Abstract
gp78, also known as the tumor autocrine motility factor receptor, is a transmembrane protein whose expression is correlated with tumor metastasis. We establish that gp78 is a RING finger-dependent ubiquitin protein ligase (E3) of the endoplasmic reticulum (ER). Consistent with this, gp78 specifically recruits MmUBC7, a ubiquitin-conjugating enzyme (E2) implicated in ER-associated degradation (ERAD), through a region distinct from the RING finger. gp78 can target itself for proteasomal degradation in a RING finger- and MmUBC7-dependent manner. Importantly, gp78 can also mediate degradation of CD3-δ, a well-characterized ERAD substrate. In contrast, gp78 lacking an intact RING finger or its multiple membrane-spanning domains stabilizes CD3-δ. gp78 has thus been found to be an example of a mammalian cellular E3 intrinsic to the ER, suggesting a potential link between ubiquitylation, ERAD, and metastasis.
Cytosolic and nuclear proteins are targeted for proteasomal degradation by the addition of multiubiquitin chains. The specificity of this process is largely conferred by ubiquitin (Ub) protein ligases (E3s). E3s interact directly or indirectly with substrate and mediate transfer of Ub from Ub-conjugating enzymes (E2s) to target proteins where isopeptide linkages are formed. Two major E3 classes have been identified. Homologous to E6-AP C terminus (HECT) domain E3s accept Ub from E2, themselves forming thiol-ester intermediates with Ub. RING finger E3s bind E2 and apparently mediate the direct transfer of Ub from E2 to substrate (reviewed in refs. 1–3).
Ubiquitylation also plays essential roles in targeting of proteins for retrotranslocation and proteasomal targeting from the endoplasmic reticulum (ER) by processes collectively known as ER-associated degradation (ERAD). ERAD serves to degrade misfolded or otherwise functionally denatured proteins. Elucidation of its details has important implications for many diseases, including cystic fibrosis, neurodegenerative disorders, α1 antitrypsin deficiency, and tyrosinase deficiency (reviewed in refs. 4–6). Additionally, ERAD has homeostatic functions in regulating hydroxymethylglutaryl-CoA reductase (7) as well as unassembled, but otherwise apparently native, components of multisubunit cell surface receptors, such as the T cell antigen receptor (TCR) CD3-δ subunit (8). Ubiquitylation is an obligate step in ERAD that appears to be required for retrotranslocation to the cytosol and proteasomal degradation (refs. 9–12 and references therein). The details by which retrograde movement and proteasomal targeting occur and the means by which Ub is conjugated to sites on proteins that are not normally exposed to the cytosol remain to be fully understood (4, 13).
Much of what is known about ubiquitylation in ERAD derives from Saccharomyces cerevisiae. Two yeast E2s that associate with the ER, Ubc6p and Ubc7p, play roles in ERAD, with Ubc7p most frequently implicated (refs. 9, 12, and 14, and references therein). Ubc6p has a C-terminal hydrophobic anchor that localizes it to the ER membrane (14). Ubc7p has no intrinsic characteristics that predict membrane association. The recruitment of Ubc7p to the ER is instead accomplished by association with Cue1p, a small N-terminal anchored ER protein (15). A single yeast ER resident E3 implicated in ERAD, Hrd1p or Der3p, has been identified. This E3 has the capacity to function with Ubc7p (7, 9, 16, 17). The substrates targeted for degradation by this E3 are varied in structure, and there is little evidence of direct E3–substrate binding.
Murine orthologs of Ubc6p and Ubc7p (MmUBC6 and MmUBC7) have been characterized and are highly conserved relative to counterparts in other mammals (11, 18, 19). Of these MmUBC7, but not MmUBC6, is implicated in degradation of unassembled TCR subunits (11). No mammalian ERAD E3 analogous to yeast Hrd1p/Der3p has been characterized, nor has the existence of a mammalian Cue1p homolog been established.
gp78 was originally isolated as a membrane glycoprotein from murine melanoma cells and was implicated in cell migration (20). Subsequently, gp78 was identified as the tumor autocrine motility factor receptor mediating tumor invasion and metastasis (21). The message encoding gp78 has recently been shown to be widely expressed in mouse tissues, and perusal of expressed sequence tag databases suggests that this is similarly true for both normal and diseased human tissues (ref. 22 and S.F. and A.M.W., unpublished data). By using a monoclonal antibody, gp78 levels were found to be increased in a number of different human malignancies, with this correlating with metastatic potential. gp78 has been shown to be expressed on the cell surface and to exhibit colocalization with caveolin when endocytosis is arrested, with evidence for internalization and transport to the ER in a manner similar to simian virus 40 (23). Other studies suggest a substantial smooth ER distribution and association with structures that have been referred to as autocrine motility factor receptor tubules (23, 24).
Recently, the full-length cDNA for gp78 has been isolated and found to predict a 643-aa protein with at least five membrane-spanning domains (22). Notably, the region C-terminal to the last transmembrane domain includes a RING finger consensus sequence (22). We demonstrate that gp78, which we find to be largely localized to the ER, has intrinsic RING finger-dependent E3 activity. In this way it can target itself and a heterologous ERAD substrate, CD3-δ, for proteasomal degradation. Moreover, gp78 binds MmUBC7 independent of the RING finger through a region that exhibits homology to yeast Cue1p. Thus, this example of a mammalian ER membrane E3 also represents convergent evolution where functions of a yeast ERAD E3 and a docking protein are incorporated in a single human polypeptide.
Methods
cDNA Cloning and Expression of Recombinant Proteins.
To construct pGEX-gp78C and pFLAG-CMV6c-gp78C, cDNA for a human gp78 fragment corresponding to amino acids 309–643 (gp78C) was amplified from human peripheral blood leukocytes by reverse transcription–PCR (SuperScript First-Strand Synthesis System for RT-PCR, Life Technologies), and cloned into pGEX4T2 (Amersham Pharmacia) or pFLAG-CMV6c (Sigma) through EcoRI and SalI sites. pCIneo-gp78 was generated by using the full-length cDNA of human gp78 in pCR2.1 (a generous gift from Jun Yokota) by using an XbaI site. To create a plasmid encoding a gp78-GFP (green fluorescent protein) and gp78ΔTail-GFP fusion proteins, cDNAs encoding full-length gp78 and gp78 amino acids 1–308 were subcloned into pEGFP N1 (CLONTECH). Plasmid encoding glutathione S-transferase (GST)-Itch was generated by subcloning of murine Itch cDNA (a gift from Neal G. Copeland) into pGEX-KG (a gift from Brian Druker). cDNA for yeast Hrd1/Der3 corresponding to amino acids 205–551 was amplified from genomic DNA by PCR and cloned into pGEX-KG by using NcoI and XhoI sites to express GST-Hrd1p/Der3pC. The following plasmids have been previously reported: pGEX-XIAP, -CIAP2, -mdm2, -AO7T, -MmUBC6, -MmUBC7, pET15b-UbcH5B, pCI-HA-MmUBC6, and pcDNA3-myc-MmUBC7 (25–28). All mutations were generated by using the QuickChange site-directed mutagenesis kit (Stratagene). Sequences of primers used for mutagenesis are available from A.M.W. on request. Recombinant protein expression and purification were previously published (25–28).
Antibodies.
Polyclonal anti-gp78 peptide (amino acids 505–529) was generated in rabbit and affinity purified. Monoclonal anti-influenza hemagglutinin (HA), anti-Myc, and anti-GFP were from Sigma; anti-Ub was from Santa Cruz Biotechnology, and anti-calnexin was from Transduction Laboratories (Lexington, KY).
Immunofluorescence and Confocal Microscopy.
For microscopy, U2OS cells were cultured on coverslips in six-well dishes and transfected with plasmids by using Lipofectamine 2000 (Invitrogen). The double staining was done by using anti-gp78 and anti-Myc or anti-HA, and confocal microscopy was performed and data were processed as described (11).
In Vitro Binding Assay.
[35S]Methionine-labeled MmUBC7 was generated by using reticulocyte lysate-based TnT Quick coupled transcription and translation system (Promega). GST-gp78C and truncated forms were purified on glutathione-Sepharose beads. Twenty picomoles of each protein was incubated with 6 μl of [35S]MmUBC7 in binding buffer (20 mM Tris⋅HCl, pH 8.0, containing 150 mM NaCl, 0.5% Triton X-100, and 2 mg/ml BSA) for 2 h at 4°C. Beads were washed three times with binding buffer without BSA and processed for analysis as described (26).
Immunoprecipitation and Immunoblotting.
Indicated plasmids were transiently transfected in U2OS or HEK 293T cells. pEGFP N1 plasmid was cotransfected as a measure of transfection efficiency. Cells were harvested 24–30 h after transfection and lysed in RIPA buffer containing 1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 10 mg/ml phenylmethylsulfonyl fluoride. For immunoprecipitation (IP), lysates were incubated with 1 μg of Ab and 40 μl of protein A beads (Zymed) for 4 h at 4°C, beads were washed five times in buffer containing 50 mM Tris⋅HCl (pH 7.5), 0.1 M NaCl, and 0.5% Triton X-100 before processing for immunoblotting as described (27). Subcellular fractionation was done essentially as reported (29). In brief, HEK 293T cells were transfected as indicated and homogenized in 50 mM Tris⋅HCl (pH 8.0) containing 1 mM 2mercaptoethanol, 1 mM EDTA, 0.32 M sucrose, and 0.1 mM phenylmethylsulfonyl fluoride by passing through a 27-gauge syringe 20 times. Lysates were then centrifuged at 9,000 × g. Microsomes and cytosol in the supernatant were separated by centrifugation at 105,000 × g for 60 min. The fractions were further processed for immunoblotting. For cycloheximide chase experiments, 24 h after transfection HEK 293T cells were divided into thirds and incubated for the indicated times with 50 μg/ml cycloheximide. Cells were then harvested and lysed in RIPA buffer followed by SDS/PAGE and immunoblotting. For stable transfection, the indicated plasmid was transfected by calcium phosphate precipitation and selected by using 0.5 mg/ml G418 (GIBCO).
In Vitro and in Vivo Ubiquitylation.
In vitro ubiquitylation assays were performed as described (27). UbcH5B was expressed in Escherichia coli as described (25). MmUBC7 was expressed as a GST fusion protein from pGEX-KG, purified on glutathione Sepharose, and cleaved from GST by using a built-in thrombin cleavage site. For detection of ubiquitylated CD3-δ, cells were transfected as indicated and after 24 h lysed in RIPA buffer containing 10 mM iodoacetamide. Immunoprecipitates from cell lysates were processed for immunoblotting with anti-Ub followed by reprobing with anti-HA to detect CD3-δ.
Results
ER Localization of gp78 and Colocalization with ER E2s.
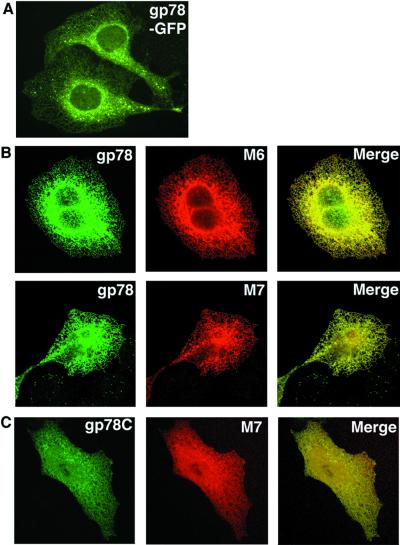
To evaluate the subcellular localization of gp78, GFP-tagged gp78 was generated. As is evident (Fig. 1A), this protein delineates primarily a lacy pattern typical of the ER. To evaluate this further, gp78 was coexpressed with two different E2s that associate with the ER membrane. MmUBC6 is a C-terminal-anchored, or type IV, membrane protein that becomes incorporated into the ER membrane posttranslationally. MmUBC7 is a nontransmembrane protein that, when transfected into cells, distributes between the ER membrane, cytosol, and the nucleus and exhibits significant ER colocalization with MmUBC6 (11). gp78 exhibits substantial colocalization with both of these E2s, consistent with an ER distribution (Fig. 1B). Because gp78 is a transmembrane protein, we reasoned that expression of a form lacking transmembrane domains (gp78C: amino acid 309–643) would alter its distribution. When coexpressed with MmUBC7, substantial colocalization persisted, although the patterns for both proteins were more diffuse than those observed when full-length gp78 was coexpressed with MmUBC7 (compare Fig. 1C to 1B Lower).
Figure 1.
Subcellular localization of gp78. (A) U2OS cells stably expressing gp78 with GFP at its C terminus (gp78-GFP) were assessed by fluorescence microscopy. (B) U2OS transiently expressing plasmids encoding gp78 and either HA-MmUBC6 (M6) or myc-MmUBC7 (M7) were assessed for colocalization by confocal microscopy after double staining with anti-gp78 and either anti-Myc or anti-HA. (C) U2OS cells transiently expressing gp78C and MmUBC7 (M7) were assessed for colocalization by confocal microscopy as in B.
In Vivo Association of gp78 and MmUBC7.
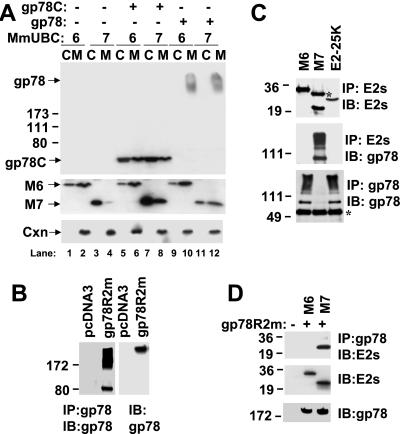
To evaluate interactions of gp78 with MmUBC6 and MmUBC7, cells transfected with plasmid encoding MmUBC6 or MmUBC7 alone or in combination with either full-length gp78 or gp78C were fractionated into cytosolic and membrane fractions (Fig. 2A). Without gp78 (lanes 1–4) MmUBC7 was found largely in the cytosolic fraction, whereas MmUBC6 was distributed between the membrane and cytosolic fractions, in accord with the posttranslational ER insertion of this protein. Wild-type (wt) gp78 (lanes 9–12), which often migrates as a high molecular weight smear because of interactions involving its membrane-spanning domains (Fig. 2B), was exclusively membrane associated, consistent with its being a polytopic membrane protein. Coexpression of gp78 did not alter distribution of MmUBC6. However, it did result in a marked recruitment of MmUBC7 from the cytosol to membrane fraction (compare lanes 3 and 4 to lanes 11 and 12). Notably, and in accord with the pattern seen on immunofluorescence, gp78C (lanes 5–8) was found membrane associated as well as in cytosolic fractions despite removal of its hydrophobic domains. Although gp78C affected neither the level nor the expression of MmUBC6, it resulted in accumulation of MmUBC7 in both membrane and cytosolic fractions (lanes 7 and 8), suggesting a functionally significant interaction between gp78 and MmUBC7.
Figure 2.
gp78 and MmUBC7 physically interact in HEK 293T cells. (A) Cells transfected with plasmids encoding gp78 or gp78C in combination with HA-MmUBC6 or Myc-MmUBC7 were fractionated into cytosolic (C) and microsomal (M) fractions and after SDS/PAGE evaluated for expression of the indicated proteins by immunoblotting. MmUBC6 (M6) and MmUBC7 (M7) were detected by using Abs against HA or Myc. Numbers on the left indicate apparent molecular weight × 10−3. Cxn, calnexin. (B) Cells were transfected as indicated. In the left two lanes, cell lysates were subject to immunoprecipitation with anti-gp78; in the right two lanes lysates from the same cells were directly resolved on SDS/PAGE. Immunoblotting (IB) was with anti-gp78. Relative amounts of gp78 migrating at its predicted molecular weight, as compared to high molecular weight complexes, is variable with IP. With direct resolution of lysates, gp78 migrates almost exclusively as a high molecular weight form. (C) Lysates from cells transfected with plasmid encoding gp78 with a mutation in a critical RING finger Cys (gp78C356G) and the indicated E2s were subjected to IP with either anti-HA (MmUBC6 and E2–25K) or anti-Myc (MmUBC7) and IB with Abs against either the epitope tags (Top) or anti-gp78 (Middle); *, IgG light chain. The same lysates were subsequently subjected to re-immunoprecipitation with anti-gp78 followed by IB, also with anti-gp78 (Bottom); *, IgG heavy chain; M6, MmUBC6; M7, MmUBC7. (D) Cells were transfected as indicated. gp78R2m is a mutant of gp78 in which predicted coordinating residues for both putative zinc binding sites have been mutated. (Top) IP with anti-gp78 followed by IB with mixture of anti-HA and anti-Myc to detect E2s. (Middle and Bottom) Immunoblots with indicated Abs of whole-cell lysates representing one-fourth of the material used for the IP presented in Top.
To further evaluate gp78 E2 interactions, a RING mutant of gp78 was coexpressed with epitope-tagged forms of either MmUBC7 or MmUBC6, or a third mammalian E2, E2–25K (48). When subjected to immunoprecipitation with Abs that recognize the N-terminal tags on these E2s, only MmUBC7 coimmunoprecipitated gp78 (Fig. 2C Top and Middle). The association between the two proteins is underscored by the depletion of gp78 from cell lysates by immunoprecipitation of MmUBC7 (Fig. 2C Bottom). It is important to note that the gp78 used was not wt, but a mutant of a crucial Cys within the RING finger, suggesting that the gp78 RING finger is dispensable for physical interactions with MmUBC7. Similar results were obtained with wt gp78 (data not shown). This RING-independent association was corroborated by expression of a double mutant of gp78 in which residues predicted to participate in the coordination of both RING finger zinc ions were mutated (gp78R2m). Immunoprecipitation of transfected gp78R2m resulted in the coimmunoprecipitation of cotransfected MmUBC7 but not MmUBC6 (Fig. 2D Top). These observations strongly suggest that despite the known importance of the RING in interactions with E2s, the interaction of MmUBC7 with the C-terminal tail of gp78 does not require an intact RING finger.
In Vitro Binding of gp78 to MmUBC7.
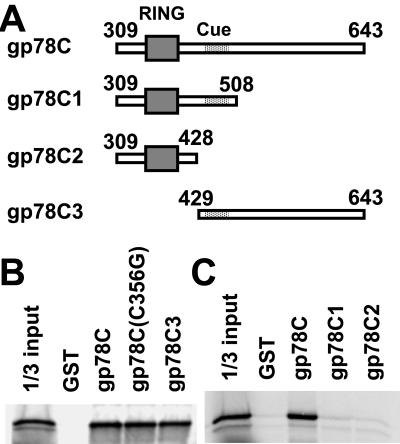
To directly evaluate interactions between gp78 and MmUBC7, GST fusions of regions of the gp78 cytoplasmic tail (Fig. 3A) were assessed for binding MmUBC7. Substantial binding was exhibited by the entire cytoplasmic tail whether the RING finger was intact or mutated [gp78C and gp78C (C356G), respectively]. In contrast, a fusion protein that included little more than the RING finger (gp78C2) exhibited no binding (Fig. 3 B and C). Ponting (30) has recently described the “Cue domain” as a ≈42-aa region of homology to yeast Cue1p found in a number of proteins, including gp78. However, a GST fusion that included both the RING finger and the Cue domain (amino acids 456–497 of gp78) was insufficient for detectable MmUBC7 binding (GST-gp78C1). On the other hand, a fusion protein that excluded the RING finger but included the entire region from amino acid 429 to the C terminus (gp78C3) bound MmUBC7 to a degree comparable to gp78C. Thus, the major site of physical interaction of gp78 with MmUBC7 is the region distal to the RING finger.
Figure 3.
In vitro analysis of gp78 interaction with MmUBC7. (A) Schematic representation of the cytoplasmic domain of gp78 (gp78C) and its truncated forms expressed as C-terminal fusions with GST and used for binding in B and C. The RING finger (amino acids 341–378) is boxed. The Cue domain (30) (amino acids 456–497) is shaded. (B and C) The indicated bead-bound GST fusions were evaluated for binding to 35S-labeled in vitro translated MmUBC7. Left lanes represent one-third of [35S]MmUBC7 used for each binding reaction.
gp78 Mediates Ubiquitylation.
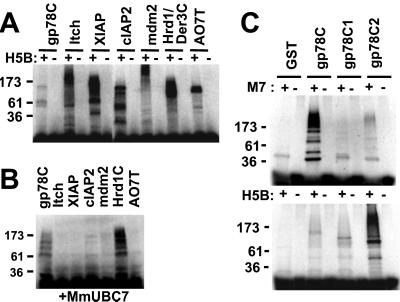
Many RING finger proteins, including some that contain little more than the RING finger, function with the core E2 UbcH5B in an in vitro ubiquitylation assay. In this assay E3s are expressed as GST fusions and serve as the primary targets for E2-mediated ubiquitylation. gp78C was assessed side by side with other recombinant E3s for ubiquitylation with UbcH5B (Fig. 4A). These E3s included a HECT (homologous to E6-AP C terminus) domain E3 (Itch) (31, 32); four other mammalian RING finger proteins; the cytoplasmic tail of the yeast ERAD E3, Hrd1p/Der3p; and gp78C. All of these proteins exhibited E2-dependent ubiquitylation although among these gp78 was by far the weakest (Fig. 4A). Strikingly, however, when MmUBC7 rather than UbcH5B was used as an E2, substantial activity was seen only with gp78 and with Hrd1p/Der3p, which has been shown to function with yeast Ubc7p (Fig. 4B) (16, 17). Neither gp78 nor Hrd1p/Der3p exhibited detectable activity with MmUBC6 (data not shown). For Hrd1p/Der3p, this is consistent with previous observations with Ubc6p.
Figure 4.
In vitro ubiquitylation of gp78. (A) In vitro ubiquitylation in the presence of E1, UbcH5B (H5B), 32P-labeled Ub, and 20 pmol of the indicated GST fusion proteins performed as described. (B) In vitro ubiquitylation using the same GST fusion proteins as in A, but MmUBC7 was used instead of UbcH5B. (C) gp78C and its truncated mutants as presented in Fig. 3A were assayed for ubiquitylation in the presence of MmUBC7 (M7) (Upper) or UbcH5B (H5B) (Lower).
Fusion proteins containing various portions of the gp78C-terminal domain were next evaluated for ubiquitylation (Fig. 4C). When compared to gp78C, truncations from the C terminus that failed to bind MmUBC7 exhibited a marked diminution in MmUBC7-mediated ubiquitylation (GST-gp78C1 and GST-gp78C2). The significance of this is underscored by the fact that with UbcH5B as the E2 (Fig. 4C Lower), gp78 ubiquitylation actually increased with elimination of most of the gp78 cytoplasmic domain (GST-gp78C2). Thus, although the gp78 RING is adequate for ubiquitylation mediated by UbcH5B, the C-terminal site responsible for binding the mammalian ERAD MmUBC7 markedly enhances ubiquitylation with this ERAD E2.
gp78 Is a RING Finger-Dependent ERAD Substrate.
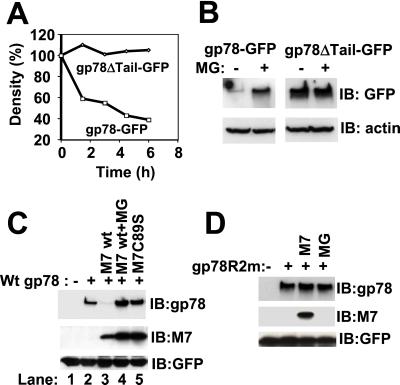
To determine whether gp78 is itself an ERAD substrate, the stability of GFP fusion of the wt (gp78-GFP) and a form of gp78 lacking the C-terminal tail (gp78ΔTail-GFP) were evaluated by pulse–chase metabolic labeling (Fig. 5A). Although the tail-less form was stable, the full-length gp78 was degraded with a half-life of ≈3 h. Degradation of wt gp78 was proteasome-dependent, as treatment with a peptide aldehyde proteasome inhibitor, MG132, markedly increased the steady-state levels of the wt but not the tail-less form (Fig. 5B).
Figure 5.
RING-, proteasome-, and MmUBC7-dependent degradation of gp78. (A) Cells stably expressing GFP-tagged wt gp78 (gp78-GFP) or GFP-tagged gp78 lacking the C-terminal tail (gp78ΔTail-GFP) were pulse labeled with [35S]methionine for 30 min followed by removal of unincorporated [35S]methionine. gp78 was then immunoprecipitated by using anti-GFP, and, after resolution on SDS/PAGE, gp78 was quantified and plotted as a function of the amount at the initiation of the chase. (B) Cells stably expressing gp78-GFP or gp78ΔTail-GFP were treated with or without MG132 (MG) for 6 h followed by immunoblotting with anti-GFP; β-actin was used as a gel loading control. (C and D) Lysates from cells transfected as indicated were processed for immunoblotting for gp78, MmUBC7 (M7), and cotransfected GFP (transfection efficiency control). Cells exposed to MG132 (MG) were treated with 20 μM for 6 h.
To determine whether MmUBC7 plays a role in the degradation of gp78, combinations of wt and inactive gp78 and MmUBC7 were coexpressed. MmUBC7 markedly decreased gp78 (Fig. 5C, compare lanes 2 and 3); however gp78 expression was efficiently recovered with MG132 (Fig. 5C, lane 4). In contrast, coexpression of catalytically inactive MmUBC7 (MmUBC7C89S) resulted in an increase in gp78, possibly by competing with binding of endogenous MmUBC7 (Fig. 5C, lane 5). Neither MmUBC7 nor MG132 altered the level of RING finger mutant gp78 (Fig. 5D). The proteasome-dependent degradation of wt gp78 was confirmed with the specific proteasome inhibitor lactacystin (data not shown). Thus, gp78 has the potential to target itself for proteasomal degradation in cells in a manner that is dependent both on its RING finger and on interactions with MmUBC7.
gp78 Targets CD3-δ for ERAD.
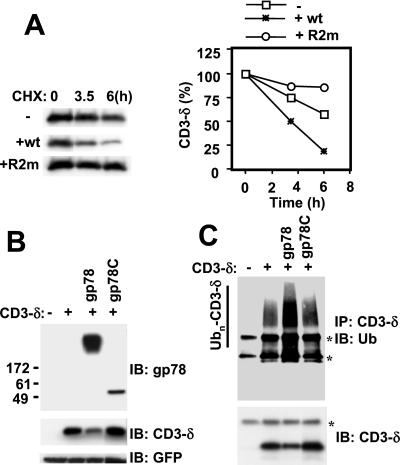
Because gp78 exhibits substantial ER localization, binds to and functions with an E2 specifically implicated in ERAD, and mediates its own degradation from the ER, this raises the question of whether it also functions to mediate degradation of a known ERAD substrate. To investigate this possibility, we examined the effect of gp78 on the TCR subunit CD3-δ. When not assembled with other receptor subunits, CD3-δ is retained in an apparently native conformation in the ER, where it is ubiquitylated and degraded (8). This degradation is proteasome-dependent and delayed by over-expression of inactive MmUBC7 (8, 11). As is evident (Fig. 6A), coexpression of wt gp78 enhanced the rate of loss of HA-tagged CD3-δ when new protein synthesis was inhibited by cycloheximide. In contrast, RING finger mutated gp78 (gp78R2m) dramatically stabilized CD3-δ. To evaluate this further, gp78C was compared to wt gp78 for effects on steady-state CD3-δ in HEK 293T cells (Fig. 6B) and in U2OS cells (Fig. 6C). In both, wt gp78 decreased CD3-δ. This decrease correlated with an increase in ubiquitylated species immunoprecipitated (Fig. 6C). In contrast, expression of gp78C, which lacks transmembrane domains, increased the levels of CD3-δ in both cell lines (Fig. 6 B and C). Collectively these observations establish that gp78 has the potential to function as an ERAD E3 for CD3-δ and that this activity is dependent on an intact RING finger and the presence of its multiple membrane-spanning domains.
Figure 6.
gp78-mediated ERAD of CD3-δ. (A) HEK 293T cells transfected with plasmid encoding HA-CD3-δ and either full-length wt gp78 or gp78R2m were treated with cycloheximide (CHX), 50 μg/ml, for the indicated times followed by evaluation of cell lysates by IB with anti-HA to detect HA-tagged CD3-δ. Bands were quantified by densitometry and plotted as a function of the amount of material when CHX was added (Right). (B) HEK 293T cells were transfected as indicated with plasmids encoding HA-CD3-δ, full-length wt gp78, and the C-terminal tail of gp78 (gp78C). After transfection (24 h), cell lysates were processed for IB with anti-gp78, anti-HA, and anti-GFP (transfection efficiency control). (C) U2OS cells transfected as in B were subject to IP with anti-HA followed by IB first with either anti-Ub (Upper) followed by reprobing of the membrane with anti-HA (Lower). *, IgG heavy chain (Upper and Lower) and light chain (Upper).
Discussion
This study demonstrates that gp78 is a RING finger-dependent Ub protein ligase that localizes primarily to the ER. This protein has the capacity to target itself and a well-characterized ERAD substrate, CD3-δ, for proteasomal degradation. gp78 is the only example to date of a mammalian E3 of the ER, and no other mammalian transmembrane receptor with in vivo E3 activity has been demonstrated. Moreover, this protein specifically interacts with an ERAD E2 through interactions involving a region C-terminal to the RING finger. Identification of gp78 as an ERAD E3 should provide a valuable tool to help understand the details of ERAD in mammalian cells. Given our observations, it now also becomes important to understand how the E3 activity of gp78 influences the metastatic potential of tumors.
Two cytoplasmic E3s, CHIP and Parkin, that also play roles in the degradation of nonmembrane proteins, have recently been shown to function in targeting specific cellular proteins for ERAD (33–36). CHIP is a U-box protein that interacts with Hsc70 and facilitates ubiquitylation of unfolded cystic fibrosis transmembrane conductance regulator as well as the glucocorticoid receptor (33, 34, 37). Among the several identified substrates for Parkin is Pael, a polytopic membrane protein that, when unfolded, is a Parkin binding partner and substrate (35). gp78 is distinguished from CHIP and Parkin in being an integral membrane protein of the ER. In this regard it most closely resembles the yeast ERAD E3 Hrd1p/Der3p with which it shares a clear preference for an E2 implicated in ERAD (16, 17). However, despite the common features their overall identity is only ≈15%, much of which is in the hydrophobic membrane-spanning domains. The RING fingers of the two proteins are similarly disparate. It will now be of interest to determine whether, despite dissimilarities, the two proteins are capable of targeting common substrates for degradation
Although gp78 enhances CD3-δ degradation and altered forms substantially delay this process, how this occurs remains to be determined. CD3-δ is distinguished from other ERAD substrates such as the cystic fibrosis transmembrane conductance regulator and TCR-α in retaining an apparently native conformation when proteasome activity is inhibited (8, 38–41). Targeting of CD3-δ for ERAD may represent a stochastic process reflective of its inability to exit the ER when not assembled as part of the multisubunit TCR. Whether the ERAD function of gp78 is restricted to itself and other substrates that are similar to CD3-δ or whether it plays a more general role in the degradation of unfolded ER proteins awaits determination.
In considering the molecular basis for the physical interaction between MmUBC7 and gp78, it is notable that this E3 includes a 42-aa region (amino acids 456–497) that has been identified by sequence analysis as being a Cue domain consensus (30). Although Cue1p, a 203-aa protein, is implicated in binding yeast Ubc7p (15), the Cue domain region of gp78 is insufficient for MmUBC7 binding, and a RING-containing construct including this area shows minimal activity with MmUBC7 despite robust activity with UbcH5B. However, when the entire region distal to the RING finger, which efficiently binds MmUBC7 and enhances activity, is aligned with Cue1p, there is 20% identity and 28% similarity over 198 aa (from 415 to 612). This suggests that interactions between this region of gp78 and MmUBC7 may be similar to those that mediate binding of Cue1p to Ubc7p. It would appear, therefore, that gp78 represents an example of convergent evolution with functions of both yeast Hrd1p/Der3p and Cue1p found within a single molecule. A notable feature of yeast Ubc7p is a 13-aa insertion within its core domain that is also found in MmUBC7. Based on the Ubc7p structure and that of the c-Cbl RING with another E2, this insertion is predicted to have a significant negative impact on physical interactions with RING fingers (3, 42, 43). Thus, one possible role for the C-terminal tail of gp78 is to increase the overall avidity of the E2–E3 interaction by providing a high-affinity binding site for MmUBC7.
gp78 was identified as the tumor autocrine motility factor (AMF) receptor with its elevated expression correlating with metastasis. Although purported to be a G protein-coupled receptor, little is actually known about its mechanism of action. Recently AMF has been shown to stimulate endothelial motility and to function as an angiogenic factor. Additionally, in endothelial cells gp78 exhibits ligand-dependent cell surface expression, suggesting that AMF plays an important role in regulating trafficking of this receptor (44). Other ER proteins also have the capacity to be expressed on the cell surface and function as receptors, as exemplified by calreticulin, which binds fibrinogen and in this way may mediate cell adhesion or promote mitogenesis (45–47). For gp78, the relationship between its E3 activity, either constitutively or in response to ligand, its subcellular distribution, and signaling properties now warrants evaluation. A particularly intriguing question for future study is whether the role of gp78 in promoting migration and metastasis is related to targeting of itself and other cellular proteins for ubiquitylation. Should this be the case, it presents the opportunity for therapeutic interventions aimed at the E3 activity or E2 interactions of this E3.
Acknowledgments
We thank Dr. Yili Yang for help in cloning gp78 cDNA, J. McNally and T. Karpova of the National Cancer Institute Core Fluorescence Imaging Facility for providing acess and advice, and Kevin Lorick and Alessandra Magnifico for helpful discussions. M.F. is a Howard Hughes Medical Institute-National Institutes of Health Research Scholar.
Abbreviations
- ER
endoplasmic reticulum
- HA
hemagglutinin
- TCR
T cell antigen receptor
- Ub
ubiquitin
- GST
glutathione S-transferase
- ERAD
ER-associated degradation
- wt
wild type
- GFP
green fluorescent protein
- IP
immunoprecipitation
- IB
immunoblotting
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Joazeiro C A, Weissman A M. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 3.Weissman A M. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino J S, Weissman A M. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plemper R K, Wolf D H. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- 6.Brodsky J L, McCracken A A. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 7.Hampton R Y, Gardner R G, Rine J. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang M, Omura S, Bonifacino J S, Weissman A M. J Exp Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordallo J, Plemper R K, Finger A, Wolf D H. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Kopito R R. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 11.Tiwari S, Weissman A M. J Biol Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- 12.Gardner R G, Swarbrick G M, Bays N W, Cronin S R, Wilhovsky S, Seelig L, Kim C, Hampton R Y. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClellan A J, Frydman J. Nat Cell Biol. 2001;3:E51–E53. doi: 10.1038/35055162. [DOI] [PubMed] [Google Scholar]
- 14.Sommer T, Jentsch S. Nature (London) 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 15.Biederer T, Volkwein C, Sommer T. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 16.Bays N W, Gardner R G, Seelig L P, Joazeiro C A, Hampton R Y. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 17.Deak P M, Wolf D H. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 18.Katsanis N, Fisher E M. Genomics. 1998;51:128–131. doi: 10.1006/geno.1998.5263. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, Wing S S. J Biol Chem. 1999;274:14685–14691. doi: 10.1074/jbc.274.21.14685. [DOI] [PubMed] [Google Scholar]
- 20.Nabi I R, Raz A. Int J Cancer. 1987;40:396–402. doi: 10.1002/ijc.2910400319. [DOI] [PubMed] [Google Scholar]
- 21.Nabi I R, Watanabe H, Silletti S, Raz A. EXS. 1991;59:163–177. doi: 10.1007/978-3-0348-7494-6_11. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu K, Tani M, Watanabe H, Nagamachi Y, Niinaka Y, Shiroishi T, Ohwada S, Raz A, Yokota J. FEBS Lett. 1999;456:295–300. doi: 10.1016/s0014-5793(99)00966-7. [DOI] [PubMed] [Google Scholar]
- 23.Benlimame N, Le P U, Nabi I R. Mol Biol Cell. 1998;9:1773–1786. doi: 10.1091/mbc.9.7.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H J, Benlimame N, Nabi I. J Cell Sci. 1997;110:3043–3053. doi: 10.1242/jcs.110.24.3043. [DOI] [PubMed] [Google Scholar]
- 25.Jensen J P, Bates P W, Yang M, Vierstra R D, Weissman A M. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 26.Lorick K L, Jensen J P, Fang S, Ong A M, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Fang S, Jensen J P, Weissman A M, Ashwell J D. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner B A, Yuan J. Nature (London) 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 30.Ponting C P. Biochem J. 2000;351:527–535. [PMC free article] [PubMed] [Google Scholar]
- 31.Perry W L, Hustad C M, Swing D A, O'Sullivan T N, Jenkins N A, Copeland N G. Nat Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 32.Qiu L, Joazeiro C, Fang N, Wang H Y, Elly C, Altman Y, Fang D, Hunter T, Liu Y C. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 33.Connell P, Ballinger C A, Jiang J, Wu Y, Thompson L J, Hohfeld J, Patterson C. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 34.Meacham G C, Patterson C, Zhang W, Younger J M, Cyr D M. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 35.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 36.Shimura H, Schlossmacher M G, Hattori N, Frosch M P, Trockenbacher A, Schneider R, Mizuno Y, Kosik K S, Selkoe D J. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 37.Ballinger C A, Connell P, Wu Y, Hu Z, Thompson L J, Yin L Y, Patterson C. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 39.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 40.Yu H, Kaung G, Kobayashi S, Kopito R R. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 41.Huppa J B, Ploegh H L. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 42.Cook W J, Martin P D, Edwards B F, Yamazaki R K, Chau V. Biochemistry. 1997;36:1621–1627. doi: 10.1021/bi962639e. [DOI] [PubMed] [Google Scholar]
- 43.Zheng N, Wang P, Jeffrey P D, Pavletich N P. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 44.Funasaka T, Haga A, Raz A, Nagase H. Biochem Biophys Res Commun. 2001;285:118–128. doi: 10.1006/bbrc.2001.5135. [DOI] [PubMed] [Google Scholar]
- 45.Gray A J, Park P W, Broekelmann T J, Laurent G J, Reeves J T, Stenmark K R, Mecham R P. J Biol Chem. 1995;270:26602–26606. doi: 10.1074/jbc.270.44.26602. [DOI] [PubMed] [Google Scholar]
- 46.Xiao G, Chung T F, Fine R E, Johnson R J. J Neurosci Res. 1999;58:652–662. doi: 10.1002/(sici)1097-4547(19991201)58:5<652::aid-jnr6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 47.Xiao G, Chung T F, Pyun H Y, Fine R E, Johnson R J. Brain Res Mol Brain Res. 1999;72:121–128. doi: 10.1016/s0169-328x(99)00188-6. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z J, Niles E G, Pickart C M. J Biol Chem. 1991;266:15698–15704. [PubMed] [Google Scholar]