Abstract
Robust and responsive, the surface of a cell is as important as its interior when it comes to mechanically regulating form and function. New techniques are shedding light on this role, and a common language to describe its properties is now needed.
Cells are now broadly appreciated to be mechanical as well as biochemical systems. They generate, transmit and respond to forces through an intricate network of mechanical components, resulting in cell movement and shape change, as well as altered signalling, modulated expression and even genomic damage. Contributions to cell mechanics from molecular motors, cytoskeletal filaments and mechanosensitive proteins have received significant attention, and the cell surface — comprising the plasma membrane and underlying cortical cytoskeleton — has emerged as a unique mechanical system capable of exerting both local and global control of cell form and function. The physical properties of the cell surface can be rapidly modulated, enabling cells to generate or accommodate changes in shape. This Perspective examines the role that cell surface mechanics plays across multiple length scales and in multiple cell types and surveys the experimental techniques that are providing new physical insight into the powerful mechanical border of cells.
Cell surface mechanics during crawling motility
The importance of cell surface mechanics is clear from the coordinated shape changes involved in crawling motility. Through extensive biochemical and genetic studies, we know many of the key cytoskeletal players in cell movement1,2. In the simplest description, assembly of actin monomers into filaments at the leading edge generates sufficient local force to displace the cell’s plasma membrane at the leading edge3–5 and motor-based contractility retracts the trailing edge at the back of the cell to generate forward motion6.
Since the plasma membrane is a deformable barrier, growth of actin filaments oriented towards the membrane during cell spreading4 and cell polarization7 can cause an increase in plasma membrane tension as the force of polymerization unfolds wrinkles in the membrane. During cell polarization this increase in plasma membrane tension acts to constrain the spread of the existing leading edge and prevents the formation of secondary fronts in chemotactic cells such as neutrophils7, providing a possible mechanism for constraining the overall size of protrusions. One way that changes in tension could regulate actin assembly at the leading edge size is purely mechanical. In support of this, a model consisting entirely of mechanical interactions between the actin cytoskeleton, myosin and the plasma membrane was found to be sufficient to predict the polarized morphologies of keratocytes8, as well as the relation between cell shape and speed5.
How else could increasing plasma membrane tension limit the expansion of the existing leading edge and prevent formation of secondary fronts? And specifically, could biochemical signalling mediated by cell surface mechanics contribute to leading edge size regulation? This question motivated a collaboration between biologists and physicists that brought together cell biology, mechanical measurements and modelling to better understand regulation of neutrophil motility. We imaged actin filament nucleation in the leading edge of cells and directly quantified plasma membrane tension with an atomic force microscope. Using a series of knock down, chemical and mechanical perturbations, we found that elevated membrane tension limits actin polymerization through phospholipase D2 (PLD2) and the mammalian target of rapamycin complex 2 (mTORC2) by an as yet uncharacterized mechanism. In the absence of this pathway, neutrophils exhibit larger leading edges, higher membrane tension and profoundly defective chemotaxis. Moreover, mathematical modelling indicated that this feedback circuit is a favourable topology to enable competition between protrusions during neutrophil polarization9. Our work, together with other recent findings10,11, highlight how biochemical signals, membrane tension and the actin cytoskeleton can collaborate to generate large-scale cellular organization.
In this Perspective, we give an overview of cell surface mechanics and its role in cellular form and function. We first discuss the composition and dynamics of the cell membrane and its underlying cortex and how these determine cell surface mechanics. We then summarize current methods to measure cell surface physical properties and clarify the terminology surrounding cell surface mechanics. Finally, we discuss future directions for research on cell surface mechanics, including how physical models could contribute to our understanding of cellular mechanics.
Molecular characterization of the cell surface
The field of mechanobiology has advanced dramatically since the early twentieth century, when D’Arcy Thompson published On Growth and Form and described how physical forces contribute to determining the size and shape of living organisms (reviewed in ref. 12). As Thompson appreciated a century ago, cell shape is the result not only of internal forces driving growth and movement but also of finely tuned cell surface mechanics. Living organisms mould the form and function of their cells and tissues by regulating the deformability and displacement of cell surfaces, achieving control of shape at multiple length scales.
The cell surface comprises the plasma membrane, the underlying cortical cytoskeleton and the ‘glue’ that binds them together — the membrane-to-cortex attachment or membrane-to-cortex adhesion energy (MCA). Their molecular origin is diverse: the plasma membrane is composed of a milieu of lipids and glycolipids with peripheral and embedded transmembrane proteins and glycoproteins. In most eukaryotic cells lacking a cell wall, including animal cells and amoebae, the cortex is composed of a thin actin network crosslinked by myosin motors and actin binding proteins, as well as networks of other filaments including septins. MCA is mediated by a layer of specialized proteins (like the ezrin, radixin, moesin (ERM) family), but also involves nonspecific frictional forces between the cortex and the plasma membrane (by, for example, electrostatic or van der Waals forces). MCA proteins generally have an F-actin binding domain and a domain that binds either directly to lipids or to proteins embedded in the membrane.
Variations in membrane, MCA or cortex composition are observed sub-cellularly, between cell types and over time. For example, cells regulate their lipid composition during the cell cycle, with consequences for both signalling and cell structure13. Moreover, MCA has been shown to differ between different cell regions14, tissue types15 and germ layers16. Such variation in MCA can be accomplished by differences in the expression level or by altering the binding activity of the MCA protein domains by cellular signalling16. The composition of the cell cortex also varies during the cell cycle and particularly between cell types. Classic examples are the surface of red blood cells and neurons, where an actin–spectrin cytoskeletal network provides mechanical resilience during capillary flow17 and touch sensation18, respectively. Mutations that interfere with the formation of this network cause severe anaemia19 and motor and cognitive disorders20, highlighting how molecular differences lead to mechanical differences.
The role of specific membrane proteins in the regulation of membrane mechanics, and vice versa, has been the focus of recent studies. BAR (Bin/amphiphysin/Rvs) protein scaffolding on membrane deformations creates a frictional barrier for lipid diffusion that is critical for processes like endocytosis. The generated deformation can be elongated by motor activity, resulting in a local increase in membrane tension that ultimately generates membrane scission21. Other membrane proteins respond to membrane tension, such as mechanosensitive channels like Piezo1, critical for responsiveness to touch in metazoans22.
Quantification of cell surface mechanics
Quantification of cell surface mechanics typically involves combining direct mechanical measurements with mathematical models to delineate the individual contributions of the plasma membrane (in-plane tension and bending rigidity), MCA (adhesion energy) and cortex (stiffness and cortical tension).
Membrane tension is a measure of the energetic cost of stretching a membrane (measured in J m–2 = N m–1). In a pure lipid bilayer/water system, it is the energy required to expand the surface area in the plane of the lipid bilayer (Tm). The molecular origin of tension is the tight packing of hydrophobic lipid molecules to avoid contact with water molecules. In cells, plasma membrane tension can appear greater than in pure lipid vesicles due to MCA and peripheral proteins that provide additional resistance to membrane deformation. MCA is defined as the free energy that is released when a unit area of membrane detaches from the cell cortex. Overall plasma membrane tension in cells is therefore approximated as the sum of Tm and MCA. It can be measured, for example, by pulling a membrane nanotube (or tether) and holding it with a constant length until it breaks by using an atomic force microscope (AFM) cantilever or an optical or magnetic tweezer23–25 (Table 1; Fig. 1). Its magnitude covers a wide range, from 3×10–6 N m–1 on apical membranes of epithelial cells to 276×10–6 N m–1 in keratocytes (reviewed in ref. 26) . To separate Tm from MCA, one can measure the tether force on cells whose membrane is detached from the cortex by actin depolymerizing agents27–29. Since cortex perturbations dramatically change overall cell morphology and thus might also affect Tm, the contribution of MCA can be more natively estimated by dynamically pulling membrane tethers at different speeds until they break, where the Tm is constant but MCA scales with the speed of tether pulling. To interpret such measurements, a classical model by Hochmuth and Sheetz24 or a more recent by Brochard-Wyart et al.30 have been used.
Table 1 |.
Methods to characterize the cell surface
| Technique | Examples | Measured quantity | Schematic | |
|---|---|---|---|---|
| Membrane tension | Tether pulling using AFM | 40× 10–12 N for HL60 cells9 | Tether rupture force | Fig. 1a |
| Interferometric particle detection using optical tweezers | 65× 10–6 N m−1 for erythrocytes44 | Power spectral density of the quadrant photodiode voltage | Fig. 1b | |
| Tether pulling using optical or magnetic tweezers | 29× 10–12 N for MEF cells45 | Tether rupture force | Fig. 1c | |
| Tether pulling using shear fluid | 86–172× 10–12 N for shear rates of 100–250 s–1 for neutrophils46 | Approach velocity, shear rate, tether length and cell size | Fig. 1d | |
| Cortical tension | AFM compression (with a flat cantilever or a bead) | 1.6× 10–3 N m–1 for HeLa cells during metaphase47 | Compressive force, contact angles of cellular deformation for a flat cantilever or deflection of the cantilever with a bead | Fig. 1e,i |
| Dual plate compression | 1.75 dyn cm–1 (or 175× 10–5 N m–1) for sea urchin eggs 40 minutes after fertilization48 | Compressive force, contact angles of cellular deformation | Fig. 1f | |
| Laser ablation | 9.15 μm min–1 orthogonal outward velocity in the anterior cortex of the one-cell C. elegans embryo49 | Response of cortex to laser ablation | Fig. 1g | |
| Micropipette aspiration | 0.035 dyn cm–1 (or 3.5× 10–5 N m–1) for passive blood granulocytes38 | Aspiration pressure and cellular deformation | Fig. 1h | |
|
Surface elasticity and viscoelasticity |
AFM indentation | 855 Pa for HL60 cells50 | Deflection of the cantilever | Fig. 1i |
| Brillouin microscopy | 5.41–8.06 GPa for plants ECM51 and 2.78 GPa for the nuclear envelope52 |
Brillouin peak shift and width | Fig. 1j | |
| Magnetic twisting cytometry | 10–11–10–8 Pa m–1 for a broad range of cancer cells lines53 | Bead displacement to a twisting magnetic field with different frequencies | Fig. 1k | |
| Cortical thickness | Electron microscopy | Tens of nm (ref. 54) | Thickness of the cortical layer | Fig. 1l |
| Fluorescence microscopy | 186 nm for the cortex of mitotic HeLa cells55 | Fluorescence peak distance | Fig. 1m | |
| Bending rigidity | Interferometric particle detection using optical tweezers | 2.8× 10–19 N m (or 67.6kBT) for erythrocytes44 | Power spectral density of the quadrant photodiode voltage or standard deviation of the distribution of fluctuation amplitudes | Fig. 1b |
| Micropipette aspiration | 10–19 N m for red blood cells or lipid bilayers23 and 1–2× 10–18 N m for cell types with a simple cortex56 | In the low tension regime, the slope of the area dilation versus the logarithm of the tension | Fig. 1h | |
| Flicker spectroscopy | 5× 10–13 erg (or 0.5× 10–19 N m) for erythrocytes57 | Shape fluctuations of vesicles from time series of optical microscopy snapshots | Fig. 1n | |
| Micropipette aspiration combined with optical tweezers | 2.7× 10–19 N m for neutrophils24 | The slope of the equilibrium tube force with the square root of the tension | Fig. 1c,h | |
| Membrane viscosity | Molecular rotors and flippers combined with FLIM | 270 cP (or 0.27 Pa s) in SKOV cells at 23 °C (ref. 58) | Fluorescence | Fig. 1o |
| Interferometric particle detection using optical tweezers | 81× 10–3 Pa s for erythrocytes44 | Power spectral density of the quadrant photodiode voltage | Fig. 1b |
Fig. 1 |. Schematics of methods to characterize the cell surface.
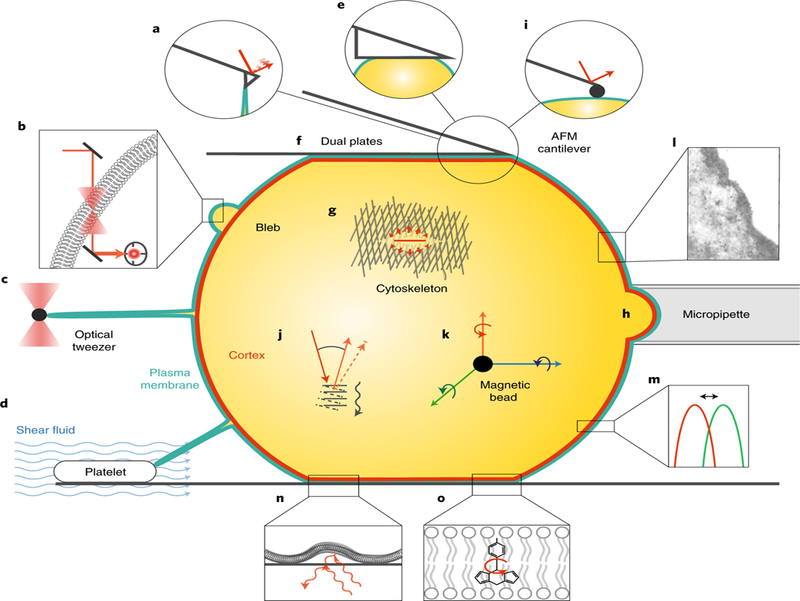
Understanding how cell shape is controlled at the molecular level requires a combination of quantitative experiments and physical modelling to relate the microscopic organization and dynamics of cortical and membrane components to global mechanical properties. See Table 1 for an overview of current techniques for quantifying cell surface mechanical properties of single cells, corresponding to the schematics pictured here. For techniques relevant to tissue mechanics and active force generation, we refer the reader to excellent recent reviews59–62. a, Tether pulling using AFM. b, Interferometric particle detection using optical tweezers. c, Tether pulling using optical or magnetic tweezers. d, Tether pulling using shear fluid. e, AFM compression with a flat cantilever. f, Dual plate compression. g, Laser ablation. h, Micropipette aspiration. i, AFM indentation. j, Brillouin microscopy. k, Magnetic twisting cytometry. l, Electron microscopy. m, Fluorescence microscopy. n, Flicker spectroscopy. o, Molecular rotors and flippers combined with FLIM. Panel l reproduced from ref. 54, EMBO.
Lipid bilayer membranes resist both extension, shear and bending. Thus, three moduli characterize membranes: the area stretch modulus (also known as the area compressibility modulus or area expansion modulus), the shear modulus and the bending rigidity. The area stretch modulus is a measure of the stiffness of the membrane under tension (measured in N m–1). At high tension values, and once most thermal fluctuations are smoothed out, its value is 0.1–1 N m–1 for various types of lipid bilayers and about 0.45 N m–1 (450 dyn cm–1) for red blood cells31. Cell membranes are thus quite resistant to extension and often treated as inextensible. When subjected to shear stresses in the plane of the membrane, a pure lipid bilayer behaves essentially as a liquid, thus the shear modulus is very small for membranes, on the order of 10–6 N m–1. The bending rigidity is a measure of the energy associated with its bending (measured in kBT or N m). In general, membranes made from lipids with unsaturated acyl chains have a lower bending modulus than membranes made from lipids with saturated acyl chains. Moreover, cholesterol increases the bending rigidity. Several methods have been developed to measure bending rigidity (Table 1; Fig. 1; reviewed in ref. 32).
The cell cortex tightly interacts with the plasma membrane. Cortical tension is the force per unit length exerted on a piece of the cortex by the cortical network around it (measured in J m–2 = N m–1). Tension gradients result in local contractions and drive cell deformations. Molecularly, cortical tension is governed by actomyosin contractility (reviewed in ref. 33) and cortical actin architecture34. The cell cortex undergoes rapid turnover, making it both mechanically rigid and highly plastic. Providing further complexity, the cell cortex is composed of actin filaments polymerized by both Arp2/3-complex activators as well as formins35. The resulting filaments might have different lengths and have different effective turnover rates and this underlies differential mechanical properties36,37. A wide range of experimental techniques have been used to measure cortical tension (Table 1; Fig. 1). Its magnitude varies considerably between cell types, ranging from approximately 10–5 N m–1 in blood granulocytes38 to 10–3 N m–1 in D. discoideum cells39,40.
The dynamic plasticity of the cell surface is essential for cells to rapidly change shape, move and exert forces. Interestingly, cell surface mechanics often have opposing effects on cell shape: cytoskeletal assembly and disassembly is intimately linked with membrane shape changes of the plasma membrane and organelles27,41, but the ability of the cytoskeleton to influence membrane shape changes is physically limited by in-plane membrane tension and MCA3. Signalling feedback loops have been identified between membrane tension and actin polymerization9,10. On the other hand, specifically perturbing membrane or cortical mechanics does not necessarily affect the other42. This highlights how we are only beginning to understand the logic of how cell surface mechanics affect signalling, cell shape and movement, and how that logic might be context dependent. This is due, at least in part, to limited information on the mechanical properties of the cell surface. Multidisciplinary approaches and specific experimental tools to measure and control cellular forces over multiple time, length and force scales are needed to understand how cellular morphogenesis, which drives complex cell behaviour such as cell motility and division, arises from membrane and cortical mechanics.
Future research on cell surface mechanics
The complexity, variability and importance of cell surface mechanics remain to be uncovered in many cellular processes. Three trends will help to account for its continued importance. First, a growing range of experimental techniques and mathematical models are being developed to allow measurement of cell surface mechanical properties (Table 1; Fig. 1). While many still require specialized skills and instruments, they are becoming more widely available. Second, a common language to describe cell surface mechanics is emerging. While errors in terminology and even units persist, the inclusion of mechanics in methods and discussions will move the field to a more careful quantification of cell surface properties. Third, understanding cell behaviour is often incomplete without consideration of cell surface mechanics. In this manner, efforts to explain biology will continue to demand investigation of cell mechanics, both internal and at the surface, to explain cell form and function.
Of course, many challenges remain: identifying molecular determinants of cell surface mechanics continues to be a challenge. Compounding this, recent studies highlight the importance of not only the composition but also the specific architecture of actin networks for their mechanical properties34,43. Moreover, bending modulus and MCA may also vary locally due to spatial variations in lipid composition and MCA proteins. This complicates efforts to obtain cell surface properties from measurements at one location on the cell. Some of those challenges can be addressed with the help of physical models and numerical simulations. Studies such as Chugh et al.34 exemplify how computational models can be used to identify the physical mechanisms responsible for the emergence of a given regime of cell surface mechanics.
If regulation of cell surface mechanics is important for proper cell function, then disruption of cell surface mechanics could be responsible for some pathologies. This raises the intriguing question of whether mechanical investigations of the cell membrane and cortex will reveal new molecular targets and offer new opportunities for intervention. Our challenge going forward is to understand mechanistically how, when and why cell surface mechanics choices are made.
Acknowledgements
We thank M. Bergert for comments on the manuscript and the schematics in Fig. 1. We acknowledge the financial support of the European Molecular Biology Laboratory (A.D.-M.), the NIH through GM114671 (D.A.F.), GM114344 (D.A.F) and GM118167 (O.D.W.), and the Chan Zuckerberg Biohub (D.A.F.).
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Mayor R & Etienne-Manneville S e front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol 17, 97–109 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol 36, 103–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raucher D & Sheetz MP Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol 148, 127–136 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gauthier NC, Fardin MA, Roca-Cusachs P & Sheetz MP Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proc. Natl Acad. Sci. USA 108, 11467–11472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keren K et al. Mechanism of shape determination in motile cells. Nature 453, 475–480 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchison T & Cramer L Actin-based cell motility and cell locomotion. Cell 84, 371–379 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Houk AR et al. Membrane tension maintains cell polarity by con ning signals to the leading edge during neutrophil migration. Cell 148, 175–188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozlov MM & Mogilner A Model of polarization and bistability of cell fragments. Biophys. J 93, 3811–3819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diz-Muñoz A et al. Membrane tension acts through PLD2 and mTORC2 to limit actin network assembly during neutrophil migration. PLoS Biol 14, e1002474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsujita K, Takenawa T & Itoh T Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat. Cell Biol 17, 749–758 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Lieber AD, Schweitzer Y, Kozlov MM & Keren K Front-to-rear membrane tension gradient in rapidly moving cells. Biophys. J 108, 1599–1603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iskratsch T, Wolfenson H & Sheetz MP Appreciating force and shape — the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol 15, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atilla-Gokcumen GE et al. Dividing cells regulate their lipid composition and localization. Cell 156, 428–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J & Sheetz MP Membrane tether formation from blebbing cells. Biophys. J 77, 3363–3370 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi K, Yonemura S, Matsui T & Tsukita S Immuno uorescence detection of ezrin/radixin/moesin (ERM) proteins with their carboxyl-terminal threonine phosphorylated in cultured cells and tissues. J. Cell Sci 112, 1149–1158 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Link V et al. Identi cation of regulators of germ layer morphogenesis using proteomics in zebra sh. J. Cell Sci 119, 2073–2083 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Bennett V & Baines AJ Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev 81, 1353–1392 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Krieg M, Dunn AR & Goodman MB Mechanical control of the sense of touch by β-spectrin. Nat. Cell Biol 16, 224–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaetani M, Mootien S, Harper S, Gallagher PG & Speicher DW Structural and functional efects of hereditary hemolytic anemia-associated point mutations in the alpha spectrin tetramer site. Blood 111, 5712–5720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda Y et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet 38, 184–190 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Simunovic M et al. Friction mediates scission of tubular membranes scafolded by BAR proteins. Cell 170, 172–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis AH & Grandl J Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans EA Bending elastic modulus of red blood cell membrane derived from buckling instability in micropipet aspiration tests. Biophys. J 43, 27–30 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochmuth FM, Shao JY, Dai J & Sheetz MP Deformation and ow of membrane into tethers extracted from neuronal growth cones. Biophys. J 70, 358–369 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charras G & Paluch E Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol 9, 730–736 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Sens P & Plastino J Membrane tension and cytoskeleton organization in cell motility. J. Phys. Condens. Matter 27, 273103 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Sheetz M Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol 2, 392–396 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Sun M et al. Multiple membrane tethers probed by atomic force microscopy. Biophys. J 89, 4320–4329 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun M et al. e efect of cellular cholesterol on membrane-cytoskeleton adhesion. J. Cell Sci 120, 2223–2231 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Brochard-Wyart F, Borghi N, Cuvelier D & Nassoy P Hydrodynamic narrowing of tubes extruded from cells. Proc. Natl Acad. Sci. USA 103, 7660–7663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waugh R & Evans EA ermoelasticity of red blood cell membrane. Biophys. J 26, 115–131 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimova R Advances in Colloid and Interface Science. Adv. Colloid Interface Sci 208, 225–234 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Salbreux G, Charras G & Paluch E Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol 22, 536–545 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Chugh P et al. Actin cortex architecture regulates cell surface tension. Nat. Cell Biol 19, 689–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bovellan M et al. Cellular control of cortical actin nucleation. Curr. Biol 24, 1628–1635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritzsche M, Erlenkamper C, Moeendarbary E, Charras G & Kruse K Actin kinetics shapes cortical network structure and mechanics. Sci. Adv 2, e1501337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suarez C & Kovar DR Internetwork competition for monomers governs actin cytoskeleton organization. Nat. Rev. Mol. Cell Biol 17, 799–810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans E & Yeung A Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration. Biophys. J 56, 151–160 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai J, Ting-Beall HP, Hochmuth RM, Sheetz MP & Titus MA Myosin I contributes to the generation of resting cortical tension. Biophys. J 77, 1168–1176 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz EC, Neuhaus EM, Kistler C, Henkel AW & Soldati T Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J. Cell Sci 113, 621–633 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Ledesma MD & Dotti CG Membrane and cytoskeleton dynamics during axonal elongationand stabilization. Int. Rev. Cytol 227, 183–219 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Diz-Muñoz A et al. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol 8, e1000544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bieling P et al. Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Betz T, Lenz M, Joanny J-F & Sykes C ATP-dependent mechanics of red blood cells. Proc. Natl Acad. Sci. USA 106, 15320–15325 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pontes B et al. Membrane tension controls adhesion positioning at the leading edge of cells. J. Cell Biol 216, 2959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidtke DW & Diamond SL Direct observation of membrane tethers formed during neutrophil attachment to platelets or P-selectin under physiological ow. J. Cell Biol 149, 719–730 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer-Friedrich E, Hyman AA, Jülicher F, Muller DJ & Helenius J Quanti cation of surface tension and internal pressure generated by single mitotic cells. Sci. Rep 4, 137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoneda M & Dan K Tension at the surface of the dividing sea-urchin egg. J. Exp. Biol 57, 575–587 (1972). [DOI] [PubMed] [Google Scholar]
- 49.Mayer M, Depken M, Bois JS, Jülicher F & Grill SW Anisotropies in cortical tension reveal the physical basis of polarizing cortical ows. Nature 467, 617–621 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Rosenbluth MJ, Lam WA & Fletcher DA Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophys. J 90, 2994–3003 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elsayad K et al. Mapping the subcellular mechanical properties of live cells in tissues with uorescence emission-Brillouin imaging. Sci. Signal 9, rs5 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Antonacci G & Braakman S Biomechanics of subcellularstructures by non-invasive Brillouin microscopy. Sci. Rep 6, 37217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coughlin MF et al. Cytoskeletal stifness, friction, and uidity of cancer cell lines with diferent metastatic potential. Clin. Exp. Metastasis 30, 237–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanakam F, Albrecht R, Eckerskorn C, Matzner M & Gerisch G Myristoylated and non-myristoylated forms of the pH sensor protein hisactophilin II: intracellular shuttling to plasma membrane and nucleus monitored in real time by a fusion with green fluorescent protein. EMBO J 15, 2935–2943 (1996). [PMC free article] [PubMed] [Google Scholar]
- 55.Clark AG, Dierkes K & Paluch EK Monitoring actin cortex thickness in live cells. Biophys. J 105, 570–580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhelev DV, Needham D & Hochmuth RM Role of the membrane cortex in neutrophil deformation in small pipets. Biophys. J 67, 696–705 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fricke K, Wirthensohn K, Laxhuber R & Sackmann E Flicker spectroscopy of erythrocytes. A sensitive method to study subtle changes of membrane bending stifness. Eur. Biophys. J 14, 67–81 (1986). [DOI] [PubMed] [Google Scholar]
- 58.López-Duarte I, Vu TT, Izquierdo MA, Bull JA & Kuimova MK A molecular rotor for measuring viscosity in plasma membranes of live cells. Chem. Commun 50, 5282–5284 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Campàs O A toolbox to explore the mechanics of living embryonic tissues. Semin. Cell Dev. Biol 55, 119–130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugimura K, Lenne PF & Graner F Measuring forces and stresses in situ in living tissues. Development 143, 186–196 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Polacheck WJ & Chen CS Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roca-Cusachs P, Conte V & Trepat X Quantifying forces in cell biology. Nat. Cell Biol 19, 742–751 (2017). [DOI] [PubMed] [Google Scholar]


