Abstract
Chemotherapy with carboplatin and paclitaxel is the standard treatment for ovarian cancer patients. Although most patients initially respond to this treatment, few are cured. Resistance to chemotherapy is the major cause of treatment failure. We applied a quantitative proteomic approach based on ICAT/MS/MS technology to analyze tissues harvested at primary debulking surgery before the initiation of combination chemotherapy in order to identify potential naive or intrinsic chemotherapy response proteins in ovarian cancers. We identified 44 proteins that are overexpressed, and 34 proteins that are underexpressed in the chemosensitive tissue compared to the chemoresistant tissue. The overexpressed proteins identified in the chemoresistant tissue include 10 proteins (25.6%) belonging to the extracellular matrix (ECM), including decorin, versican, basigin (CD147), fibulin-1, extracellular matrix protein 1, biglycan, fibronectin 1, dermatopontin, alpha-cardiac actin (smooth muscle actin), and an EGF-containing fibulin-like extracellular matrix protein 1. Interesting proteins identified as overexpressed in the chemosensitive tissue include gamma-catenin (junction plakoglobin) and delta-catenin, tumor suppressor p53-binding protein 1 (53BP1), insulin-like growth factor-binding protein 2 (IGFBP2), proliferating cell nuclear antigen (PCNA), annexin A11, and 53 kDa selenium binding protein 1. Integrative analysis with expression profiling data of eight chemoresistant tissues and 13 chemosensitive tissues revealed that 16 proteins showed consistent changes at both the protein and the RNA levels. These include P53 binding protein 1, catenin delta 1 and plakoglobin, EGF-containing fibulin-like extracellular matrix protein 1 and voltage-dependent anion-selective channel protein 1. Our results suggest that chemotherapy response may be determined by multiple and complex system properties involving extracellular–matrix, cell adhesion and junction proteins.
Introduction
Ovarian cancer ranks fourth in cancer mortality among women in the United States. There are 22,430 new cases of ovarian cancer in 2007 and about 15,280 deaths from this diseasse (American Cancer Society Statistics for 2007). Most cases are advanced at diagnosis, as ovarian cancer typically does not cause symptoms until having metastasized outside the ovaries. Platinum compounds, given as either cisor carboplatins, combined with paclitaxels, are the standard treatment for nearly all women diagnosed with ovarian cancer. Although most patients initially respond to this treatment, few are cured (Hall et al., 2004). Resistance to chemotherapy is the major cause of treatment failure. Resistance to cisplatin occurs in roughly one-third of women during primary treatment and in all patients treated for recurrent diseases (Vasey, 2003). Despite the treatment, fewer than 25% of the women with ovarian cancer experience progression-free survival of more than 4 years (McGuire et al., 1996). Currently, resistance can only be determined retrospectively after patients have gone through the treatment and experienced the burdens and toxicities of ineffective therapies. Outcomes for women with ovarian cancers could be improved by the discovery of biomarkers capable of identifying resistant tumors. In addition, a better understanding of chemotherapy resistance may lead to the development of more effective therapies.
Proteins that are differentially expressed in chemotherapy resistant and sensitive ovarian cancer cells are likely to be involved in pathways that modulate the sensitivity of ovarian cancers to cisplatins or paclitaxels, and therefore, are logical candidates as markers to monitoring treatment response or as novel therapeutic targets to overcoming chemotherapy resistance. Mass-spectrometry-based quantitative proteomics, such as the isotope-coded affinity tags (ICAT) labeling (Gygi et al., 1999) combined with liquid chromatography (LC)—tandem mass spectrometer (MS/MS), allow global identification and quantification of proteins in complex samples and are well suited for discovering potential biomarkers for human diseases (Aebersold et al., 2005; Aebersold and Mann, 2003). We and others have used ovarian cancer cell line models such as IGROV1 and GROV1-CP cells to identify proteins related to chemotherapy responses (Le Moguen et al., 2007; Stewart et al., 2006). However, to our knowledge, a direct proteomics analysis using ovarian cancer tissues from chemotherapy-resistant and chemotherapy-sensitive tissues has not yet been performed, probably due to difficulties in obtaining necessary tissues with clinically defined chemotherapy response statuses as lengthy follow-up is needed to ascertain responsiveness to chemotherapy drugs. Therefore, in this study, we conducted quantitative proteomic analysis of tissues from chemotherapy sensitive and resistant patients.
Materials and Methods
Cancer tissues
The tissues were harvested at primary debulking surgery before the initiation of combination chemotherapy with carboplatin and paclitaxel. Institutional review board approval was obtained for the acquisition of tissues. One chemosensitive tissue (serous histology, stage IIIC ovarian cancer, progression-free interval of 23 months) and one chemoresistant tissue (serous histology, stage IIIC ovarian cancer, progression-free interval of 0 months) were used for the study.
ICAT/MS/MS and data analysis
Protein isolation and ICAT/MS/MS analysis were performed as we described previously (Stewart et al., 2006). In brief, for ICAT labeling, 1 mg of proteins each from the chemoresistant and chemosensitive tissue were denatured with 6 M urea and 0.05% SDS, and then immediately reduced with 5 mM tributylphosphine. The tissue proteins were labeled with second-generation ICAT reagents (acid cleavable ICAT), either in light (12C for cisplatin sensitive) or in heavy (13C for cisplatin resistant) isotopes (Applied Biosystems, Foster City, CA). Equal amounts of the two labeled samples were combined and digested by trypsin (Promega, Madison, WI). ICAT-labeled peptides were subsequently purified by cation-exchange chromatography and avidin-affinity chromatography. Peptide mixtures were analyzed by microcapillary HPLC-electrospray ionization (ESI)-MS/MS using an ion-trap mass spectrometer (LCQ-DecaXP, ThermoFinnigan, Ringoes, NJ) as previously described (Yi et al., 2003). MS/MS spectra obtained were searched against the IPI human database (version v3.17.) using the SEAQEST algorithm (Eng et al., 1994). The SEQUEST parameters included ICAT labeling on cysteine and phosphorylation on serine, threonine, and tyrosine, and oxidation for methionine. PeptideProphet (Keller et al., 2002) and ProteinProphet (Nesvizhskii et al., 2003) were used for statistical analysis of the proteomics data. The ASAPRatio program (Li et al., 2003) was used to identify differentially expressed proteins. To calculate the false discovery rate, we used the perl script (www.matrixscience.com/downloads/decoy.pl.gz) to randomize the database entries. We chose the random option of the perl script (i.e., decoy.pl [–random] input.fasta [output.fasta]), for which the output entries are random sequences with the same average amino acid composition as the input database. The false discovery rate (FDR) was calculated by the formula Ndecoy/Ncombined, where Ndecoy (namely FP, false positives) is number of matches to the entries from decoy database and Ncombine (namely FP + TP, false positives + true positives) is number of matches to the combined database.
Results
ICAT/MS/MS analysis of chemosensitive and chemoresistant ovarian cancer tissues
We analyzed one chemosensitive tissue (serous histology, stage IIIC ovarian cancer, progression-free interval of 23 months) and one chemoresistant tissue (serous histology, stage IIIC ovarian cancer, progression-free interval of 0 months). The tissues were harvested at primary debulking surgery before the initiation of combination chemotherapy with carboplatin and paclitaxel. Therefore, the differentially expressed proteins we identified could reveal naive or intrinsic chemotherapy-responsive proteins.
Using the PeptideProphet program (Keller et al., 2002) with a cutoff score of 0.9, we identified 1,413 unique peptides (Supplementary Table 1; See online supplementary material at www.liebertonline.com) that potentially correspond to 621 proteins. The data were further analyzed by the Protein Prophet program (Nesvizhskii et al., 2003). Peptides that correspond to more than a single protein in the sequence database were apportioned among all corresponding proteins, and a minimal protein list sufficient to account for the observed peptide assignments was derived using the expectation-maximization algorithm (Nesvizhskii et al., 2003). Using a Protein Prophet cutoff score of 0.9, which corresponds to a 0.8% overall false-positive rate, we identified 502 proteins (or protein groups) in the tissue samples (Supplementary Table 2). The false discovery rate calculated by the decoy database search is 0.87%, which is similar to what was estimated from the Protein Prophet program.
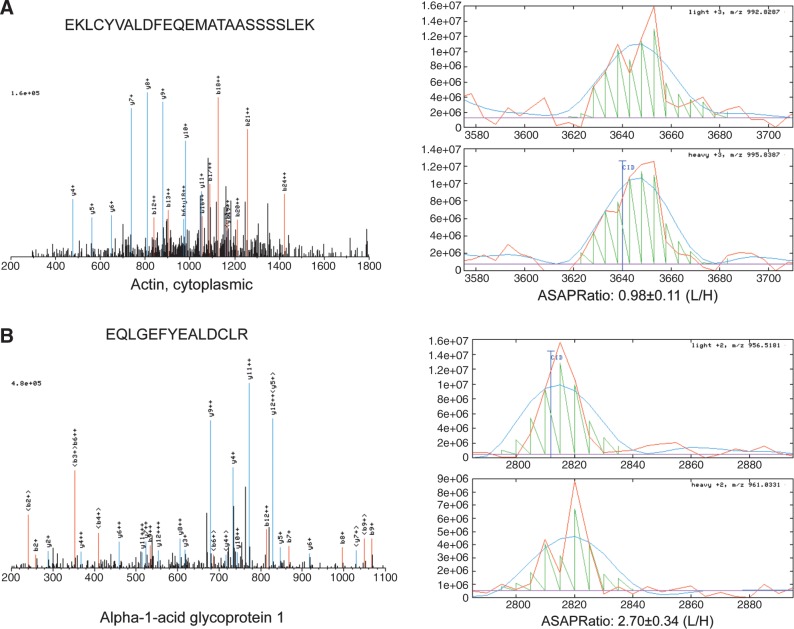
We used the ASAPRatio program (Li et al., 2003) to identify differentially expressed proteins. ASAPRatio program utilizes the signals recorded for the different isotopic forms of peptides of identical sequences and applies statistical methods including Savitzky-Golay smoothing filters, statistics for weighted samples, and Dixon's test for outliers, to evaluate protein abundance ratios and their associated errors (Li et al., 2003). We obtained quantitative data on 493 proteins. The median ICAT ratio (ASAPRatio) for these 493 proteins is 0.96 and the mean ratio is 1.18. The ratio for cytoplasmic actins (beta and gamma actin) (IPI000021439, IPI000021440), proteins that should show no difference between chemosensitive and chemoresistant tissues, is 0.98. Proteins with expression ratios of at least twofold were considered differentially expressed. We filtered out keratin (a common contaminant from sample handling), immunoglobins, and HLA antigens (the latter being two proteins that are likely associated with blood contamination in the tissues). We identified 44 proteins that were overexpressed, and 34 proteins that were underexpressed in the chemosensitive tissue compared with the chemoresistant tissue (Table 1). Figure 1 shows examples of quantification (ASAPRatio) of two proteins, the cytoplasmic actin and alpha-1-acid glycoprotein 1.
Table 1.
Differentially Expressed Proteins Identified by ICAT/MS/MS
| Protein (rep ID) | Protein probability | ASAPratio mean (R/S) | ASAPratio SD | Description |
|---|---|---|---|---|
| Proteins with ASAPRatio <0.5 | ||||
| Nucleic Acid Binding | ||||
| IPI00438229a | 1 | 0.14 | 0.03 | Transcription intermediary factor 1-beta |
| IPI00737736 | 0.99 | 0.19 | 0.02 | PREDICTED: similar to High mobility group protein 1 |
| IPI00025057a | 1 | 0.32 | 0.06 | Double-stranded RNA-specific adenosine deaminase |
| IPI00328343a | 1 | 0.34 | 0.07 | Spliceosome RNA helicase BAT1, 26-kDa protein, HLA-B associated transcript 1 |
| IPI00007163 | 0.95 | 0.34 | 0.06 | U6 snRNA-associated Sm-like protein LSm7 |
| IPI00299573a | 0.99 | 0.36 | 0.14 | 60S ribosomal protein L7a, 31-kDa protein |
| IPI00025447a | 1 | 0.39 | 0.09 | Elongation factor 1-alpha 1 |
| IPI00306332 | 1 | 0.47 | 0.06 | 60S ribosomal protein L24 |
| IPI00419880a | 1 | 0.47 | 0.06 | 30 kDa protein, 40S ribosomal protein S3a |
| IPI00028888a | 1 | 0.48 | 0.04 | Heterogeneous nuclear ribonucleoprotein D0 |
| IPI00023785a | 0.99 | 0.48 | 0.05 | Probable ATP-dependent RNA helicase DDX17 |
| IPI00220528 | 1 | 0.5 | 0.12 | Small nuclear ribonucleoprotein F |
| IPI00167949 | 1 | 0.47 | 0.13 | Interferon-induced GTP-binding protein Mx1 |
| IPI00163782a | 1 | 0.49 | 0.09 | far upstream element-binding protein |
| Chaperone/chaperonin | ||||
| IPI00304925a | 1 | 0.22 | 0.08 | heat-shock 70-kDa protein |
| IPI00290770a | 1 | 0.34 | 0.07 | Chaperonin containing TCP1, subunit 3 |
| IPI00013890a | 1 | 0.49 | 0.14 | 14-3-3 protein sigma, SFN protein |
| Catenins | ||||
| IPI00182469a | 0.99 | 0.45 | 0.09 | Catenin delta-1 |
| IPI00554711 | 1 | 0.47 | 0.06 | Junction plakoglobin (gamma catenin) |
| Others | ||||
| IPI00382682 | 0.94 | 0.3 | 0.06 | Putative matrix cell adhesion molecule-3 |
| IPI00414320a | 1 | 0.22 | 0.09 | Annexin A11 |
| IPI00027175a | 0.99 | 0.39 | 0.17 | sorcin |
| IPI00029778a | 1 | 0.44 | 0.11 | Tumor suppressor p53-binding protein 1 |
| IPI00305719a | 1 | 0.31 | 0.07 | 53 kDa protein, selenium binding protein 1 |
| IPI00297284 | 1 | 0.5 | 0.16 | Insulin-like growth factor-binding protein 2 |
| IPI00021700 | 1 | 0.5 | 0.08 | Proliferating cell nuclear antigen |
| IPI00644497 | 1 | 0.24 | 0.41 | Hypothetical protein DKFZp686O16217 |
| IPI00005721a | 1 | 0.26 | 0.05 | PREDICTED: similar to Neutrophil defensin 1 |
| IPI00102821 | 1 | 0.32 | 0.08 | PACAP protein |
| IPI00305383 | 0.98 | 0.32 | 0.04 | Ubiquinol-cytochrome c reductase complex core protein 2, mitochondrial |
| IPI00328113 | 0.99 | 0.35 | 0.06 | Fibrillin-1 |
| IPI00387168a | 0.97 | 0.35 | 0.05 | Proprotein convertase subtilisin\kexin type 9 |
| IPI00550995 | 0.99 | 0.37 | 0.05 | Hypothetical protein NOP17 |
| IPI00027230 | 1 | 0.39 | 0.08 | Endoplasmin |
| IPI00465044 | 1 | 0.39 | 0.06 | Protein RCC2 |
| IPI00023647a | 0.99 | 0.39 | 0.06 | Hypothetical protein DKFZp451P021, UBE1L2 protein |
| IPI00220617a | 0.99 | 0.4 | 0.17 | Liver phosphofructokinase |
| IPI00140420 | 0.97 | 0.42 | 0.13 | Staphylococcal nuclease domain-containing protein 1 |
| IPI00003933a | 0.94 | 0.42 | 0.1 | Hydroxyacylglutathione hydrolase |
| IPI00220766 | 1 | 0.43 | 0.09 | Lactoylglutathione lyase |
| IPI00010720 | 1 | 0.45 | 0.1 | T-complex protein 1 subunit epsilon |
| IPI00297779 | 1 | 0.48 | 0.17 | T-complex protein 1 subunit beta |
| IPI00647217 | 0.96 | 0.47 | 0.09 | Superkiller viralicidic activity 2-like 2 |
| IPI00022694a | 1 | 0.49 | 0.15 | Proteasome (Prosome, macropain) 26S subunit, non-ATPase, 4 |
| Proteins with ASAPRatio >2 | ||||
| ECM/stromal proteins | ||||
| IPI00003351a | 0.98 | 2.72 | 0.34 | Extracellular matrix protein 1 |
| IPI00019906a | 0.98 | 2.05 | 0.49 | Basigin |
| IPI00008603a | 1 | 2.73 | 0.56 | Actin, alpha cardiac, Actin |
| IPI00215628a | 1 | 2.78 | 0.8 | Versican core protein |
| IPI00296534 | 1 | 2.79 | 0.75 | Fibulin-1 |
| IPI00029658a | 1 | 2.95 | 1.18 | EGF-containing fibulin-like extracellular matrix protein 1 |
| IPI00010790a | 0.98 | 2.97 | 0.67 | Biglycan |
| IPI00292130 | 0.99 | 2.99 | 0.57 | Dermatopontin |
| IPI00012119a | 1 | 5.47 | 0.68 | Decorin |
| IPI00022418a | 1 | 3.33 | 0.71 | Fibronectin |
| Others | ||||
| IPI00216308 | 1 | 1.98 | 0.27 | Voltage-dependent anion-selective channel protein 1 |
| IPI00032328a | 1 | 2 | 0.57 | Kininogen-1 |
| IPI00298267a | 1 | 2 | 0.54 | Prostaglandin G\H synthase 1 |
| IPI00298828 | 1 | 2.01 | 0.42 | Beta-2-glycoprotein 1 |
| IPI00013847 | 1 | 2.01 | 0.34 | Ubiquinol-cytochrome c reductase complex core protein I, mitochondrial |
| IPI00414717a | 0.99 | 2.01 | 0.21 | golgi apparatus protein 1, Golgi apparatus protein 1 |
| IPI00104074a | 1 | 2.04 | 0.73 | CD163 antigen isoform b, CD163 antigen isoform a |
| IPI00296913a | 1 | 2.09 | 0.59 | ADP-sugar pyrophosphatase |
| IPI00022431 | 1 | 2.5 | 0.35 | Alpha-2-HS-glycoprotein |
| IPI00022429 | 1 | 2.7 | 0.34 | Alpha-1-acid glycoprotein 1 |
| IPI00643920 | 1 | 2.85 | 0.77 | Transketolase |
| IPI00018352a | 0.96 | 2.85 | 1.72 | Ubiquitin carboxyl-terminal hydrolase isozyme L1, 16 kDa protein |
| IPI00385887 | 0.97 | 2.88 | 0.49 | Tyrosine kinase |
| IPI00022417 | 1 | 3.06 | 1.25 | Leucine-rich alpha-2-glycoprotein |
| IPI00550363a | 1 | 3.25 | 0.5 | Transgelin-2,21-kDa protein, 24-kDa protein |
| IPI00216057 | 0.92 | 3.26 | 0.48 | Sorbitol dehydrogenase |
| IPI00004471a | 1 | 3.32 | 0.65 | Splice Isoform 2 of PDZ and LIM domain protein 3 |
| IPI00022488 | 1 | 3.41 | 0.53 | Hemopexin |
| IPI00553177 | 1 | 3.43 | 0.44 | Alpha-1-antitrypsin |
| IPI00384952 | 1 | 3.79 | 0.68 | Hypothetical protein DKFZp686K04218 |
| IPI00216088 | 1 | 4 | 0.8 | Cellular retinoic acid-binding protein 2 |
| IPI00215611 | 0.98 | 4.01 | 0.48 | Cysteine-rich protein 1 |
| IPI00217966a | 1 | 6.19 | 1.68 | Lactate dehydrogenase a |
| IPI00027434a | 1 | 10.26 | 1.17 | Rho-related GTP-binding protein RhoC |
Proteins identified as a group. See Supplementary Table 2.
FIG. 1.
Representative spectra and quantitative comparison of protein expression between chemoresistant (light) and chemosensitive (heavy) ovarian tissues using the ICAT approach. The ratio of peptides with identical sequence but different stable isotope labeling was quantified using the ASAPRatio. The quantification of the proteins was based on the quantification of the corresponding peptides for each protein identified. Please note that the scales of the y-axes were set automatically by the ASAPRatio program and are different from panel to panel. (A) Cytoplasmic actin. (B) Alpha-1-acid glycoprotein 1.
Gene ontology analysis of the differentially expressed chemotherapy response proteins
We realize that some of these differentially expressed proteins may represent tumor-to-tumor heterogeneity even the tumors are of the same histological subtype (serous for both tumors we analyzed) and tumor stage (stage IIIC for both tumors we analyzed). Each protein, when it is looked individually, has a high chance of being differentially expressed due to tumor heterogeneity. However, grouping differentially expressed proteins as functional groups may help to reduce the chance, especially when the function of a group of proteins explains potential mechanism of chemotherapy response. We used the Panther classification system (http://www.pantherdb.org/) to analyze our data in order to see which gene ontology (GO) terms were enriched in the differentially expressed proteins that we identified. We used the Bonferroni correction for multiple testing. Table 2 lists the 60 terms that are significantly enriched (p < 0.05). By molecular function, the extracellular matrix (ECM) GO term is significantly enriched in the proteins that were overexpressed in the chemoresistant tissue. In contrast, two GO terms nucleic acid binding and chaperonin were significantly enriched in the proteins that were over-expressed in the chemosensitive tissue.
Table 2.
Gene Ontology Classification of Chemotherapy Response Proteins Proteins Overexpressed in Chemosensitive Tissues
| Molecular function | NCBI: H. sapiens genes (REF) | Number of protein hits | Expected number of hits | p Value |
|---|---|---|---|---|
| Chaperone | 192 | 6 | 0.34 | 0.0000 |
| Nucleic acid binding | 2783 | 16 | 4.99 | 0.0004 |
| Chaperonin | 28 | 3 | 0.05 | 0.0031 |
| Biological process | ||||
| Protein complex assembly | 71 | 4 | 0.13 | 0.0013 |
| Protein folding | 205 | 5 | 0.37 | 0.0049 |
| Protein metabolism and modification | 2996 | 14 | 5.38 | 0.0153 |
| Proteins overexpressed in chemoresistant tissues | ||||
| Molecular function | ||||
| Extracellular matrix | 367 | 8 | 0.53 | 0.0000 |
| Biological process | ||||
| Immunity and defense | 1324 | 9 | 1.92 | 0.0026 |
| Skeletal development | 122 | 4 | 0.18 | 0.0060 |
| Mesoderm development | 545 | 6 | 0.79 | 0.0178 |
| Extracellular matrix protein-mediated signaling | 61 | 3 | 0.09 | 0.0198 |
To further reduce the chance that the differentially expressed protein identified are due to small sample size and related to individual differences rather than differences in chemotherapy responses, we performed integrative analysis with a large dataset of gene expression profiling that consists of 8 chemotherapy-resistant tissues and 13 chemotherapy-sensitive tissues. Using an average ratio of 1.5-folds as the cutoff value, we identified a total of 16 proteins whose expression changes are consistent at both the protein and the RNA levels. They include 14 proteins that are underexpressed genes in chemoresistant tissues such as P53 binding protein 1, catenin delta 1, and plakoglobin (with average ratios of 0.59, 0.48, and 0.53, respectively, comparing 8 chemoresistant to 13 chemsensitive tissues) (Table 3). In addition, two interesting protein EGF-containing fibulin-like extracellular matrix protein 1 and voltage-dependent anion-selective channel protein 1 are consistently overexpressed at both the protein and the RNA levels in chemoresistant tissues compared to chemosensitive tissues (Table 3).
Table 3.
Consistent Differential Expression Changes at Both Protein and RNA Levels
| IPI number | Protein ratios | Array ratiosa | Description |
|---|---|---|---|
| IPI00438229 | 0.14 | 0.31 | Transcription intermediary factor 1-beta |
| IPI00013890 | 0.49 | 0.39 | 14-3-3 protein sigma, SFN protein |
| IPI00023785 | 0.48 | 0.51 | Probable ATP-dependent RNA helicase DDX17 |
| IPI00027175 | 0.39 | 0.67 | sorcin |
| IPI00029778 | 0.44 | 0.59 | Tumor suppressor p53-binding protein 1 |
| IPI00182469 | 0.45 | 0.49 | Catenin delta-1 |
| IPI00290770 | 0.34 | 0.29 | Chaperonin containing TCP1, subunit 3 |
| IPI00297284 | 0.5 | 0.25 | Insulin-like growth factor-binding protein 2 |
| IPI00306332 | 0.47 | 0.48 | 60S ribosomal protein L24 |
| IPI00328343 | 0.34 | 0.41 | Spliceosome RNA helicase BAT1,26-kDa protein, HLA-B associated transcript 1 |
| IPI00387168 | 0.35 | 0.59 | Proprotein convertase subtilisin\kexin type 9 |
| IPI00414320 | 0.22 | 0.36 | Annexin A11 |
| IPI00550995 | 0.37 | 0.47 | Hypothetical protein NOP17 |
| IPI00554711 | 0.47 | 0.53 | Junction plakoglobin (gamma catenin) |
| IPI00029658 | 2.95 | 1.56 | EGF-containing fibulin-like extracellular matrix protein 1 |
| IPI00216308 | 1.98 | 1.54 | Voltage-dependent anion-selective channel protein 1 |
Average of 8 Chemo_R to 13 Chemo_S ratio.
Discussion
Identification of differentially expressed chemotherapy response proteins between chemosensitive and chemoresistant ovarian cancer tissues
Using a Protein Prophet cutoff score of 0.9, which corresponds to a 0.8% overall false-positive rate, we identified 502 proteins (or protein groups) in the tissue sample (Supplementary Table 2), which can be added to the protein catalog of the ovarian cancer tissues. These 502 proteins only represented a subset of the proteome. However, they are not necessary the most abundant 502 proteins as our protocol only analyze tryptic peptides containing cysteine as the ICAT reagent labels cysteine residues, and trypsin was used for digestion. In silico digestion of the human Swiss-Prot protein database revealed that only 22.5% of tryptic peptides contained cysteine residues. Selection and enrichment of the cysteinyl peptides, as ICAT/MS/MS protocol did, provides a significant technical advantage for detecting low abundance proteins by effectively reducing sample complexity and dynamic range (Gygi et al., 2002; Liu et al., 2005). Moreover, the reduction of sample complexity is not necessary linear with protein abundance, but is dependent on how many cysteine containing tryptic peptides a protein has. We also included those protein identified by single cysteine-containing tryptic peptides as often there is only one cysteine-containing tryptic peptides in a protein in the mass range detectable by the mass spectrometry. Table 1 also includes this information so that a user can use the number of unique peptide identified as an indication of the confidence in protein identification. Recently Vaughn et al. (2006) analyzed the characteristics of cleavable isotope-coded affinity tag (cICAT)-LC-tandem mass spectrometry for quantitative proteomic studies and they found that most of the proteins were identified with single cysteine-containing peptides (<4% of the proteins were identified by more than one unique peptide in three of the six cICAT experiments). They also compared the quantification results by Western blot analysis and by cICAT-LC-MS/MS based on single peptides. They concluded that identifications based on single cICAT-labeled peptides with tryptic ends provide sufficiently reliable protein identifications and quantification information in cICAT-LC-MS/MS-based proteomic studies (Vaughn et al., 2006).
Extracellular matrix proteins are overexpressed in the chemoresistant tissue compared to the chemosensitive tissue
Among the overexpressed proteins identified in the chemoresistant tumor sample, we found 10 proteins (25.6%) in the ECM, including decorin, versican, basigin (CD147), fibulin-1, extracellular matrix protein 1, biglycan, fibronectin 1, dermatopontin, alpha-cardiac actin (smooth muscle actin), and an EGF-containing fibulin-like extracellular matrix protein 1 (Table 1). Decorin is a proteoglycan component of the cellular matrix, and a recent study suggested that overexpression of decorin in pancreatic cancer attenuated the cytostatic action of carboplatin and gemcitabine toward pancreatic cancer cells (Koninger et al., 2004). Decorin, as well as other ECM genes (collagen XI and VI, cartilage linking protein 1), was also shown to be overexpressed in cisplatin-resistant ovarian cancer cells by SAGE analysis (Sherman-Baust et al., 2003).
Elevated levels of ECM proteins such as laminin-gamma2, collagen types I and III, fibronectin, syndecan-1, glypican-1, versican, and hyaluronan, and its receptor CD44 have all been associated with a poor prognosis of ovarian cancers (Ricciardelli and Rodgers 2006). For example, a high percentage of strong stromal versican (chondroitin sulfate proteoglycan 2) staining was associated with reduced 5-year survival rates of ovarian cancer patients (44 vs. 32%; p = 0.032) (Voutilainen et al., 2003). Basigin is an ECM metalloproteinase inducer (EMMPRIN), overexpressed in ovary tumors, and might be a prognostic marker for ovarian carcinomas (Davidson et al., 2003; Jin and others 2006). Fibulin 1 is an estrogen-regulated protein that has increased expression in the stroma of human ovarian epithelial tumors (Roger et al., 1998). Dermatopontin is a tyrosine-rich acidic ECM protein that was shown to enhance tumor growth. A mouse dermatopontin in transgenic mice under the control of the rat probasin promoter showed prostate intraepithelial neoplasia at the age of 11 months, whereas the control mice did not (Takeuchi et al., 2006).
In vertebrates there are three main groups of actin isoforms, alpha, beta, and gamma. The beta and gamma actins coexist in most cell types as components of the cytoskeleton and as mediators of internal cell motility, and the alpha actins are found in muscle tissues. We identified alpha actin, but not beta and gamma actins, as being overexpressed in the chemoresistant tissue. Globular alpha actin (G-actin) can be polymerized to form a structural filament (F-actin) in the form of a two-stranded helix. One can postulate that alpha actins could form structure filaments, and therefore reduce the penetrations of chemo drugs to cancer cells. Further experimentation is necessary to test this hypothesis.
Potential causes of drug resistance in solid tumors include genetically determined factors expressed in individual cells as well as factors related to the solid tumor environment. The latter is relevant when considering the need for drugs to penetrate into tumor tissue in order to achieve a lethal concentration in all of the tumor cells. Tannock et al. (2002) showed that the penetration of cisplatin, paclitaxel, and other chemo drugs through multiple cell layers of tumor tissue grown on collagen-coated semiporous Teflon membranes was much slower compared with penetration through the Teflon support membrane alone. They suggested that the limited penetration of anticancer drugs through tumor tissue might be an important cause of clinical resistance of solid tumors to chemotherapy. Sherman-Baust et al. (2003) showed that remodeling of the ECM through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Our data further support the hypothesis that chemoresistance is associated with increased expression of ECM proteins, which potentially limits the penetration of anticancer drugs through the ECM to the tumor cells.
When we compared the protein expression changes with the gene expression changes from 8 chemoresistant and 13 chemosensitive tissues, we found that the ECM/stromal proteins that we identified as overexpressed in chemoresistant tissues at protein levels are not overexpressed at RNA levels at the 1.5-fold cutoff value, suggesting that the difference in abundance of these ECM proteins maybe more likely related to protein turnover rate of the ECM proteins rather than new protein synthesis that requires new RNA synthesis.
Other interesting proteins that were overexpressed in the chemoresistant tissues include cellular retinoic acid-binding protein 2, Rho-related GTP-binding protein RhoC, CD163 and several glycoproteins (Table 1). The functional roles of these proteins in the chemotherapy response remain to be investigated.
Chaperone and nucleic acid binding proteins are enriched in the proteins overexpressed in the chemosensitive tissue compared to the chemoresistant tissue
By GO molecular functions, chaperone and nucleic acid binding GO terms are enriched in the proteins overexpressed in the chemosensitive tissue (Table 2). Cisplatin acts to kill cells by binding to DNAs and interfering with the cellular DNA repair mechanism (Jamieson and Lippard, 1999). A few of the nucleic acid binding proteins identified are cancer markers themselves. For example, DeSouza et al. (2005) identified HNPRD as a marker over expressed by 4.62-fold in endometrial cancer compared to normal tissues using a combination of iTRAQ (Wiese et al., 2007) and cICAT approaches combined with multidimensional liquid chromatography and tandem mass spectrometry (DeSouza et al., 2005). HNPRD binds to AU-rich elements contained in the 3′ untranslated region of many short-lived mRNAs and controls the stability of AU-rich element-containing mRNAs, including mRNAs encoding proto-oncogenes, cytokines, or other signaling molecules (Nagata et al., 1999). HNPRD overexpression leads to tumorigenesis in transgenic mice (Gouble et al., 2002).
We also identified three chaperon proteins that are overexpressed in the chemosensitive tissue (Table 1). They are the 70 kDa heat-shock protein (Hsp70), chaperonin containing TCP1 subunit 3, and 14-3-3 protein sigma. Two of them, chaperonin containing TCP1 subunit 3 and 14-3-3 protein sigma, are also differentially expressed at RNA levels comparing 8 chemoresistant to 13 chemosensitive tissues (Table 3). These chaperon proteins were previously reported to be associated with ovarian cancers. For example, Akahira et al. (2004) showed that decreased expression of 14-3-3 sigma was associated with advanced disease in human epithelial ovarian cancer. The roles of TCP1 and 14-3-3 protein sigma in chemotherapy responses remain to be investigated. Our result that heat-shock protein 70 Kda (Hsp70) is overexpressed in the chemosensitive tissue is different from the general belief that overproduction of heat-shock proteins fosters resistance to chemotherapeutic agents and irradiation. Although our observation could be an exception, the exact role of Hsp70 in modulating the chemotherapy response needs to be revisited. Different heat-shock proteins may have different specific functions. For example, inhibition of Hsp90 has been used to combat various forms of cancer, and the induction of Hsp70 has been used to help the recovery of patients from diseases such as ischemic heart disease, diabetes, and neurodegeneration (Soti et al., 2005).
Gamma-catenin (junction plakoglobin) and delta-catenin are overexpressed in the chemosensitive tissue compared to the chemoresistant tissue
Interestingly, we identified two catenins, gamma-catenin (junction plakoglobin) and delta-catenin, to be overexpressed in the chemosensitive tissue. These two proteins are also differentially expressed at RNA levels comparing 8 chemoresistant to 13 chemosensitive tissues (Table 3). The plakoglobin gene localizes on chromosome 17q21 and is subjected to loss of heterozygosity in breast and ovarian cancers. Davidson et al. (2000) showed that E-cadherin and alpha-, beta-, and gamma-catenin protein expression is upregulated in ovarian carcinoma cells from serous effusions and metastatic lesions. Nuclear gamma-catenin expression was associated with serous histology and poor differentiation (Voutilainen et al., 2006). Liang et al. (2004) showed reduced levels of gamma-catenin in two resistant cancer cell lines, human KB epidermoid adenocarcinoma cells (KB-CP) and human BEL7404 hepatoma cells (7404-CP). Their result is consistent with our observation that gamma-catenin was overexpressed in chemosensitive tissue. Catenins are linked to transmembrane protein cadherins. Gamma-catenin (junction plakoglobin) and delta-catenin are both junction plaque proteins (Franke et al., 1989; Keirsebilck et al., 1998). These two catenins may regulate the adherens junction, consisting of E-cadherin and members of the catenin family, thereby increasing drug uptake and chemosensitivity. Consistent with this, we also identified a cell adhesion protein (putative matrix cell adhesion molecule-3) that is overexpressed in chemosensitive tissue (Table 1). Therefore, cell adhesion and cell junction proteins might play roles in modulating chemosensitivity of ovarian cancer cells.
Other interesting proteins that we identified as overexpressed in the chemosensitive tissue include tumor suppressor p53-binding protein 1 (53BP1), insulin-like growth factor-binding protein 2 (IGFBP2), proliferating cell nuclear antigen (PCNA), annexin A11, and 53-kDa selenium binding protein 1. Among them, 53BP1, IGFBP2, annexin A11 are also differentially expressed at RNA levels comparing eight chemoresistant to thirteen chemosensitive tissues (Table 3).
It is worth noting that the p53 gene itself has been thoroughly studied in ovarian cancers and in chemotherapy responses (Kigawa et al., 2001). It was shown that nonresponders (chemoresistant) to platinum-based chemotherapy had a higher frequency of p53 mutations than responders (chemosensitive) had (83% for nonresponders vs. 16% for responders) (Kigawa et al., 2001). Introduction of the p53 gene into cells markedly enhanced the sensitivity to cisplatin and cisplatin-induced apoptosis. The combination treatment using a recombinant adenovirus carrying a wild-type p53 gene (AxCAp53) and cisplatin showed significantly greater growth suppression of ovarian cancer cells in an ovarian cancer xenograft model, compared with treatments of either AxCAp53 or cisplatin alone. More recently, Yan et al. (2006) showed that overexpression of PTEN upregulated p53 content and increased the sensitivity of chemoresistant cells to cisplatin-induced apoptosis independent of the PI3K/Akt pathway. We have identified p53 binding protein 1 (53BP1) as overexpressed in the chemosensitive tissue. If it is confirmed by further study, this protein may be useful as a novel therapeutic target to increase chemosensitivity of ovarian cancer cells.
Conclusions
It is our point of view that response to chemotherapy involves systems properties at multiple levels in each individual patient, which include liver drug metabolic rate, kidney drug clearance rate, stromal cell barriers to drug penetrance and tumor cell themselves. Well-established mechanisms of cisplatin's action are that cisplatin and carboplatin form DNA interstrand and intrastrand crosslinks, and thus cause DNA damage and result in apoptosis of cells. Mechanisms of cisplatin resistance identified so far include alterations in cellular drug transport, enhanced DNA repair, and enhanced intracellular detoxification involving glutathione and metallothionein proteins (Agarwal and Kaye 2003; Gosland et al., 1996). Paclitaxel (Taxol) blocks cell-cycle progression through centrosomal impairment, induction of abnormal spindle formations and suppression of spindle microtubule turnover (Abal et al., 2003). Mechanisms of paclitaxel resistance include tubulin mutations, tubulin isotype selection, and posttranslational modifications, and dysfunction of other regulatory proteins (Orr et al., 2003). Our results showed that ECM and cell adhesion and junction proteins might all play important roles in modulating chemotherapy responses. Our data provide novel insights to understanding the mechanism of chemotherapy response, and may offer guidance to future experimentation and to developing novel approaches to overcome chemotherapy resistance in ovarian cancer cells.
We admit that a major weakness of this investigation is that the sample size is limited by the limited availability of well-defined specimen, as it is difficult to obtain naïve chemotherapy-sensitive and -resistant tissues. Most ovarian cancers demonstrate partial responses to chemotherapy, probably due to the existence of mixed sensitive and resistant cells in the primary tumors. We have tried hard to obtain a truly chemosensitive tissue with progression-free interval of 23 months (serous histology, stage IIIC ovarian cancer) and one chemoresistant tissue with immediate (progression-free interval of 0 months) cancer progression despite of chemotherapy. Among the differentially expressed proteins that we identified, it is possible that some of them may be due to interperson heterogeneity. However, the identification of multiple differentially expressed proteins that belong to the same functional group (e.g., same GO Terms) strongly suggest a functional association with disease phenotype rather than random tissue heterogeneity. In addition, many of the proteins we identified have been previously reported in association with ovarian cancer and chemotherapy response as we discussed in the previous sections, confirming their roles in chemotherapy response. Furthermore, we compared the proteomics changes with transcriptomics data consisting of 8 chemoresistant and 13 chemosensitive tissues and identified that a subset of the proteins changed at both the protein and the RNA levels, suggesting those changes are probably true changes relevant to the biology chemotherapy response (Table 3). Further experimentation will be necessary to determine whether the differentially proteins we identified have causal roles in chemotherapy response, which can be used as novel drug targets, or whether they are merely associated with chemotherapy response, which can be used as biomarkers to monitor chemotherapy response for ovarian cancers.
Acknowledgments
This publication was made possible by grant 5P50CA 083636, 1P50GM076547, 5U54CA119347 from NIH, and a grant from Marsha Rivkin Center for Ovarian Cancer Research, and grant 2006AA02Z4A2, 2006DFA32950, 2007DFC30360 from MOST, China.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Abal M., Andreu J.M., and Barasoain I. (2003). Taxanes: microtubule and centrosome targets, and cell cycle dependent mechanisms of action. Curr Cancer Drug Targets 3, 193–203 [DOI] [PubMed] [Google Scholar]
- Aebersold R., and Mann M. (2003). Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- Aebersold R., Anderson L., Caprioli R., Druker B., Hartwell L., and Smith R. (2005). Perspective: a program to improve protein biomarker discovery for cancer. J Proteome Res 4, 1104–1109 [DOI] [PubMed] [Google Scholar]
- Agarwal R., and Kaye S.B. (2003). Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer 3, 502–516 [DOI] [PubMed] [Google Scholar]
- Akahira J., Sugihashi Y., Suzuki T., Ito K., Niikura H., Moriya T., et al. (2004). Decreased expression of 14-3-3 sigma is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin Cancer Res 10, 2687–2693 [DOI] [PubMed] [Google Scholar]
- Davidson B., Berner A., Nesland J.M., Risberg B., Berner H.S., Trope C.G., et al. (2000). E-cadherin and alpha-, beta-, and gamma-catenin protein expression is up-regulated in ovarian carcinoma cells in serous effusions. J Pathol 192, 460–469 [DOI] [PubMed] [Google Scholar]
- Davidson B., Goldberg I., Berner A., Kristensen G.B., and Reich R. (2003). EMMPRIN (extracellular matrix metalloproteinase inducer) is a novel marker of poor outcome in serous ovarian carcinoma. Clin Exp Metastasis 20, 161–169 [DOI] [PubMed] [Google Scholar]
- Desouza L., Diehl G., Rodrigues M.J., Guo J., Romaschin A.D., Colgan T.J., et al. (2005). Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J Proteome Res 4, 377–386 [DOI] [PubMed] [Google Scholar]
- Eng J., McCormack A., and Yates J. (1994). An approach to correlate tandem mass special data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 [DOI] [PubMed] [Google Scholar]
- Franke W.W., Goldschmidt M.D., Zimbelmann R., Mueller H.M., Schiller D.L., and Cowin P. (1989). Molecular cloning and amino acid sequence of human plakoglobin, the common junctional plaque protein. Proc Natl Acad Sci USA 86, 4027–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosland M., Lum B., Schimmelpfennig J., Baker J., and Doukas M. (1996). Insights into mechanisms of cisplatin resistance and potential for its clinical reversal. Pharmacotherapy 16, 16–39 [PubMed] [Google Scholar]
- Gouble A., Grazide S., Meggetto F., Mercier P., Delsol G., and Morello D. (2002). A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res 62, 1489–1495 [PubMed] [Google Scholar]
- Gygi S.P., Rist B., Gerber S.A., Turecek F., Gelb M.H., and Aebersold R. (1999). Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17, 994–999 [DOI] [PubMed] [Google Scholar]
- Gygi S.P., Rist B., Griffin T.J., Eng J., and Aebersold R. (2002). Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J Proteome Res 1, 47–54 [DOI] [PubMed] [Google Scholar]
- Hall M.D., Amjadi S., Zhang M., Beale P.J., and Hambley T.W. (2004). The mechanism of action of platinum(IV) complexes in ovarian cancer cell lines. J Inorg Biochem 98, 1614–1624 [DOI] [PubMed] [Google Scholar]
- Jamieson E.R., and Lippard S.J. (1999). Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev 99, 2467–2498 [DOI] [PubMed] [Google Scholar]
- Jin J.S., Yao C.W., Loh S.H., Cheng M.F., Hsieh D.S., and Bai C.Y. (2006). Increasing expression of extracellular matrix metalloprotease inducer in ovary tumors: tissue microarray analysis of immunostaining score with clinicopathological parameters. Int J Gynecol Pathol 25, 140–146 [DOI] [PubMed] [Google Scholar]
- Keirsebilck A., Bonne S., Staes K., Van Hengel J., Nollet. F., Reynolds A., et al. (1998). Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics 50, 129–146 [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., and Aebersold R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kigawa J., Sato S., Shimada M., Takahashi M., Itamochi H., Kanamori Y., et al. (2001). p53 gene status and chemosensitivity in ovarian cancer. Hum Cell 14, 165–171 [PubMed] [Google Scholar]
- Koninger J., Giese N.A., Di Mola F.F., Berberat P., Giese T., Esposito I., et al. (2004). Overexpressed decorin in pancreatic cancer: potential tumor growth inhibition and attenuation of chemotherapeutic action. Clin Cancer Res 10, 4776–4783 [DOI] [PubMed] [Google Scholar]
- Le Moguen K., Lincet H., Marcelo P., Lemoisson E., Heutte N., Duval M., et al. (2007). A proteomic kinetic analysis of IGROV1 ovarian carcinoma cell line response to cisplatin treatment. Proteomics 7, 4090–4101 [DOI] [PubMed] [Google Scholar]
- Li X.J., Zhang H., Ranish J.A., and Aebersold R. (2003). Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem 75, 6648–6657 [DOI] [PubMed] [Google Scholar]
- Liang X.J., Shen D.W., and Gottesman M.M. (2004). Down-regulation and altered localization of gamma-catenin in cisplatin-resistant adenocarcinoma cells. Mol Pharmacol 65, 1217–1224 [DOI] [PubMed] [Google Scholar]
- Liu T., Qian W.J., Chen W.N., Jacobs J.M., Moore R.J., Anderson D.J., et al. (2005). Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: the human mammary epithelial cell proteome. Proteomics 5, 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire W.P., Hoskins. W.J., Brady M.F., Kucera P.R., Partridge E.E., Look K.Y., et al. (1996). Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med 334, 1–6 [DOI] [PubMed] [Google Scholar]
- Nagata T., Kurihara Y., Matsuda G., Saeki J., Kohno T., Yanagida Y., et al. (1999). Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0. J Mol Biol 287, 221–237 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., and Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- Orr G.A., Verdier-Pinard P., McDaid H., and Horwitz S.B. (2003). Mechanisms of Taxol resistance related to microtubules. Oncogene 22, 7280–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardelli C., and Rodgers R.J. (2006). Extracellular matrix of ovarian tumors. Semin Reprod Med 24, 270–282 [DOI] [PubMed] [Google Scholar]
- Roger P., Pujol P., Lucas A., Baldet P., and Rochefort H. (1998). Increased immunostaining of fibulin-1, an estrogen-regulated protein in the stroma of human ovarian epithelial tumors. Am J Pathol 153, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman-Baust C.A., Weeraratna A.T., Rangel L.B., Pizer E.S., Cho K.R., Schwartz D.R., et al. (2003). Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell 3, 377–386 [DOI] [PubMed] [Google Scholar]
- Soti C., Nagy E., Giricz Z., Vigh L., Csermely P., and Ferdinandy P. (2005). Heat shock proteins as emerging therapeutic targets. Br J Pharmacol 146, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.J., White J.T., Yan X., Collins S., Drescher C.W., Urban N.D., et al. (2006). Proteins associated with Cisplatin resistance in ovarian cancer cells identified by quantitative proteomic technology and integrated with mRNA expression levels. Mol Cell Proteomics 5, 433–443 [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Suzuki M., Kumagai J., Kamijo T., Sakai M., and Kitamura T. (2006). Extracellular matrix dermatopontin modulates prostate cell growth in vivo. J Endocrinol 190, 351–361 [DOI] [PubMed] [Google Scholar]
- Tannock I.F., Lee C.M., Tunggal J.K., Cowan D.S., and Egorin M.J. (2002). Limited penetration of anticancer drugs through tumor tissue: a potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res 8, 878–884 [PubMed] [Google Scholar]
- Vasey P.A. (2003). Resistance to chemotherapy in advanced ovarian cancer: mechanisms and current strategies. Br J Cancer 89 (Suppl 3), S23–S28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn C.P., Crockett D.K., Lim M.S., and Elenitoba-Johnson K.S. (2006). Analytical characteristics of cleavable isotope-coded affinity tag-LC-tandem mass spectrometry for quantitative proteomic studies. J Mol Diagn 8, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutilainen K., Anttila M., Sillanpaa S., Tammi R., Tammi M., Saarikoski S., et al. (2003). Versican in epithelial ovarian cancer: relation to hyaluronan, clinicopathologic factors and prognosis. Int J Cancer 107, 359–364 [DOI] [PubMed] [Google Scholar]
- Voutilainen K.A., Anttila M.A., Sillanpaa S.M., Ropponen K.M., Saarikoski S.V., Juhola M.T., et al. (2006). Prognostic significance of E-cadherin-catenin complex in epithelial ovarian cancer. J Clin Pathol 59, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese S., Reidegeld K.A., Meyer H.E., and Warscheid B. (2007). Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 [DOI] [PubMed] [Google Scholar]
- Yan X., Fraser M., Qiu Q., and Tsang B.K. (2006). Overexpression of PTEN sensitizes human ovarian cancer cells to cisplatin-induced apoptosis in a p53-dependent manner. Gynecol Oncol 102, 348–355 [DOI] [PubMed] [Google Scholar]
- Yi E.C., Lee H., Aebersold R., and Goodlett D.R. (2003). A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun Mass Spectrom 17, 2093–2098 [DOI] [PubMed] [Google Scholar]