2 years old), while the most affected age group was 16 to 30 years old (64.9%), and it seemed to relate to the population of male IDUs (Table 2). IDUs were also dominant among most ethnic groups, except Chinese, among whom heterosexual transmission was more dominant (61.3%). Based on the retrospective data about the initiation period of risk behaviors from 194 subjects (Table 3), the most common risk factor prior to 1991 was sexual transmission. However, in the period from 1991 to 1995, the number of IDUs started to increase, followed by a dramatic increase of IDUs among male subjects. Interestingly, data from 2001 and afterward showed a decreasing number of IDUs, whereas the ratio of heterosexual transmission among female subjects was increasing.
Table 1.
Epidemiological and Clinical Characteristics of 208 HIV-1 Seropositive Indonesian Subjects, Recruited in 2007, in Jakarta, Indonesiaa
| Characteristics | Male (%) | Female (%) | Total (%) |
|---|---|---|---|
| Gender | 155 (74.5)b | 53 (25.5)b | 208 (100) |
| Age group (years) | |||
| <16 | 3 (1.9) | 1 (1.9) | 4 (1.9) |
| 16–30 | 98 (63.2) | 37 (69.8) | 135 (64.9) |
| 31–45 | 48 (31.0) | 12 (22.6) | 60 (28.8) |
| >45 | 6 (3.9) | 3 (5.7) | 9 (4.3) |
| Risk factors | |||
| IDU | 91 (58.7) | 4 (7.5) | 95 (45.7) |
| Heterosexual | 30 (19.4) | 41 (77.4) | 71 (34.1) |
| Mix | 21 (13.4) | 6 (11.3) | 27 (13.0) |
| MSM | 4 (2.6) | 0 (0) | 4 (1.9) |
| MTCT | 3 (1.9) | 1 (1.9) | 4 (1.9) |
| Tattoo | 3 (1.9) | 0 (0) | 3 (1.4) |
| Transfusion | 2 (1.3) | 0 (0) | 2 (1.0) |
| Unknown | 1 (0.6) | 1 (1.9) | 2 (1.0) |
| Ethnics | |||
| Javanese | 35 (22.6) | 16 (30.2) | 51 (24.5) |
| Betawi | 40 (25.8) | 8 (15.1) | 48 (23.1) |
| Chinese | 25 (16.1) | 6 (11.3) | 31 (14.9) |
| Sundanese | 21 (13.5) | 7 (13.2) | 28 (13.5) |
| Manado | 8 (5.2) | 6 (11.3) | 14 (6.8) |
| Batak | 10 (6.5) | 1 (1.9) | 11 (5.3) |
| Minang | 7 (4.5) | 3 (5.7) | 10 (4.8) |
| Ambon | 4 (2.6) | 2 (3.8) | 6 (2.9) |
| Bali | 1 (0.6) | 1 (1.9) | 2 (1.0) |
| Dayak | 2 (1.3) | 0 (0.0) | 2 (1.0) |
| Others | 2 (1.3) | 3 (5.7) | 5 (2.5) |
| Education | |||
| Uneducated | 4 (2.6) | 1 (1.9) | 5 (2.4) |
| Primary school | 1 (0.6) | 0 (0.0) | 1 (0.5) |
| Secondary school | 12 (7.7) | 1 (1.9) | 13 (6.2) |
| High school | 113 (72.9) | 41 (77.4) | 154 (74.0) |
| Professional school | 1 (0.6) | 3 (5.7) | 4 (1.9) |
| University | 23 (14.8) | 7 (13.2) | 30 (14.4) |
| Graduate school | 1 (0.6) | 0 (0.0) | 1 (0.5) |
| Hospital | |||
| Private | 26 (16.8) | 11 (20.8) | 37 (17.8) |
| Public | 129 (83.2) | 42 (79.2) | 171 (82.2) |
| Treatment status | |||
| ARV naive | 56 (36.1) | 32 (60.4) | 88 (42.3) |
| ARV treated | 99 (63.9) | 21 (39.6) | 120 (57.7) |
| CD4 count at enrollment (cells/mm3) | |||
| <200 | 91 (58.7) | 26 (49.1) | 117 (56.2) |
| 201–500 | 51 (32.9) | 19 (35.8) | 70 (33.7) |
| >500 | 13 (8.4) | 8 (15.1) | 21 (10.1) |
| Initial disease stagec | |||
| AIDS | 133 (85.8) | 29 (54.7) | 162 (77.9) |
| ARC | 16 (10.3) | 8 (15.1) | 24 (11.5) |
| HIV | 6 (3.9) | 16 (30.2) | 22 (10.6) |
| History of opportunistic infection | |||
| Tuberculosis | 106 (68.4) | 19 (35.8) | 125 (60.1) |
| Oropharyngeal candidiasis | 94 (60.6) | 19 (35.8) | 113 (54.3) |
| Chronic diarrhea | 76 (49.0) | 13 (24.5) | 89 (42.8) |
| Toxoplasmosis | 33 (21.3) | 4 (7.5) | 37 (17.8) |
| Cytomegalovirus infection | 26 (16.8) | 3 (5.7) | 29 (13.9) |
IDU, injecting drug use; mix, subjects with both IDU and heterosexual risk factors; MSM, male having sex with male; MTCT, mother-to-child transmission; Transfusion, blood transfusion; ARV, antiretroviral; ARC, AIDS-related complex.
Percentage from total number of subjects (n = 208). Other percentage are from each column group (female and male groups).
Initial disease stage is subject's infection stage at the time of HIV diagnosis.
Table 2.
Distribution of Risk Factor among Age Groupa
| Risk factor group | |||||||
|---|---|---|---|---|---|---|---|
| Age group (years old) | IDU n (%) | Heterosexual n (%) | Mix n (%) | MSM n (%) | MTCT n (%) | Others n (%) | Total |
| <16 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (100) | 0 (0.0) | 4 |
| 16–30 | 75 (55.6) | 37 (27.4) | 20 (14.8) | 2 (1.5) | 0 (0.0) | 1 (0.7) | 135 |
| 31–45 | 20 (33.3) | 26 (43.3) | 7 (11.7) | 2 (3.3) | 0 (0.0) | 5 (8.3) | 60 |
| >45 | 0 (0.0) | 8 (88.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (11.1) | 9 |
IDU, injecting drug use; Heterosexual, heterosexual transmission; Mix, subjects with both IDU and heterosexual risk factors; MSM, male having sex with male; MTCT, mother-to-child transmission; Others, other risk factors such as blood transfusion, tattoo, and unknown transmission.
Table 3.
Distribution of Subjects in Each Period of Risk Behavior Initiationa
| Risk factor group | ||||||||
|---|---|---|---|---|---|---|---|---|
| Period | IDU n (%) | Heterosexual n (%) | Mix n (%) | MSM n (%) | MTCT n (%) | Tattoo n (%) | Transfusion n (%) | Total |
| <1991 | 2 (13.3) | 9 (60.0) | 0 (0.0) | 2b (13.3) | 0 (0.0) | 1 (6.7) | 1 (6.7) | 15 |
| 1991–1995 | 11 (61.1) | 3 (16.7) | 3 (16.7) | 0 (0.0) | 0 (0.0) | 1 (5.6) | 0 (0.0) | 18 |
| 1996–2000 | 66c (61.1) | 24d (22.2) | 18 (16.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 108 |
| 2001–2005 | 10 (22.2) | 24e (53.3) | 4 (8.9) | 2 (4.4) | 4 (8.9) | 1 (2.2) | 0 (0.0) | 45 |
| >2005 | 0 (0.0) | 8f (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 8 |
IDU, injecting drug use; Heterosexual, heterosexual transmission; Mix, subjects with both IDU and heterosexual risk factors; MSM, Male having sex with male; MTCT, Mother-to-child transmission; Transfusion, blood transfusion.
These include one subject infected with HIV-1 subtype B.
These include two subjects infected with HIV-1 subtype B, with 97% (64 of 66) male subjects.
These include 45.8% (11 of 24) female subjects.
These include 70.8% (17 of 24) female subjects.
These include 87.5% (7 of 8) female subjects.
Of patients 56.3% were in the late stage of HIV infection when they were diagnosed, suggesting the possibility of earlier exposure to HIV. In the private hospital, the number of patients initially diagnosed in earlier stages (HIV or ARC) was greater than in public hospitals, (37.8% and 18.7%, respectively). Antiretroviral (ARV) treatment seemed effective; the number of subjects in AIDS stages decreased from 84.2% initially to 24.2% at enrollment in the treated group (n = 120). The median ARV therapy duration was 8 months (minimum 1 month and maximum 84 months).
At the time of sample collection, the median of CD4 cell counts from all subjects (n = 208) was 174 cells/μ1 (minimum 1 cells/μl and maximum 997 cells/μl). Lower CD4 median cell counts were found in ARV-naive cases (n = 88): 58 cells/μl with a minimum of 1 cell/μl and a maximum of 997 cells/μl, and higher CD4 median cell counts were found among treated cases (n = 120): 205 cells/μl with a minimum of 4 cells/μl and maximum of 896 cells/μl.
Only 65 of 208 subjects were examined for coinfection with hepatitis C virus (HCV), and 38.5% of them had positive anti-HCV antibody. As for coinfection with hepatitis B (HB) virus, HB antigen was examined in 55 subjects, and 30.9% of them were positive. HB coinfection was found in almost every risk group; however, hepatitis C coinfection was found mostly in IDUs (p = 0.000) (Table 4).
Table 4.
Hepatitis B and/or Hepatitis C Coinfection with HIV-1 Among Risk Factor Groupsa
| Risk factor group | ||||||||
|---|---|---|---|---|---|---|---|---|
| IDU n (%) | Heterosexual n (%) | Mix n (%) | MSM n (%) | MTCT n (%) | Tattoo n (%) | Transfusion n (%) | Total | |
| Hepatitis B | 9 (52.8) | 2 (11.8) | 4 (23.5) | 1 (5.9) | 0 (0.0) | 1 (5.9) | 0 (0.0) | 17 |
| Hepatitis C | 21 (84.0) | 0 (0.0) | 4 (16.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 25 |
Of 208 subjects, 55 were tested for HBs antigen and 65 were examined for antihepatitis C virus antibody. IDU, injecting drug use; Heterosexual, heterosexual transmission; Mix, subjects with both IDU and heterosexual risk factors; MSM, male having sex with male; MTCT, mother-to-child transmission; Transfusion, blood transfusion.
Subtype distribution and recombinant analysis
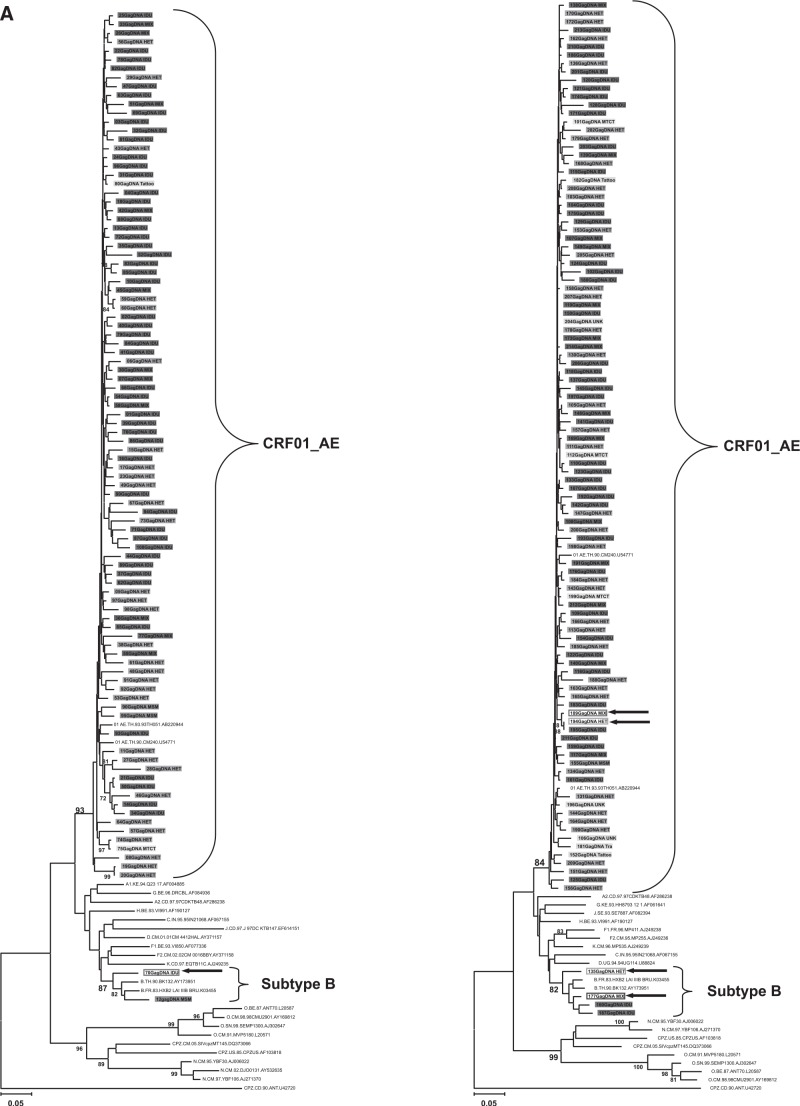
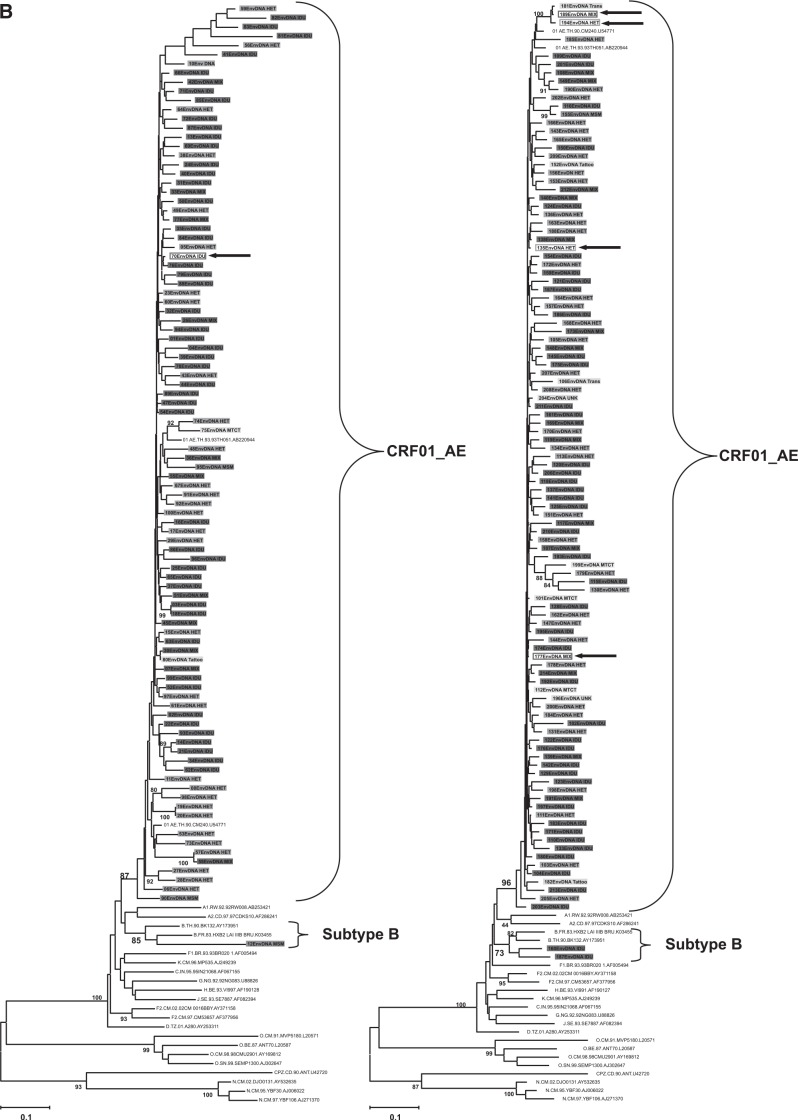
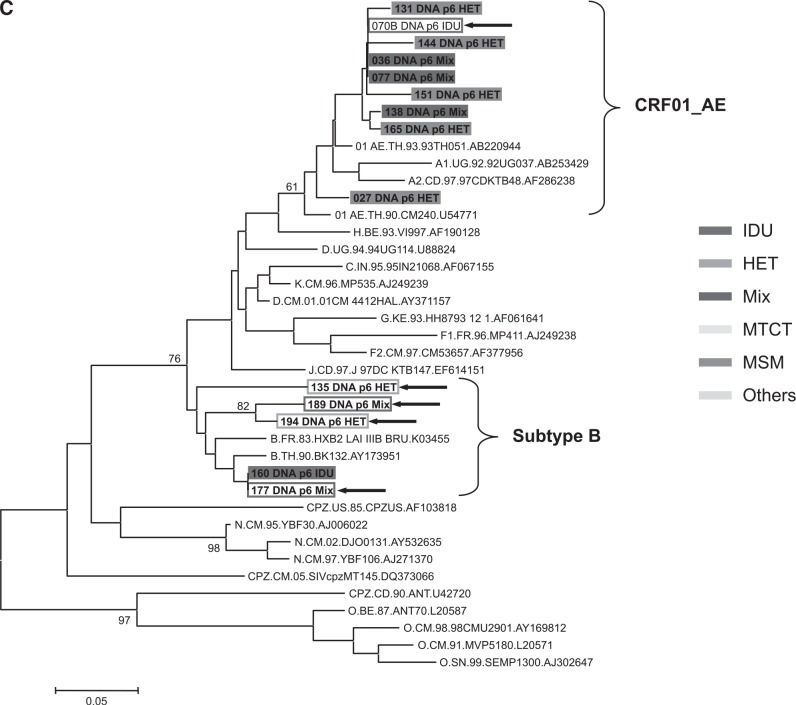
The genetic subtype of 208 subjects was determined by phylogenetic trees based on nucleotide sequences of gag p17 (396–411 bp), env C2V3 (321–357 bp), and p6 (159–162 bp) genes; 200 subjects (96.2%) were CRF01_AE and three subtype Bs (1.4%) were identified. Of the remaining five samples (2.4%), three samples (07IDJKT070, 07IDJKT135, and 07IDJKT177) indicated different subtypes between the gag p17 and env C2V3 regions: subtype B and CRF01_AE, respectively, and two samples (07IDJKT189 and 07IDJKT194) showed subtype B in the p6 gene and CRF01_AE in other regions (Fig. 1).
FIG. 1.
Phylogenetic tree analysis based on the nucleotide sequences of gag p17 (A), env C2V3 (B), and gag p6 (C) regions obtained from various risk groups. Left panels in (A) and (B) include sample numbers 1 to 100 and right panels in (A) and (B) include sample numbers 101 to 214, with reference sequences obtained from the Los Alamos National Laboratory HIV Database (http://hiv-web.lanl.gov/), respectively. (C) consists of five recombinant forms and nine randomly chosen samples. Each color represents risk factor groups. Open rectangular boxes and arrows indicate five recombinant forms with subtype discordance among the gag p17, env C2V3, and gag p6 genes. IDU, intravenous drug use; HET, heterosexual transmission; Mix, subjects with both IDU and heterosexual risk factors; MSM, males having sex with males; MTCT, mother-to-child transmission; Others, other risk factors such as blood transfusion, tattoo, and unknown transmission.
CRF01_AE dominated in all risk factor groups. Three samples identified with subtype B came from one MSM and two IDUs, and all of them initiated their risky behavior before the year 2000 (Table 3).
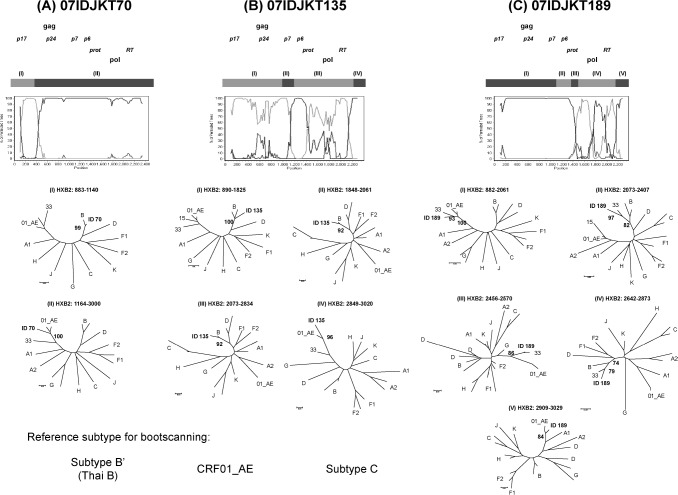
Of the five possible recombinant forms, three samples (07IDJKT070, 07IDJKT135, and 07IDJKT189) were successfully amplified, cloned, and sequenced for gag p17-RT regions (2.3 kbp). Bootscanning analysis revealed different breakpoint numbers and positions (Fig. 2), suggesting, at the very least, intersubtype recombination between CRF01_AE and Subtype B′. 07IDJKT070 showed one breakpoint separating Thai B′ in gag p17 and CRF01_AE in the rest of the following regions, while 07IDJKT135 indicated a more complex mosaic pattern with three identified breakpoints. 07IDJKT189 showed a recombination pattern very similar to CRF33_01B from Malaysia.14
FIG. 2.
Bootscanning analysis of three recombinant forms. In all, 2.3-kb gene sequences from p17 to part of RT were analyzed. Subtype B Thai variant (AY173951) and CRF01_AE (U54771) were used as putative parental strains, with subtype C (AF067155) as the outgroup. All samples showed different breakpoint number and positions. (A) 07IDJKT70 mainly consisted of CRF01_AE, while (B) 07IDJKT135 harbored a more complicated mosaic pattern. (C) 07IDJKT189 showed a pattern similar to CRF33_01B previously reported from Kuala Lumpur, Malaysia. Reference sequences were used for phylogenetic tree construction, including HXB2 and subtype B′ (Thai variant) for the subtype B group reference sequence. 01_AE, CRF01_AE; 33, CRF33_01B; 15, CRF15_01B.
Discussion
The number of HIV patients in Indonesia, especially in Jakarta, is steadily increasing. UNAIDS reported the estimated prevalence of people living with HIV in Indonesia increased from 0.1% in 2001 to 0.2% in 2007, and it was followed by a large increase in the estimated number of women living with HIV from 10,000 in 2001 to 54,000 in 2007. Another issue of concern is the high percentage of HIV prevalence among IDUs, which accounted for 52.4% in 2007.3
This study showed male IDUs from younger generations as dominant subjects, similar to other reports from Indonesia2 and UNAIDS.3 However, in the heterosexual risk group, female subjects were dominant, and more than half of them were diagnosed with HIV infection during the screening process after their spouses or sex partners were diagnosed with HIV. Therefore, these females visited hospitals at a relatively early stage and had higher CD4 counts, with most of them enrolled as ARV-naive cases. Of this female heterosexual group 73.2% had HIV-positive IDU or ex-IDU spouses or sex partners, suggesting HIV cross-spread from male IDUs to their sex partners. This was also supported by the situation previously reported in Indonesia,1 which showed a low percentage of condom use in both commercial and noncommercial sexual intercourse.
The number of HIV patients in Indonesia started to increase steadily from 1991, and in this period most cases were from the MSM and heterosexual risk groups. The dynamics of HIV-1 transmission began in 1999, when IDU started to dominate as the main risk factor,2 consistent with our data regarding the initiation of risk behavior. The decreasing number of IDUs newly initiated since 2001 is likely related to successful anti-drug abuse campaigns in Indonesia. Yet the ratio of patients infected by heterosexual transmission remains high. This suggests that knowledge regarding cross-spreading HIV infection may still be low and safer sexual practice remains uncommon. Phylogenetic analysis in our study showed that the samples from different risk groups clustered closely to each other in the gag, env, and p6 regions, which also suggests HIV cross-spreading between IDU and heterosexual groups.
In this study, the lack of understanding of HIV infection was indicated by the large number of subjects who were already in the AIDS stage when they were diagnosed, low CD4 counts among ARV-naive subjects, and a high occurrence of opportunistic infections; in addition, it also seemed to be affected by the socioeconomic status of subjects. Furthermore, people living in metropolitan or urban areas have easier access to illicit drugs and free, unsafe sex is more common, especially among the younger generation,1 and even though most subjects in this study have at least a high school education, most of them still persist in using unsterilized needles despite their understanding of the risk.
Sharing unsterilized needles can lead to other problems, such as HCV and/or HBV coinfection. HCV can be spread efficiently through exposure to contaminated blood or blood products, thus the HCV coinfection rate among IDUs is high.24 Our study showed 84% of 25 subjects with HCV co-infection were IDUs, suggesting a high HCV coinfection rate among IDUs in Indonesia. Greater effort on the part of government, NGOs, and all concerned parties is needed to provide more information regarding HIV transmission, including prevention and treatment, especially to females and the young adult generation.
Subtype B and CRF01_AE were introduced relatively independently among two risk groups at the beginning of the HIV-1 endemic in Southeast Asia.5,6 However, recent studies in these countries show that cases of subtype B are decreasing, and CRF01_AE is taking over as the dominant subtype in all risk groups.4,8 In Indonesia, the first study focusing on subtype distribution showed 12 subtype B (63.2%) and 7 CRF01_AE cases among 19 HIV-positive samples collected in 1993 from high-risk volunteers.19 The second study reported 2 subtype B (12.5%) and 14 CRF01_AE (87.5%) cases among the 24 positive sera collected in 1999–2000 from an STD clinic in Papua.20 The current study conducted in 2007 from various risk groups in Jakarta indicated that 96.2% of 208 subjects were infected with CRF01_AE in every risk group. Taken together, the molecular epidemiological shift as observed in Thailand is considered to have occurred in Indonesia as well, to which interconnections between different risk groups might contribute. Moreover, we believe that CRF01_AE might be the founder among Indonesian IDUs, based on the high prevalence of CRF01_AE among IDUs in this study, and due to data indicating that 12 of 13 IDUs who initiated their risk behavior in the earlier phase of the HIV epidemic in Indonesia were infected with CRF01_AE (data not shown).
The increasing prevalence of CRF01_AE/B intersubtype recombinants in Southeast Asia was represented in molecular epidemiological studies in Thailand and Malaysia.8,11 In a study conducted in 1999–2000 in Thailand, no pure subtype B and an increasing number of recombinants showing various chimeric patterns between subtype B and CRF01_AE were reported among 38 seroconverters.8 A study conducted in 2004 in Malaysia showed evidence of changing HIV-1 molecular epidemiology toward the predominance of CRF01_AE/B intersubtype recombinants among IDUs.11 In our study, five (2.4%) of 208 samples showed subtype discordance between three different genes suggesting CRF01_AE/B unique recombinant forms. Their number was higher than subtype B, however, although the decreasing number of subtype B, pure subtype B is still circulating, which might contribute to the generation of CRF01_AE/B recombinants in Indonesia, as well as in Thailand or Malaysia. Interestingly, three of them, which were confirmed as true recombinants, showed different mosaic patterns (Fig. 2), even though they consist mostly of CRF01_AE and subtype B, the two main circulating subtypes in Southeast Asia, including Indonesia. Furthermore, two of them were not related to either current reference subtypes or the strains identified recently in Thailand and Malaysia,13–18 suggesting that they were generated in Indonesia. These unique recombinants came from different risk groups, which implies the dissemination of these recombinant forms into the general population. In this study, we also identified a recombinant form (01IDJKT189), which indicates a close pattern to CRF33_ 01B, previously reported in Malaysia. As neighboring countries, people frequently travel between these countries, and with a similar sociocultural background, might make the spreading of HIV, including CRF33_01B, highly likely.
Continuous study is necessary to determine the direction the dynamics of HIV molecular epidemiology will take and how it can affect the current and future global HIV/AIDS situation in Indonesia.
Sequence Data
GenBank accession numbers of nucleotide sequences reported in this article are AB468161–AB468365 for gag p17 sequences, AB468366–AB468572 for env C2V3 sequences, AB468573–AB468575 for gag-pol 2.3-kbp sequences, and AB468576–AB468586 for gag p6 sequences.
Acknowledgments
We thank Dr. Gunawan Martinus, Dr. Dyah Waluyo, and the staff at Cipto Mangunkusumo National Hospital, Kramat 128 Hospital, and Sulianti Saroso Infectious Disease Hospital for their help during specimen collection. We thank Dr. Wataru Habano for technical advice and Ms Kumi Furusawa for technical assistance. This work was supported by grants-in-aid for Advanced Medical Science Research by the Ministry of Science, Education, Sports, and Culture of Japan, a grant from the Imai Memorial Trust for AIDS Research, and a fellowship from the Takeda Science Foundation. Last but not least, we are grateful to all the patients who have participated in this study and their families. This work is dedicated to them.
Disclosure Statement
No competing financial interests exist.
References
- 1. Riono P. and Jazant S. The current situation of the HIV/AIDS epidemic in Indonesia: AIDS Educ Prev 2004;16(3Suppl. A):78–90 [DOI] [PubMed] [Google Scholar]
- 2. Directorate General of CDC: Statistic of HIV/AIDS cases in Indonesia: Reported up to April 2008. Jakarta (Indonesia): Ministry of Health Republic of Indonesia, 2008. Available from http://www.aidsindonesia.or.id
- 3. UNAIDS/WHO. AIDS epidemic update 2008. Geneva, Switzerland: WHO/UNAIDS;2008. Available from http://www.unaids.org
- 4. Ruxrungtham K, Brown T, and Phanupak P: HIV/AIDS in Asia. Lancet 2004;364:69–82 [DOI] [PubMed] [Google Scholar]
- 5. Ou CY, Takebe Y, Weniger BG, et al. : Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet 1993;341:1171–1174 [DOI] [PubMed] [Google Scholar]
- 6. Weniger BG, Takebe Y, Ou CY, and Yamazaki S: The molecular epidemiology of HIV in Asia. AIDS 1994;(8 Suppl. 2):S13–28 [PubMed] [Google Scholar]
- 7. Verachai V: HIV infection among substance abusers in Thanyarak Institute on Drug Abuse, Thailand, 1987–2002. J Med Assoc Thai 2005;88:76–79 [PubMed] [Google Scholar]
- 8. Tovanabutra S, Beyrer C, Sakkhachornphop S, et al. : The changing molecular epidemiology of HIV type 1 among Northern Thai drug users, 1999 to 2002. AIDS Res Hum Retroviruses 2004;20:465–475 [DOI] [PubMed] [Google Scholar]
- 9. Hudgens MG, Longini IM, Jr, Vanichseni S, et al. : Subtype-specific transmission probabilities for human immunodeficiency virus type 1 among injecting drug users in Bangkok, Thailand. Am J Epidemiol 2002;155:159–168 [DOI] [PubMed] [Google Scholar]
- 10. Malim MH, Emerman M: HIV-1 sequence variation: Drift, shift, and attenuation. Cell 2001;104:469–472 [DOI] [PubMed] [Google Scholar]
- 11. Tee KK, Saw TL, Pon CK, et al. : The evolving molecular epidemiology of HIV type 1 among injecting drug users (IDUs) in Malaysia. AIDS Res Hum Retroviruses 2005;21: 1046–1050 [DOI] [PubMed] [Google Scholar]
- 12. Motomura K, Kusagawa S, Kato K, Nohtomi K, Lwin HH, Tun KM, et al. : Emergence of new forms of human immunodeficiency virus type 1 intersubtype recombinants in Central Myanmar. AIDS Res Hum Retroviruses 2000;16: 1831–1843 [DOI] [PubMed] [Google Scholar]
- 13. Tovanabutra S, Watanaveeradej V, Viputtikul K, et al. : A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses 2003;19:561–567 [DOI] [PubMed] [Google Scholar]
- 14. Tee KK, Li XJ, Nohtomi K, et al. : Identification of a novel circulating recombinant form (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr 2006;43:523–529 [DOI] [PubMed] [Google Scholar]
- 15. Tovanabutra S, Kijak GH, Betrer C, et al. : Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in Northern Thailand. AIDS Res Hum Retroviruses 2007;23:829–833 [DOI] [PubMed] [Google Scholar]
- 16. Wang B, Lau KA, Ong LY, et al. : Complex pattern of the HIV-1 epidemic in Kuala Lumpur, Malaysia: Evidence for expansion of circulating recombinant form CRF33_01B and detection of multiple other recombinants. Virology 2007;367: 288–297 [DOI] [PubMed] [Google Scholar]
- 17. Lau KA, Wang B, Kamarulzaman A, Ng KP, and Saksena NK: Near full-length sequence analysis of a unique CRF01_AE/B recombinant from Kuala Lumpur, Malaysia. AIDS Res Hum Retroviruses 2007;23:1139–1145 [DOI] [PubMed] [Google Scholar]
- 18. Lau KA, Wang B, Kamarulzaman A, Ng KP, and Saksena NK: Continuous crossover(s) events of HIV-1 CRF01_AE and B subtype strains in Malaysia: Evidence of rapid and extensive HIV-1 evolution in the region. Curr HIV Res 2008;6:108–116 [DOI] [PubMed] [Google Scholar]
- 19. Porter KR, Mascola JR, Hupudio H, Ewing D, VanCott TC, Anthony RL, et al. : Genetic, antigenic and serologic characterization of human immunodeficiency virus type 1 from Indonesia. J Acquir Immune Defic Syndr Hum Retrovirol 1997;14:1–6 [DOI] [PubMed] [Google Scholar]
- 20. Foley B, Donegan E, Silitonga N, Wignall FS, Busch MP, and Delwart EL: Importation of multiple HIV type 1 strains into West Papua, Indonesia. AIDS Res Hum Retroviruses 2001; 17:1655–1659 [DOI] [PubMed] [Google Scholar]
- 21. Naganawa S, Sato S, Nossik D, Takahashi K, Hara T, Tochikubo O, et al. : First report of CRF03_AB recombinant HIV type 1 in injecting drug users in Ukraine. AIDS Res Hum Retroviruses 2002;18:1145–1149 [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Dudley J, Nei M, and Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599. (Publication PDF at http://www.kumarlab.net/publications). [DOI] [PubMed]
- 23. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, and Ray SC: Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73(1):152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sulkowski MA: Viral hepatitis and HIV coinfection. J Hepatol 2008;48:353–367 [DOI] [PubMed] [Google Scholar]