Abstract
Hantaviruses cause hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS), which infects over 200,000 people worldwide. Sin Nombre virus (SNV) and Andes virus (ANDV) cause the most severe form of HCPS with case fatality ratios of 30–40%. There are no specific therapies or vaccines for SNV. Using high throughput flow cytometry, we screened the Prestwick Compound Library for small molecule inhibitors of the binding interaction between UV-inactivated and fluorescently labeled Sin Nombre virus (SNVR18) particles, and decay accelerating factor (DAF) expressed on Tanoue B cells. Eight confirmed hit compounds from the primary screen were investigated further in secondary screens that included infection inhibition, cytotoxicity, and probe interference. Antimycin emerged as a bona fide hit compound that inhibited cellular infection of the major HCPS (SNV) and HCPS (Hantaan) causing viruses. Confirming our assay’s ability to detect active compounds, orthogonal testing of the hit compound showed that antimycin binds directly to the virus particle and blocks recapitulation of physiologic integrin activation caused by SNV binding to the integrin PSI domain.
Keywords: hantavirus, high throughput screen, drug discovery, integrin activation, PAC-1, PSI domain, cyclic AMP, Rap1, mechanotransduction, G protein, flow cytometry
INTRODUCTION
Hantaviruses are an important group of emerging pathogenic viruses and represent a paradigm for zoonotic airborne transmission. Sin Nombre virus (SNV) is a Category A pathogen that causes the most severe form of HCPS with case fatality ratios of 30–50%.1 Tissue tropism involves the vascular endothelium of the heart, kidney, lung, and lymphoid organs where dysregulation of the endothelial barrier function, edema, and focal hyaline membranes are central features.2–4 There is no approved, effective therapy and treatment of severe disease is supportive, including the use of extracorporeal membrane oxygenation (ECMO).5 The ability of a virus to break the species barrier critically depends on its capacity to enter human cells efficiently. Inhibiting viral entry allows blocking the pathogen before it can take control of the host cell and represents a promising strategy for therapeutic anti-viral intervention.
The hantavirus genome comprises three segments of negative sense ssRNA, which are coated with multiple copies of the nucleoprotein. A lipid bilayer membrane envelope in which two glycoproteins, Gn and Gc are localized covers the virus.4 These envelope glycoproteins are necessary for virus attachment to entry receptors and fusing with endocytic vesicles once inside the host cell. Pathogenic hantaviruses, including Andes (ANDV), SNV, Hantaan (HTNV), Puumala (PUUV), Seoul Virus, and New York-1 (NY-1) have been reported to use αvβ3 and αIlbβ3 integrins for host cell entry that leads to productive infection. The hantavirus engages the PSI domain of the β3 integrin subunit of low-affinity conformers of αvβ3 and αIIbβ3 integrins, to enter host cells.6 The effect of hantavirus engagement of the PSI domain on integrin activity is not well understood. One long-held view is that hantaviruses bind to inactive integrins and maintain them in an inactive state.7 However, recent single-molecule atomic force microscopy experiments from our lab have shown that SNV engagement of the integrin PSI domain induces higher affinity for its cognate RGD ligands. We further demonstrated that bent conformation low-affinity β3 integrins interact in cis with the G protein-coupled receptor P2Y2R via an RGD sequence in the first extracellular loop of P2Y2R.8 As a result hantavirus engagement of the integrin-P2Y2R complex stimulates a force-dependent integrin outside-in signaling cascade. Outside-in signaling requires a surface-anchored ligand, which can support traction force upon its exertion.8 The cis binding to P2Y2R provides the requisite fixed scaffold for outside-in signaling. In this way the physiologically activated (inside out) and extended conformation integrin αIlbβ3 is recognized by PAC-1.9 PAC-1 is an IgM antibody that selectively binds to the fibrinogen-binding site of activated αIIbβ3.10 Others have shown that pathogenic hantaviruses also recognize the glycosylphosphatidyl-inositol - linked protein, CD55/decay-accelerating factor (DAF), which they use as an effector for integrin activation during cell entry.11,12 DAF is 70-kDa member of the complement system, which is often exploited by various pathogens to gain entry into host cells.13, 14 Although the mechanism that links the interaction of hantaviruses with DAF and integrin-mediated cell entry is currently not well understood; it is known that crosslinking of DAF by proteins or viruses stimulates cell signaling cascades upstream of integrin activating pathways.13, 15
In this study, we tested whether the combination of targeting DAF binding inhibitors in primary screens and the PSI domain in subsequent tests could be used to identify hits and and their molecular mechanism of action. As an effector for cellular infection by multiple pathogens, DAF is an attractive target for discovery campaigns of small molecules that inhibit binding between DAF and its pathogenic ligands. DAF is expressed in a wide range of cells including Tanoue B suspension cells, which are amenable to homogeneous flow cytometry assays.12, 16 Homogeneous flow cytometry, uses small sample volumes and allows resolution of free and bound (virus), and obviates a wash step. High signal to background of individual cells in suspension is possible as compared to adherent cells in the virus or antibody-based assays. Finally, flow cytometry is readily amenable to multiplexing.
Accordingly, we developed a flow cytometry-based high throughput screen (HTS) assay for small molecule inhibitors of UV-inactivated and fluorescently labeled SNV (SNVR18), and DAF expressed in β3 integrin-null Tanoue B cells.16 We conducted a primary HTS campaign of the Prestwick chemical library. We tested candidate compounds in orthogonal screens for probe interference, as well as inhibition of infection and cytotoxicity. To validate a potential mechanism of action of our hit compound, antimycin, we assayed the compound for its ability to interfere with SNV-induced integrin signaling cascade. We first applied cAMP Difference Detector in situ (cADDis) BacMam fluorescent biosensor kit17 to test the effect of antimycin on SNV induced cyclic AMP production in CHO-A24 BacMam transformed cells. Second, we tested the effect of antimycin on PAC-1 staining of SNV activated cells.9
MATERIALS AND METHODS
Materials. 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(carboxyfluorescein) (fluorescein PE) and dioleoyl-L-a-phosphatidylcholine (DOPC), cholesterol, and egg sphingomyelin were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). 5 μm diameter glass beads were obtained in dry form from Duke Scientific Corp (Palo Alto, CA). Octadecyl Rhodamine B chloride (R18), and 5-octadecanoylaminofluorescein (F18) were purchased from Molecular Probes (Eugene, OR) and used without further purification. We have previously characterized the spectroscopy of these probes.18 Phosphate-buffered saline (PBS) was purchased from Mediatech, Inc, Herndon, VA). Dimethyl sulfoxide (DMSO) and Sephadex G-50 were purchased from Sigma. TRIS (10 mM or 25 mM Tris, 150 mM NaCl, pH 7.5) and HHB (30 mM HEPES, 110 mM NaCl, 10 mM KCl, 1 mM MgCl26H2O and 10 mM glucose, pH 7.4) buffer, and Hanks Balanced Saline Solution (HBSS) (4mM NaH2CO3, 0.4mM MgSO4, 1 mM CaCl2 or 1 mM MgCl2) were prepared under sterile conditions and stored in 50 ml tubes at 20°C.
Cell Culture
Tanoue B cells and Vero E6 cells were maintained in either sterile filtered RPMI media, or minimum essential media (MEM). All media contained 10% heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin, 100 μg/mL streptomycin, 10 mM HEPES, pH 7.4, 20 μg/mL ciprofloxacin, 2 mM L-glutamine, at 37°C in a CO2 water-jacketed incubator of 5% CO2 and 95% air (Forma Scientific, Marietta, OH). CHO A24 cells were grown in DMEM/F12 HAM mixture (Gibco, Life Technologies) with 2 mM L-Glutamine and 1 mg/mL G418 (Sigma). CHO C3 cells were grown in F-12K media (Cellgro, Mediatech), 100 units/mL Penicillin/Streptomycin, 2 mM L-glutamine. All cell lines are tested for the presence of mycoplasma at every cell passage using commercial kits such as MycoAlert™ mycoplasma detection kit (ww.lonza.com).
Production of SNV
Stocks of SNV particles were propagated in Vero E6 cells as previously described.19 The typical yield is ≈1010 particles/ml in 15ml aliquots. The particle to infectious unit ratio is 104 to 1.16 The binding assays on which the HTS screen is based does not distinguish between infectious and non-infectious particles. The 15 ml stocks are concentrated to 1.5mL. The virions are then rendered non-infectious by a 15 sec dose of UV light at 254-nm as described.19 The neutralization of irradiated supernatants is confirmed by a focus reduction assay before transfer, with documentation from the BSL-3 laboratory as required by UNM Biosafety. The structural integrity of UV-irradiated SNV is checked by immunoblotting of key protein components (viz: nuclear N proteins, and glycoprotein components Gc and Gn) measured against nonirradiated samples.19 We also use transmission electron microscopy (TEM) to show that the UV treatment leaves virions physically intact with an average diameter of 193 nm.16
Fluorescent labeling and quantifying of SNVR18
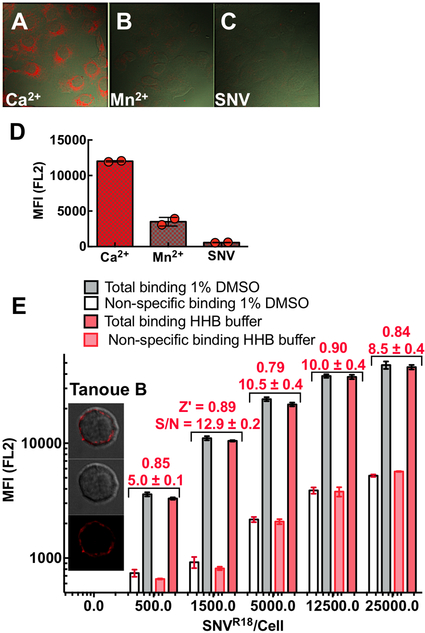
Hantaviruses consist of a nucleocapsid surrounded by a lipid bilayer membrane-derived from membrane lipids of the host cells. As such, SNV is amenable to staining by fluorescent derivatives of lipophilic lipid probes such as octadecyl rhodamine (R18).16 We use a combination of fluorescence and absorption measurements to quantify purified and fluorescently labeled particles, SNVR18. On average, our preparations of labeled virions are usually 500 μl stocks containing 108SNVR18/μl, each bearing ≈ 1×104 R18 probes/SNV. We typically establish a signal dynamic range and variation (Z’ score) for each batch of labeled SNVR18 by titrating SNVR18 particles to 104 cells in 10μl volumes in a plate assay (Fig. 1).
Figure 1.
Confocal microscopy images show differential binding of SNVR18 to Vero E6 cells in HHB/0.1% HSA buffer in the presence of A. 1.5 mM Ca2+ which supports an integrin bent conformation low-affinity state, B. 1 mM Mn2+ activates integrins and induces an extended integrin conformation. C. Excess unlabeled SNV used to block SNVR18. D. Flow cytometry measurements of SNVR18 binding to Vero E6 cells in the presence Ca2+, Mn2+, and excess unlabeled SNV. 100,000 Vero E6 /well, were incubated with 1500 SNVR18/cell at 37°C under mild vortexing for 15 mins. The cells were then washed 3x in the buffer, then either removed from the bottom of the wells by 0.025% Trypsin/0.0526 mM EDTA for flow cytometric measurement or fixed in 2% PFA at 0°C for 30 mins before mounting in Vector Shield containing 1X DAPI for microscopic imaging. E. Equilibrium binding of SNVR18 titers to 10,000 Tanoue B cells in a 384 well plate. Quadruplicate measurements were performed for each condition to determine the signal to background ratio and Z’ values for each titer. The Z’-score values for each condition were comparable, here Z’ scores for +DMSO are shown. Error bars represent ±SD.
Virus Probe Validation
The median fluorescence for each well was plotted vs. plate position. Before engaging in the HT screen of the Prestwick Compound library (PCL), we tested the suitability of our virus preparations in a pilot assay measuring reproducibility as previously described.16 We tested the virus for specific binding to Vero E6 cells and Tanoue B cells using confocal microscopy imaging and flow cytometry. For Vero E6 cells we used Mn2+ to block specific binding of SNV to low-affinity integrins.16 Using a plate-based assay, we assessed signal to background levels using increasing concentrations of fluorescently labeled SNV (SNVR18) to a fixed number of cells, while using excess unlabeled SNV as a blocking agent. We assessed the dynamic signal range and data variation associated with these measurements concerning Z’ factors:20
Positive controls are neat SNVR18, and negative controls are samples pre-blocked with 10X unlabeled SNV. An assay with Z’ scores ranging from 0.5 to 1.0 is considered as excellent, whereas a Z’ score of 0 reflects a “yes/no” assay.
HTS assay for specific SNVR18/ DAF interactions in Tanoue B cells
We performed HTS of the 1200 compound PCL distributed in four, 384-well plates. A total volume of 5 μl Tanoue B cells at a concentration of 2×106 cells/mL in HHB buffer was distributed to each well of a 384-well polystyrene plate except columns 23 and 24.
10 μl HHB buffer was added to columns 23 and 24 for washes and to aid in well-identification for analysis. In column 2 unlabeled SNV (10x conc. excess) was added as a positive control for inhibition of SNVR18 binding (2.5 μL/well). The library compounds (10 μM final concentration) were transferred to columns 3–22 using a 100 nL pin tool. The four plates were incubated at room temperature for 30 min. The SNVR18 was then added to columns 1 and 3–22 at 3×107 SNVR18/well in 5 μL (1.5×1010 SNVR18/mL), which is a 3000-fold excess of virus compared to cells. This virus to cell ratio was optimal for S/N (Fig. 1E). The plate was returned to the vortex mixer for an additional 25 min at 1000 rpm. After 25 min the plate was sampled, and samples delivered into an Accuri flow cytometer with HyperCyt automation and analyzed using HyperView software for automated well identification and cell analysis. The median fluorescence value from wells in columns 1 and 2 was used to calculate a Z’ factor.
Confirmatory dose response
The selected hit compounds were screened in a 10-point dose response assay (0.1μM- 20 μM) in the same way as the library screen. Each well, including the controls, had a constant 1% DMSO concentration, which maintained continuous light scatter characteristics of cells. The EC50 values were all in the 10 μM range.
Cell viability assays
We tested the hit compounds for toxicity using the Cell Titer Glo® kit from Promega. This package assesses the level of ATP found in viable cells. Cells were subjected to a titrated concentration range of 0.1 μM – 20 uM of hit compounds over 24 hrs. Toxicity ≥ 50% toxicity was used to exclude the hit from further advancement. Toxicity was used as a factor in triaging compounds in the context of its relationship to the compound’s IC50.
Infection Assays
We assessed the ability of the compounds to prevent entry of live hantaviruses (SNV and HTNV) into Vero E6 cells in a BSL3 laboratory setting. This assay uses a qRT-PCR readout based upon TaqMan technology, which we have used extensively.19, 21 It has previously been optimized for SNV, ANDV, and HTNV using a multiplicity of infection (MOI) of 0.01 and a readout 48 hr after infection. Primers and probes for viral S segment have already been developed and evaluated for each virus.19, 21 We plated Vero E6 cells (5×104/well) in a 48-well plate, and the following day, we infected separate wells in triplicate with each of the two viruses in the presence or absence of 10 μM concentrations of the HTS screen hit compound. After a 48 hr incubation, we prepared supernatant RNA using viral RNA preparation kits (Qiagen) and subjected the RNA to TaqMan analysis as previously described.19, 21
Alternatively, infectivity is analyzed by standard Western blot analysis of N-protein expression.9 Briefly, for Western blot SNV N protein was detected with αSNV/N, (hyperimmune rabbit anti-SN virus N protein) used at a 1:1000 dilution with a secondary antibody (Peroxidase AffiniPure Goat Anti-Rabbit IgG; used at 1:1000 dilution) from Jackson Immuno Research Laboratories (cat# 111-035-003, lot# 104668). We treated the nitrocellulose membrane with an HRP substrate from Pierce, SuperSignal ® West Pico (Product # 0034077) before imaging. Quantitative analysis of the gel was performed using a BioRad Molecular Imager, ChemiDoc XRS+ equipped with Image Lab Software 4.1. We verified equal loading by detecting of calnexin with anti-calnexin mABs (clone H-70 used at 1:500 dilution; from Santa Cruz) or β-actin on the same membrane with the anti-β-actin (clone AC-74 used at 1:2000 from Sigma).
Pertussis toxin
150,000 CHO A24 cells were plated onto a 48 well plate for 24 hours and then treated with 100ng/ml pertussis toxin (PTX) and incubated for ~18 hrs at 37°C and then used in infection or GTPase activity assays.
GTPase effector trap flow cytometry assay (G-trap)
This assay has been previously described.9, 22 PTX-treated and mock-treated CHO A24 were serum-starved overnight, in a 48 well plate with 50,000 cells in each well. After incubating the cells with SNVR18 for 5 min cells in 100μl media at 37°C, we lysed the cells with 100μL ice-cold RIPA buffer. Lysates were kept cold at all times to limit hydrolysis of active GTPases. For each sample, 50 μl was used to analyze for GTP loading of Rap1. 10,000 beads functionalized with the Rap1 effector protein RAL GDS were used to capture Rap1GTP from the cell lysate as previously described.9, 22
cyclic AMP assay
We used an Upward (U0200G) cAMP Difference Detector in situ (cADDis) BacMam fluorescent biosensor kit purchased from Montana Molecular (www.montanamolecular.com)17 to assay cyclic AMP production in CHO-A24 cells after exposure to SNV. The sensor application involves a two-step process supplied by the manufacturer. It consists of inoculating cells plated in a 96 well plate with a baculovirus transduction system in appropriate media. The assay can be tested after two days. We optimized the assay for our system by titrating the sensor expression and measuring the response of the positive control (e.g., forskolin) relative to our SNV assay. It is essential to perform the optimization process regarding the corresponding reaction between the positive control and the test of interest (e.g., SNV). This is because excess sensor expression, which is optimal for the positive control might be undesirable for the assay target as it limits response compared to low-expressing effectors as in the case of SNV. We plated BacMam-transduced cells in a 48 well plate (48,000 cells/well) and tested the cells for SNV- induced cAMP production using a BioTek Synergy 2-plate reader. Cell activity inhibitors were added to target wells and allowed to incubate for 30 min. We recorded baseline fluorescence readings of normal and transformed cells for 5 min before adding SNV and forskolin. We performed triplicate readings for each condition. We measured the cellular response as Δfluorescence pre- and post-activation.
PAC-1 binding assay
We incubated CHO-A24 cells (1,000 cells/μl) in HHB buffer containing 1:20 dilution PAC-1, a monoclonal IgM antibody that selectively binds to physiologically activated integrin αIIbβ3.23 To test candidate compounds for inhibition of integrin activation, we added 10 μM aliquots of each compound to 20 μl volumes of the CHO-24 cell suspensions for 30 min. Negative control samples were treated with 1% DMSO. We then added SNV aliquots (5,000 SNVR18/cell) to the cell samples maintained at 37°C for 5 min and then quenched cell activation with ice-cold buffer. The samples were incubated for 30 min to enable PAC-1 binding to active-conformation integrins to reach equilibrium. The samples were centrifuged once and resuspended in 50 μl HHB buffer and then read on an Accuri C6 flow cytometer.
RESULTS
Assay validation
To identify compounds that inhibit binding of SNV18 in an HTS campaign, we first established the specificity16 and affinity of binding of SNVR18 to DAF- functionalized beads and Tanoue B cells.12 Figure 1 shows SNVR18 binding to Vero E6 cells expressing αvβ3 integrin and DAF. SNV binds to the PSI domain of low-affinity integrins.6 Thus, treatment of cells with Mn2+cations, which induce an extended conformation blocks SNV binding.6, 16 Here we show that treatment of cells with 1.5 mM Mn2+ inhibits SNVR18 binding to Vero E6 cells by ≥ 70% (Fig. 1B). We also show that unlabeled SNV efficiently inhibits SNVR18 from binding to Vero E6 cells (Fig. 1D). Because the library compounds are solubilized in DMSO, which is cytotoxic above 1% concentration by volume, we tested whether 1% DMSO had adverse effects on our assay in a simulated HTS setting. In a 384 well plate, titration of SNVR18 particles to DAF-expressing Tanoue B cells yielded comparable Z’ scores of 0.8 – 0.9 across a 50 fold range of virus particle titers in standard cell media and 1%DMSO buffer. Although non-specific binding appeared to increase with increasing titers of SNVR18, the signal to background levels (S/N in Fig 1E) increased from 5 at the lowest SNVR18 titer to ~13 before decreasing to ~ 9 fold at the highest titer of SNVR18.
Primary HTS
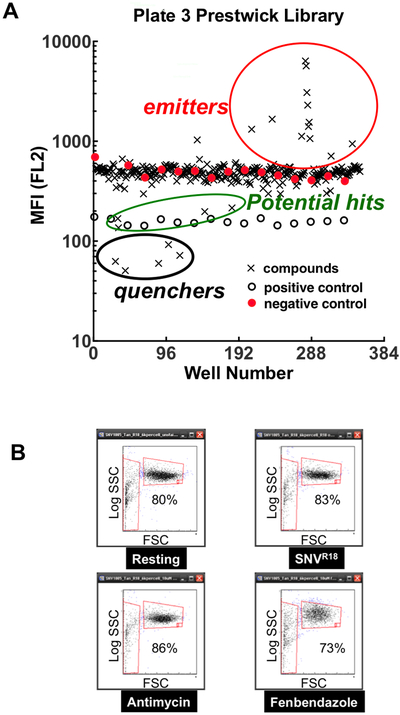
The median fluorescence for each well was plotted vs. plate position (Fig. 2A). The average Z’ score for the plates was ~0.5, indicating that the assay had appropriate statistical reliability for HTS. As shown, the library compounds comprising fluorescent, light emitting compounds, and compounds that quenched SNVR18 fluorescence emission below the positive control were not considered. We calculated the percent inhibition based on the following equation: %Inhibition=100[1-((MFItest - MFIblocked)/(MFIunblocked-MFIblocked))] where, MFIi is the median fluorescence intensity of cells in wells containing test compounds, blocked control wells and unblocked controls, respectively. From these screens, 24 compounds were hits; inhibiting the binding response of the virus to the Tanoue B cells at >70%. We also analyzed forward and side scattergrams (FSC and SSC in Fig. 2B) for clear indications of cytotoxicity. Dead or dying cells are expected to become smaller (reduced FSC) or more granular (increased SSC). In general, the scattergrams for the vehicle (DMSO only) treated cells, and noncytotoxic compounds displayed ≥ 80%events in the gate associated with healthy cells. We observed an increase in log SSC and a decrease in the percentage of the population of viable cells, resulting from treatment with cytotoxic compounds (e.g., 73% viable cells treated with Fenbendazole in Fig 2B).
Figure 2.
High throughput screen of the Prestwick Compound Library (PCL). A. A plot of the median fluorescence intensity (MFI) emitted in the FL2 fluorescence channel (570 nm, excitation at 488 nm) from one run of plate 3 of the PCL. The compound-containing test wells are indicated as crosses. Negative (closed circles) and positive (open circles) are as shown. B. Qualitative determination of short-term cytotoxic effects of drug targets on cell viability measured by flow cytometric scattergrams. Plots of side (SSC) versus forward scatter (FSC) of cells after 30 min exposure to potential hit compounds. Left vertical gate shows dead cells, and horizontal gate shows viable cells. Fenbendazole treated cells show a decrease in the percentage of viable cells. Fenbendazole is also shown to cause an increase in SSC, typically an indication of increased cell granularity.
We performed dose-response measurements by testing the effects of compounds at 10 concentrations ranging in a half-log dilution series from nM to μM range. Eight compounds were confirmed (gossypol, trifluoperazine, niclosamide, nifedipine, tocopherol, fenbendazole, antimycin, and tolfenamic acid) by showing reduced SNVR18 fluorescence in a dose-dependent manner. We fitted the data to a dose-response inhibition curve with a variable slope in GraphPad Prism Software Version 6.0h. IC50 values ranged from 0.5 μM to 40 μM (Supplemental Figure S1.
Confirming the activity of hit compounds
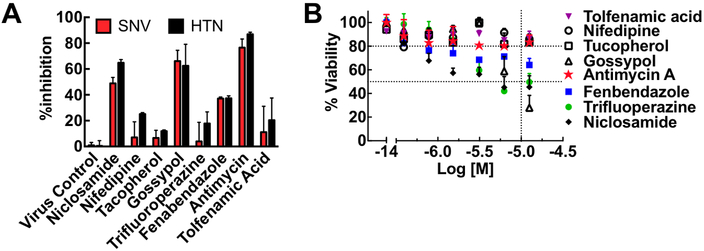
Testing for infection inhibition and cytopathic effects. We next tested the candidate compounds for infection inhibition and cytotoxicity. Vero E6 cells were incubated with the eight candidate compounds for 30 min before infection with 0.1 moi Sin Nombre and Hantaan virus inocula. Results indicated that three compounds inhibited the infection of Vero E6 cells by SNV and HTN: niclosamide (~60% SNV; ~70%HTN inhibition), gossypol ~(70% SNV and HTN), and antimycin (≥80% SNV and HTN) (Fig. 3A). However, the five other compounds failed to inhibit infection significantly. Four of the compounds inhibited infection by ≤20%, whereas, Fenbendazole inhibited infection by nearly 40%, which is below our set threshold for acceptability in our primary screen. We then subjected Vero E6 cells to doses of compounds spanning a concentration range of 0.1 μM – 20 uM over 24 hr in a 96 well plate, to test cells for viability after long-term exposure to hit compounds. We determined toxicity using the Cell Titer Glo® kit from Promega. Fig. 3B shows the concentration-dependent effects of the eight compounds on cell viability. Niclosamide (~50% viability), and gossypol (~30% viability) were notably cytotoxic. Fenbendazole (~70%) was moderately cytotoxic with increasing concentration of the drugs. Antimycin, nifedipine, and tocopherol had minimal cytopathic effects after 24 hours at the limiting concentration of the HTS assay.
Figure 3.
Cytotoxic hits yield false positive results on Hantavirus infectivity. A. A plot of % inhibition of infection by Sin Nombre (SNV) and Hantaan virus (HTN) measured by a TaqMan assay as previously described.19, 21 Target compounds were used at 10 μM. B. A plot of cell viability versus concentration of hit compounds after 24 hr exposure to compounds. Cell viability was determined using a Promega™ CellTiter 96™ Aqueous One Solution Cell Proliferation Assay following the manufacturer’s protocol (see text for details). Error bars represent ±SD for duplicate measurements
Distinction of fluorescence quenching from competitive virus displacement
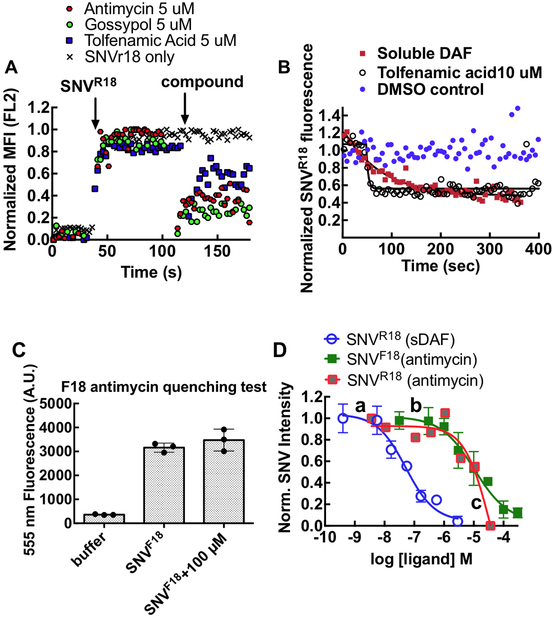
Having identified antimycin as an active inhibitor of infection with minimal direct cytotoxicity, we performed additional competitive binding experiments to investigate its mechanism of action. We tested the compounds for probe quenching and determined that the compound characteristics were consistent with molecules that formed non-fluorescent complexes or aggregates upon interacting with R18. The quenching mechanism is commonly described as static quenching24 (Supplemental Material Fig S2). Thus, a decrease of cell-associated fluorescence in our screening assay could feasibly have resulted from quenching of SNV-bound R18 rather than (or in addition to) competitive displacement of the virus by the test compound, a false positive result. Quenching can be distinguished from competitive displacement by the kinetics with which fluorescence is diminished. We therefore used soluble DAF (sDAF) to compete off the SNVR18 from DAF-functionalized beads as a positive control for the kinetics of competition binding. Over a 3 min time course of analysis on a BD FACsan flow cytometer, Antimycin and two other of the hit compounds (Gossypol, Tolfenamic acid) promoted a precipitous drop (koff ~ 0.25 s−1) in bead fluorescence (Fig. 4A, 4B). By contrast, beads treated with 10μM soluble DAF displayed a single exponential decay curve of SNVR18 (koff ~ 0.01 s−1, Fig. 4B). The significant discrepancies in fluorescence loss kinetics suggested that static quenching confounded the competition-binding assays.
Figure 4.
Investigation of fluorescence reporter interference by candidate hit compounds. A. Real-time quenching of SNVR18 fluorescence by candidate compounds as they associate rapidly with envelope membranes of the R18-tagged virus. 50,000 Tanoue B cells suspended in 200 μl HHB buffer were analyzed on a flow cytometer for a 30-second baseline. SNVR18 aliquots were added to the tube. Samples were read ~60 sec, then 1μl aliquots of hit compounds were added. Readings were resumed for 90 sec. Data were converted to ASCII using IDLeQuery software B. Comparing loss of SNVR18 fluorescence from DAF-Fc functionalized beads treated with 10 μM soluble DAF (positive control for competitive dissociation;(koff = 0.01s-1) and tolfenamic acid (likely quenching kq = 0.25s−1). C. SNV labeled with 5-octadecanoylaminofluorescein (F18) is refractory to fluorescence quenching by antimycin. Three cuvettes: 1) buffer only, 2) 10,000 SNVF18 particles suspended in 200 μl HHB buffer, 3) SNVF18 and 100 μM antimycin were analyzed of a spectrofluorometer. D. Quality control check on competitive inhibition curves for SNV binding to DAF on Tanoue B cells and DAF functionalized beads. Equilibrium binding of SNVR18/F18 to 10,000 DAF functionalized beads or 10,000 Tanoue B cells in 10 μL sample tubes. Samples volumes were increased to 50 μl and then analyzed on a flow cytometer. Data were fit to a competition binding curve (Y=Bottom + (Top-Bottom)/(1+10^((LogIC50-X)*HillSlope))). a) Competition binding curve between SNVR18 on DAF functionalized beads and various concentrations of soluble DAF (sDAF), representing a prototypical competition binding isotherm (IC50 = 50 nM). b) Binding curve for antimycin against SNVF18 bound to Tanoue B cells (IC50 =10.0μM). The SNVF18 binding curve shows similar characteristics to the sDAF curve. c) Binding curve for antimycin against SNVR18 bound to Tanoue B cells. The curve deviates from a standard competitive equilibria binding curve (see text). Binding measurements were made by flow cytometry with fluorescence excitation at 488 nm. R18 fluorescence emission intensity was detected in the FL2 channel (570 nm), and F18 fluorescence emission intensity in the FL1 channel (533 nm) Error bars represent triplicate ±SD for triplicate measurements..
Electrostatic attraction and aromatic stacking interactions are often associated with static quenching.24 R18 is a positively charged fluorophore; we, therefore, tested whether SNV particles labeled with the negatively-charged 5-octadecanoylaminofluorescein (F18) were refractory to quenching by antimycin. We first established that antimycin does not quench SNVF18 fluorescence by mixing antimycin with SNVF18 (Fig. 4C). We then compared the dose-response curves for antimycin in binding competition with SNVR18 and SNVF18 to Tanoue B cells using a flow cytometer (Fig. 4D). We used soluble DAF titrations to compete with SNVR18 in a binding assay to DAF-functionalized beads, as a standard control for competition binding.12 The data were fit to a competition binding curve (Y=Bottom + (Top-Bottom)/(1+10^((LogIC50-[ligand])*HillSlope))) using GraphPad Prism 6.0h. None of the parameters were fixed, the unfixed Hillslopes were not significantly different (~ −0.8), indicating single site binding. Fixing the Hillslopes to unity did not significantly change the fit parameters. The SNVF18 antimycin curve was characteristically similar to the sDAF curve but shifted to the right due to lower affinity for antimycin (IC50 = 50 nM for sDAF vs. 1C50 = 10μM for antimycin). Normal competition curves characteristically show that the dose-response concentration ranges of the target ligands span more than two orders of magnitude25 as indicated for sDAF versus SNV18 and antimycin versus SNVF18. Conversely, the effective concentration range of antimycin titrations used in the antimycin-SNVR18 curve was relatively narrow suggesting that the process did not represent an actual competition binding curve, but one confounded by probe quenching.
SNV binding to low-affinity integrin stimulates cyclic AMP production
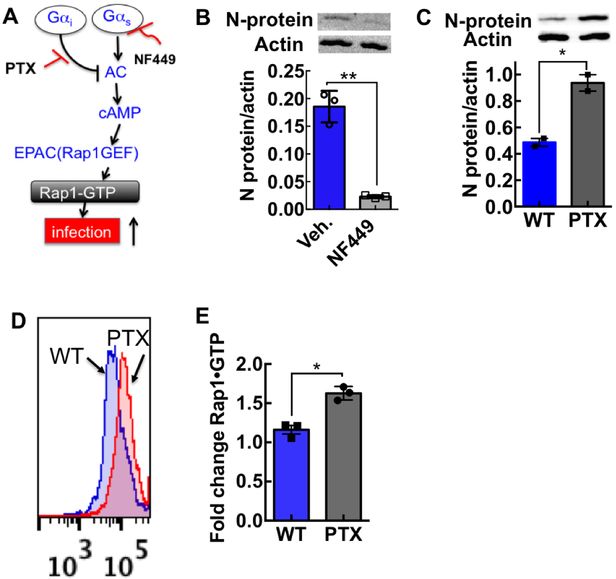
We have recently shown that SNV engagement of the integrin PSI domain involves the transduction of tensile force transmitted through the integrin.9 Others have shown that force-stimulated integrins might recruit stress-dependent signaling molecules to focal adhesions including cyclic AMP by activating Gαs at the site of force application.26 For our purposes, Gαs stimulation results in integrin activation, by a pathway involving cAMP and the exchange protein activated by cAMP (Epac) and Rap1.27 As a rationale for establishing a cyclic AMP assay for assessing the mechanism of action of our inhibitor compounds, we first tested whether SNV binding to the integrin PSI domain activated the Gαi/Gαs signaling axis (Fig. 5A).28 These assays used DAF-null CHO-A24 cells, stably expressing integrins αIIbβ3 and P2Y2R receptors.9 We then used a Gαs-selective antagonist NF44929 to determine whether Gαs signaling was necessary for SNV infection of CHO-A24 cells. We used a Western blot to measure SNV N-protein expression as described in the methods. We established loading controls of the gels by using a typical standard housekeeping gene such as β-actin on the same membrane. Quantitative analysis of the gel was performed using a BioRad Molecular Imager. NF449 reduced the viral N-protein by >80% relative to vehicle-treated cells, indicating that Gαs signaling was essential for infection. (Fig 5B) After that, we tested whether Gαi signaling was involved in SNV infectivity by using pertussis toxin (PTX) to block Gαi activity. Inhibition of Gαi signaling with PTX increased viral N-protein 2-fold relative to control cells (Fig. 5C). As a matter, of course, we then compared GTP-loading of Rap1, in PTX and non-PTX treated cells 5 min after exposure to UV killed SNVR18. The nearly two-fold increase in Rap1-GTP in PTX-treated cells was comparable to the rise in infection (Fig. 5D&E) indirectly showing that cAMP stimulation was a decisive factor in the infection of CHO-A24.
Figure 5. Cyclic AMP stimulation potentiates SNV infectivity.
A. Pertussis toxin (PTX) treatment of CHO-A24 cells suggests that Gαi and Gαs acting through cyclic adenosine monophosphate (cAMP) regulate SNV infectivity. Gαs signaling up-regulates adenylate cyclase (AC) upstream of cAMP. cAMP stimulates the exchange protein activated by cAMP (Epac)27 a guanine nucleotide exchange factor (GEF) for Rap1.27 Rap 1 is a small GTP binding protein, which is an effector for integrin activation.39 Gαi signaling blocks Gαs activity upstream of AC. Pertussis toxin (PTX) inhibits Gαi inhibitory function. B. NF449, a Gαs specific antagonist, inhibits SNV infectivity. CHO-A24 cells were treated with 65 μM NF449 for 10 min before infection. C. PTX treatment induces a nearly two-fold increase in progeny SNV in CHO- A24 cells. 150,000 CHO A24 cells were plated onto a 48 well plate for 24 hours and then treated with 100 ng/ml PTX and incubated for ~18hrs at 37°C and then used in infection. CHO-A24 cells were treated with 100ng/ml PTX for 18 hours before infection. Viral N-protein was measured using a Western blot as previously described.9, 12 Quantitative analysis of the gel was performed using a BioRad Molecular Imager, ChemiDoc XRS+ equipped with Image Lab Software 4.1. We verified equal loading by detecting of β-actin on the same membrane with the anti-β-actin (clone AC-74 used at 1:2000 from Sigma). D. Panel inserts show fluorescence histograms of active Rap1 measured in wild-type and PTX treated cells. E. A ratiometric plot of SNV activated GTPases in PTX-treated cells and WT untreated cells measured at 5 min after activation. Values are means ± SD of triplicate measurements.
Antimycin inhibits cAMP stimulation by directly binding to SNV
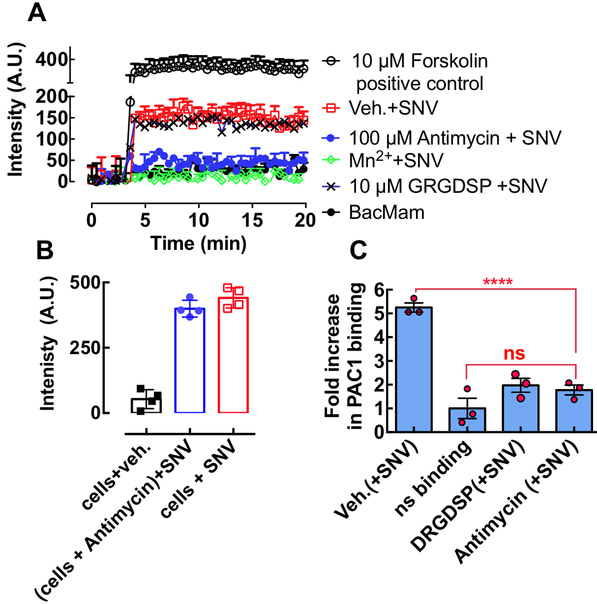
We measured SNV-induced cAMP production in 48,000 CADDis-transformed CHO-A24 cells under various conditions (Fig. 6A). As shown, SNV binding to low-affinity β3 integrins stimulated cAMP signaling (red squares). Preincubating antimycin with SNVR18 and adding to the cells (100 μM antimycin + SNV in Fig 6A) inhibited stimulation of cAMP more effectively than incubating antimycin with cells before the addition of SNVR18 (Fig. 6 B; also see Supplemental Figure S3 for kinetic data). This result suggested that antimycin binds directly to the virus and not to the cell receptor. We treated other wells with 1.5 mM Mn2+ and 10 μM GRGDSP followed by SNV. We used Mn2+ as a positive control for inhibition of SNV binding to integrins.6, 16 Also, Mn2+ was applied to determine whether Mn2+-induced integrin activation caused cAMP production. The data show that Mn2+, which prevented SNV binding to the cells6, 16 did not affect cAMP production. We used the soluble fibronectin hexapeptide GRGDSP to disrupt the putative cis interaction of αIIbβ3 with RGDP2Y2R in CHO-A24 cells. Interestingly, GRGDSP did not affect SNV-stimulated cAMP response, indicating that cAMP stimulation is not integrin binding to an anchored ligand.
Figure 6.
A. A plot of Green fluorescence of CHO-A24 cells expressing an Upward cAMP Difference Detector in situ (CADDis) sensor, responding to 10 μM forskolin (positive control for cAMP activity) and SNVR18 alone and mixed with 100 μM Antimycin, 1.5 mM Mn2+ and GRGDSP. Error bars represent ±SD for triplicate measurements. B. The plot compares endpoint cyclic AMP signals produced in antimycin treated cells (no wash) and untreated cells (see Fig. S3 for real-time assay). C. SNVR18 binding to αIIbβ3 recapitulates physiologic integrin activation indicated by PAC-1 binding. The graph shows a plot of fold increase over resting cells in PAC-1 staining of CHO-A24 cells after exposure to SNVR18. 5,000 CHO-A24 cells were suspended in 20 μl media containing 1:20 dilution PAC-1 antibody. Select tubes were treated for 30 min with 10 μM GRDGSP. SNVR18 samples were premixed with antimycin to block SNVR18 binding to cells. To induce PAC-1 staining SNV (5,000 particles/cell) were added to the tubes and incubated with virus for 5 min. Cell activation was terminated by placing tubes in ice-cold water. PAC1 was allowed to equilibrate for 30 min. Cells were washed once and read on a flow cytometer. Error bars represent standard error of 3 triplicate measurements. Statistical significance of the difference between the vehicle (Veh) and compound-treated samples was determined by Dunnett multiple comparison t-tests.
Testing antimycin for inhibition of full αIIbβ3 integrin activation
SNV binds to the PSI domain of low-affinity integrins interacting in cis with P2Y2R and stimulates outside-in signaling. Outside-in signaling requires a surface-anchored ligand, such as P2Y2R which can support traction force upon its exertion.8 The resulting changes in conformation to full activation allow the integrin’s ligand binding site to be recognized by PAC1, an integrin activation-dependent IgM antibody.9 Exposure of CHO-A24 cells to SNV stimulated a 5-fold increase in PAC-1 staining above non-specific background (Veh. and ns binding in Fig. 6B). Antimycin preincubated with SNV inhibited PAC-1 staining (Antimycin in Fig. 6B), whereas preincubating antimycin with cells produced irreproducible results (data not shown). GRGDSP (GRGDSP (+SNV) in Fig 6B) inhibited PAC1 staining unlike cAMP (Fig. 6A). Thus this result reveals that cAMP stimulation is an upstream signaling event relative to signaling associated with the integrin conformational change required for the molecular recognition of PAC1.9
DISCUSSION
We demonstrate that a BSL3 pathogen, SNV, can be produced and inactivated in sufficient quantities of known concentration12, 16 for use in an HTS campaign. We have performed HTS of the 1200 compound PCL with a homogeneous flow cytometry-based assay to identify compounds that can inhibit Sin Nombre virus binding to DAF. We have identified one compound, antimycin. Antimycin blocked binding of SNV to integrin β3-null but DAF-replete Tanoue B cells. Antimycin inhibited infection of Vero cells that express DAF and integrin β3. Antimycin inhibits integrin-associated cAMP production and molecular recognition by PAC1, an IgM antibody that binds to activated integrins. Therefore, the results suggest that antimycin’s molecular mechanism of action is interacting with the hantavirus’s receptor-binding Gn/Gc proteins, which in turn interferes with the virus’s ability to bind cellular receptors.
Antimycin is a specific inhibitor of respiration and is the active component found in the commercial fish poison Fintrol™. The value of antimycin as a candidate compound is based on the known, structural details of its interaction with counter-structures associated with its bioactivity.30, 31 Thus, such information could be used in SAR optimization as well as identification of structural homologs, which might serve as templates for optimization. Our secondary receptor blocking assays are carried out in a short time frame that rules out the possibility of cytopathic effects.
It is worth noting that antimycin shares common features with small antiviral compounds that inhibit infection by partitioning into the envelope membranes of diverse viruses such as influenza and hepatitis C. These compounds block infectivity of unrelated viruses by inhibiting fusion of the viral envelope membrane to the host cell.32, 33 While antimycin associates with the viral envelope and thus quenches R18, like the other hits found in our study, antimycin blocks SNV binding to cells. In contrast to the other HTS hit compounds that quenched R18 but did not inhibit infection.
The most significant aspect of our study is the development of secondary homogenous cell suspension assays to screen for small molecule inhibitors of SNV-induced integrin activation. These tests are compatible with flow cytometry and 96 or 384 well plate format, and therefore amenable to HTS configuration. Integrin activation plays a critical role in many physiological processes, including activation of the immune response, inflammation, hemostasis, thrombosis, cell migration, and tumor cell invasion. Thus, discovery campaigns to understand the signaling pathways and their modulation are important steps in controlling the diseases associated with dysregulated integrin activity.34–36
It is important to note that the concept of SNV-induced integrin activation post-occupancy of the PSI domain of low-affinity integrins is novel, and has only been recently described.9 The canonical switchblade model of physiological integrin activation postulates that resting integrins assume a low-affinity bent conformation for their ligand and upon full activation, integrins acquire an extended structure with an open headpiece.37 Integrin αIIbβ3 thus activated by SNV recognizes the PAC1 antibody.9 The intermediate signaling steps that occur between SNV engagement and molecular recognition of PAC-1 by the fully active integrin have not been explored. In this study, we show that cAMP production is an early downstream consequence of SNV engagement. cAMP is usually activated downstream of ligand-occupied heterotrimeric G protein-coupled receptors (GPCRs) that associate with the Gαs subunit. In this setting active Gαs (GTP bound) can induce the plasma membrane-bound adenylyl cyclase to catalyze the conversion of ATP to cAMP.38 Direct binding of cAMP to Rap1-guanine nucleotide exchange factor (Rap1-GEF) Epac1 provides a Gαs dependent mechanism for SNV-induced physiologic integrin activation.39, 40 Our study relies on inhibition assays of Gαi and Gαs to support the idea that, the stimulation of cAMP accumulation is linked to Gαs activation.26 In sum, our study exploits two mechanically transduced signal transduction steps, cAMP accumulation and change in integrin conformation to validate the HTS discovery target, antimycin, as a bona fide inhibitor of SNV interaction with cell entry receptors.
We propose to exploit lessons from this study to extend the original HTS screen to a two color HTS screen. Thus, instead of using Tanoue B cells for a primary screen, we propose to use CHO-A24 cells, where SNV binding to the PSI domain of the integrin can be measured directly in one fluorescence channel (FL2) in tandem with integrin activation as reported by PAC-1 staining in an alternate fluorescence channel (FL1). Cell-associated SNV fluorescence would monitor direct inhibition of SNV binding, and modulation of PAC-1 staining could suggest a mechanism of action. High-throughput flow cytometry would be the platform of choice for this activity.
Supplementary Material
Acknowledgments:
This work was supported by NIH Grants, R21NS066429, MH084690, UNM Department of Pathology and School of Medicine RAC award.
References
- 1.Epidemiology Hjelle B. and diagnosis of hantavirus infections In: Rose BD, (ed.). UpToDate. Wellesley, MA: Wolters Kluwer, 2014. [Google Scholar]
- 2.Zaki SR, Greer PW, Coffield LM, et al. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am J Pathol. 1995, 146, 552–579. [PMC free article] [PubMed] [Google Scholar]
- 3.Jonsson CB, Figueiredo LTM and Vapalahti O. A Global Perspective on Hantavirus Ecology, Epidemiology, and Disease. Clinical Microbiology Reviews. 2010, 23, 412–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaheri A, Strandin T, Hepojoki J, et al. Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol. 2013, 11, 539–550. [DOI] [PubMed] [Google Scholar]
- 5.Wernly JA, Dietl CA, Tabe CE, et al. Extracorporeal membrane oxygenation support improves survival of patients with Hantavirus cardiopulmonary syndrome refractory to medical treatment. European Journal of Cardio-Thoracic Surgery. 2011, 40, 1334–1340. [DOI] [PubMed] [Google Scholar]
- 6.Raymond T, Gorbunova E, Gavrilovskaya IN, et al. Pathogenic hantaviruses bind plexin-semaphorin-integrin domains present at the apex of inactive, bent alphavbeta3 integrin conformers. Proc Natl Acad Sci U S A. 2005, 102, 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackow ER and Gavrilovskaya IN. Hantavirus regulation of endothelial cell functions. Thromb Haemost. 2009, 102, 1030–1041. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Ju L, Rushdi M, et al. Receptor-mediated cell mechanosensing. Mol Biol Cell. 2017, 28, 3134–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bondu V, Wu C, Cao W, et al. Low-affinity binding in cis to P2Y2R mediates force-dependent integrin activation during hantavirus infection. Mol Biol Cell. 2017, 28, 2887–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shattil SJ, Hoxie JA, Cunningham M, et al. Changes in the platelet membrane glycoprotein IIb.IIIa complex during platelet activation. J Biol Chem. 1985, 260, 11107–11114. [PubMed] [Google Scholar]
- 11.Krautkramer E and Zeier M. Hantavirus causing hemorrhagic fever with renal syndrome enters from the apical surface and requires decay-accelerating factor (DAF/CD55). J Virol. 2008, 82, 4257–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buranda T, Swanson S, Bondu V, et al. Equilibrium and Kinetics of Sin Nombre Hantavirus Binding at DAF/CD55 Functionalized Bead Surfaces. Viruses-Basel. 2014, 6, 1091–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyne CB and Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006, 124, 119–131. [DOI] [PubMed] [Google Scholar]
- 14.Diaz F and Rodriguez-Boulan E. Open sesame! Coxsackieviruses conspire to trespass the tight junctional gate. Dev Cell. 2006, 10, 151–152. [DOI] [PubMed] [Google Scholar]
- 15.Bozym RA, Morosky SA, Kim KS, et al. Release of intracellular calcium stores facilitates coxsackievirus entry into polarized endothelial cells. PLoS Pathog. 2010, 6, e1001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buranda T, Wu Y, Perez D, et al. Recognition of decay accelerating factor and alpha(v)beta(3) by inactivated hantaviruses: Toward the development of high-throughput screening flow cytometry assays. Anal Biochem. 2010, 402, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tewson PH, Martinka S, Shaner NC, et al. New DAG and cAMP Sensors Optimized for Live-Cell Assays in Automated Laboratories. J Biomol Screen. 2016, 21, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buranda T, Wu Y, Perez D, et al. Real-time partitioning of octadecyl rhodamine B into bead-supported lipid bilayer membranes revealing quantitative differences in saturable binding sites in DOPC and 1:1:1 DOPC/SM/cholesterol membranes. J Phys Chem B. 2010, 114, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescott J, Ye C, Sen G, et al. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J Virol. 2005, 79, 15007–15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang JH, Chung TD and Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 21.Botten J, Mirowsky K, Kusewitt D, et al. Persistent Sin Nombre virus infection in the deer mouse (Peromyscus maniculatus) model: sites of replication and strand-specific expression. J Virol. 2003, 77, 1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buranda T, Basuray S, Swanson S, et al. Rapid parallel flow cytometry assays of active GTPases using effector beads. Anal Biochem. 2013, 144, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coller BS and Shattil SJ. The GPIIb/IIIa (integrin alphaIIbbeta3) odyssey: a technology-driven saga of a receptor with twists, turns, and even a bend. Blood. 2008, 112, 3011–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2 ed New York: Plenum Press, 1999. [Google Scholar]
- 25.Motulsky HJ and Mahan LC. The kinetics of competitive radioligand binding predicted by the law of mass action. Mol Pharmacol. 1984, 25, 1–9. [PubMed] [Google Scholar]
- 26.Alenghat FJ, Tytell JD, Thodeti CK, et al. Mechanical control of cAMP signaling through integrins is mediated by the heterotrimeric Galphas protein. J Cell Biochem. 2009, 106, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bos JL. Epac: a new cAMP target and new avenues in cAMP research. Nat Rev Mol Cell Biol. 2003, 4, 733–738. [DOI] [PubMed] [Google Scholar]
- 28.Lefkowitz RJ. G protein-coupled receptors and their regulation. Faseb Journal. 1997, 11, A873–A873. [Google Scholar]
- 29.Hohenegger M, Waldhoer M, Beindl W, et al. Gsalpha-selective G protein antagonists. Proc Natl Acad Sci U S A. 1998, 95, 346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulley H and Mohammad R. Small-molecule inhibitors of Bcl-2 protein. Drugs of the Future. 2004, 29, 369–381. [Google Scholar]
- 31.Strong FM, Dickie JP, Loomans ME, et al. The Chemistry of Antimycin-A .9. Structure of the Antimycins. Journal of the American Chemical Society. 1960, 82, 1513–1514. [Google Scholar]
- 32.Colpitts CC, Ustinov AV, Epand RF, et al. 5-(Perylen-3-yl)ethynyl-arabino-uridine (aUY11), an arabino-based rigid amphipathic fusion inhibitor, targets virion envelope lipids to inhibit fusion of influenza virus, hepatitis C virus, and other enveloped viruses. J Virol. 2013, 87, 3640–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hakobyan A, Galindo I, Nanez A, et al. Rigid amphipathic fusion inhibitors demonstrate antiviral activity against African swine fever virus. J Gen Virol. 2017, 99, 148–156. [DOI] [PubMed] [Google Scholar]
- 34.Ley K, Rivera-Nieves J, Sandborn WJ, et al. Integrin-based therapeutics: biological basis, clinical use and new drugs. Nat Rev Drug Discov. 2016, 15, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao JT and Davis MJ. The roles of integrins in mediating the effects of mechanical force and growth factors on blood vessels in hypertension. Curr Hypertens Rep. 2011, 13, 421–429. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Vootukuri S, Shang Y, et al. RUC-4: a novel alphaIIbbeta3 antagonist for prehospital therapy of myocardial infarction. Arterioscler Thromb Vasc Biol. 2014, 34, 2321–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo BH, Carman CV and Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007, 25, 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassone-Corsi P The cyclic AMP pathway. Cold Spring Harb Perspect Biol. 2012, 4, a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagarrigue F, Kim C and Ginsberg MH. The Rap1-RIAM-talin axis of integrin activation and blood cell function. Blood. 2016, 128, 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shattil SJ, Kim C and Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010, 11, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.