Abstract
We show that the clinical phenotype associated with BRD4 haploinsufficiency overlaps with Cornelia de Lange syndrome (CdLS) – most often caused by mutation of NIPBL. More typical CdLS was observed with a de novo BRD4 missense variant, which retains the ability to co-immunoprecipitate with NIPBL but which binds poorly to acetylated histones. BRD4 and NIPBL display correlated binding at super-enhancers and appear to co-regulate developmental gene expression.
Keywords: BRD4, NIPBL, Cohesin, Cornelia de Lange syndrome, Enhancer, De Novo Mutation, Super-enhancer
Super-enhancers are clustered cis-regulatory elements (CRE) controlling genes important for cell type specification. Super-enhancers are molecularly defined as genomic intervals with high levels of H3K27 acetylation and binding of both BRD4 and the mediator complex1. A critical role for cohesin in super-enhancer function has been recently reported 2. Acute depletion of cohesin resulted in disruption of higher-order chromatin structure and disordered transcription of genes predicted to be under super-enhancer control. The most widely studied human cohesinopathy is Cornelia de Lange syndrome (CdLS), a severe multisystem neurodevelopmental disorder, which is associated with a generalized deregulation of developmental genes3,4. Typical CdLS is caused by heterozygous or mosaic loss-of-function (LOF) mutations in the gene encoding NIPBL. NIPBL is recruited to sites of double-strand breaks in DNA5 and functions as a transcriptional activator6 but it is best known for its role in loading the cohesin complex onto DNA and is required for cohesin-mediated loop extrusion and TAD formation7,8 9. Causative mutations in the genes encoding the core components of the cohesin ring (SMC1A, SMC3, RAD21)10–12 and the SMC3 deacetylase HDAC813 have been identified in CdLS-like conditions. However, individuals with de novo mutations in the chromatin associated/modifying proteins ANKRD1114,15, KMT2A4 and AFF416, which have no known association with cohesin can also present with CdLS-like disease.
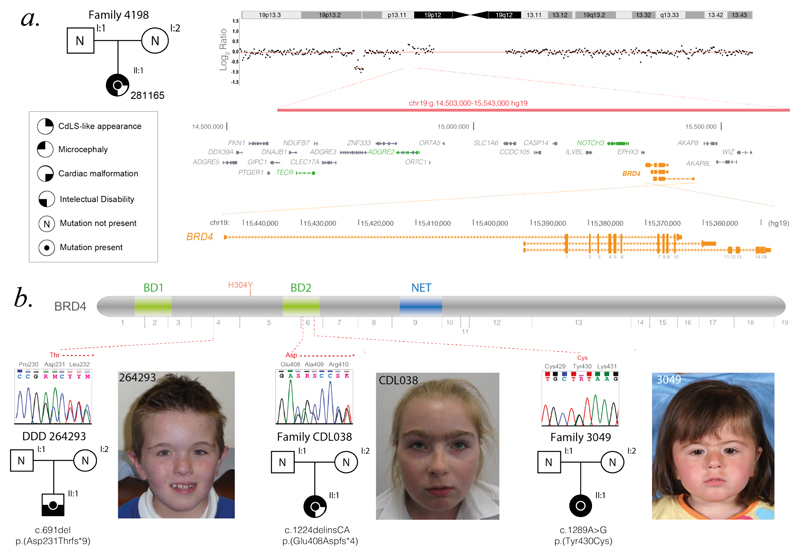
To identify novel disease loci we studied 92 individuals with CdLS in whom no plausibly diagnostic variants could be identified in the known causative genes. In this group we identified 2/92 (2.2%) individuals with de novo mutations affecting BRD4. The first individual had a CdLS-like condition and a heterozygous 1.04 Mb deletion encompassing BRD4 (plus 28 other protein-coding genes (Family 4198; DECIPHER 281165)) (Fig. 1A, Supplemental Fig. 1&2). Targeted re-sequencing in the remaining 91 affected individuals identified an individual, with more typical CdLS, who has a de novo missense mutation located in the second bromodomain (BD2) of BRD4 (3049; NM_058243.2 c.1289A>G, p.(Tyr430Cys))(Fig. 1B, Supplemental Fig. 5). Subsequently two affected individuals who were not part of the original cohort were identified with de novo frame-shift mutations in BRD4. The first of these was identified via ongoing screening of individuals with CdLS-like disorders (CDL038 NM_058243.2 c.1224delinsCA p.(Glu408Aspfs*4) ; Fig. 1B). The second indel variant was discovered through analysis of trio whole exome sequencing data generated by the Deciphering Developmental Disorders study 17 (DDD; DECIPHER 264293 NM_058243.2 c.691del p.(Asp231Thrfs*9); Fig. 1B). The latter individual was recruited to DDD on the basis of intellectual disability, mild short stature and a ventricular septal defect but had not been suspected to have CdLS (Supplemental Table 1). On review of 7 reported heterozygous multigenic deletions encompassing BRD4 we found a significant phenotypic overlap with CdLS (Supplemental Fig. 3; Supplemental Table 1) with at least 2/7 fulfilling the established CdLS diagnostic criteria18. Taken together; these data support BRD4 haploinsufficiency as the likely genetic mechanism for the CdLS-like phenotype.
Figure 1. BRD4 mutations in CdLS and CdLS-like disorders.
A. Pedigree drawing of proband with CdLS-like disorder associated with a de novo 1.04 Mb microdeletion of 19p (red bar), the location of which is shown on the Log2 ratio plot of the array-based comparative genomic hybridization. Below on the left is a key to the symbols using in the pedigree images. To the right of this is the RefSeq gene content of the deleted region with the location of BRD4 indicated in orange. The genes colored green are known disease genes associated with phenotypes that are not consistent with the clinical presentation in this case. Details of each is given in the supplementary notes. B. Pedigree drawings and facial photographs of probands with intragenic mutations of BRD4, with cartoon of BRD4 protein indicating the position of each of the variants in relation to the first bromodomain (BD1), the second bromodomain (BD2) and the N-terminal extra terminal domain (NET). The position of an inherited p.His304Tyr variant (orange text) reported in a single family with inherited cataracts is indicated and discussed in supplementary notes.
It has been previously reported that mice carrying heterozygous LOF mutations in Brd4 show marked early postnatal mortality, severe prenatal onset growth failure, abnormalities of the craniofacial skeleton and reduced body fat19; all features common in CdLS. Brd4 homozygous null embryos die soon after implantation. Heterozygous LOF mutations in only 12 other non-imprinted autosomal mouse genes have both postnatal lethality and postnatal growth retardation recorded as features in the Mouse Genome Informatics database (MGI); one of these being Nipbl 20 (Supplemental Fig. 4A-C). 4 of these 13 haploinsufficient mouse genes have been implicated in super-enhancer function (Brd4, Nipbl, Chd7 and Crebbp)1,21 (Supplemental Fig. 4E).
BRD4, is a member of the bromodomain and extraterminal domain (BET) protein family with tandem bromodomains that ‘read’ acetylated lysine marks on chromatin. BRD4 binds mostly to hyper-acetylated genomic regions that encompass promoters and enhancers and BRD4 levels are particularly high at super-enhancers22. BRD4 regulates transcription elongation by paused RNA polymerase II (Pol II) via mediating the release of Cdk9 activity, which results in phosphorylation of serine 2 of Pol II C-terminal domain (CTD). Tyr430, the residue substituted in individual 3049 with the more typical CdLS phenotype (Fig. 1B), lies within the third alpha helix (αB) of the second bromodomain (BD2) of BRD4; close to the recognition site that mediates binding to acetylated lysine23. p.Tyr430Cys (Y430C) is a non-conservative amino acid substitution which could plausibly impair the binding of BRD4 to acetyl lysine. Indeed, compared to wild-type BD1 and BD2, a tagged BRD4 BD2 containing the Y430C mutation shows reduced binding to acetylated histone peptides in vitro (Fig. 2A). In mouse BRD4 the “human equivalent” missense variant would be p.Tyr431Cys; the difference in amino acid numbering is the result of an “extra” proline in the poly-proline repeat (position 215-217 in the human protein; Supplemental Fig. 6). To avoid confusion we will use Brd4Y430C as the mouse variant designation; we introduced this variant onto both alleles (Brd4Y430C/Y430C) of mouse embryonic stem cell (mESC) lines by Cas9-induced homology directed repair (HDR). BRD4 immuno-precipitation (IP) in Brd4Y430C/Y430C mESC shows impaired binding to acetylated histones (H3K9ac and K3K27ac) (Fig. 2B, Supplemental Fig. 18).
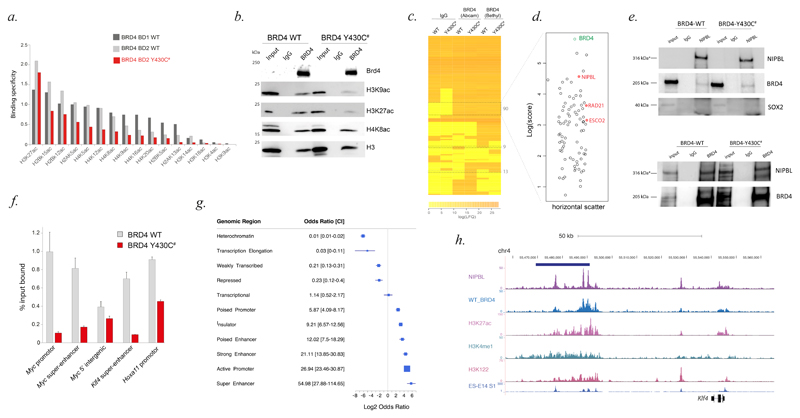
Figure 2. Binding of BRD4 wild-type and BRD4 p.Tyr430Cys variant to histone and non-histone proteins.
A. Specificity of binding to acetylated histone tail peptides of wild-type BRD4 Bromodomain 1 (BRD4 BD1 WT), wild-type BRD4 Bromodomain 2 (BRD4 BD2 WT), and BRD4 p.Tyr430Cys mutant BD2 (BRD4 BD2 Y430C). B. Cropped immunoblots of endogenous BRD4 IPs and rabbit normal IgG (control) from Brd4 wild-type and Brd4Y430C/Y430C mESCs. Input =1% of mESC nuclear extract. Antibodies detect BRD4, H3K9ac, H3K27ac, H4K8ac and H3. C. Heatmap of the label-free mass spectrometry quantitative output values (average of triplicates) assigned to each protein following IP from BRD4 wild-type (WT) and BRD4-Y430C (MUT) mESC using IgG only control or Abcam/Bethyl antibody against BRD4. D. A plot of the log Andromeda scores assigned to the 90 proteins which are absent in the IgG controls and present in both cell lines using both BRD4 antibodies. Horizontal scatter aids visibility of each open circle and has no data correlate. E. Cropped immunoblot of reciprocal IP using BRD4 and NIPBL antibodies in Brd4 wild-type and Brd4Y430C/Y430C mESCs. Antibodies detect BRD4, NIPBL and SOX2. F. Percentage (%) input bound for BRD4 ChIP-qPCR across genomic regions in WT and BRD4 Y430C mutant MEFs (error bars = standard error of the mean from n=2 biological replicates). G. Forest plot of the log2 odds ratio with confidence intervals (CI) of different functional genomic categories within intersecting regions from BRD4 and NIPBL mESC ChIP. H. UCSC Genome Browser graphic showing colocalisation of BRD4 and NIPBL ChIPseq peaks over the super enhancer (blue bar) at Klf4 locus. H3K27ac, H3K4me1, H3K122ac and super enhancer tracks are previously published.
Label-free quantitative (LFQ) mass spectrometry (MS) following IP was performed using two different BRD4 antibodies on lysates from Brd4Y430C/Y430C and wild-type mESCs (Supplemental Table 4). This detected 1,082 proteins present in BRD4 IP from both cell lines, 90 of which were absent in all IgG controls (Fig. 2C). Of these, BRD4 was the top hit with three of the remaining 89 proteins being NIPBL, Rad21 (core cohesin ring component) and Esco2 (SMC3 acetylase) (Fig. 2D, Supplemental Fig. 7). Other subunits of cohesin (SMC1A, SMC3, STAG2, PDS5A, PDS5B) also showed evidence of enrichment (Supplemental Fig. 8). The association of BRD4 with NIPBL was replicated using LFQ MS on an independent Brd4Y430C/Y430C mESC line created using the same genome editing protocol. Reciprocal IPs using antibodies to NIPBL and SMC3 confirmed the BRD4 interaction with both NIPBL and the core cohesin ring (Fig. 2E, Supplemental Fig. 9&20). In mESC Brd4Y430C/Y430C shows a similar level of NIPBL association to wild-type Brd4, suggesting that this interaction is unlikely to be mediated via co-binding to acetylated chromatin (Fig. 2E, Supplemental Fig. 19).
In order to further assess the functional consequences of the BRD4 missense variant we generated F0 mouse embryos following zygote injections of reagents to induce Cas9-mediated HDR. As judged from digital sectioning from optical projection tomography, the morphology of Brd4Y430C/+ and Brd4Y430C/Y430C F0 mouse embryos is indistinguishable from wild-type embryos. We also generated apparently non-mosaic F0 embryos homozygous for a 15bp in-frame deletion (NM_020508.4 c.1288_1302del; p.(Cys430_Asn434del; Supplemental Fig. 10); designated as Brd4C429_N433del/ C429_N433del to maintain consistency with human nomenclature) showing significant growth restriction at 13.5 dpc as their only obvious phenotype (Supplemental Fig. 11). We derived mouse embryonic fibroblasts (MEF) from 13.5 dpc F0 Brd4Y430C/Y430C, Brd4C429_N433del/ C429_N433del and control mouse embryos. The MEFs lines initially established from Brd4C429_N433del/ C429_N433del embryos did not survive in long-term culture, but we were able to generate sufficient cells for western blot analysis. Both wild-type and Brd4Y430C/Y430C MEF lines expressed comparable levels of BRD4 protein. However, in Brd4C429_N433del/ C429_N433del MEFs the BRD4 band was undetectable (Supplemental Fig. 12) using an antibody raised against a peptide representing amino acid numbers 1312-1362 in the mature peptide. The apparent null status of these cells may be the result of rapid degradation of the abnormal protein or an artifact due to change in the epitope. The latter may also explain the survival of these homozygous embryos past implantation.
BRD4 Chromatin IP (ChIP) from Brd4Y430C/Y430C MEF showed reduced binding to the promoters and super-enhancers of known BRD4 targets compared with wild-type MEF (Figure 2F). To assess regions of common binding, we performed BRD4 ChIP-seq on wild-type mESC and compared this to publicly accessible NIPBL and BRD4 ChIP-seq data from mouse (Supplemental Fig. 13) and human (Supplemental Fig. 14) ESCs. We used the intersection of the BRD4-bound and NIPBL-bound genomic intervals from the ChIP-seq data to create a set of high-confidence shared binding sites. By comparing different functional genomic categories, mESC super-enhancers show the highest level of enrichment with heterochromatin being the least enriched (Figure 2G&H, Supplemental Fig.15). To look for any common functional effect on gene expression we then generated array-based transcriptome data from control, Brd4Y430C/Y430C and Nipbl+/- MEFs (Supplemental Fig. 16). Of the >18000 genes probed on the microarray, 3049 have a transcription start site within 1Mb of a defined MEF super-enhancer (Super Enhancer Archive). These super-enhancer-associated genes showed significantly higher level of differential expression in both Brd4 (p = 0.002) and Nipbl (p = 0.006) mutant cells compared to genes that are not super-enhancer associated. There is also a significant overlap in the specific genes that show differential expression in both BRD4 and NIPBL mutant MEFs (Supplemental Fig. 17).
CdLS can be considered a transcriptomopathy4, presumed to result from loss of cohesin-dependent chromatin loops or a cohesin-independent NIPBL-mediated transcriptional activity6. Our identification of de novo heterozygous loss of function mutations in BRD4 in a CdLS-like disorder, together with the functional genomic data presented above, suggests that CdLS may be more specifically defined as a disorder of super-enhancer function. Delineation of any direct or indirect physical interaction and/or functional co-dependency of BRD4 with NIPBL can now reasonably become a topic of investigation.
Online Methods Section
Methods
Patient ascertainment
All the clinical research activity relating to this report has been in accordance with World Medical Association Declaration Of Helsinki on the Ethical Principles For Medical Research Involving Human Subjects. The research was conducted using protocols approved by UK multicenter ethics committees under the references; 04:MRE00/19 (MRC HGU) and 10/H0305/83 (DDD). Two of the affected individuals (4198 II:1 & 3049 II:1 Figure 1) are part of a larger cohort of patients with a diagnosis of CdLS or possible CdLS, referred by experienced clinical geneticists or pediatricians to the MRC Human Genetics Unit for research genetic analysis12. The third (CDL038) was referred to the DNA Diagnostic Laboratory in NHS Lothian with a CdLS-like disorder. The final affected individual (264293) was identified using the trio whole exome sequence data generated by the Deciphering Developmental Disorders study.
Array comparative genomic hybridization
Array comparative genomic hybridization (aCGH) was performed using the Nimblegen 135k microarray platform (Roche Nimblegen) as described previously21. Results were compared with the Database of Genomic Variants and polymorphic CNVs excluded.
Droplet digital PCR
A pair of oligonucleotide primers and the matching 5′ FAM-labelled Universal Probe Library (UPL) probe (# 25) (Roche) were designed to target coding exon 17 of the BRD4 gene using ProbeFinder software version 2.50 (Roche).
Each 20 μl ddPCR reaction consisted of 40 ng of genomic DNA, 1X ddPCR SuperMix for probes (No dUTP) (Bio-Rad Inc.), forward and reverse primers at 1 μM each, UPL probe #25 at 250 nM, and 1X 5′ VIC-labelled RNase P TaqMan Copy Number Reference assay (Thermo Fisher Scientific). Droplet generation using the QX200 droplet generator (Bio-Rad Inc.) followed by amplification, 95°C for 10 minutes, 40 cycles of 94°C for 30 seconds and 57°C for 60 seconds, and a final incubation at 95°C for 10 minutes, were performed as per manufacturer’s instructions (Bio-Rad Inc.). Following completion of the PCR, plates were read using the QX200 droplet reader (Bio-Rad Inc.). Analysis of droplet counts, amplitudes and DNA copy number were performed with QuantaSoft software (Bio-Rad Inc.) for channel 1 = FAM and channel 2 = VIC.
Mutation analysis by DDD Trio Exome Sequencing, Ion AmpliSeq PCR-Ion PGM, and Sanger sequencing
As part of a DDD Complementary Analysis Protocol #35 VCF files on the first 4293 trios with whole exome sequence were searched for candidate de novo mutations in BRD4. Only one possible de novo disruptive variant was identified in BRD4. This variant was validated as de novo using the approach mentioned below. No other plausible cause for the developmental disorder was apparent on trio based whole exome analysis.
An AmpliSeq panel encompassing the coding exons of BRD4 and nine other candidate genes was designed using the Ion AmpliSeq Designer tool (Life Technologies, IAD41056). Library preparation and sequencing on the Ion PGM platform (Life Technologies), followed by sequence alignment and variant calling on software NextGENe version 2.3.3 (Soft Genetics) were performed as described previously12. A total of 92 individuals were screened, who had previously scored as negative for mutations in NIPBL, SMC1A, SMC3, HDAC8 and RAD21, and large-scale genomic deletions/duplications. The same panel also applied to subsequent clinical referrals to the NHS DNA diagnostic laboratory in Edinburgh was used to identify one further de novo heterozygous loss of function mutation in an individual who had a CdLS-like phenotype.
Any significant variants were confirmed by Sanger sequencing and analysed using Mutation Surveyor software version 3.30, as described previously21. The BRD4 sequence identifier, NC_000019.10 was used in the analysis. Sequence variant nomenclature is reported according to the BRD4 transcript variant, NM_058243. Primer sequences and PCR conditions are available upon request.
Plasmids, expression and purification of proteins
Human BRD4 BD1 and BD2 plasmids were kindly gifted by Prof Stefan Knapp (Nuffield Department of Clinical Medicine, Oxford). Proteins were expressed and purified at the Edinburgh Protein Production Facility (EPPF) as described previously22.
Site-directed Mutagenesis
The point mutation c.1289A>G, predicted to result in the protein variant p.Tyr430Cys (Y430C) was introduced into the BRD4-BD2 and FLAG-mBRD4 constructs using the QuikChange II XL Site-directed Mutagenesis kit (Agilent Technologies) following the manufacturer's instructions. The presence of the desired mutations was confirmed by Sanger sequencing.
CRISPR/Cas9 construct design
Guide RNA (gRNAs) 1 and 2 were designed across p.Tyr430 using online tool DNA 2.0. The wild-type and mutant repair templates (chr17:32,220,150-32,220,271; GRCm38) were synthesized by IDT as 122 bp UltramerssODN bearing the desired sequence change. For genome editing in mouse embryonic stem cells (mESCs) gRNAs 1 and 2 were cloned into PX461 (Addgene plasmid #48140) and PX462 (Addgene plasmid #62987) respectively. For genome editing in mouse embryos both gRNAs were cloned into PX461 and the full gRNA template sequence was amplified from the resulting PX461 clone using universal reverse primer and T7 tagged forward primers. The gRNA PCR template was used for in vitro RNA synthesis using T7 RNA polymerase (NEB), and the RNA template subsequently purified using RNeasy mini kit (Qiagen) purification columns. Cas9n mRNA was procured from Tebu Bioscience.
Genome editing in mouse embryonic stem cells (mESCs)
To generate mESCs carrying the p.Tyr430Cys missense variant in BRD4, 46C cells were cotransfected with gRNAs 1 and 2 (0.5 μg/ml) and the mutant repair template (0.5μg/2ml) using Lipofectamine® 3000 Transfection Reagent (ThermoFisher) as per the manufacturer’s instructions. After 48 hours, successfully transfected cells were selected for: firstly by puromycin treatment, and subsequently by FACS based on GFP expression. Resulting GFP and puromycin positive cells were plated at 500 cells/10cm2. After 1 week, colonies were picked and plated in duplicate as 1 colony/well of a 96 well plate. Genomic DNA was extracted from the colonies and sequenced by Sanger sequencing. Wild-type clones and clones homozygous for the p.Tyr430Cys variant were expanded and frozen for later use.
Genome editing in mouse embryos and generation of mouse embryonic fibroblast (MEFs)
To generate mouse embryos carrying the p.Tyr430Cys variant in BRD4, injections were performed in single cell mouse zygotes. Injection mix contained Cas9 mRNA (50 ng/μl), gRNAs 1 and 2 (25 ng/μl) and each repair template DNA (75 ng/μl). The embryos were later harvested for analysis at 13.5 dpc stage of embryonic development. MEFs were isolated from limbs of individual E13.5 embryos by mincing in 1 ml of medium (DMEM, 10% FCS, 50 U/ml penicillin and 50 mg/ml streptomycin,). Resulting suspensions were grown at 37°C, 5% CO2 and 3% O2, and non-adherent cells removed after 24 hours. MEFs from embryos with unedited Brd4 alleles, clean homozygous knock-in for p.Tyr430Cys in Brd4 (Brd4Tyr430Cys/Tyr430Cys and homozygous knock-in for an in-frame deletion (Brd4Cys430_Asn434del/Cys430_Asn434del) alleles were used for further experimentation.
Generation of heterozygous loss-of-function Nipbl MEFs
Mice with Nipbl floxed allele (a kind gift from Heiko Peters, University of Newcastle) were crossed with Cre745 mice (a kind gift from DJ Kleinjan, University of Edinburgh), containing a CAGGS-Cre construct in which Cre recombinase is under control of a chicken b-actin promoter to excise Nipbl exon 1. Embryos were collected at 13.5 dpc. MEFs were isolated from heterozygous Nipbl knockout embryo limbs by mincing in 1 ml of medium (DMEM, 10% FCS, 50 U/ml penicillin and 50 mg/ml streptomycin,). Resulting suspensions were grown at 37°C, 5% CO2 and 3% O2, and non-adherent cells removed after 24 hours.
Histone tail peptide arrays
A modified histone peptide array (Active motif, #13005) experiment was performed as described previously23. Briefly, the array was blocked in TBST buffer (10 mM Tris/HCl pH 8.3, 0.05% Tween-20, 150 mM NaCl) containing 5% non-fat dried milk at 4°C overnight. The membrane was washed with TBST for 5 min, and incubated with 10 ηM purified His-tagged BRD4 BD1 or wild-type (WT) and p.Tyr430Cys (Y430C) BD2 domains, at room temperature (RT) for 1 hour in interaction buffer (100 mM (0.5 μg/3 ml) KCl, 20 mM HEPES pH 7.5, 1 mM EDTA, 0.1 mM DTT, 10% glycerol). After washing in TBST, the membrane was incubated with mouse α-His (Sigma, H1029, 1:2,000 dilution in TBST) for 1 hour at RT. The membrane was then washed 3 times with TBST for 10 min each at RT, and incubated with horseradish peroxidase conjugated α-mouse antibody (1:10,000 in TBST) for 1 hour at RT. The membrane was submerged in ECL developing solution (Pierce, #32209), imaged (Image-quant, GE Healthcare) and the data quantified using array analyzer software (Active motif).
Nuclear extract co-immunoprecipitation
30 × 106 wild-type and p.Tyr430Cys (Y430C) BRD4 mESCs were trypsinised, pelleted and resuspended in 5 ml ice-cold swelling buffer (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5mM DTT, Complete Mini EDTA-free protease inhibitor (Roche)) for 5 minutes on ice. Nuclei were pelleted by centrifugation at 2,000 rpm for 5 minutes at 4°C. The resulting nuclear pellets were sonicated in 2 ml RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, benzonase and Complete Mini EDTA-free protease inhibitor (Roche)), using a Bioruptor® Plus sonication device (Diagenode) at 4°C, 30 seconds on, 30 seconds off. It was noted that prolonged (1 hour) exposure to the detergents in RIPA buffer affected the interactions of BRD4 as measured by mass spectrometry. Nuclear extracts were cleared by centrifugation at 13,000 rpm for 10 minutes at 4°C. Protein A Dynabeads (life technologies) were blocked prior to antibody coupling by washing 3 times with 5% BSA in PBS. Antibodies were coupled to the beads at 5 mg/ml by rotation for 1 hour at 4°C. Equivalent nuclear protein amounts were incubated with antibody coupled beads for 1 hour at 4°C. Beads were washed and pulled down proteins analysed by mass spectrometry or western blot. Antibodies used: BRD4 (Bethyl A301-985A100), SMC3 (Bethyl 0300-060A), NIPBL (Bethyl A301-779A) and normal rabbit IgG (Santa-Cruz, sc-2027).
Western blots
For western blot analysis beads were washed 5 times with RIPA buffer, bound proteins eluted by boiling in 1X NuPage LDS buffer (ThermoFisher Scientific) with 1X NuPage reducing agent (ThermoFisher Scientific) for 5 minutes and separated on a 3-8% tris-acetate gel (reciprocal BRD4/SMC3/NIPBL IPs and MEF cell lysates) or 4-12% bis-tris gel (BRD4 IPs for acetylated histone binding) (ThermoFisher Scientific). Following electrophoresis, proteins were transferred to nitrocellulose membranes (ThermoFisher Scientific) using iBlot 2 Dry Blotting System (ThermoFisher Scientific) for 7 minutes (when probing for proteins <250 kDa only) or to PVDF membranes by wet transfer for 90 minutes (when probing for proteins >250 kDa) and incubated with primary antibodies overnight at 4°C. Membranes were washed 3 times in TBST and probed with HRP-conjugated secondary antibody (anti-Rb/anti-goat, 1:10,000) for 1 hour at RT. After 3 more washes in TBST, membranes were incubated with Pierce™ ECL Western Blotting Substrate (ThermoFisher Scientific) for 5 minutes and imaged using (Image-quant, GE Healthcare). Antibodies used: BRD4 (Bethyl A301-985A100, 1:3,000), SMC3 (Bethyl 0300-060A, 1:1,000), H3K27ac (Genetex GTX128944, 1:1,000), H4K8ac (Abcam ab15823, 1:1,000), H3K9ac (Abcam, ab10812, 1:500), H3 (Abcam, ab1791, 1:5,000), NIPBL (Bethyl A301-779A, 1:1000), SOX2 (Abcam ab97959, 1:1000) Actin-b (Abcam ab8229, 1:500).
Mass spectrometry
For analysis by mass spectrometry, beads were washed 3 times with Tris-saline buffer, and excess buffer removed. Immunoprecipitations were digested on beads, desalted and analysed on a Q-Exactive plus mass spectrometer as previously described24. Proteins were identified and quantified by MaxLFQ25 by searching with the MaxQuant version 1.5 against the Mouse proteome data base (Uniprot). Modifications included C Carbamlylation (fixed) and M oxidation (variable). Bioinformatic analysis was performed with the Perseus software suite.
Chromatin immunoprecipitation-quantitative PCR
Primary MEFs isolated from 13.5 dpc embryos were cultured for 3-4 passages in DMEM media supplemented with 15% FCS, 1% Pen/Strep, L-Glutamine, non-essential amino acids and Sodium pyruvate. Cells were harvested by trypsinizing and fixed immediately with 1% formaldehyde (Thermo Fisher Cat. 28906) (25°C, 10 min) in PBS, and stopped with 0.125 M Glycine. Chromatin immunoprecipitation (ChIP) was performed as described previously26. Briefly, cross linked cells were re-suspended in Farnham lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, Complete Mini EDTA-free protease inhibitor (Roche)) for 30 minutes and centrifuged at 228 g for 5 minutes at 4°C. Nuclei were resuspended in RIPA buffer (1X PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS (filtered 0.2 -0.45 micron filter unit) + Complete Mini EDTA-free protease inhibitor (Roche)) and sonicated using a Bioruptor® Plus sonication device (Diagenode) at full power for 40 minutes (30 seconds on, 30 seconds off) to produce fragments of 100-500 bp. 3 μg of each antibody was incubated with Protein A Dynabeads (ThermoFisher Scientific, 10001D) in 5 mg/ml BSA in PBS on a rotating platform at 4°C for two hours. An arbitrary concentration of 50 μg chromatin was incubated with antibody bound Dynabeads in a rotating platform at 4°C for 16 hours. Beads were washed 5 times (5 minutes each) on a rotating platform with cold LiCl wash buffer (100 mM Tris pH 7.5, 500 mM LiCl, 1% NP-40, 1% Sodium deoxycholate) and one time with RT TE buffer. ChIP complexes were eluted with elution buffer (1% SDS, 0.1 M NaHCO3) and Input and ChIP samples were incubated at 65°C for 5 hours to reverse the crosslinks. 2 μl of RNase A (20 mg/ml) was added and samples were incubated at 37°C for 1 hour before 2 μl of Proteinase K was added and samples were incubated for 2 hours at 55°C. DNA was purified using QIAquick PCR Purification Kit (Qiagen, Cat. 28104), and analysed by qPCR for primers described in Supplementary Table 2. Antibodies used: rabbit IgG (Santa Cruz sc-2025), BRD4 (Bethyl A301-985A100).
Chromatin immunoprecipitation-sequencing (ChIP-Seq)
Wild-type mESCs were cultured in GMEM media supplemented with 10% FCS, 1% Pen/strep, L-glutamine, non-essential amino acids, sodium pyruvate and 1,000 U/ml LIF. Cells were harvested by trypsinizing and fixed immediately with 1% formaldehyde (Thermo Fisher Cat. 28906) (25°C, 10 minutes) in PBS. This reaction was quenched with 0.125 M Glycine. ChIP was carried out as above. After purification, DNA was eluted in 20 μl and libraries were prepared for ChIP and input samples as previously described27. Samples were sequenced at BGI (Hong Kong; 50-base single-end reads) using the HiSeq 4000 system (Illumina).
Transcriptome analysis
RNA was extracted from Brd4Tyr430Cys/Tyr430Cys and Nipbl heterozygous null MEFs using the RNeasy Mini Kit (Qiagen) as per the manufacturer’s instructions. 1 μg of RNA was hybridised to a SurePrint G3 Mouse GE 8x60K microarray (Agilent; G4852A) and scanned on a Nimblegen scanner as described previously28.
Quantile normalisation and background correction (method normexp) of the microarray data was carried out using the bioconductor package limma29. Gene level expression was calculated by averaging the probes signal that mapped to the same gene (Gene Symbols mapped to Probe identifiers obtained from GEO, GPL13912, Agilent-028005 SurePrint G3 Mouse GE 8x60K Microarray). The normalised signal for technical replicate samples was averaged prior to Differential expression (DE) analysis.
Transcriptome analysis statistics
Gene level differential expression was conducted using the bioconductor package limma29. Briefly, a linear model was fitted to each gene. Then empirical Bayes moderation was applied to the linear model fit to compute moderated t-statistics, moderated F statistic, and log-odds of differential expression. The Benjamini & Hochberg method was used to correct the p-values for multiple testing. Genes were identified as significantly differentially expressed if the FDR q value < 0.1.
To test whether genes (n= 3049/19113) with a transcription start site within 1Mb of MEF Super Enhancers (SE) are more highly ranked relative to other expressed genes in terms differential expression (t-statistic) we performed a Mean-rank Gene Set Test (geneSetTest, bioconductor package limma).
A hypergeometric test was performed on the differentially expressed gene sets for Brd4 and Nipbl to determine if DE genes were significantly enriched between the two groups.
ChIP-seq analysis
Bowtie 2 (version 2.2.6) was used to map reads to mouse (mm9) and human (hg19) genomes (options bowtie2-align-s --wrapper basic-0)30. To calculate the correlation of NIPBL and BRD4 with other histone modifications (see Supplementary Table 3), the correlation of the ChIP-seq binding profiles across the genome was calculated. DeepTools (version 2.3.5) multiBamSummary was used to calculate the coverage of mapped reads in 150 bp sequential bins across the mm9/hg19 genome (options --binSize 150bp, --ignoreDuplicates,--black, ListFileName,--extendReads 150,–mappingquality 30)31. Genomic bins within Blacklisted regions and chrX and chrY were excluded from the analysis. Genomic bins were also restricted to regions of open chromatin using DNase I hypersensitive sites identified by the ENCODE project32 (see Supplementary Table 3).
The genome wide coverage matrix was imported into R and Pearson's R was calculated. Correlation scores were visualised as a heatmap using the R package pheatmap (options; euclidean distance and complete clustering method) 33.
Peak calling
To call BRD4 and NIPBL bound regions we used the MACS peak caller (2.1.1). For BRD4 peaks we used the parameters broadPeaks and an FDR cut-off of 0.1.
For NIPBL we used the public ChIP-Seq dataset NIPBL in V6.5 (C57BL/6-129) murine ES cells (GEO ID, GSM560350) and, the accompanying whole cell extract dataset (GEO ID, GSM56035) as background.
For peak calling we used MACS with the parameters narrowPeaks and an FDR cut off of 0.1. To perform intersections on genomic ranges, such as peaks regions, we used bedtools intersect (2.26.0)34.
Any peaks that intersected with the mm9 genome blacklist regions or mapped to non-canonical chromosomes were removed from subsequent analysis.
Genomic region enrichment
To determine the preference of co-localised NIPBL and BRD4 binding to specific chromatin states we performed fisher enrichment analysis on a chromatin state map in mouse embryonic stem cells (ChromHMM, mm9). This state map has annotated the genome into six major chromatin states including; active promoter, poised promoter, strong enhancer, poised or weak enhancer, insulator, repressed, transcribed and heterochromatin.
In addition we looked at enrichment of co-localised NIPBL and BRD4 binding sites with Super Enhancers (SE) regions found in mESC line E14 using data from SEA: Super-Enhancer Archive35.
We used the consensus set of SE regions from two mES E14 replicates to define SE regions.
Genomic region enrichment statistics
To calculate enrichment of peaks we used bedtools (2.26.0) fisher test. The fishers odds ratio was converted to Log2 scale and plotted using R forest plot package.
Supplementary Material
Acknowledgements
We thank the CdLS Foundation of UK and Ireland and particularly the families of the affected children for their time and support for the research. GO, MA, HB, WAB, PM, DRF were funded by the MRC University Unit award to the University of Edinburgh for the MRC Human Genetics Unit. AvK’s work was supported by a Carnegie Trust Research Incentive Grant 70382. The Deciphering Developmental Disorders study presents independent research commissioned by the Health Innovation Challenge Fund (grant HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant WT098051). The views expressed in this publication are those of the authors and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research through the Comprehensive Clinical Research Network.
Footnotes
URLs
Database of Genomic Variants; http://dgv.tcag.ca/dgv/app/home
Ion AmpliSeq Designer tool; http://www.ampliseq.com
Blacklisted regions; https://sites.google.com/site/anshulkundaje/projects/blacklists
ChromHMM, mm9; https://github.com/gireeshkbogu/chromatin_states_chromHMM_mm9
Super Enhancer Archive; http://www.bio-bigdata.com/SEA/
pheatmap: Pretty Heatmaps (2015) v1.0.8. https://CRAN.R-project.org/package=pheatmap
Author Contributions
WAB, MMP and DRF conceived the study. DRF, WAB and MMP wrote the manuscript. All authors have read and commented on the manuscript. GO, MA, HB, NC, MMP and DDD generated the molecular biology and animal model data. AvK generated and analysed the mass spectrometry data. FJS, EW, AR, SMP provided expert clinical interpretation and details of the phenotype for each affected individual. AB performed the metanalysis of the reported deletion cases. JR provided expert technical advice and cel reagents. GRG performed the genomic and transcriptomic informatic analysis.
URLs
Super Enhancer Archive; http://www.bio-bigdata.com/SEA/
DECIPHER; http://decipher.sanger.ac.uk
UCSC Genome Browser; https://genome.ucsc.edu
Mouse Genome Informatics database; http://www.informatics.jax.org
Deciphering Developmental Disorders Study; https://www.ddduk.org
The authors delcare that they have no competing financial interests
Data Accessibility Statement
Results of array-based comparative genomic hybridization from individual II:1 (family 4198) are available on DECIPHER database (ID 281165). The DDD trio-based exome data used to identify the de novo frame-shift mutation in individual 264293 is available from European Genome-phenome Archive (EGA) under accession EGAD00001001848
References
- 1.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao SSP, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watrin E, Kaiser FJ, Wendt KS. Gene regulation and chromatin organization: relevance of cohesin mutations to human disease. Curr Opin Genet Dev. 2016;37:59–66. doi: 10.1016/j.gde.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Yuan B, et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest. 2015;125:636–651. doi: 10.1172/JCI77435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bot C, et al. Independent mechanisms recruit the cohesin loader protein NIPBL to sites of DNA damage. J Cell Sci. 2017;130:1134–1146. doi: 10.1242/jcs.197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuin J, et al. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 2014;10:e1004153. doi: 10.1371/journal.pgen.1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haarhuis JHI, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707.e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 9.Schwarzer W, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deardorff MA, et al. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet. 2012;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil-Rodriguez MC, et al. De novo heterozygous mutations in SMC3 cause a range of Cornelia de Lange syndrome-overlapping phenotypes. Hum Mutat. 2015;36:454–462. doi: 10.1002/humu.22761. [DOI] [PubMed] [Google Scholar]
- 12.Musio A, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 13.Deardorff MA, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansari M, et al. Genetic heterogeneity in Cornelia de Lange syndrome (CdLS) and CdLS-like phenotypes with observed and predicted levels of mosaicism. J Med Genet. 2014;51:659–668. doi: 10.1136/jmedgenet-2014-102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parenti I, et al. Broadening of cohesinopathies: exome sequencing identifies mutations in ANKRD11 in two patients with Cornelia de Lange-overlapping phenotype. Clin Genet. 2016;89:74–81. doi: 10.1111/cge.12564. [DOI] [PubMed] [Google Scholar]
- 16.Izumi K, et al. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet. 2015;47:338–344. doi: 10.1038/ng.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deciphering DDS. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kline AD, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143A:1287–1296. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- 19.Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawauchi S, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Discov. 2017;7:322–337. doi: 10.1158/2159-8290.CD-16-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno T, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vollmuth F, Blankenfeldt W, Geyer M. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J Biol Chem. 2009;284:36547–36556. doi: 10.1074/jbc.M109.033712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerth-Kahlert C, et al. Mol Genet Genomic Med. 2013;1:15–31. doi: 10.1002/mgg3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filippakopoulos P, et al. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradeepa MM, et al. PLoS Genet. 2012;8(5):e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turriziani B, et al. Biology (Basel) 2014;3(2):320–32. doi: 10.3390/biology3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox J, et al. Mol Cell Proteomics. 2014;9:2513–26. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DS, et al. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 30.Pradeepa MM, et al. Nature Genetics. 2016;48:681–686. doi: 10.1038/ng.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Illingworth R, et al. Genes Dev. 2015;29(18):1897–902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, et al. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg S. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez F, et al. Nucleic Acids Res. 2016;44(W1):W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ENCODE Project Consortium. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR, Hall IM. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, et al. Nucleic acids research. 2016;44(D1):D172–D179. doi: 10.1093/nar/gkv1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.