Abstract
Aberrant glutamatergic signaling has been implicated in altered metabolic activity in many cancer types including malignant melanoma. Previously, we have illustrated the role of metabotropic glutamate receptor 1 (GRM1) in neoplastic transformation of melanocytes in vitro and spontaneous metastatic melanoma in vivo. In this study, we showed that autocrine stimulation constitutively activates the GRM1 receptor and its downstream mitogenic signaling. GRM1-activated (GRM1+) melanomas exhibited significantly increased expression of glutaminase (GLS), which catalyzes the first step in the conversion of glutamine to glutamate. In cultured GRM1+ melanoma cell lines, CB-839, a potent, selective, and orally bioavailable inhibitor of GLS, suppressed cell proliferation while riluzole, an inhibitor of glutamate release, promoted apoptotic cell death in vitro and in vivo. Combined treatment with CB-839 and riluzole treatment proved to be superior to single agent treatment, restricting glutamate bioavailability and leading to effective suppression of tumor cell proliferation in vitro and tumor progression in vivo.
Hyperactivation of GRM1 in malignant melanoma is an oncogenic driver, which acts independently of canonical melanoma proto-oncogenes, BRAF or NRAS. Overall, these results indicate that expression of GRM1 promotes a metabolic phenotype that supports increased glutamate production and autocrine glutamatergic signaling, which can be pharmacologically targeted by decreasing glutamate bioavailability and the GLS-dependent glutamine to glutamate conversion.
Keywords: melanoma, GRM1, mGluR1, glutamate, glutaminase, CB-839, cancer, metabolism
INTRODUCTION
Melanoma is the most aggressive type of skin cancer and its incidence is on the rise worldwide, accounting for almost 10,000 deaths every year [1, 2]. Although surgically curable at early stages, late stage melanoma is difficult to treat due to genomic tumor variability and therapy resistance [3]. The constitutive activation of the mitogen activated protein kinase pathway is frequently altered in melanoma and initially responsive to targeted treatment [4]. However, disease relapse and tumor progression impedes long-term survival [5]. Newer immunotherapies including CTLA-4 or PD-1 blockade initially display great therapeutic efficacies in patients with advanced melanoma [6, 7]. However, almost all patients acquire resistance to these therapies after varying periods of time [8, 9]. Therefore, there is a clear need for improved combinatorial treatments to combat melanoma.
Utilizing a transgenic mouse model [10], we have established the genotype and tumorigenicity of aberrant expression of metabotropic glutamate receptor 1 (GRM1) in melanoma [11]. Glutamate is one of the most abundant amino acids in the human body and the predominant excitatory neurotransmitter in the vertebrate central nervous system. In addition to glutamate being the natural ligand of GRM1, our interest in exploring the consequences of altered glutaminolytic glutamate production is based on previous studies demonstrating that increased resistance to targeted therapy is a result of augmented glutamine dependency in melanoma cells [12, 13]. Consistent with this, we have demonstrated that a reduction in expression or function of GRM1 resulted in a decrease in melanoma cell proliferation in vitro and tumor burden in vivo [14]. Riluzole is an FDA approved drug for amyotrophic lateral sclerosis (ALS), a disease that is, at least in part, mediated by the pathological accumulation of glutamate. We pioneered preclinical studies with riluzole, which blocks glutamate release and leads to decreased melanoma cell growth in vitro and tumor progression in vivo [15, 16]. The exact mechanism of how riluzole inhibits the release of glutamate is unknown, but functionally, riluzole acts as an indirect antagonist of GRM1.
GRM1-expressing melanoma cells release excess glutamate into the extracellular environment to warrant constitutive activation of the receptor [17]. Glioma cells use glutamate as an autocrine or paracrine signal to promote cellular migration and invasion [18]. Glioma cells release excess glutamate through a cystine-glutamate antiporter system (xCT), which causes the excitotoxic death of neurons and permits tumor cell expansion [19, 20]. Excessive release of glutamate by transformed glioma cells has also been linked to more aggressive growth compared to parental cells [21]. The brain is a common metastatic site for secondary tumors to arise in metastatic melanoma [22]. In addition, enhanced glutamate release has been observed in both breast cancer and prostate cancer cells [23, 24].
Glutamate is the most abundant and multifaceted biomolecule that plays a fundamental role in multiple metabolic processes and signaling in human cells. The vital role of glutamine metabolism in cancer cell proliferation suggests that glutaminolytic enzymes feeding into the tricarboxylic acid (TCA) cycle could be appealing targets for therapy. It has been shown that GLS (glutaminase), an important glutaminolytic enzyme involved in the conversion of glutamine to glutamate, has elevated activity in tumors and is positively correlated with transformation and oncogenesis [25–28]. These findings brought about the design and development of CB-839, a potent, selective, and orally bioavailable GLS inhibitor. Recent studies have shown that CB-839 exhibits anti-proliferative activity in vitro against a panel of triple-negative breast cancer (TNBC) cell lines, as well as in vivo breast cancer models, suggesting that GLS inhibition could lead to therapeutic benefit in patients with TNBC and other glutamine-dependent tumors [29]. In addition, CB-839 is well tolerated in preclinical studies in mice, with no weight loss or toxicity observed [29]. Recent reports also suggest that combining potent GLS inhibitors with other targeted therapies increases the durability of therapeutic responses in a variety of cancers [30, 31]. These results prompted us to investigate a novel therapeutic approach to target glutaminolytic glutamate production and utilization in GRM1+ melanoma through combined actions of CB-839 and riluzole.
MATERIALS AND METHODS
Reagents, antibodies and western immunoblots
CB-839 (PubChem CID: 71577426) and riluzole (PubChem CID: 5070) were purchased from Selleckchem, Houston, TX). CB-839 and riluzole were dissolved in dimethyl sulfoxide (DMSO, Fisher Scientific) as 50 mM and 100 mM stock solutions, respectively, and used in treatments at the indicated concentrations. Anti-GLS antibody was purchased from Novus Biologicals (NBP1–58044, Littelton, CO). Monoclonal anti-α-tubulin antibody was purchased from Sigma Aldrich (T6074, St. Louis, MO). Anti-GRM1 antibody was purchased from Lifespan BioSciences (LS-C354444, Seattle, WA).
In order to prepare protein lysates, media was removed and cells were washed once with ice-cold phosphate buffered saline (PBS) [32]. After removal of PBS, extraction buffer was added directly to the plates and cells were collected with a cell scraper. Cell extracts were incubated on ice for 20 min. Cell debris was removed by centrifugation at 14,000 rpm at 4 ºC for 20 min, supernatant was collected and stored at −80 ºC. 25 μg of protein was routinely used for Western immunoblot analysis. The intensity of the protein bands on the blots was quantified and performed using the NIH-approved ImageJ software.
Cell lines, cell culture reagents and conditions
C8161, C81–61, UACC903, and 1205Lu were provided by Drs. Mary J.C. Hendrix (Children’s Memorial Research Center, Chicago, IL), Jeffrey Trent (The Translational Genomics Research Center, Phoenix, AZ), and Meenhard Herlyn (Wistar Institute, Philadelphia, PA) about 20 years ago and have been kept in our liquid nitrogen tank. These cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). hTERT/CDKR24C/p53DD (AR7119; immortalized normal human melanocytes) cells were provided by Dr. David Fisher (Harvard Medical School, Boston, MA) and maintained in Medium 254 with human melanocyte growth supplements (M-254, Invitrogen, Carlsbad, CA) [33]. All cell lines were maintained at 37 ºC in a humidified 5% CO2 incubator. We maintain a large number of frozen vials after the cells were tested and confirmed by Hoechst stain (Sigma-Aldrich) to be free of mycoplasma contamination. When there are less than two vials left in the liquid nitrogen tank, the cells are thawed out and the Hoechst mycoplasma stain test is repeated, and numerous vials are frozen down again. The most recent testing on these cells was performed in September 2017. We routinely try not to keep the cells in culture for more than 30 days. All cells are less than 7 passages from the time we obtained the cells. Cell line authentication was not conducted.
For metabolite quantification experiments, 100,000 cells per well were seeded in replicate (n=6) in 6-well plates (657160, Greiner Bio-One, Kremsmünster, Germany) in DMEM (10–017, Corning Cell-Gro, Manassas, VA) supplemented with 10% FBS, 1% Penicillin-Streptomycin (30–002-CI, Corning Cell-Gro, Manassas, VA), and 1% MEM Non-Essential Amino Acids (25–025-C, Corning Cell-Gro, Manassas, VA). 24 hours following seeding, media was aspirated and replaced with MEM (Corning Cell-Gro, Manassas, VA) supplemented with 1 g/L D-Glucose (0188, Amresco, Solon, OH), 2 mM L-Glutamine (G3126, Sigma Merck, Darmstadt, Germany), 10% FBS, and 1% MEM Vitamins (25–020-CI, Corning Cell-Gro, Manassas, VA).
Constructing melanoma cell lines with altered GRM1 expression
The stable human melanoma cell line C8161 TetR siGRM1 B22–20 clone (C8161si) was generated and maintained in 1 µg/ml blasticidin and 10 µg/ml hygromycin [15]. Induction of siGRM1 was carried out by incubating the cells with 10 ng/ml of doxycycline for 4 days. A stable human melanoma cell line C81–61 GRM1–6 (C81–61OE) clone that expresses elevated GRM1 levels compared to parental cell lines was selected with 10 µg/ml blasticidin [34].
TGS melanoma model of ectopic expression of GRM1
The original TG-3 strain (transgenic founder 3 with Grm1+) was crossed with hairless SKH-1 mice to arrive at the TGS strain—brother-sister littermates have been mating for the past 18 years. TG-3 mice were established as a result of a classic case of insertional mutagenesis that led to the ectopic expression of Grm1 in melanocytes. TG-3 mice spontaneously develop metastatic melanoma with 100% penetrance. SKH-1 is an uncharacterized/non-pedigreed hairless strain of mice. The goal was to make the pigmented lesions visible on the TGS mice. Genotypes of TGS mice were performed as described for TG-3 [11].
Tumorigenicity assays
The C8161 xenograft study was performed according to our Institutional Animal Care and Use Committee approved protocol. C8161 cells were harvested by trypsinization and resuspended in PBS (107 cells/ml). 5–6-week-old immunodeficient nude male and female mice were subcutaneously injected with 106 C8161 cells in each dorsal flank. Tumor growth was monitored weekly with a vernier caliper and calculated with the formula (d2 x D/2) [35]. Once tumor volumes reached 10 to 20 mm3, mice were divided into four treatment groups and received vehicle (DMSO), riluzole (10 mg/kg), CB-839 (200 mg/kg), or the combination of riluzole (10 mg/kg) and CB-839 (200 mg/kg) by oral gavage, daily. The experiment was terminated when the xenografts in the vehicle group reached maximum permitted size.
MTT cell proliferation/viability assays
Cell proliferation was ascertained using MTT reagent [17]. All cell lines were cultured in RPMI-1640 medium, which contains 0.02 g/L of glutamate, supplemented with 10% FBS. Briefly, each cell line was cultured in 96-well culture plates (~2500 cells per well) followed by treatment with vehicle (DMSO), CB-839 or/and riluzole at varying concentrations. At indicated time points, the number of viable cells was determined by measuring absorbance (at 560 nm with a reference wavelength of 750 nm) using a 96-well plate reader (Infinite M200 Tecan USA, Durham, NC).
Glutamate quantification
Glutamate concentration in the conditioned media was measured after 0, 2 or 4 days in culture with glutamate-free MEM using the Glutamine/Glutamate Determination Kit (GLN1, Sigma-Aldrich) according to the manufacturer’s instructions. The determination of L-glutamate was done by measuring the dehydrogenation of L-glutamate to α-ketoglutarate accompanied by reduction of NAD+ to NADH. The conversion of NAD+ to NADH was determined by measuring absorbance at 340 nm using a 96-well plate reader (Infinite M200 Tecan USA, Durham, NC). The amount of NADH is proportional to the amount of glutamate in each sample.
Metabolite extraction
Following 24 hours incubation in supplemented MEM, 5 μL of supernatant containing conditioned media was transferred to micro centrifuge tubes (MT-0200-BC, Biotix, San Diego, CA) with 1 mL of cold [−20 °C (253 K)] extraction buffer consisting of 50% methanol (A452, Fisher Scientific, Fair Lawn, NJ) in ultrapure (18.2 MΩ x cm) water with 20 µM L-Norvaline (N7627 Sigma Merck, Darmstadt, Germany) and 20 µM DL-Norleucine (N1398, Sigma Merck, Darmstadt, Germany) and dried by vacuum centrifugation in a speedvac concentrator (DNA120OP115, Savant, Thermo Fisher Scientific, Waltham, MA) overnight. The remaining media was aspirated and the cells washed quickly with cold 0.9% sodium chloride in ultrapure water (Amresco) and placed on ice. To each well, 1 mL of cold extraction buffer was added, the cells scraped on ice and the entire solution was then transferred to a pre-chilled micro centrifuge tube. Tubes were then frozen in liquid nitrogen, thawed, and placed in a digital shaking dry bath (8888–0027, Thermo Fisher Scientific, Waltham, MA) set to 1100 rpm for 15 min at 4° C (277K). Samples were then centrifuged for 15 min at 4 °C (277 K) and 12500 g in a refrigerated centrifuge (X1R Legend, Sorvall, Thermo Fisher Scientific, Waltham, MA) using a fixed-angle rotor (F21–48×1.5, Sorvall, Thermo Fisher Scientific, Waltham, MA). Supernatants were transferred to new micro centrifuge tubes and dried by vacuum centrifugation overnight.
Metabolite derivatization
Dried, extracted cell samples or media supernatants were derivatized by addition of 20 μL of 2.0% methoxyamine-hydrochloride in pyridine (MOX, TS-45950, Thermo Fisher Scientific, Waltham, MA) followed by 90 min incubation in a digital shaking dry bath at 30 °C (303 K) and 1100 rpm. Then 90 μL of N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA, 394866–10X1ML, Sigma Merck, Darmstadt, Germany) was added and samples incubated at 37 °C (310 K) and 1100 rpm for 30 min before centrifugation for 5 min at 14,000 rpm and 4 °C. The supernatant was transferred to an auto sampler vial (C4000LV3W, Thermo Fisher Scientific, Waltham, MA) with screw cap (C5000–53B, Thermo Fisher Scientific, Waltham, MA) for analysis by gas chromatography (GC, TRACE 1310, Thermo Fisher Scientific, San Jose, CA) coupled to a triple-quadruple GC mass spectrometry system (QQQ GCMS, TSQ8000EI, TSQ8140403, Thermo Fisher Scientific, San Jose, CA).
GCMS
Samples were analyzed on a QQQ GCMS system equipped with a 0.25 mm inner diameter, 0.25 μm film thickness, 30 m length 5% diphenyl/95% dimethyl polysiloxane capillary column (Trace GOLD TG-5MS, 26098–1420, Thermo Fisher Scientific, Waltham, MA) and run under electron ionization at 70 eV. The GC was programed with an injection temperature of 250 °C (523 K) and split less injection volume of 1.0 μL. For media samples, a 1:20 split injection was used. The GC oven temperature program started at 50 °C (323 K) for 1 min, rising to 300 °C (573 K) at 10 K/min with a final hold at this temperature for 6 min. The GC flow rate with helium carrier gas (HE, HE 5.0UHP, Praxair, Danbury, CT) was 1.2 mL/min. The transfer line temperature was set at 290 °C (563 K) and ion source temperature at 295 °C (568 K). A range of 50–600 m/z was scanned with a scan time of 0.25 s.
Metabolomics data processing
Metabolites were identified using Trace Finder (Version 3.3, Thermo Fisher Scientific, Waltham, MA) based on in-house libraries of metabolite retention times and fragmentation patterns. Identified metabolites were quantified using the selected ion count peak area for specific mass ions, and standard curves generated from reference standards run in parallel. Peak intensities were normalized for extraction efficiency using L-norvaline as an internal standard. The mean and standard deviation for each quantified metabolite was calculated for each cell line and treatment condition. A univariate t-test was used to compare means for each metabolite and cell line.
Statistical analysis
Statistical significance of experimental data was calculated using the Stat Plus software (Version 6.2.30) on Microsoft Excel® evaluated by the appropriate t-test or ANOVA depending on variables. The statistical significance was set at a p value of *p<0.05, **p<0.01 or ***p<0.001.
RESULTS
Elevated circulating plasma glutamate levels in a Grm1-driven melanoma model
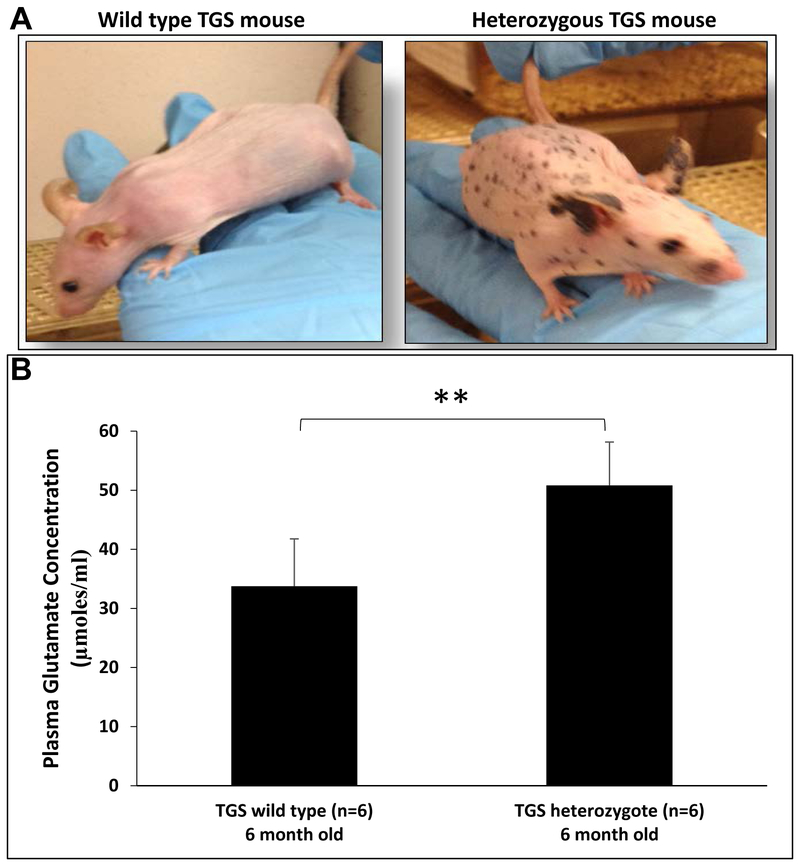
We derived TGS mice from crosses between melanoma-prone TG-3 [11, 36–38] with hairless SKH-1. Onset and progression of the pigmented lesions are very similar in TG-3 and TGS mice; in the absence of hair the pigmented lesions are readily visible in TGS mice. Homozygous TGS mice that harbor two copies of the disrupted endogenous Grm1 gene succumb to large tumor burden by 4–5 months old; thus, are not included in our studies. Heterozygous Grm1+/− TGS mice with only one copy of the disrupted endogenous Grm1 are viable, show highly pigmented tumors and bear large tumor burden by 11–12 months of age, indicating that Grm1 signaling stimulates melanomagenesis. An image addressing the visual (phenotypic) difference between heterozygous a TGS mouse and a wild type TGS mouse is shown in Figure 1A. Comparison of glutamate levels in circulating blood plasma between 6-month old heterozygous TGS and wild type (no disrupted Grm1) mice, heterozygous TGS mice showed elevated glutamate levels (Figure 1B), suggesting aberrant Grm1 expression may promote an increase in its natural ligand, glutamate, in circulation to ensure constitutive activation of Grm1 receptor; similar observation was made in in vitro culture cells [17].
Figure 1: (A) Phenotypes of wild type and heterozygous TGS genotypes:
Pigmented lesions on the skin of heterozygous TGS mice are easily visualized compared to the WT without any copies of the disrupted endogenous metabotropic glutamate receptor 1 (Grm1) gene. Homozygous and heterozygous TGS mice are indistinguishable in the progression of the disease; the major difference is the onset of the disease, 2-3 months for homozygous TGS, and 5-7 months for heterozygous TGS. (B) Elevated circulating glutamate levels in plasma isolated from heterozygous TGS mice: Glutamate concentration in plasma isolated from wild type (6 month old) or heterozygous TGS mice (6 month old) was measured using the Glutamate Determination Kit (GLN1, Sigma-Aldrich) according to the manufacturer’s instructions. Data are given as μmoles of glutamate per mL of plasma and represented as mean ± standard deviation (n=6). Student’s t-test was used to calculate statistical significance. **p value < 0.01.
Elevated GLS detected in GRM1+ human melanoma cell lines
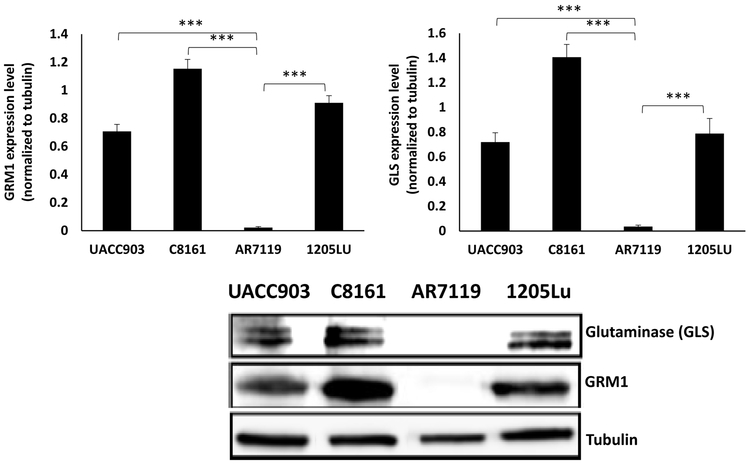
Ectopic expression of GRM1 is sufficient to induce cellular transformation in vitro and spontaneous melanoma development in vivo [11]. To investigate a possible relationship between GRM1 expression, altered glutamate bioavailability and glutaminase (GLS), we first confirmed GRM1 expression in C8161, UACC903 and 1205Lu human melanoma cells, plus immortalized normal human melanocytes, hTERT/CDKR24C/p53DD (AR7119). C8161 is a malignant human melanoma cell line that expresses wild type B-Raf proto-oncogene, serine/threonine kinase (BRAF). UACC903 and 1205Lu are other malignant melanoma cell lines that harbor the BRAFV600E mutation. C8161, UACC903, and 1205Lu demonstrated significantly elevated levels of GRM1 and GLS compared to hTERT/CDKR24C/p53DD cells with almost undetectable GRM1 and much lower GLS expression (Figure 2).
Figure 2: A link between GLS and GRM1 levels in human melanoma cells:
Western immunoblots of metabotropic glutamate receptor 1 (GRM1) and glutaminase (GLS) on protein lysates from three different human melanoma cell lines (C8161, UACC903 and 1205Lu) and one normal immortalized melanocyte cell line, hTERT/CDKR24C/p53DD (AR7119). Lysates were probed with the indicated antibodies. GLS expression correlated with GRM1 levels. Quantification of the intensity of GRM1 and GLS bands normalized to tubulin is displayed above the gel on a bar graph. Data is expressed as a mean ± standard deviation of three independent experiments. Student’s t-test was used to calculate statistical significance. ***p value < 0.05.
GLS inhibition reduces proliferation/viability of GRM1+ human melanoma cells
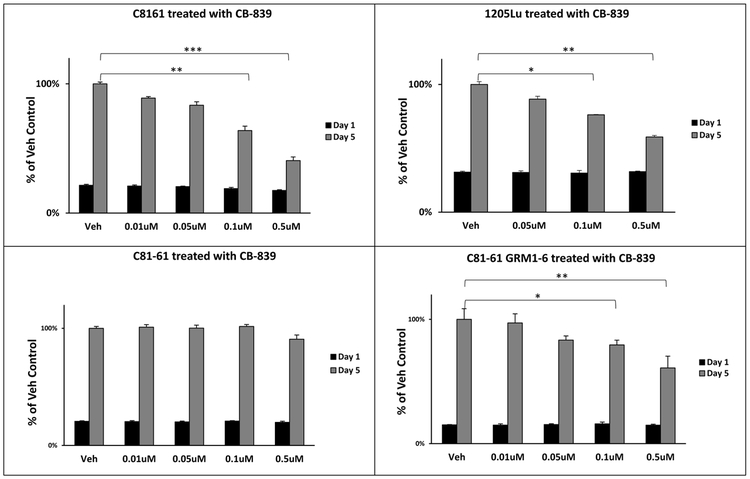
In in vitro tetrazolium-based proliferation/viability assays all three GRM1-expressing human melanoma cells, C8161, 1205Lu, and UACC903 displayed modest efficacy in suppressing cell growth in the presence of CB-839 as compared with the control vehicle (DMSO) group regardless of their BRAF genotypes (Figure 3 and Supplementary Figure 1A). It is noteworthy that a considerably higher concentration of CB-839 (10–50 μM) is required to observe a reduction in UACC903 cell proliferation, likely due to the presence of other driver mutations including BRAFV600E (Supplementary Figure 1A). Furthermore, to determine if GRM1 expression modulates the responsiveness to GLS inhibition, exogenous human GRM1 cDNA was introduced into an early stage melanoma cell line, C81–61, which does not express endogenous GRM1 (see profiling data below). Characterization of several GRM1-expressing C81–61 clones confirmed that these clones were transformed and tumorigenic [34]. We compared the growth rate of the parental C81–61 cell line to the C81–61OE cell line in the presence of CB-839. A marked reduction in the cell proliferation of C81–61OE was seen with 0.5 μM CB-839 as compared to the vehicle (DMSO) control (Figure 3). Strikingly, very minute if any changes were detected in growth of the parental C81–61 cells with analogous treatment conditions (Figure 3). These results suggest that GRM1 expression may influence the responsiveness of melanoma cells to GLS inhibition.
Figure 3: Inhibition of GLS reduces proliferation of GRM1-expressing melanoma cells:
MTT cell viability/proliferation assays were performed on GRM1-expressing C8161, 1205Lu and C81-61OE (C81-61 GRM1-6) cell lines, and GRM1-negative C81-61 cells. Treatment conditions for all cells were vehicle (DMSO) or CB-839 at 0.01, 0.05, 0.1 and 0.5 μM. Each time point and concentration represents a mean ± standard deviation of four independent measurements of the absorbance. A one-way ANOVA test with Bonferroni’s post-hoc analysis was used to calculate statistical significance between experimental and control groups. *p value < 0.05, **p value < 0.01, and ***p value < 0.001.
Combinatorial treatment with CB-839 and riluzole leads to enhanced inhibition of GRM1+ melanoma cell proliferation
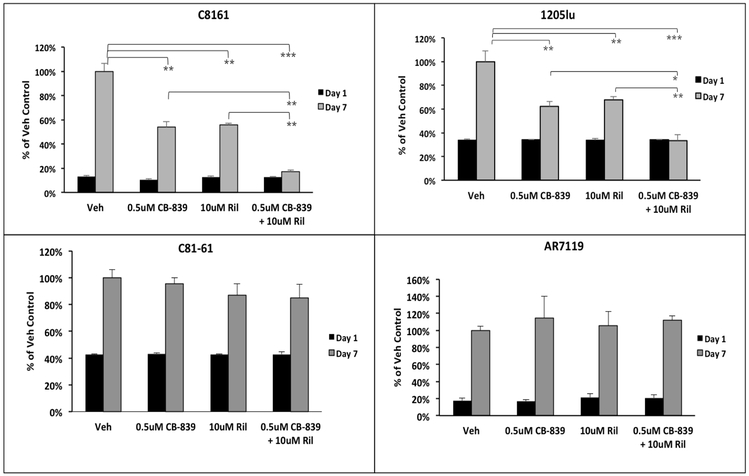
Suppressive effects of riluzole on GRM1+ melanoma cell proliferation were reported earlier [17, 39, 40]. Here, the consequences of including both CB-839 and riluzole on cell growth of two GRM1-expressing human melanoma cell lines were investigated. As shown in Figure 4, C8161 and 1205Lu cells were treated for 7 days with 0.5 μM CB-839, 10 μM riluzole or 0.5 μM CB-839 + 10 μM riluzole. Treatment with either CB-839 or riluzole reduced C8161 cell proliferation by ~40%, while combining both CB-839 and riluzole led to an ~85% decrease when compared to vehicle treated control cells. 1205Lu cells also displayed significant reduction in cell proliferation in the presence of both CB-839 and riluzole as compared to either agent alone (Figure 4). As mentioned above, a higher dose of CB839 and riluzole was needed to diminish cell proliferation in UACC903 cells (Supplementary Figure 1B) similar to our earlier observations [41]. The GRM1-negative C81–61 and hTERT/CDKR24C/p53DD (AR7119) cells were used as negative controls – these cells did not respond to riluzole plus CB-839 treatment (Figure 4), confirming that GRM1 expression is required to be responsive to these compounds at the indicated doses. Furthermore, increasing evidence illustrates that the presence of a mutation in BRAF oncoprotein frequently makes some cancer cells less responsive to various targeted treatments [42]. Taken together, our results suggest that CB-839 combined with riluzole can enhance the anti-proliferative properties of GRM1+ human melanoma cells, and that higher doses are needed for some BRAF-mutated cells.
Figure 4: Enhanced suppression of proliferation of GRM1-expressing human melanoma cells with CB-839 and riluzole:
MTT cell viability/proliferation assays were performed on GRM1-expressing C8161 and 1205Lu cells. GRM1-negative C81-61 and AR7119 cells were used as controls. Since AR7119 cells do not readily take up the tetrazolium MTT reagent, the trypan blue exclusion assay was performed on these cells as an alternative. The treatment conditions were vehicle (DMSO), CB-839 or/and riluzole at 0.5 μM and 10 μM respectively. Each time point and concentration represents a mean ± standard deviation of four independent measurements of the absorbance. A two-way ANOVA test with Bonferroni’s post-hoc analysis was used to calculate statistical significance between experimental and control groups. *p value < 0.05, **p value < 0.01, and ***p value < 0.001.
CB-839 treatment leads to inhibition of glutamate release from GRM1+ human melanoma cells
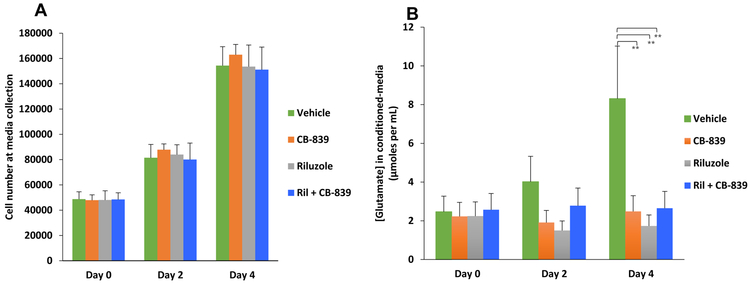
Inclusion of riluzole in cultured media modulated the amount of glutamate released by melanoma cells [17]. To determine the consequences on the level of glutamate released by GRM1+ melanoma cells upon treatment with CB-839 only or riluzole + CB-839, C8161 cells were plated in glutamate-free MEM media followed by collection of conditioned media at days 0, 2 and 4. We plated different number of C8161 cells so at time of collecting the conditioned-media samples, the cell numbers were very similar among the different days (Figure 5A). In parallel we also performed cell viability/cell proliferation MTT assays to ensure that the treated cells were viable, as the levels of glutamate release were determined. We showed that extracellular glutamate levels were significantly reduced in the conditioned culture media isolated from CB-839, riluzole or CB-839 + riluzole treated C8161 cells compared to the untreated cells (Figure 5B).
Figure 5: CB-839 treatment leads to inhibition of glutamate release in GRM1+ human melanoma cells:
Human melanoma C8161 cells were assessed for the amount of glutamate they release into the extracellular medium after treatment with CB-839, riluzole or CB-839 + riluzole. (A) Different number of cells was plated such that comparable numbers of cells were present at time of sample (conditioned medium) collection. The bar graph represents the number of viable cells during sample collection at day 0, day 2 or day 4. (B) Concentrations of extracellular glutamate within each treatment group are shown. Statistical analysis was performed between control (vehicle) and treated pairs to show significance. Each bar represents mean ± standard deviation, n=3. **p value < 0.01 and ns = no significance.
Concurrent administration of CB-839 and riluzole diminishes in vivo tumorigenicity
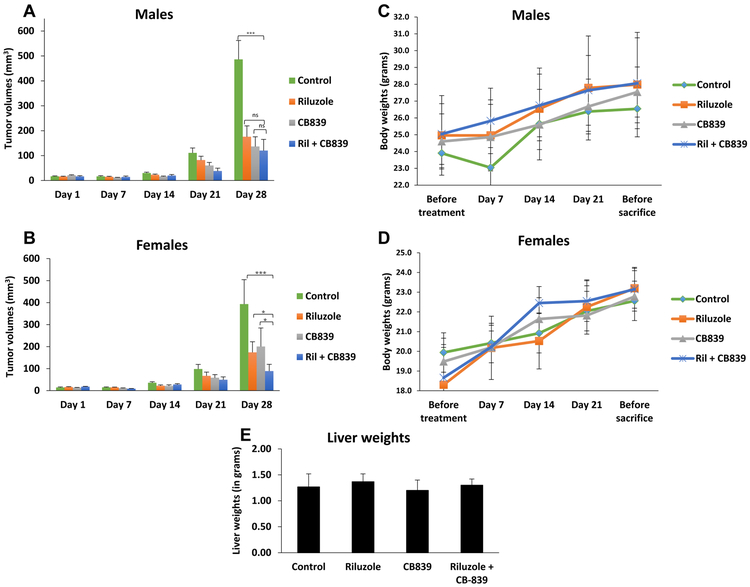
Next, to confirm our in vitro anti-tumorigenic potential in GRM1+ melanoma cells upon combining CB-839 with riluzole, we conducted in vivo experiments on established C8161 xenografts. C8161 cells were inoculated into the dorsal flanks of immunodeficient nude mice and when the tumor volumes reached approximately 10 to 20 mm3, the mice were randomly divided into 4 treatment groups: vehicle (DMSO), riluzole (10 mg/kg), CB-839 (200 mg/kg), or the combination of riluzole (10 mg/kg) and CB-839 (200 mg/kg) by oral gavage, daily. In support of the encouraging in vitro findings, we showed considerable suppression of tumor progression in mice treated with the combination of CB-839 plus riluzole as evident by the decrease in tumor volumes (Figures 6A and 6B). Interestingly, CB-839 plus riluzole mediated enhanced suppression of tumor growth in female mice (p<0.05) but only moderate in male mice (p=0.113) when compared to either agent alone. It is worth noting that C8161 cells were isolated from a female human patient and recent reports support the importance of the cell lines’ gender when screening for anti-cancer agents [43]. In addition, our co-workers have previously demonstrated the prevalent cross-talks between GRM1 and estrogen signaling [44] thereby providing some clarification on the sex-biased differences observed in this study. Most importantly, all of the treatment groups did not significantly affect the body and liver weights of the mice (Figures 6C, 6D and 6E), highlighting that these compounds are not toxic and well tolerated even when administered together. Absence of liver toxicity in all treatment groups was confirmed by hematoxylin and eosin (H&E) staining of liver tissue followed by exhaustive histopathological evaluation of the slides (Supplementary Figure 2).
Figure 6: In vivo xenograft tumorigenicity assay:
Xenografts were established in male (n=20) and female (n=20) mice using C8161 cells. The groups were control (vehicle: DMSO + PBS), riluzole (10 mg/kg), CB-839 (200 mg/kg), or the combination of riluzole (10 mg/kg) and CB-839 (200 mg/kg). All agents were given daily by p.o. gavage. All tumor-bearing mice were euthanized after 28 days due to tumor burden in the control (vehicle) group. A two-way ANOVA test with Bonferroni’s post-hoc analysis was used to calculate statistical significance between all treated pairs. Each bar represents volumes of tumors (mean ± standard error), n=5 for each of the 4 treatment groups for males (A) or females (B). Body weight of male (C) and female (D) mice was monitored throughout the course of treatment administration. Liver weights (n=3 for each group) were measured upon termination of the experiment (E). *p value < 0.05, ***p value < 0.001, and ns = no significance.
Modulation of GRM1 alters the intracellular production of glutaminolytic and glycolysis metabolites in human melanoma cells
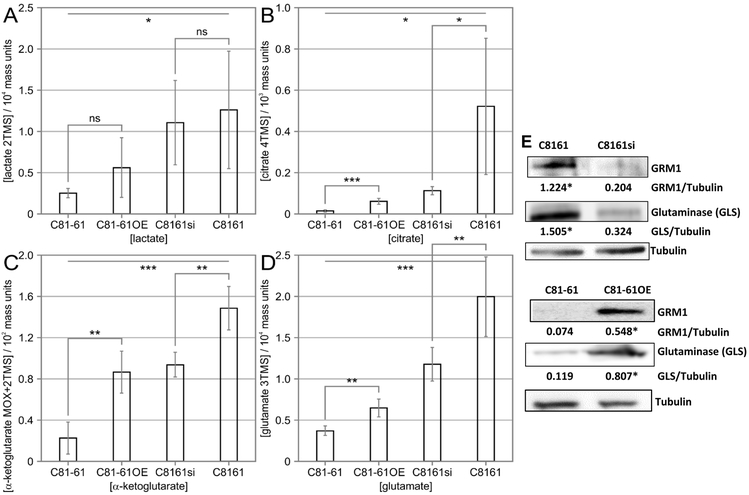
We next asked whether modulation of GRM1 expression affects the intracellular levels of key glutaminolytic and glycolytic metabolites. We analyzed both overexpression of GRM1 in a GRM1 low background (parental C81–61 and C81–61OE) and suppression of GRM1 in a GRM1 high background (parental C8161 and C8161si). While manipulation of GRM1 expression levels failed to alter intracellular lactate concentration (Figure 7A), higher levels of GRM1 correlated with significantly increased levels of intracellular citrate, α-ketoglutarate, and glutamate (p<0.01) (Figures 7B-D). This indicates that GRM1 expression does not increase lactate fermentation but does increase levels of TCA cycle intermediates. The increased intracellular pool size of glutamate could be a direct result of increased conversion of glutamine into glutamate via the activity of GLS. To determine whether modulating GRM1 expression affects the level of GLS, we assayed GLS protein levels by western blot. Consistent with our observed elevated glutamate concentrations, cells with higher levels of GRM1 also had higher levels of GLS protein (Figure 7E). These results suggest that GRM1 expression increases glutamate production by increasing GLS expression.
Figure 7: Modulation of GRM1 alters the intracellular production of glutaminolytic and glycolytic metabolites in human melanoma cells:
The intracellular concentrations of lactate (A), citrate (B), α-ketoglutarate (C), and glutamate (D) were detected in C81-61, C81-61OE (C81-61 GRM1-6), C8161 and C8161si (C8161 TetR siGRM1 B22-20) cells by GCMS analysis. Data represent the mean of six independent reads ± standard deviation. (E) Modulations in GRM1 and subsequent changes in glutaminase protein levels in C81-61, C81-61OE, C8161 and C8161si cells were determined by Western blot. Tubulin was used as the loading control. Numerical values indicating quantified intensity of GRM1 and GLS bands normalized to tubulin are displayed below the gel. Data is expressed as a mean ± standard deviation of three independent experiments. Paired, homoscedastic Student’s t-test was used to calculate statistical significance with p value threshold levels. *p value < 0.05, **p value < 0.01, ***p value < 0.001, and ns = no significance.
DISCUSSION
Metabotropic glutamate receptor 1 (GRM1) is an oncogenic driver in the neuroectodermal-derived lineage of melanocytes [45]. Aberrant glutamatergic signaling activates mitogenesis and melanomagenesis independent from canonical mitogen activated protein kinase signaling [4]. The high frequency of ectopic GRM1 expression in melanoma and its signaling cascades implicated in cellular transformation has made GRM1 a principal research target to improve treatment of melanoma. In this study, the role of GRM1 in modulating glutamate bioavailability in melanoma cells was explored. Our results suggest that GRM1 expression promotes a metabolic phenotype that supports increased glutamate production and autocrine glutamatergic signaling. Glutamatergic signaling through GRM1 promotes expression of GLS increasing the conversion of glutamine into glutamate. Melanoma cells heavily depend on anaplerosis via glutamine [46, 47]. GRM1+ melanoma cells upregulate GLS and support increased levels of glutamate. Excess amounts of intracellular glutamate are transported to the extracellular environment, where it serves as trigger for the GRM1 receptor. In neuronal cell lineages, cytoplasmic glutamate is exported via vesicular glutamate transporters or cystine-glutamate exchangers [48].
We demonstrate elevated glutamate levels in systemic circulation of heterozygous TGS mice (which harbor only one copy of the disrupted Grm1) compared to that of wild type TGS mice (no disrupted Grm1). The abundant availability of the endogenous ligand for GRM1 leads to constitutive activation of the GRM1 receptor, further promoting cell proliferation and metabolism pathways. To break such positive feedback circuits, we sought after different pharmacological vulnerabilities of glutamate signaling and metabolism. While treatment with the GRM1-specific inhibitor riluzole resulted in a significant proliferative disadvantage of tumor cell growth, it is insufficient as single treatment. In contrast, combined treatment of riluzole and CB-839 resulted in substantial tumor cell death. The combined inhibition of glutamatergic signaling via riluzole and of GLS activity via CB-839 efficiently reduced the conversion of glutamine to glutamate and interrupted GRM1 activation. As a consequence extracellular glutamate was reduced thus lowering the availability of the natural ligand of GRM1. Such rational combination of two complementary drug-targeting approaches disrupted the ability to circumvent individual blockages yielding a robust response, as evidenced by the enhanced reduction of tumor progression. While inhibition of glutamatergic signaling by decreasing glutamate release via riluzole or inhibition of GLS activity via CB-839 resulted in reduced cell proliferation as well as diminished tumor growth, the combination of both compounds was most effective. We also expected to see increased reduction of glutamate in the conditioned media after co-treatment with riluzole and CB-839 but this was not the case. Tumor cells have the ability to compensate for GLS inhibition and can overcome glutamate deprivation under such conditions including through increased anaplerosis, for example by asparagine synthase [49]. This compensatory mechanism may have contributed to intracellular glutamate production and release into the extracellular milieu. Alternatively, other glutamate transport pathways may have been altered to counterbalance the reduced glutamate levels due to burden exhibited by the combinatorial treatment [50]. Additional metabolic flux analysis would assist in revealing the source of glutamate before and after treatment with riluzole plus CB-839. Taken together, combined targeting of glutamate production and its eventual release showed highest efficacy in vitro and in vivo.
The role of glutamine metabolism in cancer cells is well established. However, it is less clear how its role is influenced by the tumor microenvironment, which is often subject to nutrient and oxygen shortage [51]. Recently, we reported that one of the consequences of aberrant GRM1 signal transduction is downstream activation of the hypoxia-induced transcription factor 1, HIF-1α, which promotes angiogenesis even in normoxic conditions [34]. Glutamine metabolism is strongly coupled to HIF-1α activity and enhanced in tumors [52]. Metabolomics data show that pool sizes of TCA cycle intermediates are increased by GRM1 expression, while glycolytic intermediates are unaffected. This further suggests that GRM1 signaling is tightly connected to glutamine metabolism. Recent reports in ovarian cancer cells demonstrate that GLS inhibition enhances the effectiveness of chemotherapy [53] and also improves the efficacy of other targeted therapies [30, 31], suggesting the critical role of targeting GLS in an attempt to improve overall patient response. These insights, combined with our data, support the rationale to target glutaminolytic glutamate bioavailability by combining riluzole with CB-839 in order to combat GRM1+ human neoplasia.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Findings demonstrate that targeting glutaminolytic glutamate bioavailability is an effective therapeutic strategy for GRM1-activated tumors.
ACKNOWLEDGEMENTS
This research has been funded in part by a grant from New Jersey Health Foundation and Veterans Administration Research award 101BX003742 to Suzie Chen (SC). Raj Shah (RS) is thankful for the support of the Bristol-Myers Squibb Co. Graduate Research Fellowship. SC and RS are grateful for the support of the NIEHS T32 Training Grant in Environmental Toxicology (ES007148). SC and RS also appreciate the support of the New York Society of Cosmetic Chemists. Fabian V. Filipp (FVF) is grateful for the support by grants CA154887 from the National Institutes of Health, National Cancer Institute, GM115293 NIH Bridges to the Doctorate, NSF GRFP Graduate Research Fellowship Program, CRN-17–427258 by the University of California, Office of the President, Cancer Research Coordinating Committee, and the Science Alliance on Precision Medicine and Cancer Prevention by the German Federal Foreign Office, implemented by the Goethe-Institute, Washington, DC, USA, and supported by The Federation of German Industries (BDI), Berlin, Germany. We would like to thank Andrew Boreland, Mengying Zhu and Xueting Wang for assistance in performing some of the pilot experiments. We appreciate the kind generosity of Dr. Allison Isola for sharing blood plasma samples of TGS wild type and heterozygous mice. We also value the help of Dr. Kenneth Reuhl for his in-depth evaluation of the liver H&E slides.
Footnotes
The authors declare no potential conflict of interest
REFERENCES
- 1.Siegel V, Adding patient education of skin cancer and sun-protective behaviors to the skin assessment screening on admission to hospitals. Medsurg Nurs, 2012. 21(3): p. 183–4. [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2016. CA: A Cancer Journal for Clinicians, 2016. 66(1): p. 7–30. [DOI] [PubMed] [Google Scholar]
- 3.Sabel MS, Liu Y, and Lubman DM, Proteomics in Melanoma Biomarker Discovery: Great Potential, Many Obstacles. International Journal of Proteomics, 2011. 2011: p. 181890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipp FV, Precision medicine driven by cancer systems biology. Cancer Metastasis Reviews, 2017. 36(1): p. 91–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zecena H, et al. , Systems biology analysis of mitogen activated protein kinase inhibitor resistance in malignant melanoma. BMC Systems Biology, 2018. 12: p. 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DB and Sosman JA, Therapeutic Advances and Treatment Options in Metastatic Melanoma. JAMA Oncol, 2015. 1(3): p. 380–6. [DOI] [PubMed] [Google Scholar]
- 7.Bucheit AD and Davies MA, Emerging insights into resistance to BRAF inhibitors in melanoma. Biochemical Pharmacology, 2014. 87(3): p. 381–389. [DOI] [PubMed] [Google Scholar]
- 8.Poulikakos PI, et al. , RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature, 2011. 480(7377): p. 387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulikakos PI and Rosen N, Mutant BRAF melanomas--dependence and resistance. Cancer Cell, 2011. 19(1): p. 11–5. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, et al. , Development of early melanocytic lesions in transgenic mice predisposed to melanoma. Pigm. Cell Res, 2000. 13: p. 158–164. [DOI] [PubMed] [Google Scholar]
- 11.Pollock PM, et al. , Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia. Nat Genet, 2003. 34(1): p. 108–12. [DOI] [PubMed] [Google Scholar]
- 12.Baenke F, et al. , Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells. Molecular Oncology, 2016. 10(1): p. 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez-Davies JE, et al. , Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. Journal of Translational Medicine, 2015. 13(1): p. 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin SS, et al. , Oncogenic activities of metabotropic glutamate receptor 1 (Grm1) in melanocyte transformation. Pigment Cell Melanoma Res, 2008a. 21(3): p. 368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wangari-Talbot J, et al. , Functional effects of GRM1 suppression in human melanoma cells. Mol Cancer Res, 2012. 10(11): p. 1440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall BA, et al. , Disruption of GRM1-mediated signalling using riluzole results in DNA damage in melanoma cells. Pigment Cell Melanoma Res, 2014. 27(2): p. 263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namkoong J, et al. , Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Research, 2007. 67(5): p. 2298–305. [DOI] [PubMed] [Google Scholar]
- 18.Lyons SA, et al. , Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res, 2007. 67(19): p. 9463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sontheimer H, A role for glutamate in growth and invasion of primary brain tumors. J Neurochem, 2008. 105(2): p. 287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye ZC, Rothstein JD, and Sontheimer H, Compromised glutamate transport in human glioma cells: reduction-mislocalization of sodium-dependent glutamate transporters and enhanced activity of cystine-glutamate exchange. J Neurosci, 1999. 19(24): p. 10767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takano T, et al. , Glutamate release promotes growth of malignant gliomas. Nat Med, 2001. 7(9): p. 1010–5. [DOI] [PubMed] [Google Scholar]
- 22.Xie TX, et al. , Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res, 2006. 66(6): p. 3188–96. [DOI] [PubMed] [Google Scholar]
- 23.Seidlitz EP, et al. , Cancer cell lines release glutamate into the extracellular environment. Clin Exp Metastasis, 2009. 26(7): p. 781–7. [DOI] [PubMed] [Google Scholar]
- 24.Koochekpour S, et al. , Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res, 2012. 18(21): p. 5888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lora J, et al. , Antisense glutaminase inhibition decreases glutathione antioxidant capacity and increases apoptosis in Ehrlich ascitic tumour cells. European Journal of Biochemistry, 2004. 271(21): p. 4298–4306. [DOI] [PubMed] [Google Scholar]
- 26.Cairns RA, Harris IS, and Mak TW, Regulation of cancer cell metabolism. Nat Rev Cancer, 2011. 11(2): p. 85–95. [DOI] [PubMed] [Google Scholar]
- 27.Wang JB, et al. , Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell, 2010. 18(3): p. 207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filipp FV, et al. , Glutamine-fueled mitochondrial metabolism is decoupled from glycolysis in melanoma. Pigment Cell Melanoma Res, 2012. 25(6): p. 732–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross MI, et al. , Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther, 2014. 13(4): p. 890–901. [DOI] [PubMed] [Google Scholar]
- 30.Momcilovic M, et al. , Targeted inhibition of EGFR and glutaminase induces metabolic crisis in EGFR mutant lung cancer. Cell reports, 2017. 18(3): p. 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie C, et al. , Inhibition of mitochondrial glutaminase activity reverses acquired erlotinib resistance in non-small cell lung cancer. Oncotarget, 2016. 7(1): p. 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen-Solal KA, et al. , Progressive appearance of pigmentation in amelanotic melanoma lesions. Pigment Cell Res, 2002. 15(4): p. 282–9. [DOI] [PubMed] [Google Scholar]
- 33.Garraway LA, et al. , Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature, 2005a. 436(7047): p. 117–22. [DOI] [PubMed] [Google Scholar]
- 34.Wen Y, et al. , Activation of the glutamate receptor GRM1 enhances angiogenic signaling to drive melanoma progression. Cancer Res, 2014. 74(9): p. 2499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stepulak A, et al. , NMDA antagonist inhibits the extracellular signal-regulated kinase pathway and suppresses cancer growth. Proc Natl Acad Sci U S A, 2005. 102(43): p. 15605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohtani Y, et al. , Metabotropic glutamate receptor subtype-1 is essential for in vivo growth of melanoma. Oncogene, 2008. 27: p. 7162–7170. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, et al. , Spontaneous melanocytosis in transgenic mice. J. Invest. Dermatol, 1996. 106: p. 1145–1150. [DOI] [PubMed] [Google Scholar]
- 38.Zhu H, et al. , Development of heritable melanoma in transgenic mice. J. Invest. Dermatol, 1998. 110: p. 247–252. [DOI] [PubMed] [Google Scholar]
- 39.Le MN, et al. , The Glutamate Release Inhibitor Riluzole Decreases Migration, Invasion and Proliferation of Melanoma Cells. The Journal of investigative dermatology, 2010. 130(9): p. 2240–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isola AL, et al. , Exosomes released by metabotropic glutamate receptor 1 (GRM1) expressing melanoma cells increase cell migration and invasiveness. Oncotarget, 2018. 9(1): p. 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HJ, et al. , Glutamatergic pathway targeting in melanoma; single agent and combinatorial therapies. Clin Cancer Res, 2011. 17: p. 7080–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spagnolo F, et al. , BRAF-mutant melanoma: treatment approaches, resistance mechanisms, and diagnostic strategies. OncoTargets and therapy, 2015. 8: p. 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nunes LM, et al. , The gender of cell lines matters when screening for novel anti-cancer drugs. Aaps j, 2014. 16(4): p. 872–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolfi SC, et al. , Abstract 4457: Regulation of GRM1 by estrogen receptor in breast cancer. Cancer Research, 2014. 74(19 Supplement): p. 4457–4457. [Google Scholar]
- 45.Filipp FV, et al. , Frontiers in pigment cell and melanoma research. Pigment Cell Melanoma Res, 2018. 31(6): p. 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott DA, et al. , Comparative Metabolic Flux Profiling of Melanoma Cell Lines: BEYOND THE WARBURG EFFECT. The Journal of Biological Chemistry, 2011. 286(49): p. 42626–42634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filipp FV, et al. , Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment cell & melanoma research, 2012. 25(3): p. 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schallier A, et al. , Region- and age-specific changes in glutamate transport in the AbetaPP23 mouse model for Alzheimer’s disease. J Alzheimers Dis, 2011. 24(2): p. 287–300. [DOI] [PubMed] [Google Scholar]
- 49.Krall AS, et al. , Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nature Communications, 2016. 7: p. 11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin CS, et al. , The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat Commun, 2017. 8: p. 15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayers JR and Vander Heiden MG, Famine versus feast: understanding the metabolism of tumors in vivo. Trends in biochemical sciences, 2015. 40(3): p. 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metallo CM, et al. , Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature, 2012. 481(7381): p. 380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masamha CP and LaFontaine P, Molecular targeting of glutaminase sensitizes ovarian cancer cells to chemotherapy. J Cell Biochem, 2018. 119(7): p. 6136–6145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.