Abstract
Rare diseases pose a global challenge, in that their collective impact on health systems is considerable, whereas their individually rare occurrence impedes research and development of efficient therapies. In consequence, patients and their families are often unable to find an expert for their affliction, let alone a cure. The tide is turning as pharmaceutical companies embrace gene therapy development and as serviceable tools for the repair of primary mutations separate the ability to create cures from underlying disease expertise. Whereas gene therapy by gene addition took decades to reach the clinic by incremental disease-specific refinements of vectors and methods, gene therapy by genome editing in its basic form merely requires certainty about the causative mutation. Suddenly we move from concept to trial in 3 years instead of 30: therapy development in the fast lane, with all the positive and negative implications of the phrase. Since their first application to eukaryotic cells in 2013, the proliferation and refinement in particular of tools based on clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) prokaryotic RNA-guided nucleases has prompted a landslide of therapy-development studies for rare diseases. An estimated thousands of orphan diseases are up for adoption, and legislative, entrepreneurial, and research initiatives may finally conspire to find many of them a good home. Here we summarize the most significant recent achievements and remaining hurdles in the application of CRISPR/Cas technology to rare diseases and take a glimpse at the exciting road ahead.
Key Points
| Accelerated molecular characterization of rare disease cases and the advent of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) technology promise to enable rapid therapy development for many rare genetic diseases. |
| The adoption of editing technology reduces the time from conception to evaluation of advanced therapy approaches compared to gene addition, encouraging an unprecedented number of research groups and studies to focus on rare diseases. |
| As CRISPR/Cas-based tools are customized to tackle rare diseases in clever ways, the results reveal and help address remaining unknowns and obstacles in the clinical translation of the new technology, including those concerning efficiency, specificity, delivery, immunity, preservation of stemness, and avoiding malignant transformation. |
Rare Diseases: Towards Curative Treatments
Rare Diseases and Rarer Disease Expertise
Rare diseases are common and ubiquitous, and for decades rare therapies were anything but well done. As this introductory sentence employs puns commonly used in the rare disease literature, its two parts summarize the double challenge faced by society and patients, respectively. First, it is a widely accepted estimate that globally between 6000 and 8000 individually rare diseases across all therapeutic areas affect between 8 and 10% of the population and therefore taken together pose a widespread and pervasive challenge [1, 2]. Second, rare disease patients are frequently treated suboptimally, if they receive treatment at all, because few clinicians have the opportunity to build up disease expertise for rare diseases. Consequently, many a rare disease patient does not receive a definite diagnosis or is initially misdiagnosed due to often missing molecular (causative) information for the disease in question, clinical features that overlap with other, more common afflictions, and limited global access by the treating physicians to relevant data [2–4]. Many rare diseases have early or prenatal onset, making prompt and appropriate treatment all the more important. However, where even diagnosis is difficult, disease management often relies on ad hoc symptomatic treatments or repurposed medicines developed for other diseases, so that an estimated 94% of rare diseases are without an approved treatment [5, 6]. This problem is exemplified by the ongoing search for effective treatments for two relatively prominent rare diseases, Shwachman–Diamond syndrome and cystic fibrosis, as reviewed by Bezzerri and Cipolli [7] and by Cabrini [8], respectively, in this issue of Molecular Diagnosis and Therapy. Moreover, a cure is only available where stem cell or organ transplantation is feasible for the tissues affected and where a compatible donor can be identified [9–12]. Accordingly, the average standard of care for rare disease patients is suboptimal, at high long-term cost to national health budgets and individual health. In response, governments try to incentivize investment in rare disease drug development by introducing orphan drug legislation that lowers regulatory thresholds for clinical translation [13] and invest in the collaboration of international rare disease experts, such as through the International Rare Diseases Research Consortium (IRDiRC) [14]. Specialty international networks of rare disease expertise, including the nascent European Reference Networks (ERNs) or other international networks complement this effort [2, 15]. Likewise, rare disease expertise is pooled by establishing government-funded or co-funded information portals, such as the Genetic and Rare Diseases Information Center (GARD) run by the US National Center for Advancing Translational Sciences [16, 17] and the European Orphanet [18], complemented by academic disease-specific databases [19, 20] and patient portals such as the National Organization for Rare Disorders (NORD) and EURORDIS (Rare Diseases Europe) [21, 22]. Most rare diseases are not and cannot be in the standard medical curriculum, but at ever increasing speed the knowledge base increases and becomes available to anyone willing to look. This is particularly true for the assumed vast majority of classifiable rare diseases that have a genetic cause, with key developments the rapid discovery of novel causative mutations for rare diseases by massively parallel sequencing [23] and adoption of global variant database and nomenclature standards to make the information universally accessible and interpretable [14, 19]. While one of the major contributions of rapid variant discovery to reducing the global burden of disease is seen in rare disease prevention [24], it likewise provides the foundation for stratified if not personalized rare disease treatments by gene therapy.
Teething Troubles of Gene Therapies for Rare Diseases
As reviewed by us and others in this issue of Molecular Diagnosis and Therapy [25, 26], gene therapy for rare diseases has achieved remarkable successes for several rare diseases, including curative treatments. As a prerequisite for success, robust and clearly defined clinical trial design is imperative in order to deal with the unique challenges inherent to rare diseases. These challenges include small and highly fragmented study populations and the difficulty of setting clear clinical endpoints, often in the absence of control groups, without in-depth understanding of the disease state, and in the face of high heterogeneity of the disease phenotype and further complications unique to rare diseases [27]. Despite these difficulties, several successful trials have already resulted in orphan drug designations for rare diseases as an important milestone on the road to market approval. For instance, market approval was achieved for Glybera to treat lipoprotein lipase deficiency (EMEA/H/C/002145) in 2012, for Strimvelis® to treat adenosine-deaminase-linked severe combined immunodeficiency (ADA-SCID) (EMEA/H/C/003854) in 2016 [2], and for Luxturna™ to treat mutation-associated retinal dystrophy in the USA in late 2017 and in the European Union (EU) in late 2018 (EMEA/H/C/004451) [29]. Glybera has since been withdrawn from the market for lack of profitability, indicating a persistent and worrisome problem in the provision of ultra-rare disease drugs despite high development cost and uncertain reimbursement models, in the case of Glybera exacerbated by limited safety and efficacy data that prevented full market penetration [30, 31]. By contrast, Strimvelis® and Luxturna™ have both provided substantial benefits to patient cohorts in the absence of adverse events after years of follow-up, but their continued provision will once more depend on their profitability. In view of the economic lessons learnt [30], gene therapy companies will thus preferentially seek market approval of drugs for more prominent rare diseases. In consequence, and unless other funding mechanisms are developed, the current trend holds the danger that a wealth of competing treatments will become available for more prominent rare diseases, whereas a multitude of ultra-rare diseases will be left out. Unless development costs are vastly reduced, gene therapy for those diseases would then merely and sadly remain an academic exercise. One factor that will make translation more affordable is the maturation of the corresponding regulatory framework at national and international level [32], which already provides a level of certainty about the ground rules for translation and facilitates compliance across jurisdictions and harmonization of the development and approval process. Moreover, curative treatments, such as Strimvelis® and Luxturna™, are usually based on permanent gene addition, which provides trans-acting components or a functional copy of the defective gene, either as an integrating transgene or, for non-dividing cells, as a stable episome [25]. Despite the apparent simplicity of the concept, years if not decades of study of disease and regulatory mechanisms are usually required in order to achieve both efficient delivery and stable physiological expression of the therapeutic transgene. For many rare diseases, required insights into the underlying disease mechanism are insufficient to allow such development, even if funding were available.
Gene Therapy Version 2.0
As more rare disease therapies based on gene addition advance from clinical trials toward market approval, a new wave of treatments based on gene editing technology are being developed, moving from preclinical studies to their first clinical trials. A slow development at first, based on zinc finger nucleases and other hard-to-design tools, the advent of transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) RNA-guided nucleases (RGNs) has led to recently accelerated development and a veritable explosion of tools and rare disease targets [25]. However, as permanent gene addition by genome integration has inherent risks, such as those of insertional mutagenesis, so does gene editing. First, target recognition is error-prone to some degree, so that any editing application may introduce changes at hard-to-predict off-target sites, dependent on similarities to the on-target recognition sequence. For each editing tool, the risk of off-target modifications therefore needs to be assessed comprehensively early on and potentially eliminated by changing the tool or recognition site. Second, the pre-existing humoral and cell-mediated adaptive immunity to Cas9, as a common environmental antigen, poses the risk of triggering widespread immune responses in patients receiving CRISPR/Cas9-based gene therapy, at least in in vivo applications of these tools [33–35]. The third and fourth risks relate to gene editing by designer nucleases being universally based on the introduction of double-strand breaks (DSBs) in the genome as triggers of repair by the efficient but imprecise non-homologous end-joining (NHEJ) mechanism or the inefficient but precise homology-directed repair (HDR) mechanism [25]. Specifically, efficient induction of DSBs may cause P53-mediated apoptosis for cycling cells [36–38], leading to a principally but not widely proven enrichment for P53-deficient, and thus more cancer-prone, cells by DSB-mediated editing. Additionally, and of still more acute concern, even single DSBs may lead to wide-ranging deletions or chance recombination events involving the on-target site [39], and as long as the frequency and pattern of such events is not clarified, application of gene editing to rare diseases that are also manageable with palliative treatments may be premature. While the risk of off-target activity cannot be eliminated at the design stage but rather only assessed later on, at least any problems relating to DSBs may be avoided by a new class of editing tools: base editors. Based on the CRISPR/Cas platform and allowing certain targeted base substitutions by chemical base editing, base editors act independent from DSBs and thus without elevating the risk of chromosomal rearrangements or cancer-prone cells [40, 41].
Importantly, and although gene editing tools may be employed in a variety of other strategies, the option of targeting and correcting the causative mutation is a conceptual landmark shift compared to gene addition. Faithfully correcting causative mutations, though inefficient at present, is in principle the most universally applicable gene editing strategy, which moreover does not presuppose any prior knowledge of molecular disease mechanisms for therapy development. However, it is also the approach that will have the greatest problems creating revenue, as mutation-specific therapies have the same overheads as more universal approach but will have more limited patient numbers. CRISPR/Cas technology will at least help minimize development cost, as this new type of editing tools can be designed—and in many cases applied—with great ease. Conceptually, therefore, the possibilities for rare disease therapy development with these new tools and methods are near limitless and, assuming that safety concerns can be addressed, they may have tremendous impact on treatment prospects for all genetically defined rare diseases. In the following sections we summarize to what extent CRISPR/Cas-based therapeutic approaches have already been applied to rare diseases and which key improvements are required for clinical translation.
Rare Repair: Employing the Molecular Toolkit
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Gene Therapy Studies in Numbers
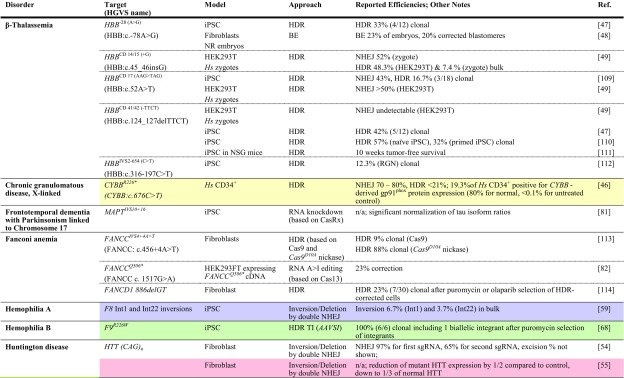
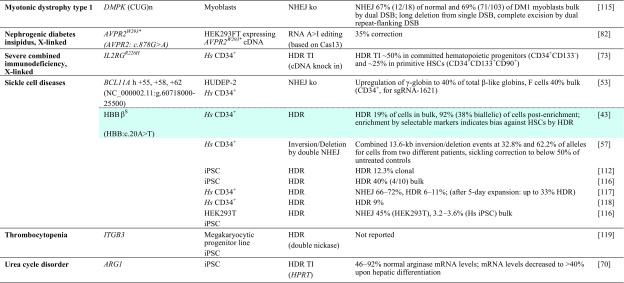
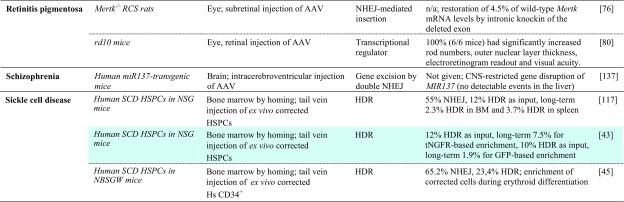
Presently only 25 registered clinical trials concern CRISPR/Cas technology, 19 of them targeting malignancies, two targeting infectious diseases, and four targeting rare diseases. The latter all concern β-hemoglobinopathies, with three aiming to treat patients (ClinicalTrials.gov identifier NCT03655678 [141] and NCT03745287 [142] by CRISPR Therapeutics Ltd. for β-thalassemia and sickle cell disease [SCD], respectively, and NCT03728322 [143] by Allife Medical Science and Technology Co. Ltd. for β-thalassemia) and one aiming to use SCD focus groups to gauge knowledge, attitudes, and beliefs about clinical application of CRISPR/Cas technology (NCT03167450 [144]). The focus group study and its testing the waters for CRISPR/Cas application in a patient population with exemplary palliative care amongst rare diseases indicate that numbers and representation of rare diseases in clinical trials may be about to change. To date, the number of research papers citing CRISPR in connection with ‘gene therapy’ has passed the 16,000 mark and in connection with ‘rare disease’ has passed the 1000 mark (Google Scholar), which even at this relatively early stage precludes truly comprehensive coverage of the field. This section therefore focuses on key studies that serve to illustrate current progress, be it in vitro in primary patient-derived cells and cellular disease models (Table 1), animal rare disease models (Table 2), or chimeric models with in vitro and in vivo data (Tables 1, 2 with correspondingly colored entries for data in both systems).
Table 1.
Achievements based on RNA-guided nuclease technology towards therapy development for rare diseases in human patient-derived cells
Reports are sorted by disease, then by gene and disease-causing mutation. All iPSCs are of human patient-derived origin. Colored fields indicate correspondence with studies in animal models in Table 2
ARG1 arginase 1, BE base editor, bulk population-wide percentages after treatment, without enrichment or antibiotic selection of corrected cells, cDNA complementary DNA, clonal percentages amongst pre-selected antibiotic-resistant clones, DM1 myotonic dystrophy type 1, DSB double-strand break, HDR homology-directed repair, HGVS Human Genome Variation Society, Hs Homo sapiens, HUDEP-2 human umbilical cord blood-derived erythroid progenitor cell line, iPSC induced pluripotent stem cell, ITGB3 integrin beta-3, ko knockout, mRNA messenger RNA, n/a not available, NHEJ non-homologous end-joining, NR embryos embryos created by fusion of thalassemic fibroblast nuclei with in vitro matured enucleated oocytes, NSG NOD (non-obese diabetic)-SCID (severe combined immunodeficient)-Il2rg-/-, TI targeted integration, zygotes tripronuclear zygotes
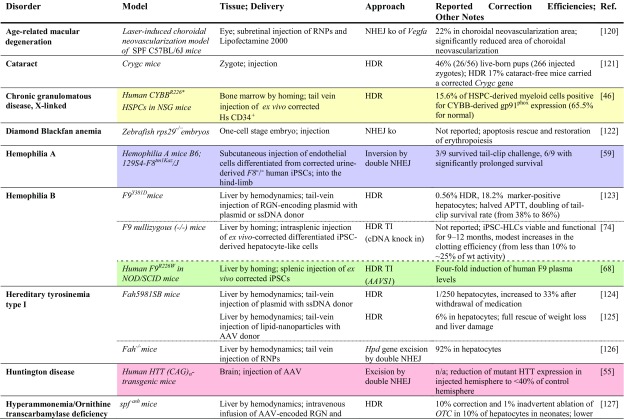
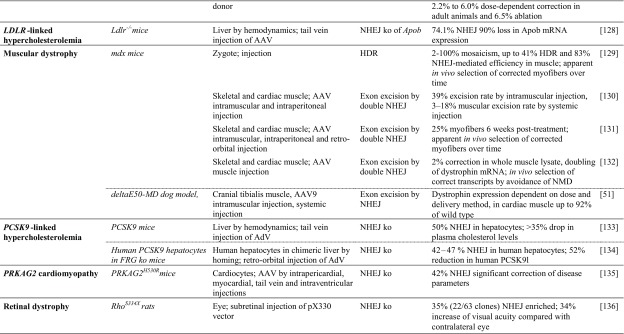
Table 2.
Achievements based on RNA-guided nuclease technology towards therapy development for rare diseases in in vivo studies
Studies listed are based on chimeric or transgenic animal disease models. Reports are sorted by disease, then by model. Percentage of ‘correction’ refers to alleles. Colored fields indicate correspondence with in vitro studies in patient-derived cells in Table 1
AAV adeno-associated virus, AdV adenovirus, APTT activated partial thromboplastin time; cDNA complementary DNA, CNS central nervous system, FRGFah–/–Rag2–/–Il2rg–/–, GFP green fluorescent protein, HDR homology-directed repair, Hs Homo sapiens, HSPC hematopoietic stem and progenitor cell, iPSC induced pluripotent stem cell, ko knockout, LDLR low density lipoprotein receptor, MIR137 microRNA 137, mRNA messenger RNA, n/a not available, NHEJ non-homologous end-joining, NOD non-obese diabetic, NSG NOD (non-obese diabetic)-SCID (severe combined immunodeficient)-Il2rg-/-, OTC ornithine transcarbamylase, PRKAG2 protein kinase AMP-activated non-catalytic subunit gamma 2, RNPs ribonucleoprotein particles, SCID severe combined immunodeficient, SPF specific pathogen-free, ssDNA single-stranded DNA, TI targeted integration, tNGFR truncated nerve growth factor receptor
Mutation-Specific Precision Repair
Correction of the causative mutation would restore the genotype of normal or at least carrier individuals (Fig. 1a). Whereas the approach may be substituted for suitable point mutations by the nascent base editing technology over time, at present it usually relies on DSB-activated HDR and is thus limited by the correspondingly low efficiency in primary cells. Notwithstanding this difficulty, the approach has been adopted for countless rare diseases for proof of principle and in exceptional studies has reached or approached clinically relevant levels of efficiency. Of the numerous studies targeting specific HBB mutations in SCD and β-thalassemia [42], a recent publication by Dever et al. [43] stands out (Tables 1, 2, turquoise). In the study, CD34+ cells from SCD patients were electroporated with ribonucleoprotein particles (RNPs) and provided with an adeno-associated virus (AAV) donor template in order to correct the causative sickling mutation. At already high 19% HDR efficiency in bulk culture, the study employed selectable markers for fluorescence-activated cell sorting to reach 92% HDR efficiency in sorted cells. Using truncated nerve growth factor receptor (tNGFR) as the selectable marker for sorting, subsequent analyses in NSG mice allowed a long-term chimerism of 7.5% corrected cells from an initial 12%, indicating that a substantial percentage of corrected and engrafted cells were long-term repopulating cells. Moreover, the study noted a bias against long-term repopulating cells in HDR-positive cells, in line with results for zinc-finger nuclease (ZFN)-mediated HDR elsewhere [44]. By contrast, an independent study recently published as preprint demonstrated over 20% correction of the HBBE6V (sickle cell disease) mutation in patient-derived HSCs engrafted in the bone marrow of NBSGW mice after CRISPR/Cas9-mediated HDR-based editing. It further demonstrated substantial percentages of HDR-corrected cells in the erythroid compartment of the bone marrow, in part through in vivo selection of corrected cells [45]. Similarly encouraging results were also achieved by De Ravin et al. [46] for X-linked chronic granulomatous disease (Tables 1 and 2, yellow), who employed electroporation of RNP and a single-stranded oligodeoxynucleotide (ssODN) donor to achieve up to 21% HDR in all alleles. Functional analyses in vitro after myeloid differentiation revealed significant correction of CYBB-derived gp91phox expression by repair of the CYBB premature stop codon, from background levels in negative controls up to a quarter of the percentage of gp91phox-positive cells seen in normal controls. These results were mirrored in vivo in NSG mice, where 15.6% of human myeloid cells were gp91phox-positive; once more, a quarter of the percentage seen for normal cells (65.5%). Myeloid-lineage differentiation led to an enrichment of gp91phox-positive cells compared with other lineages, indicating the therapeutic benefit for cell survival and a possible reduction of requirements for therapeutic bone marrow chimerism upon clinical application. Of note for many diseases with a high level of allelic heterogeneity, different mutations in compound heterozygote patients of recessive diseases may be corrected with the same RGN and donor template, as demonstrated by Xie et al. [47] for two β-thalassemia mutations in the HBB gene: one in its promoter, and one in exon 2. Using a donor template spanning both mutations and a single RGN to target intron 1 of HBB, the authors corrected both mutations in induced pluripotent stem cells (iPSCs) and established functional correction after erythroid differentiation. β-Thalassemia has also served as a model disease to pioneer repair by base editing [48], which gave 23% base editing efficiency for the HBB−78(A>G) mutation, and repair in human tripronuclear zygotes based on HDR [49]. The latter study indicated a high error rate and highlighted the risk of off-targeting highly sequence-similar paralogs, which in this case led to off-targeting of the HBD gene. For many target sites such difficulties can be addressed by alternative guide RNAs and, increasingly, by alternative Cas molecules with differing protospacer adjacent motif (PAM) requirements that would help exploit minor sequence differences between paralogs. Overall, mutation-specific precision repair has seen vast improvements in efficiency and precision in long-term repopulating cells recently, to a point where its application corrects potentially therapeutic percentages of cells, in particular where therapeutic correction confers a selective advantage in chimeric or animal disease models.
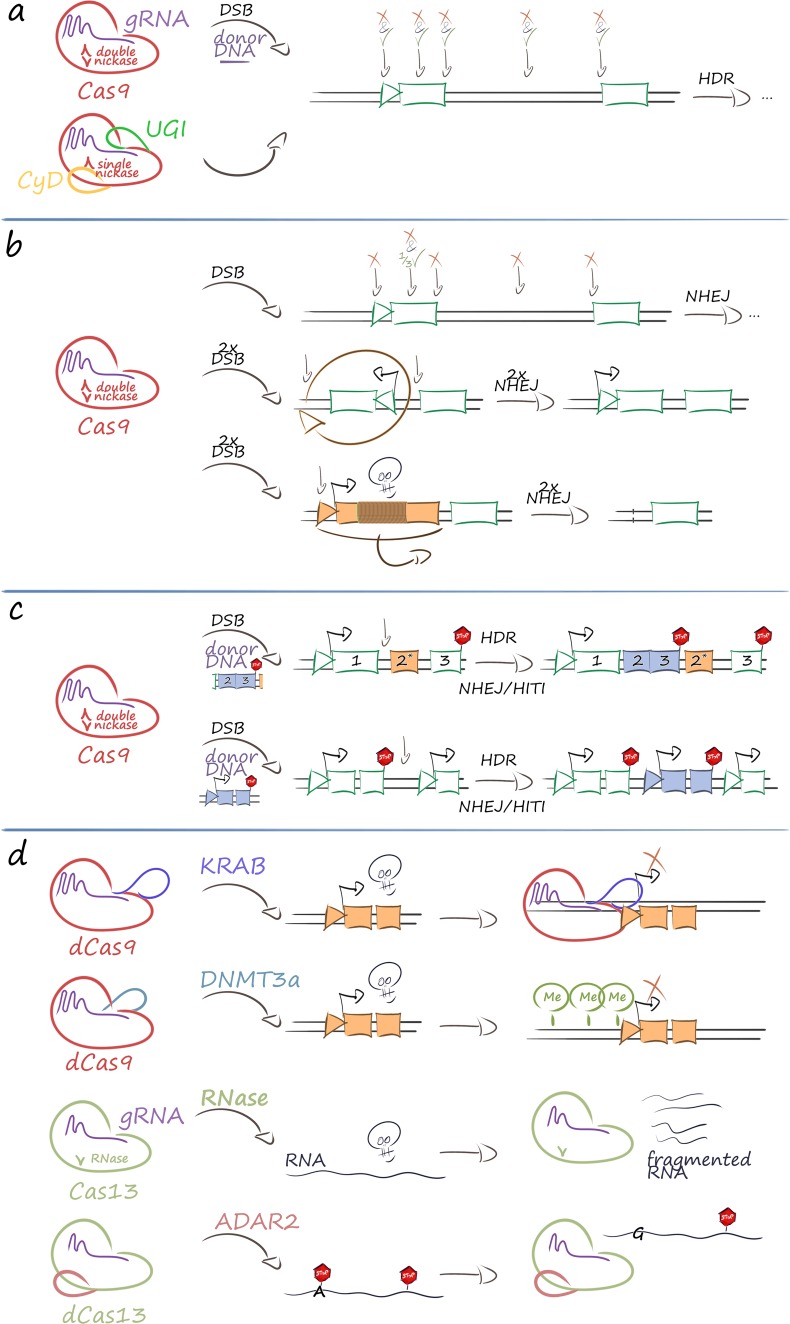
Fig. 1.
Therapeutic strategies based on CRISPR/Cas-based tools. The therapeutic use of CRISPR/Cas ribonucleoprotein complexes is wide-ranging and in its present application falls into four main categories. a Mutation-specific precision editing of any suitable nucleotide can be performed either by the original DNA endonuclease (double nickase) Cas9 molecule and HDR-mediated repair or by DNA base editors. This can be used to correct disease-causing mutations (green tick) or introduce disabling sequence changes (red cross). The figure illustrates as exemplary positions (from left to right) changes in the promoter region, the coding sequence, the splice donor site, deep intronic regulatory sequences, and the splice acceptor site. b Sequence disruption and rearrangement can be achieved by double nickases and NHEJ-based mechanisms. Top: sequences can be disrupted by a single DSB in the promoter region, the coding sequence, the splice donor site, deep intronic regulatory sequences, and the splice acceptor site. For some applications the statistical one-third of in-frame insertion and/or deletion (indel) events for changes in the open reading frame may be corrective and exploited for therapy. Middle: pathogenic inversions in the genome can be reversed by two flanking DSBs and NHEJ-mediated re-ligation (shown). Alternatively, inversion or excision can disable pathogenic or unwanted regulatory regions (not shown). Bottom: deletions for the removal of pathogenic or unwanted regulatory regions can likewise be achieved by two flanking DSBs and NHEJ-mediated re-ligation (shown), but has also been demonstrated by an appropriately spaced pair of single nickases (not shown). c Targeted integration in the genome can be achieved by a single DSB and, depending on the precision required at the junctions, can employ HDR-mediated repair or NHEJ-mediated HITI. Top: insertion of a cDNA under control of an endogenous promoter is one application of targeted integration; integration in intronic sequences of the mutated gene will allow repair of mutations downstream from the DSB. Bottom: insertion of expression cassettes into safe harbor loci, such as an intergenic region (shown) or the intron of an endogene (not shown) that will allow stable transgene expression, can be used for safe gene addition. d Modulation of gene expression can be performed at multiple levels and may employ, from top to bottom, deactivated Cas9 fused to transcriptional regulators (e.g., in order to dynamically repress expression of pathogenic genes), deactivated Cas9 fused to epigenome modifiers (e.g., in order to permanently repress expression of pathogenic genes by DNA methylation), Cas13-like RNA-guided RNA nucleases (e.g., in order to dynamically knock down mRNA of pathogenic genes), and nuclease-deficient Cas13-like fusions to chemical RNA-base modifiers (e.g., in order to repair nonsense mutations by A>G conversion). For clarity, epigenetic modifiers for DNA demethylation, histone acetylation, histone deacetylation, and others are not shown. Likewise, RNA base conversion for disease-causing missense mutations (such as demonstrated Ala>Trp conversion) is not shown. Throughout, the skull indicates a pathogenic gene or molecule and the STOP sign indicates a translation termination (nonsense) codon. ADAR2 adenosine deaminase acting on RNA 2 [82], cDNA complementary DNA, CRISPR/Cas clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas), CyD cytidine deaminase domain for C>U conversion in the single-stranded DNA loopout, currently with precision of ≤ 2 base pairs, dCas9 deactivated Cas9 without endonuclease activity, DNMT3a catalytic domain of DNA methyltransferase 3 alpha for DNA methylation and potentially persistent repression of gene expression for affected promoters [78], DSB double-strand break, gRNA guide RNA, HDR homology-directed repair, HITI homology-independent targeted integration, indel insertion and/or deletion, KRAB catalytic domain of Krüppel-associated box epigenetic repressor [80], mRNA messenger RNA, NHEJ non-homologous end-joining, UGI uracil DNA glycosylase inhibitor domain to prevent base excision repair and removal of base edit
Therapy by Disruption or Genome Rearrangement
Generally lower efficiency and the apparent bias against long-term repopulating cells by HDR-based repair [43, 44] favor NHEJ-based approaches for suitable targets. Random frameshifts, disruption of toxic sequences, and genome rearrangements for deletion or inversion can be achieved by NHEJ-mediated repair and applied for therapeutic effect (Fig. 1b).
Many rare disease cases are caused by frameshift or premature stop mutations, and where the frequency of random indels leading to open reading frame (ORF) repair is sufficient, both types of mutations can be bridged by random NHEJ-based disruption. Initial proof of principle for this approach was established in Duchenne muscular dystrophy (DMD) [50] based on TALEN technology, where approximately one-third of disruption events led to in-frame correction of the dystrophin gene, an outcome that at sufficiently high efficiency of delivery to target cells would be fully or partially therapeutic in DMD patients. Recent progress made in the field of gene editing in DMD is reflected in a study where AAV vectors were used to deliver CRISPR/Cas9 components intramuscularly and systemically in a canine large-animal model of DMD [51]. The study demonstrated high efficacy of single-cut genome editing for restoration of dystrophin expression, reaching up to ≈ 80% of normal levels in some muscles after 8 weeks. Interestingly, in cardiac muscle, dystrophin levels in the dog receiving the highest dose reached 92% of normal. The same approach, based again on RGN technology, also succeeded in correcting mutation and phenotype in cell lines holding a naturally occurring (XCGD) frameshift mutation in the CYBB transgene, with similar potentially therapeutic targeting rates also for CYBB in its normal genomic DNA (gDNA) context [52]. With 24% frequency of frameshift mutations for CYBB XCGD and similar contribution of frameshift mutations to epidemiology for X-linked severe combined immunodeficiency (X-SCID, IL2Rγ gene), Wiskott-Aldrich Syndrome (WAS, WASP gene), ADA-SCID (ADA gene), and β-thalassemia (HBB gene), this led the authors to argue that a quarter of patients with these disorders may be treatable with the same approach [52].
Although knockout of gene function or regulatory sequences by a single RGN is mainly used for functional studies, the approach has also been applied towards therapeutic use. The possibly most significant application to date is once again in the context of β-hemoglobinopathies, where Canver et al. [53] achieved high-level induction of the endogenous HBB paralogs HBG1 and HBG2. The study employed a total of over 700 RGNs to pinpoint the erythroid enhancer of the γ-globin repressor BCL11A, and identified a highly γ-globin-inducing RGN target in one of three candidate regions. The resulting γ-globin expression potentially compensates for absence of HBB in β-thalassemia and has an additional anti-sickling effect in SCD, so that the approach may be widely applicable across β-hemoglobinopathies. The study went on to demonstrate action in SCD, using integrating lentiviral vectors (LVs) for proof of concept, in primary CD34+ cells of SCD patients and tripled the number of γ-globin-positive cells in the process. Approaches such as the one described require fundamental insights into molecular mechanisms of disease and relevant disease modifiers, but potentially cover a range of diseases or at least of different primary mutations in the case of allelic heterogeneity. They are thus more widely applicable than mutation-specific approaches and therefore more likely to be chosen for clinical translation.
Other studies have used pairs of nucleases instead of single RGNs in order to achieve potentially therapeutic outcomes. This was applied by two studies for Huntington disease [54, 55], possibly representative also of other dominant trinucleotide repeat disorders. Both studies employed patient-derived fibroblasts and haplotype-specific RGNs in order to disable selectively the mutant HTT allele. In the first case the entire mutant HTT gene was excised with two haplotype-selective RGNs, whereas in the second study, promoter-proximal allele-specific RGNs and a universal RGN in intron 1 of HTT were used to inactivate the mutant allele. In a further study, two CRISPR/Cas9 nickases either side of the HTT trinucleotide repeat expansion were used for its excision, although frameshift creation and employment of an allele-independent targeting approach would inactivate both HTT alleles in case of bi-allelic events [56]. In another study for SCD therapy development, different-sized deletions/inversions in the HBB locus were introduced in erythroid cells in order to gauge their potential to induce γ-globin by changing the expression dynamics of the locus. The most effective RGN combination, giving a 13.6 kb inversion/deletion, was then tried in primary CD34+ cells of two SCD patients, which combined with green fluorescent protein (GFP)-based selection resulted in 32.8% and 62.2% inversion/deletion allele frequency, respectively, and a 50% reduction of sickling under low oxygen pressure as a mark of phenotypic correction [57]. In another study with a focus on cystic fibrosis, three different deep intronic mutations with impact on CFTR splicing were excised by pairs of RGNs, an approach that resulted in restoration of correct splicing in a minigene functional assay [58]. While these studies did not use technology suitable for clinical application, they served to provide solid proof of concept for paired RGN application and NHEJ-based therapy for rare diseases. Importantly, Park et al. [59] (Tables 1, 2, purple) applied pairs of RGNs designed to revert disease-causing inversion events in the F8 gene from two different hemophilia A patients. Applying electroporation of RGNs as RNPs into urine-derived iPSC cells, the study achieved inversion efficiencies in bulk populations of 6.7% for a 1 kb inversion and of 3.7% for a 22 kb inversion. Transplantation of these cells into an F8 hemophilia A mouse model resulted in improvement of survival time and in three of nine mice even achieving overall survival upon bleeding injury.
To conclude, NHEJ-mediated repair, although more efficient than HDR, has more limited applications, as disruption, excision, or rearrangement may not be suitable therapeutic approaches for many mutations and rare diseases. However, where the causative mutation is suitable for repair by disruption or excision, or where insights into molecular mechanisms allow the deactivation of negative disease modifiers for therapy of multiple causative mutations, efficient strategies based on NHEJ might be preferable to those based on HDR for clinical translation.
Targeted Integration
There are many cases where precise repair and disruption may both be unsuitable therapeutic approaches. First, many rare diseases with a high level of allogeneic heterogeneity cannot be addressed by modulating disease modifiers, unlike β-hemoglobinopathies. In those cases, therapy needs to cover a range of the correspondingly rare individual mutations in order to become applicable to a larger number of patients and make therapy development practicable as a commercial undertaking. Second, and a more fundamental consideration, molecular therapies for diseases caused by large deletions usually require the reintroduction of the lost genetic material. In both instances, targeted integration (TI) of expression cassettes or gene fragments would be therapeutic, given sufficiently high transfer and integration rates, and would minimize the risk of insertional mutagenesis.
A major application for TI is based on so-called safe harbor loci as integration sites that allow stable expression of the integrated transgene while not being of essence in the affected cell lineages. Moreover, this strategy can reuse effective and safe RGNs for the addition of different transgenes to the same site, which lowers the development effort for individual therapies. Besides the AAV integration site 1 (AAVS1) locus in human hematopoietic stem and progenitor cells (HSPCs) [60], the HPRT gene locus has also been employed as a safe harbor locus for TI in human cells [61], as has the chemokine (CC motif) receptor 5 (CCR5) [62], and the popular murine ROSA26 locus has also found its ortholog in the human genome [63]. TI strategies that disrupt HPRT function can additionally enrich modified cells by selection against non-integrants with 6-thioguanine in vitro [64] and, with mixed results, in vivo [64, 65]. In a deviation from the safe harbor principle, co-targeting of the HPRT locus can also be used for chemical selection in order to enrich for editing events at independent loci, which have a high level of co-occurrence [61]. Whereas AAVS1 and HPRT are used for insertion of autonomous expression cassettes, ROSA26 and CCR5 can be employed to achieve expression of transgenes from the endogenous control elements of the locus [63, 66, 67]. Regarding the AAVS1 site, in a seminal study employing ZFNs for gene therapy of X-SCID, fine-tuning of conditions allowed between 3 and 6% efficiency of HDR-based integration in primitive to early HSPCs [44]. TI at the AAVS1 site was also exploited in a study for hemophilia B therapy development, which employed urine-derived hemophilia B iPSCs for in vitro and in vivo evaluation [68] (Tables 1, 2, green). In short-term follow-up in immunodeficient mice, iPCS-derived cells with AAVS1 integration of a human elongation factor 1α (EF1α)-promoter-driven F9 complementary DNA (cDNA) achieved eight-fold induction of serum F9 levels compared to integration-free control cells. Recently, conditions were refined for RGN-mediated AAVS1-TI, and it was found that excluding Cas9 expression from G1 phase of the cell cycle significantly enhanced HDR-mediated integration at the locus [69]. The HPRT locus served as safe harbor for therapy in iPSCs of a patient with ARG1-linked urea cycle disorder [70]. The strategy targeted exon 1 of HPRT for integration of a construct holding both a puromycin resistance expression cassette and an ARG1 cDNA driven by the EF1α promoter. This allowed selection of HPRT disruption and of puromycin resistance to give between 46 and 92% of normal arginase messenger RNA (mRNA) levels in undifferentiated iPSCs; this was decreased, however, to between 27 and 39% upon hepatic differentiation. Finally, the murine ROSA26 locus was recently used for TI after adenoviral delivery of RGNs and a human α-1-antitrypsin HDR donor into the liver of wild-type mice [71], which achieved overall allele frequencies in the liver of between 5 and 26% as a tentative basis for therapy of human α-1-antitrypsin deficiency. Of note, AAVS1 encodes the phosphatase 1 regulatory subunit 12C (PPP1R12C), also known as the myosin binding subunit 85 (MBS85), and, as for CCR5, the exact importance of the gene encoded at this locus is not yet known [62]. Many transgenes can interfere with splicing of PPP1R12C transcripts [67], and a study investigating TI at the AAVS1 locus detected a change in cellular contractile force, implying that at least for the treatment of myopathies, AAVS1 may not be an inert safe harbor locus after all [72]. Therefore, studies to date on what might be more generally called ‘standard loci for TI’ indicate the scope for reapplying the corresponding tools in a modular fashion for safe integration, selection, or ubiquitous expression in therapy development for rare diseases, while also highlighting that suitability of each locus needs to be examined on a case-by-case basis.
A further application for TI is integration in the mutant locus itself, in order to exploit endogenous transcriptional control elements and, for gain-of-function mutations, to inactivate a potentially toxic mutant while adding a functional copy. The former strategy was pursued in the original ZFN-based study for X-SCID in parallel to using the AAVS1 safe harbor locus and in a follow-up study was also extended to RGN-mediated integration into the IL2RGR226H mutant gene [73]. Using RNP electroporation and AAV delivery of the donor template, the latter study achieved 50% reporter-positive committed hematopoietic progenitors (CD34+CD133−) and 25% reporter-positive primitive hematopoietic stem cells (CD34+CD133+CD90+). Both studies used the endogenous locus and promoter to drive cDNA expression of a partial IL2RG cDNA, but in one of many improvements to the previous study [44], the more recent work employs a cDNA and integration site downstream of exon 1 (instead of exon 4), suitable for repair of any causative IL2RG mutation from exon 2 onwards [73]. A similar approach was also employed in a study for haemophilia B, where CRISPR/Cas9 and an HDR donor DNA were used to knock a full-length human F9 cDNA into exon 1 of the F9 endogene in iPSCs from two hemophilia B patients. These corrected iPSCs were differentiated into hepatocyte-like cells (HLCs) and transplanted into hemophilic mice, where they achieved engraftment, expansion, and production of F9 at therapeutic levels 6–9 months after transplantation [74].
Studies on TI based on RGNs are still infrequent, but bound to proliferate with the continuously improving efficiency of the approach. The original concept of efficient TI is dependent on RGN application with high general HDR efficiency, which amongst other improvement strategies may be increased five-fold by silencing of NHEJ components [75]. However, the more recent approach of homology-independent TI (HITI) [76] relies on NHEJ and may increase TI efficiencies considerably for many applications. With ongoing development and wide scope for therapeutic application [77], HITI as an approach for higher-efficiency TI may therefore turn out to be an important addition to the rare disease therapy toolkit.
Modulation of Gene Expression
A plethora of tools based on CRISPR/Cas technology have already been developed that allow control of gene expression, and transcriptional regulators as well as RNA editors have already been used towards rare disease therapy. Both technologies have the clear disadvantage that for curative therapy development permanent integration or lifelong repeat application would be required, even though AAV-based approaches can show persistence of expression over several months. The recent development of epigenome editing tools to our knowledge still awaits its application to rare diseases, but results in modifications that appear permanent and therefore potentially suitable for curative therapy. This is the case at least for DNA modification, with as yet conflicting results for the persistence of histone modifications and their causative effect on gene repression [78].
Potential disease-specific application of transcriptional modulation was demonstrated for PCSK9-linked familial hypercholesterolemia in a study that employed a nuclease-deactivated Cas9 (dCas9) linked to the Krüppel-associated box epigenetic repressor (KRAB) motif and achieved significant knockdown of Pcsk9 mRNA in mouse liver [79]. The study further showed maintenance of significant knockdown after double-hit delivery by AAV vectors for at least 168 days, and also a transient peak of treatment-specific liver damage, as measured by elevated alanine transaminase (ALT) serum levels. Moreno et al. [80] employed a split Cas9 system based on two AAV vectors in order to increase targeting capacity for transcriptional regulation and demonstrated its utility in a murine model of retinitis pigmentosa. Using a KRAB fusion as a transcriptional repressor and a VP64 + rTA fusion as transcriptional activator, the study achieved effective reprogramming of photoreceptor types in vivo first in normal mice and then in the rd10 murine model of autosomal recessive retinitis pigmentosa. In the latter, transcriptional repression of Nrl effectively converted rod- to cone-like receptors, prevented photoreceptor degeneration, and 6 weeks after injection gave significantly higher visual acuity for all eyes injected.
Therapeutic relevance of RNA knockdown was demonstrated for frontotemporal dementia with parkinsonism using the compact Cas13d-derived CasRx system [81]. AAV delivery of CasRx to patient-derived iPSCs significantly reduced the pathologically elevated ratio of the 4R and 3R isoforms of tau (encoded by MAPT) to 50% of mock-treated control value, while healthy controls showed 20–30% of control value. The same study also demonstrated absence of double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) cleavage as a safety feature, and the ability of CasRx to process an array of multiple guide RNAs for multiplexed targeting of endogenous RNA from the same construct.
A landmark study by Cox et al. [82] employed a deactivated Cas13 (dCas13) fused to an adenosine deaminase acting on RNA 2 (ADAR2) catalytic domain in order to repair pathogenic mutations at the RNA level. Effective A>G base conversion and reversion of nonsense mutations was demonstrated for pathogenic transcripts implicated in X-linked nephrogenic diabetes insipidus (AVPR2 W293* at up to 35% efficiency) and Fanconi anemia (FANCCW506* at up to 22% efficiency) as full-length transcripts in transiently transfected HEK293T cells, and was also detected above 20% efficiency for an additional nine of 34 partial transcripts tested, including nonsense mutations of MEPCP2 (Rett syndrome), ADGRV1 (Usher syndrome type 2C), AHI1 (Joubert syndrome 3), PRKN (Parkinson disease 2), COL3A1 (Ehlers-Danlos syndrome type 4), BRCA1 (breast–ovarian cancer, familial 1), MYBPC3 (primary familial hypertrophic cardiomyopathy), APC (familial adenomatous polyposis 1), and BMPR2 (primary pulmonary hypertension).
Evidently, CRISPR/Cas technology has moved beyond gene editing and allows flexible and precise modification of the epigenome and the transcriptome. Its versatility and developments such as the use of dCas9 or small Cas13 derivatives in combination with additional functional domains will accelerate basic research into transcriptional, epigenome, and RNA-level regulation of gene expression, besides contributing tools for translational medicine. The early findings discussed here showcasing such modular use of CRISPR/Cas technology demonstrate that therapy of rare diseases will have its fair share of benefit, but that certain aspects such as efficiency and permanence of treatment still need to be addressed.
Using the Right Tool for the Job
The versatility and ease of use of CRISPR/Cas technology and its derivatives have speedily led to the development of a still expanding battery of diverse tools and approaches to tackle disease. In this currently fast-moving field, a key dilemma for researchers working on therapy development for rare diseases is their need to initiate their work in the present with a definite choice of tools and approaches for their disease of interest, despite the risk that their choice might appear outdated in the near future.
The unique molecular, pathophysiological, and clinical characteristics of the disease of interest will be a determining factor in the choice of therapeutic strategy, where such knowledge exists. However, and towards eventual translation to the clinic, consideration of the number of potentially treatable patients with a given approach also needs to be given adequate weight in the choice of treatment strategy. Should a mutation-specific approach be employed, should a targeted insertional approach be employed instead, in order to cover a range of disease-causing alleles, or might the latter also be achieved by manipulation of mutation-independent disease modifiers? Other than for large deletions or rearrangements as causes of disease, mutation-specific precision repair will be universally applicable as a treatment approach, wherever patient numbers and efficiency of correction are suitable. For loss-of-function mutations, knock-in of functional cDNA or of gene fragments in the defective endogene or targeted gene addition of expression cassettes in safe harbor loci might be feasible approaches. For gain-of-function mutations, knock-in into and deactivation of the defective endogene would once again be suitable, as would disease-allele-specific disruption, transcriptional inactivation, or RNA knockdown. Where the required nucleotide change is suitable, the quickly evolving approach of DNA base editing might provide a safer option than DSB-based precision repair, whereas RNA base editing might provide transient and safer alternatives to DNA precision repair or disruption instead.
The molecular strategy, however, is only one of the choices to be made. For delivery of CRISPR/Cas tools, selecting in vivo or ex vivo application and the choice of vector type and administration routes will depend on clinical phenotypes and accessibility and type of affected tissues. Choice of timing, too, is of great importance when treating chronic and progressive diseases, so as to maximize therapeutic benefit while minimizing risks. The treatment delivery timepoint is crucial but often uncertain, as gene editing applied too late may not be able to repair established damage, whereas premature application, such as of gene silencing CRISPR/Cas treatments, may lead to unexpected and deleterious gene dysfunctions [83, 84]. More importantly, each study has to calculate the risks cautiously and independently for each patient, in order to define the acceptable level of risk in the pursuit of a cure. Finally, informative trial design, suitable manufacturing of reagents, and the potential for commercialization all need to be considered, before even the most ingenious molecular strategy may translate into a successful therapy for rare disease patients.
Rare Trends: Clinical Translation for Rare Diseases is Taking Off
From Shakedown to Maiden Voyage
There are still many unknowns in the therapy development for rare diseases based on CRISPR/Cas technology. With the field developing at a staggering pace, it is unclear how well current cutting-edge technology in terms of off-target effects, efficiencies of delivery, and efficiencies of cell-autonomous correction will stand up to comparison 10 or even 2 years down the line. After all, safety standards and assumptions from the early days of gene addition, made by professionals no less diligent than those involved in gene editing today, appear naïve in retrospect. The correspondingly risky application of the fledgling CRISPR/Cas technology has thus been spearheaded by application to cancer, and the first-in-man Chinese CRISPR/Cas application to a lung cancer patient in 2016, only 3 years after the technology was first used in eukaryotic cells, is already history [85]. Meanwhile, researchers and clinicians dealing with less acute diseases rightly chose to err on the side of caution. Whereas the current generation of RGN therapeutics for ex vivo application is fully characterized for off-target activity and can be delivered efficiently in virus- and DNA-free fashion, new studies continue to uncover potentially problematic aspects of DSB-related technologies. Application in vivo faces additional challenges of delivery and safety, some of which will be hard to address in murine or chimeric models. Differences in size, longevity, cytokine microenvironments, and specific DNA and RNA sequences are only some parameters that may interfere with meaningful analysis and interpretation of results and will necessitate research in larger models, non-human primates or, for what remains unknown once all preclinical tests are passed, clinical trials.
Flight Plan
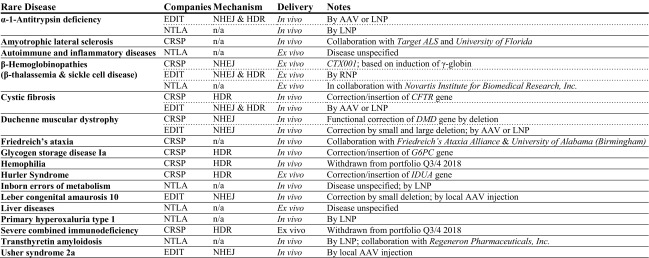
With the advent of CRISPR/Cas and the ongoing progress of gene therapy clinical trials in general, companies involved in rare disease therapy development have multiplied over the last years [86], and with the accessibility of the technology, even crowdfunding for smaller biotech companies is considered an option [87]. The three leading companies in the field of CRISPR/Cas pharmaceuticals, CRISPR Therapeutics, Intellia Therapeutics, and Editas Medicine, 4 years after their initial funding and 2 years after going on the stock exchange, are pushing forward ambitious programs for rare disease therapy development in the USA and Europe. The product pipelines for non-malignant, non-infectious disorders read like a who-is-who of prominent rare diseases, with ongoing addition and removal of candidate diseases for early-stage development (Table 3). To date, and as a first for CRISPR/Cas application to rare diseases, CRISPR Therapeutics is recruiting for a clinical phase I/II trial for β-hemoglobinopathies with its CTX001 product in Germany and the UK (NCT03655678 [141]), with an initial cohort of 12 patients, extendable up to 45. A suspension put on the trial by the US Food and Drug Administration (FDA) was lifted on 10 October 2018 [88], a development that, while opening up parallel trial initiation in the USA, indicates persistent uncertainties about the safety of the technology and the dissemination of related information.
Table 3.
Rare disease product pipelines of leading companies based on clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) technology
Only product development for non-malignant diseases is listed, sorted by rare disease. Information has been extracted from the product pipelines given on the company websites of CRISPR Therapeutics [138], Editas Medicine [139], and Intellia Therapeutics [140], last update 22 November 2018. Recent changes in the portfolio, as indicated, suggest ongoing readjustments of early-stage targets, which may also affect current portfolio content or result in additional disease targets in the future
AAV adeno-associated virus, CRSP CRISPR Therapeutics, EDIT Editas Medicine, HDR homology-directed repair, LNP lipid nanoparticle, n/a not available, NHEJ non-homologous end-joining, NTLA Intellia Therapeutics, RNP ribonucleoprotein particle
Ethical Turbulence
Although CRISPR/Cas-related research publications for rare diseases are proliferating, commercial exploitation of promising approaches is making it increasingly difficult to survey and critically assess cutting-edge translational progress. As development switches from academic laboratories to biotech and pharmaceutical companies, so does dissemination of results from peer-reviewed articles to business statements and announcements on websites. Therefore, the good news for patients that investment in gene therapy is booming [89] has the flipside that increasingly frequent conflicts of interest in research interfere with unbiased dissemination of methods and results, endangering current governance structures and possibly long-term trust in the technology [90–92]. As a prominent example, preprint publication in early 2018 of the pre-existing population-wide immunity against Cas9 proteins [33–35] prompted market upheaval in CRISPR/Cas-related stocks and the declaration that the problem had already been investigated and found to be no impediment to therapeutic application [93, 94]. Be that as it may, early dissemination of related findings would have been better for overall trust in the technology and in the integrity of those involved in their commercial development. Similarly, full disclosure of methodology and results for analyses of off-target and recombination events would help establish trust in the technology and the safety of prospective trials, given that it is unclear whether such analyses have taken into account the latest developments in the field. For instance, off-target assessment often relies on amplification of areas closely flanking the sites of interest, and such assessment would miss long-distance recombination events recently found to be triggered even by single-DSB approaches [39]. The ongoing withdrawal of frontline developments from peer review and from independent experimental confirmation is therefore a concrete problem ethically as well as—it turns out—commercially. Whereas secrecy is therefore a major concern of the field, another still more pressing concern is the erosion of ethical boundaries in the clinical application of CRISPR/Cas technology [95]. Recent reports of the birth of CRISPR/Cas-edit twins, engineered to prevent infection by HIV from their father [96], indicate an absence of checks and balances in the application of CRISPR/Cas at one if not more institutions equipped to apply the technology to human embryos, and a possibly wider and worrying loss of perspective in the assessment of risks and benefits for human life.
Budget Travel
Gene addition approaches by ex vivo therapy are costly to implement, and drug companies struggle to switch business models for inherited diseases from lifelong drug-based management to one-off curative treatments. A contributing factor to the recent sale of GlaxoSmithKline’s gene therapy portfolio to start-up company Orchard Therapeutics might therefore have been the significant price tag of potential treatments. For the approved orphan drug Strimvelis® for ADA-SCID, which is based on retroviral gene addition, this is a significant €594,000 per patient [97]. However, beyond the suffering prevented by this curative treatment, the cumulative cost of palliative treatment is several-fold this cost. What is more, non-viral ex vivo delivery for RGNs, and in particular in vivo delivery, might make one-shot therapies easier and cheaper to implement. A significant development here is the potential transformation of gene therapy centers from good manufacturing practice (GMP) containment facilities to closed-circuit GMP devices in a clean room, at least for disorders treatable by HSPC transplantation. Addition of an in-line lentiviral transduction module to the Miltenyi CliniMACS Prodigy® device for isolation of HSPCs in a recent Fanconi anemia trial more than halved the personnel and time requirements for treatment [98]. An alternative module for electroporation of RNPs and HDR donors allows analogous simplification of infrastructure and procedures also for CRISPR/Cas delivery [98]. The machine would make a clinical gene therapy program for suitable disorders widely affordable and traditional GMP facilities at times obsolete [99]. Another emerging technology, in utero gene therapy, may change not only the cost and accessibility of rare disease treatments, but would moreover increase efficiency of treatment and expand the range of disorders to which gene therapy may be applicable [100]. The risk of inadvertent germline transmission and the inability to predict possible success in human application from experimental models are concerns en route to clinical translation, but the landmark drop in required vector amounts and treatment cost, and, more fundamentally, the applicability to early-onset and prenatally lethal disorders, would be another game changer for gene therapy. Ongoing research into possible complications of treatment [101], improvements to efficiency [102], and successful tests in non-human primates, such as those for treatments of hemophilia B [103] and Gaucher disease [104], may help overcome bioethical and regulatory hurdles in implementing the technique for rare diseases.
Not a Trip, But a Journey
It therefore seems that rare diseases and CRISPR/Cas technology are going places together, albeit with choices and uncertainties ahead and with the constant need to maintain an open scientific culture and safeguard ethical boundaries for its clinical application. CRISPR/Cas-based technology, newly derived tools, and ongoing auxiliary improvements to delivery methods, including GMP-in-a-box and in utero delivery, have great potential to transform and democratize gene therapy for rare diseases. Mutation-specific cures may thus become affordable, even for small patient cohorts, and allow restoration of normal or carrier genotype as the most immediate and effective therapy. Increasingly efficient manipulation of adult stem cells has already made them the safe substrate of choice for much of CRISPR/Cas-based therapy development for rare diseases and sidelined the approach of editing and clonally selecting iPSCs, which still pose safety concerns for clinical application [105]. Ongoing improvements to efficiencies of CRISPR/Cas-based tools and approaches and refined use of inert human markers, such as tNGFR [43], may further endorse the use of adult stem cells and will encourage still wider adoption of CRISPR/Cas editing technology. In parallel, increasing affordability and application of massively parallel sequencing not only accelerates the discovery of new therapy targets for rare diseases, but applied to CRISPR/Cas also helps delineate off-target and recombination events associated with treatment. It appears that the most comprehensive currently available analysis methods allow us to fully characterize existing CRISPR/Cas-based tools for DNA-level efficiency and safety, while a switch to the most efficient of the high-specificity RGN variants [106–108] will likely become routine for new commercial research and development. Gene editing as version 2.0 of gene therapy has been accelerated enormously by the young CRISPR/Cas technology. Precision editing, targeted disruption, rearrangement and integration, and modulation of gene expression based on the CRISPR/Cas platform already offer staggering possibilities for gene therapy development and, given the current impetus, the repertoire of tools and approaches for rare disease gene therapy will continue its rapid expansion. As further CRISPR/Cas-based gene therapies enter clinical trials for rare diseases, versions 2.1 and higher will always be just around the corner.
Funding
The authors were co-funded by Eurobank Cyprus Ltd (Scholarship 33173114), TELETHON Cyprus, the EU Erasmus+ program, and the European Union’s Seventh Framework Program for Research, Technological Development and Demonstration under Grant agreement no. 306201 (THALAMOSS).
Conflict of interest
Panayiota Papasavva, Marina Kleanthous, and Carsten W. Lederer have no conflicts of interest to disclose. No writing assistance was utilized in the production of this review.
References
- 1.Szajner P, Yusufzai T. Introducing rare diseases. Rare Dis. 2013;1:e24735. doi: 10.4161/rdis.24735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moliner AM, Waligora J. The European Union policy in the field of rare diseases. Adv Exp Med Biol. 2017;1031:561–587. doi: 10.1007/978-3-319-67144-4_30. [DOI] [PubMed] [Google Scholar]
- 3.Westermark K, Holm BB, Soderholm M, Llinares-Garcia J, Riviere F, Aarum S, et al. European regulation on orphan medicinal products: 10 years of experience and future perspectives. Nat Rev Drug Discov. 2011;10:341–349. doi: 10.1038/nrd3445. [DOI] [PubMed] [Google Scholar]
- 4.Kliegman RM, Ruggeri BE, Smith MM. The team-based approach to undiagnosed and rare diseases. Pediatr Clin North Am. 2017;64:17–26. doi: 10.1016/j.pcl.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Pavan S, Rommel K, Mateo Marquina ME, Höhn S, Lanneau V, Rath A. Clinical practice guidelines for rare diseases: the Orphanet Database. PLoS One. 2017;12:e0170365. doi: 10.1371/journal.pone.0170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin CP, Cutillo CM, Lau LPL, Jonker AH, Rath A, Julkowska D, et al. Future of rare diseases research 2017-2027: an IRDiRC perspective. Clin Transl Sci. 2018;11:21–27. doi: 10.1111/cts.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezzerri V, Cipolli M. Shwachman-Diamond syndrome: molecular mechanisms and current perspectives. Mol Diagn Ther. 2018 doi: 10.1007/s40291-018-0368-2. [DOI] [PubMed] [Google Scholar]
- 8.Cabrini G. Innovative therapies for cystic fibrosis: the road from treatment to cure. Mol Diagn Ther. 2018 doi: 10.1007/s40291-018-0372-6. [DOI] [PubMed] [Google Scholar]
- 9.Chabannon C, Kuball J, Bondanza A, Dazzi F, Pedrazzoli P, Toubert A, et al. Hematopoietic stem cell transplantation in its 60 s: a platform for cellular therapies. Sci Transl Med. 2018;10:eaap9630. doi: 10.1126/scitranslmed.aap9630. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein DL, Lobritto S, Iuga A, Remotti H, Schiano T, Fiel MI, et al. Lysosomal acid lipase deficiency allograft recurrence and liver failure- clinical outcomes of 18 liver transplantation patients. Mol Genet Metab. 2018;124(1):11–19. doi: 10.1016/j.ymgme.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Choi K-A, Choi Y, Hong S. Stem cell transplantation for Huntington’s diseases. Methods. 2018;133:104–112. doi: 10.1016/j.ymeth.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Steward CG, Jarisch A. Haemopoietic stem cell transplantation for genetic disorders. Arch Dis Child. 2005;90:1259–1263. doi: 10.1136/adc.2005.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall AK, Carlson MR. The current status of orphan drug development in Europe and the US. Intractable Rare Dis Res. 2014;3:1–7. doi: 10.5582/irdr.3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Dunnen JT. Sequence variant descriptions: HGVS nomenclature and Mutalyzer. Curr Protoc Hum Genet. 2016;90:7.13.1–7.13.19. doi: 10.1002/cphg.2. [DOI] [PubMed] [Google Scholar]
- 15.Robinson HM. Increasing the involvement of diverse populations in genomics-based health care—lessons from haemoglobinopathies. J Community Genet. 2017;8:311–318. doi: 10.1007/s12687-017-0327-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis J, Snyder M, Hyatt-Knorr H. Marking 15 years of the genetic and rare diseases information center. Transl Sci Rare Dis. 2017;2:77–88. doi: 10.3233/TRD-170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CTSA Principal Investigators. Shamoon H, Center D, Davis P, Tuchman M, Ginsberg H, et al. Preparedness of the CTSA’s structural and scientific assets to support the mission of the National Center for Advancing Translational Sciences (NCATS) Clin Transl Sci. 2012;5:121–129. doi: 10.1111/j.1752-8062.2012.00401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: The orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 19.Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 20.Kountouris P, Lederer CW, Fanis P, Feleki X, Old J, Kleanthous M. IthaGenes: an interactive database for haemoglobin variations and epidemiology. PLoS One. 2014;9:e103020. doi: 10.1371/journal.pone.0103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NORD—National Organisation for Rare Disorders. https://rarediseases.org. Accessed 9 Sept 2018.
- 22.EURORDIS—Rare Diseases Europe. https://www.eurordis.org. Accessed 13 May 2018.
- 23.Wright CF, FitzPatrick DR, Firth HV. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet. 2018;19:253–268. doi: 10.1038/nrg.2017.116. [DOI] [PubMed] [Google Scholar]
- 24.Cram DS, Zhou D. Next generation sequencing: coping with rare genetic diseases in China. Intractable Rare Dis Res. 2016;5:140–144. doi: 10.5582/irdr.2016.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patsali P, Kleanthous M, Lederer CW. Disruptive technology: CRISPR/Cas-based tools and approaches. Mol Diagn Ther. 2019 doi: 10.1007/s40291-019-00391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghiaccio V, Chappell M, Rivella S, Breda L. Gene therapy for beta-hemoglobinopathies: milestones, new therapies and challenges. Mol Diagn Ther. 2019 doi: 10.1007/s40291-019-00383-4. [DOI] [PubMed] [Google Scholar]
- 27.US Food and Drug Administration. Guidance for industry: considerations for the design of early-phase clinical trials of cellular and gene therapy products. 2015. https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-bio-gen/documents/document/ucm564952.pdf. Accessed 13 Feb 2019.
- 28.Aiuti A, Roncarolo MG, Naldini L. Gene therapy for ADA-SCID, the first marketing approval of an ex vivo gene therapy in Europe: paving the road for the next generation of advanced therapy medicinal products. EMBO Mol Med. 2017;9:737–740. doi: 10.15252/emmm.201707573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Commission. Marketing authorisation (EU/1/18/1331) for “Luxturna - voretigene neparvovec,” granted on 22.11.2018. http://ec.europa.eu/health/documents/community-register/2018/20181122142655/dec_142655_en.pdf. Accessed 19 Feb 2019.
- 30.Yu TTL, Gupta P, Ronfard V, Vertès AA, Bayon Y. Recent progress in European advanced therapy medicinal products and beyond. Front Bioeng Biotechnol. 2018;6:1–8. doi: 10.3389/fbioe.2018.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touchot N, Flume M. Early insights from commercialization of gene therapies in Europe. Genes (Basel). 2017;8:78. doi: 10.3390/genes8020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halioua-Haubold CL, Peyer JG, Smith JA, Arshad Z, Scholz M, Brindley DA, et al. Regulatory considerations for gene therapy products in the US, EU, and Japan. Yale J Biol Med. 2017;90:683–693. [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner DL, Amini L, Wendering DJ, Burkhardt L-M, Akyüz L, Reinke P, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2019;25(2):242–248. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 34.Wagner DL, Amani L, Wendering DJ, Reinke P, Volk H-D, Schmueck-Henneresse M. High prevalence of S. pyogenes Cas9-specific T cell sensitization within the adult human population-A balanced effector/regulatory T cell response. bioRxiv. 2018 [Google Scholar]
- 35.Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25(2):249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, et al. P53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 37.Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 39.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, et al. Base editing with a Cpf1–cytidine deaminase fusion. Nat Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 41.Plosky BS. CRISPR-mediated base editing without DNA double-strand breaks. Mol Cell. 2016;62:477–478. doi: 10.1016/j.molcel.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Lederer CW, Kleanthous M. Beta testing: preclinical genome editing in β-globin disorders. Cell Gen Ther Insights. 2015;1:231–242. [Google Scholar]
- 43.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magis W, DeWitt MA, Wyman SK, Vu JT, Heo S-J, Shao SJ, et al. In vivo selection for corrected β-globin alleles after CRISPR/Cas9 editing in human sickle hematopoietic stem cells enhances therapeutic potential. bioRxiv. 2018 [Google Scholar]
- 46.De Ravin SS, Li L, Wu X, Choi U, Allen C, Koontz S, et al. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci Transl Med. 2017;1:eaah3480. doi: 10.1126/scitranslmed.aah3480. [DOI] [PubMed] [Google Scholar]
- 47.Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, et al. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang P, Ding C, Sun H, Xie X, Xu Y, Zhang X, et al. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Chen Y, Sun X, Liang P, Xu Y, Zhang X, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ousterout DG, Perez-Pinera P, Thakore PI, Kabadi AM, Brown MT, Qin X, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Ther. 2013;21:1718–1726. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sürün D, Schwäble J, Tomasovic A, Ehling R, Stein S, Kurrle N, et al. High efficiency gene correction in hematopoietic cells by donor-template-free CRISPR/Cas9 genome editing. Mol Ther Nucleic Acids. 2018;10:1–8. doi: 10.1016/j.omtn.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin JW, Kim K-H, Chao MJ, Atwal RS, Gillis T, MacDonald ME, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25:4566–4576. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dabrowska M, Juzwa W, Krzyzosiak WJ, Olejniczak M. Precise excision of the CAG tract from the huntingtin gene by Cas9 nickases. Front Neurosci. 2018;12:75. doi: 10.3389/fnins.2018.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antoniani C, Meneghini V, Lattanzi A, Felix T, Romano O, Magrin E, et al. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human β-globin locus. Blood. 2018;131:1960–1973. doi: 10.1182/blood-2017-10-811505. [DOI] [PubMed] [Google Scholar]
- 58.Sanz DJ, Hollywood JA, Scallan MF, Harrison PT. Cas9/gRNA targeted excision of cystic fibrosis-causing deep-intronic splicing mutations restores normal splicing of CFTR mRNA. PLoS One. 2017;12:1–13. doi: 10.1371/journal.pone.0184009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park C-Y, Kim DH, Son JS, Sung JJ, Lee J, Bae S, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR–Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Castaño J, Bueno C, Jiménez-Delgado S, Roca-Ho H, Fraga MF, Fernandez AF, et al. Generation and characterization of a human iPSC cell line expressing inducible Cas9 in the “safe harbor” AAVS1 locus. Stem Cell Res. 2017;21:137–140. doi: 10.1016/j.scr.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao S, Tammaro M, Yan H. Enriching CRISPR–Cas9 targeted cells by co-targeting the HPRT gene. Nucleic Acids Res. 2015;43:4–11. doi: 10.1093/nar/gkv675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 63.Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 64.Choudhary R, Baturin D, Fosmire S, Freed B, Porter CC. Knockdown of HPRT for selection of genetically modified human hematopoietic progenitor cells. PLoS One. 2013;8:e59594. doi: 10.1371/journal.pone.0059594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hacke K, Szakmary A, Cuddihy AR, Rozengurt N, Lemp NA, Aubrecht J, et al. Combined preconditioning and in vivo chemoselection with 6-thioguanine alone achieves highly efficient reconstitution of normal hematopoiesis with HPRT-deficient bone marrow. Exp Hematol. 2012;40(3–13):e3. doi: 10.1016/j.exphem.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casola S. Mouse models for miRNA expression: the ROSA26 locus. In: Monticelli S, editor. MicroRNAs and the immune system: methods and protocols. New York: Humana Press; 2010. pp. 145–163. [DOI] [PubMed] [Google Scholar]
- 67.Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 68.Lyu C, Shen J, Wang R, Gu H, Zhang J, Xue F, et al. Targeted genome engineering in human induced pluripotent stem cells from patients with hemophilia B using the CRISPR–Cas9 system. Stem Cell Res Ther. 2018;9:92. doi: 10.1186/s13287-018-0839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14:1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 70.Lee PC, Truong B, Vega-Crespo A, Gilmore WB, Hermann K, Angarita SA, et al. Restoring ureagenesis in hepatocytes by CRISPR/Cas9-mediated genomic addition to arginase-deficient induced pluripotent stem cells. Mol Ther Nucleic Acids. 2016;5:e394. doi: 10.1038/mtna.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stephens CJ, Kashentseva E, Everett W, Kaliberova L, Curiel DT. Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther. 2018;25:139–156. doi: 10.1038/s41434-018-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mizutani T, Li R, Haga H, Kawabata K. Transgene integration into the human AAVS1 locus enhances myosin II-dependent contractile force by reducing expression of myosin binding subunit 85. Biochem Biophys Res Commun. 2015;465:270–274. doi: 10.1016/j.bbrc.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 73.Schiroli G, Ferrari S, Conway A, Jacob A, Capo V, Albano L, et al. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci Transl Med. 2017;9:eaan0820. doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- 74.Ramaswamy S, Tonnu N, Menon T, Lewis BM, Green KT, Wampler D, et al. Autologous and heterologous cell therapy for hemophilia B toward functional restoration of factor IX. Cell Rep. 2018;23:1565–1580. doi: 10.1016/j.celrep.2018.03.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suzuki K, Izpisua Belmonte JC. In vivo genome editing via the HITI method as a tool for gene therapy. J Hum Genet. 2018;63:157–164. doi: 10.1038/s10038-017-0352-4. [DOI] [PubMed] [Google Scholar]
- 78.O’Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, et al. DCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res. 2017;45:9901–9916. doi: 10.1093/nar/gkx578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thakore PI, Kwon JB, Nelson CE, Rouse DC, Gemberling MP, Oliver ML, et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR–Cas9 repressors. Nat Commun. 2018;9:1674. doi: 10.1038/s41467-018-04048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moreno AM, Fu X, Zhu J, Katrekar D, Shih Y-RV, Marlett J, et al. In situ gene therapy via AAV-CRISPR–Cas9 mediated targeted gene regulation. Mol Ther. 2018;26:1–10. doi: 10.1016/j.ymthe.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(665–676):e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, et al. RNA editing with CRISPR–Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gray SJ. Timing of gene therapy interventions: the earlier, the better. Mol Ther. 2016;24:1017–1018. doi: 10.1038/mt.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shin JW, Lee J-M. The prospects of CRISPR-based genome engineering in the treatment of neurodegenerative disorders. Ther Adv Neurol Disord. 2018;11:175628561774183. doi: 10.1177/1756285617741837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 86.CBInsights. The CRISPR economy: 7 private startups pursuing the new frontier in biotech. 2017. https://www.cbinsights.com/research/crispr-startups-to-watch/. Accessed 17 May 2018.
- 87.Fumagalli DC, Gouw AM. Crowdfunding for personalized medicine research. Yale J Biol Med. 2015;88:413–414. [PMC free article] [PubMed] [Google Scholar]
- 88.Vertex Pharmaceuticals. CRISPR Therapeutics and Vertex announce FDA has lifted the clinical hold on the investigational new drug application for CTX001 for the treatment of sickle cell disease. 2018. https://investors.vrtx.com/news-releases/news-release-details/crispr-therapeutics-and-vertex-announce-fda-has-lifted-clinical. Accessed 26 Nov 2018.
- 89.Smith DM, Culme-Seymour EJ, Mason C. Evolving industry partnerships and investments in cell and gene therapies. Cell Stem Cell. 2018;22:623–626. doi: 10.1016/j.stem.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 90.Capps B. Can a good tree bring forth evil fruit? The funding of medical research by industry. Br Med Bull. 2016;118:5–15. doi: 10.1093/bmb/ldw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lundh A, Sismondo S, Lexchin J, Busuioc OA, Bero L. Industry sponsorship and research outcome (review) Cochrane Database Syst Rev. 2012;12:MR000033. doi: 10.1002/14651858.MR000033.pub2. [DOI] [PubMed] [Google Scholar]
- 92.Capps B, Chadwick R, Joly Y, Mulvihill JJ, Lysaght T, Zwart H. Falling giants and the rise of gene editing: ethics, private interests and the public good. Hum Genom. 2017;11:20. doi: 10.1186/s40246-017-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ledford H. How the immune system could stymie some CRISPR gene therapies. Nature. News explainer. 2018. http://www.nature.com/articles/d41586-018-00335-8. Accessed 13 Feb 2019.
- 94.Molteni M. Clashes over the future of gene therapy and Crispr at the US’s biggest biotech meeting. WIRED. 2018. https://www.wired.com/story/clashes-over-the-future-of-gene-therapy-at-the-uss-biggest-biotech-meeting/. Accessed 12 May 2018.
- 95.Cribbs AP, Perera SMW. Science and bioethics of CRISPR-CAS9 gene editing: an analysis towards separating facts and fiction. Yale J Biol Med. 2017;90:625–634. [PMC free article] [PubMed] [Google Scholar]
- 96.Cyranoski D, Ledford H. Genome-edited baby claim provokes international outcry. Nature. 2018;563:607–608. doi: 10.1038/d41586-018-07545-0. [DOI] [PubMed] [Google Scholar]
- 97.Financial Times. GSK sells rare disease gene therapy portfolio. 2018. https://www.ft.com/content/b8574ce8-3e24-11e8-b7e0-52972418fec4. Accessed 18 May 2018.
- 98.Adair JE, Waters T, Haworth KG, Kubek SP, Trobridge GD, Hocum JD, et al. Semi-automated closed system manufacturing of lentivirus gene-modified haematopoietic stem cells for gene therapy. Nat Commun. 2016;7:13173. doi: 10.1038/ncomms13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Engel M. Gene therapy goes global: portable device could make future cancer, HIV cures affordable. Fred Hutch. 2016. https://www.fredhutch.org/en/news/center-news/2016/10/gene-therapy-in-a-box-goes-global.html. Accessed 18 May 2018.
- 100.Witt R, MacKenzie TC, Peranteau WH. Fetal stem cell and gene therapy. Semin Fetal Neonatal Med. 2017;22:410–414. doi: 10.1016/j.siny.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 101.Shangaris P, Loukogeorgakis SP, Blundell MP, Petra E, Shaw SW, Ramachandra DL, et al. Long-term hematopoietic engraftment of congenic amniotic fluid stem cells after in utero intraperitoneal transplantation to immune competent mice. Stem Cells Dev. 2018;27:515–523. doi: 10.1089/scd.2017.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahmed SG, Waddington SN, Boza-Morán MG, Yáñez-Muñoz RJ. High-efficiency transduction of spinal cord motor neurons by intrauterine delivery of integration-deficient lentiviral vectors. J Control Release. 2018;273:99–107. doi: 10.1016/j.jconrel.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mattar CNZ, Gil-Farina I, Rosales C, Johana N, Tan YYW, McIntosh J, et al. In utero transfer of adeno-associated viral vectors produces long-term factor IX levels in a cynomolgus macaque model. Mol Ther. 2017;25:1843–1853. doi: 10.1016/j.ymthe.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Massaro G, Mattar CNZ, Wong AMS, Sirka E, Buckley SMK, Herbert BR, et al. Fetal gene therapy for neurodegenerative disease of infants. Nat Med. 2018;24:1317–1323. doi: 10.1038/s41591-018-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin S-L, Ying S-Y. Mechanism and method for generating tumor-free iPS cells using intronic MicroRNA miR-302 induction. Methods Mol Biol. 2018;1733:265–282. doi: 10.1007/978-1-4939-7601-0_22. [DOI] [PubMed] [Google Scholar]
- 106.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song B, Fan Y, He W, Zhu D, Niu X, Wang D, et al. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015;24:1053–1065. doi: 10.1089/scd.2014.0347. [DOI] [PubMed] [Google Scholar]
- 110.Yang Y, Zhang X, Yi L, Hou Z, Chen J, Kou X, et al. Naïve induced pluripotent stem cells generated from β-thalassemia fibroblasts allow efficient gene correction with CRISPR/Cas9. Stem Cells Transl Med. 2016;5:8–19. doi: 10.5966/sctm.2015-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ou Z, Niu X, He W, Chen Y, Song B, Xian Y, et al. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Sci Rep. 2016;6:32463. doi: 10.1038/srep32463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu P, Tong Y, Liu X, Wang T, Cheng L, Wang B, et al. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2-654 (C>T) mutation in beta-thalassemia-derived iPSCs. Sci Rep. 2015;5:12065. doi: 10.1038/srep12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Osborn MJ, Gabriel R, Webber BR, DeFeo AP, McElroy AN, Jarjour J, et al. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum Gene Ther. 2015;26:114–126. doi: 10.1089/hum.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Skvarova Kramarzova K, Osborn MJ, Webber BR, DeFeo AP, McElroy AN, Kim CJ, et al. CRISPR/Cas9-mediated correction of the FANCD1 gene in primary patient cells. Int J Mol Sci. 2017;18:1269. doi: 10.3390/ijms18061269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van Agtmaal EL, André LM, Willemse M, Cumming SA, van Kessel IDG, van den Broek WJAA, et al. CRISPR/Cas9-induced (CTG·CAG)n repeat instability in the myotonic dystrophy type 1 locus: implications for therapeutic genome editing. Mol Ther. 2017;25:24–43. doi: 10.1016/j.ymthe.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang X, Wang Y, Yan W, Smith C, Ye Z, Wang J, et al. Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33:1470–1479. doi: 10.1002/stem.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wen J, Tao W, Hao S, Zu Y. Cellular function reinstitution of offspring red blood cells cloned from the sickle cell disease patient blood post CRISPR genome editing. J Hematol Oncol. 2017;10:119. doi: 10.1186/s13045-017-0489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang N, Zhi H, Curtis BR, Rao S, Jobaliya C, Poncz M, et al. CRISPR/Cas9-mediated conversion of human platelet alloantigen allotypes. Blood. 2016;127:675–680. doi: 10.1182/blood-2015-10-675751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim K, Park SW, Kim JH, Lee SH, Kim D, Koo T, et al. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27:419–426. doi: 10.1101/gr.219089.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, et al. Correction of a genetic disease in mouse via use of CRISPR–Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 122.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med. 2016;8:477–488. doi: 10.15252/emmm.201506039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yin H, Song C-Q, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pankowicz FP, Barzi M, Legras X, Hubert L, Mi T, Tomolonis JA, et al. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun. 2016;7:1–6. doi: 10.1038/ncomms12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta R, Zhang M, et al. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep. 2017;7:44624. doi: 10.1038/srep44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Long C, Amoasii L, Mireault AA, Mcanally JR, Li H, Sanchez-ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2015;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR–Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang X, Raghavan A, Chen T, Qiao L, Zhang Y, Ding Q, et al. CRISPR–Cas9 targeting of PCSK9 in human hepatocytes in vivo—brief report. Arterioscler Thromb Vasc Biol. 2016;36:783–786. doi: 10.1161/ATVBAHA.116.307227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xie C, Zhang YP, Song L, Luo J, Qi W, Hu J, et al. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 2016;26:1099–1111. doi: 10.1038/cr.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, et al. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Murlidharan G, Sakamoto K, Rao L, Corriher T, Wang D, Gao G, et al. CNS-restricted transduction and CRISPR/Cas9-mediated gene deletion with an engineered AAV vector. Mol Ther Nucleic Acids. 2016;5:e338. doi: 10.1038/mtna.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.CRISPRTX. CRISPR Therapeutics—our programs. http://www.crisprtx.com/programs/pipeline. Accessed 7 Mar 2019.
- 139.Editas. Editas Medicine—areas of research. https://www.editasmedicine.com/pipeline/. Accessed 7 Mar 2019.
- 140.Intellia. Intellia Therapeutics—R&D pipeline. https://www.intelliatx.com/pipeline/. Accessed 17 May 2018.
- 141.Vertex Pharmaceuticals Incorporated, CRISPR Therapeutics. A safety and efficacy study evaluating CTX001 in subjects with transfusion-dependent β-thalassemia [ClinicalTrials.gov identifier NCT03655678]. National Institutes of Health, ClinicalTrials.gov. https://clinicaltrials.gov. Accessed 12 Feb 2019.
- 142.Vertex Pharmaceuticals Incorporated, CRISPR Therapeutics. A safety and efficacy study evaluating CTX001 in subjects with severe sickle cell disease [ClinicalTrials.gov identifier NCT03745287]. National Institutes of Health, ClinicalTrials.gov. https://clinicaltrials.gov. Accessed 12 Feb 2019.
- 143.Allife Medical Science and Technology Co., Ltd. iHSCs with the gene correction of HBB intervent subjests with β-thalassemia mutations [ClinicalTrials.gov identifier NCT03728322]. National Institutes of Health, ClinicalTrials.gov. https://clinicaltrials.gov. Accessed 12 Feb 2019.
- 144.National Human Genome Research Institute (NHGRI). Examining the knowledge, attitudes, and beliefs of sickle cell disease patients, parents of patients with sickle cell disease, and providers towards the integration of CRISPR in clinical care [ClinicalTrials.gov identifier NCT03167450]. National Institutes of Health, ClinicalTrials.gov. https://clinicaltrials.gov. Accessed 12 Feb 2019.