Abstract
In eukaryotes, double-stranded (ds) RNA induces sequence-specific inhibition of gene expression, referred to as RNA interference (RNAi). In invertebrates, RNAi can be triggered effectively by either long dsRNAs or 21- to 23-nt-long short interfering (si) duplex RNAs, acting as effectors of RNAi. siRNAs recently have been shown to act as potent inducers of RNAi in cultured mammalian cells. However, studies of RNAi activated by long dsRNA are impeded by its nonspecific effects, mediated by dsRNA-dependent protein kinase PKR and RNase L. Here, we report that the RNAi response can be induced effectively by long dsRNA in nondifferentiated mouse cells grown in culture. Transfection of dsRNA into embryonal carcinoma (EC) P19 and F9 cells results in a sequence-specific decrease in the level of proteins expressed from either exogenous or endogenous genes. dsRNA-mediated inhibition of the reporter gene also occurs in mouse embryonic stem cells. The RNAi effect is mediated by siRNAs, which are generated by cleavage of dsRNA by the RNaseIII-like enzyme, Dicer. We demonstrate that extracts prepared from EC cells catalyze processing of dsRNA into ≈23-nt fragments and that Dicer localizes to the cytoplasm of EC and HeLa cells.
In many eukaryotes, double-stranded (ds) RNA inhibits gene expression in a sequence-specific manner by triggering degradation of mRNA. This effect, referred to as RNA interference (RNAi), has been studied most extensively in Caenorhabditis elegans and Drosophila melanogaster. Posttranscriptional gene silencing (PTGS) in plants and quelling in Neurospora crassa are related to RNAi (reviewed in refs. 1–3). Genetic and biochemical studies have revealed that RNAi/PTGS is a very complex reaction, involving many different proteins of mostly undefined function (1–3). Characterization of RNAi/PTGS mutants and studies with viruses in plants have revealed that one biological function of RNAi/PTGS is to prevent transposition and invasion of foreign nucleic acids. In addition, mutations in some PTGS/RNAi-related genes have different developmental consequences in C. elegans and plants (1–3). At least some of the developmental effects are explained by recent findings that the dsRNA-processing enzyme functioning in RNAi also is involved in the biogenesis of small temporal (st) RNAs, which regulate development in C. elegans and probably other animals (4, 5). RNAi offers a way to inactivate genes of interest and, thus, provides a powerful tool to study gene function. Specific inhibition of gene activity also can be achieved by stable expression of dsRNA hairpins in transgenic lines (1–3).
Biochemical studies are beginning to unravel the mechanistic details of RNAi. This has been facilitated greatly by the demonstration that RNAi effects can be reproduced in Drosophila cell or embryo extracts (6–8). In both cells and in vitro, dsRNA first is processed into 21- to 23-nt short interfering RNAs (siRNAs), identified in plants, Drosophila, and C. elegans (8–11). The RNaseIII-like enzyme, Dicer, responsible for the generation of siRNAs has been identified recently (12). Subsequently, siRNAs seem to associate with a multicomponent nuclease, identified in Drosophila and called RISC, and guide this enzyme for sequence-specific degradation of mRNA (1, 12).
dsRNA-mediated inhibition of gene expression also has been studied in mammalian systems. Microinjection of dsRNA into mouse oocytes or early embryos results in specific inhibition of activity of both maternally and zygotically expressed proteins (13, 14). Inhibition of gene expression also has been achieved more recently in various cells lines treated with siRNAs (15, 16). However, elicitation of RNAi with long dsRNAs in cultured mammalian cells or cell extracts generally has been less successful. Although Ui-Tei et al. (17) reported specific effects for Chinese hamster ovary cells, introduction of dsRNA into many other mammalian cell lines (15, 16), or rabbit reticulocyte lysates (7), resulted mainly in a nonspecific decrease in gene expression. These failures are explained most readily by the action of two latent enzymes forming part of IFN defense pathways and activated by long dsRNA (18). The first is 2′-5′-oligoadenylate (2–5A) synthetase, which is activated by dsRNA to increase synthesis of 2–5A that is required for activation of the sequence-nonspecific RNase, RNase L (19). The second is protein kinase PKR, the active form of which phosphorylates the translation factor eukaryotic initiation factor 2α (eIF2α), leading to general inhibition of protein synthesis and cell death (20). In addition, PKR induces transcription of genes encoding interferons, which, in turn, activate synthesis of PKR and 2–5A synthetase, amplifying nonspecific effects of dsRNA (18, 20).
In this study, we use nondifferentiated mouse embryonal carcinoma (EC) cell lines F9 and P19 to investigate the RNAi response to dsRNA. In undifferentiated EC cells as in normal embryonic cells, IFN genes are refractory to induction by viral infection and dsRNA (21–24). In addition, these cells are deficient in some of the dsRNA-activated enzymes discussed above and generally are less sensitive to exogenous interferons (19, 21, 25, 26). We find that a specific RNAi response to long dsRNA can be induced in EC cells and embryonic stem (ES) cells. We demonstrate further that extracts prepared from EC cells catalyze processing of dsRNA into ≈23-nt RNAs and that the processing enzyme, Dicer, is localized in the cytoplasm of mammalian cells.
Materials and Methods
Plasmids and PCR Templates Used for RNA Synthesis.
The plasmid pβact-eGFP (27) was used as a reporter for cotransfections and to produce the double T7 promoter-tagged PCR fragments used for generating dsRNA–green fluorescent protein (GFP). Oligonucleotides used for PCR amplification, T7-ATGGTGAGCAAGGGCGAGGAGC and T7-GTACAGCTCGTCCATGCCGAG, contained the T7 promoter sequences (TTAATACGACTCACTATAGGGAGA, abbreviated as T7). The plasmid pIND-LacZ (Invitrogen) and oligonucleotides T7-ATGGGGGGTTCTCATCATCATC and T7-CTCAGGTCAAATTCAGACGGC were used for producing the PCR product that was used for generating dsRNA-LacZ. Plasmids containing cDNAs encoding mouse integrins α3 and β1 (obtained from T. Tsuji, School of Pharmaceutical Sciences, Tokyo, and S. Denda, Howard Hughes Medical Institute, San Francisco) were used for preparation of PCR templates for the synthesis of dsRNA-β1 and dsRNA-α3, using oligonucleotide pairs T7-ATGGGCCCCGGCCCCTGCCG and T7-GCCTACCTGCACCGTGTACCC and T7-ATGAATTTGCAACTGGTTTCC and T7-GCCACCTTCTGGAGAATCC, respectively. dsRNA-GFP, -LacZ, -α3, and -β1 were 727, 620, 753, and 771 bp long, respectively. They correspond to the initiator ATG-proximal regions of cDNAs. For preparation of templates for synthesis of single-stranded (ss) GFP RNAs, only one of the two oligonucleotide primers contained the T7 promoter sequence. pβact-eGFP was also used for preparation of PCR templates for the synthesis of 123-bp dsRNA. The following primer pairs were used: T7-CCACTACCTGAGCACCCAGT and GGGAGAGTACAGCTCGTCCATGCCGAG and T7-GTACAGCTCGTCCATGCCGAG and GGGAGACCACTACCTGAGCACCCAGT.
pCFP-Dicer, expressing the cyan fluorescent protein (CFP) fused with human Dicer, was constructed by transferring the full-length Dicer cDNA (unpublished results) into the pdECFP-C1 vector (kindly provided by S. Wiemann, University of Heidelberg), using Gateway cloning technology (Life Technologies, Gaithersburg, MD).
RNA Synthesis.
Cold dsRNAs were synthesized by using PCR templates and the Ambion T7 MegaScript kit, following the manufacturer's recommendations. RNA was precipitated with LiCl and resuspended in buffer TN (20 mM Tris⋅HCl, pH 7.5/50 mM NaCl). 32P-labeled dsRNAs were prepared by synthesis of individual strands, using [α-32P]UTP (specific activity, 800 Ci/mmol) and the Maxiscript kit (Ambion, Austin, TX). After purification on a PAGE-8 M urea gel, both strands were annealed by mixing equimolar amounts of RNA in buffer TN. For annealing, RNA samples were heated at 95°C for 3 min, transferred to 75°C for 30 min, and then slowly cooled over 4 h to 25°C. dsRNA preparation quality was analyzed on 1.5% agarose gels, using dsDNA and ssRNAs as markers. In addition, dsRNA was tested for resistance to digestion by RNases A and T1. dsRNAs used for transfection contained 18-nt 3′ overhangs at both termini. The labeled, 727-bp dsRNA contained 6-nt 3′ overhangs, and the 123-bp dsRNA had blunt ends.
Culture and Transfection of Cells.
F9 and P19 embryonal teratocarcinoma cell lines, obtained from P. Caroni of Friedrich Miescher Institute for Biomedical Research, were grown in DMEM containing 15% and 10% FBS, respectively. For F9 cells, 0.1% gelatin-coated plates were used. Cells were passaged every 2 days and split 24 h before the transfection. Mouse E14 ES cells were cultured on plates coated with inactivated fibroblast feeder cells in DMEM containing 10% FBS and supplemented with sodium-pyruvate, l-glutamine, nonessential amino acids, and leukemia inhibitory factor (dilution, 1:10,000; Life Technologies) (28). HeLa cells were grown as described (29). Cells (1 × 107) were transfected by electroporation with 30 μg of indicated plasmid, with or without the addition of 30 μg of indicated RNA. Optimal settings for transfection of EC and HeLa cells were 320 V and 500 μF, and, for ES cells, 250 V and 500 μF. After electroporation, cells were incubated for 10 min at 20°C and plated at a density of ≈2 × 106 cells per 10-cm plate. Medium was replaced every 24 h.
Fluorescence-Activated Cell Sorting (FACS).
The percentage of GFP-expressing cells and fluorescence intensity was assessed by FACS (FACScalibur; Becton Dickinson) analysis 48 h posttransfection. Control nontransfected cells and cells transfected with pβact-eGFP were used to gate for forward scatter and side scatter and to determine the number of GFP-positive cells and their mean fluorescence (25,000 events captured per sample).
To analyze the effects on expression of integrins, transfected GFP-positive F9 cells (≈4 × 105) were sorted by using a FACSVantage SE sorter (Becton Dickinson). Acquisition and analysis of the FACS data were performed by using cellquest software (Becton Dickinson).
In Vivo Imaging and Immunofluorescence.
For in vivo imaging, transfected cells were grown on acid-washed coverslips (precoated with 0.1% gelatin for F9 cells and inactivated fibroblast feeder cells for ES cells) in 6-well plates. Cells were imaged in DMEM Special nonautofluorescent medium (Life Technologies) at 37°C in purpose-built observation chambers (30). Indirect immunofluorescence was performed on cells growing on a polylysine-precoated coverslip as described (29). Abs used were D347 (1:100 dilution), followed by a Texas red-labeled α-rabbit Ab, and α-GFP Ab (1:400, Roche, Gipf-Oberfrick, Switzerland), followed by a FITC-labeled α-mouse Ab. When applicable, the nucleus was stained with 4′,6-diamidino-2-phenylindole (50 ng/ml). Images were analyzed by using a Leica laser-based confocal microscope (Leica, Deerfield, IL); data were collected on an Applied Vision System and processed by deconvolution.
Cell Extracts and Fractionation.
Total protein extracts were prepared with RIPA lysis buffer as described (31). To prepare cytoplasmic extracts used for dsRNA-processing assays, EC cells (≈6 × 107) were washed extensively with PBS and scraped with 1 ml of lysis buffer (50 mM Tris⋅HCl, pH 8.0/20 mM KCl/1 mM MgCl2/1 mM DTT/1× protease inhibitor mix without EDTA (Roche)]. Cells were disrupted on ice with a Dounce homogenizer (pestle B), and nuclei were spun down by centrifugation for 10 min at 2,000 rpm. The supernatant was recentrifuged at 10,000 rpm for 10 min. Protein concentration was adjusted to 10 mg/ml, and the extract was supplemented with glycerol to 10% and stored at −80°C. Nuclear and cytoplasmic fractions of EC cells were isolated as described by Verdel et al. (32), using 0.25% Triton X-100.
Protein concentration was calculated by using Bradford reagent with BSA as a standard.
Northern Analysis.
RNA was extracted from cell samples by using the RNeasy Mini Kit (Qiagen). RNA (15 μg) was separated in a 1.2% agarose-formaldehyde gel and blotted onto Hybond-N+ membrane. Hybridization was carried out as described (31), using a 32P-labeled GFP and mouse glyceraldehyde-3-phosphate dehydrogenase cDNA probes.
Adhesion Assays.
For adhesion assays, GFP-positive cells isolated by FACS were counted and their density was adjusted to 100 cells/μl in FBS-free DMEM. Aliquots of 103 cells were spotted in wells of NunClon plates (Nunc) precoated with increasing amounts of laminin or fibronectin. Other details were as described (33).
dsRNA-Processing Assay.
Assays (20 μl) contained 10 μl of cytoplasmic extract and 0.5 fmol of the 32P-labeled dsRNA in 20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/2 mM MgCl2/5 mM DTT. When assessing the ATP effect, 0.5 mM ATP, 10 mM creatine phosphate, and 0.03 mg/ml creatine kinase were added to the reaction. ATP was depleted from the extract by incubation for 20 min at 37°C with 2 mM glucose and 0.2 units/μl hexokinase (Sigma). Reactions were analyzed by PAGE-Urea as described (9).
Antibodies, Western Blots, and Immunoprecipitations.
The α-Dicer D347 Ab was obtained by immunizing rabbits (Eurogentec, Brussels) with the keyhole limpet hemocyanin-coupled peptide SNKYLDGNANKSTSDGSC of human Dicer. Abs were purified from serum by using a Sulfolink peptide-affinity column (Pierce). Western blots were performed as described (31), using α-Dicer Ab D347 at 1:1,000 dilution, α-integrin α3 Abs C-18 and N-19 (1:500, Santa Cruz Biotechnology), α-integrin β1 Ab FnR3 (1:250; ref. 34), α-GFP (1:4,000; Roche), anti-α tubulin (1:5,000: NeoMarker), α-hGAR1 (1:3,000; ref. 29), α-mitogen-associated protein kinase (1:2,000; New England Biolabs), α-Cdk2 (1:500; Santa Cruz Biotechnology), and horseradish peroxidase-conjugated secondary Abs (Amersham Pharmacia).
In immunoprecipitation experiments, Protein A-Sepharose beads (10 μl) coated with 5 μg of D347 Ab were incubated with 50 μl of EC cell cytoplasmic extracts. Aliquots of supernatants and pellets were analyzed on 8% SDS/PAGE and tested for the dsRNA-processing activity.
Results
dsRNA-Mediated Inhibition of Reporter Genes in EC Cells.
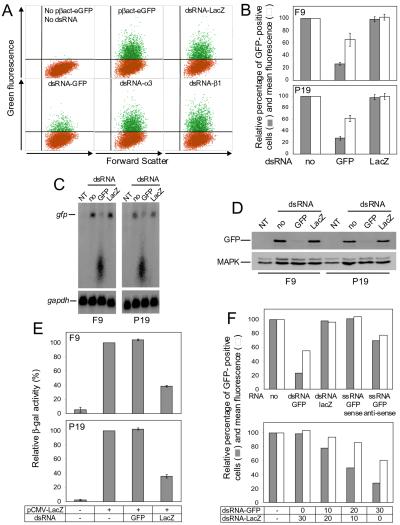
We used EC lines F9 and P19 to study the response of cells grown in culture to long dsRNA. Initially, we tested the effect of dsRNA on transient expression of reporter genes encoding the GFP and β-galactosidase (β-gal), using ≈700-bp-long, gene-specific dsRNAs (dsRNA-GFP and dsRNA-LacZ, respectively). We first investigated the effect of dsRNA-GFP on GFP expression, using dsRNA-LacZ as a control, by transfecting EC cells with the GFP-encoding plasmid pβact-eGFP alone or together with either dsRNA-GFP or dsRNA-LacZ. The number of cells expressing GFP was analyzed by FACS 48 h after transfection. About 25% of F9 (Fig. 1A) or P19 (data not shown) cells transfected with pβact-GFP were GFP-positive. Cotransfection with dsRNA-GFP decreased the fraction of fluorescent cells by more than 70%; in addition, the mean fluorescence of the remaining GFP-positive cells was reduced by 35–40%. In contrast, cotransfection of dsRNA-LacZ altered neither the number of GFP-positive cells nor their mean fluorescence (Fig. 1 A and B). Likewise, cotransfection of pβact-GFP with dsRNAs specific for integrin α3 and β1 mRNAs had no effect on GFP fluorescence (Fig. 1A; see also below). The specificity of the dsRNA-GFP on GFP expression was confirmed by fluorescence microscopy performed with living cells. Imaging of transfected EC cells indicated a decrease in GFP fluorescence in cells treated with dsRNA-GFP but not dsRNA-LacZ (data not shown). Similarly, Northern (Fig. 1C) and Western (Fig. 1D) blot analyses demonstrated decreased accumulation of GFP mRNA and its encoded protein in cells cotransfected with dsRNA-GFP but not dsRNA-LacZ.
Figure 1.
Effect of dsRNA on activity of genes expressed in transfected EC cells. (A) FACS analysis of F9 cells transfected with pβact-eGFP in the absence or presence of different dsRNAs as indicated. Fluorescent cells are in green. (B) Effect of dsRNA on the number of GFP-positive EC cells and their fluorescence intensity. Cells were transfected with pβact-eGFP in the absence or presence of dsRNA-GFP or dsRNA-LacZ. Values (means ± SD; n = 4) for the number of fluorescent cells and mean fluorescence were calculated relative to the GFP-positive cells transfected with only pβact-eGFP. (C) Northern analysis of GFP mRNA levels in cells transfected with either pβact-eGFP alone or cotransfected with dsRNA-GFP or dsRNA-LacZ. NT, nontransfected cells. (Lower) Hybridization of the same blot with the glyceraldehyde-3-phosphate dehydrogenase mRNA-specific probe. (D) Western blot analysis of GFP levels in cells transfected with either pβact-eGFP alone or cotransfected with dsRNA-GFP or dsRNA-LacZ. (Lower) Reprobing with α-mitogen-associated protein kinase Ab. (E) Effect of dsRNA on β-gal activity in extracts of cells transfected with pCMV-LacZ and indicated dsRNA. Activity was measured by using the chemiluminescent β-gal assay (Roche). Values represent averages of two independent experiments, with the range indicated. (F) Effect of GFP ssRNAs (Upper) and increasing doses of cotransfected dsRNA-GFP (Lower) on the number and fluorescence intensity of GFP-positive P19 cells. Cells were transfected with pβact-eGFP alone or in the presence of dsRNA-GFP, or dsRNA-LacZ, or ssRNA-GFP of the sense or antisense orientation. (Lower) Cells were transfected with pβact-eGFP alone or in the presence of 10, 20, or 30 μg of dsRNA-GFP supplemented with dsRNA-LacZ to a total amount of 30 μg. Values are averages from two experiments.
We also compared the effect of dsRNA-LacZ and dsRNA-GFP on the lacZ gene expression in cells transfected with the plasmid pCMV-LacZ, expressing β-gal. As shown in Fig. 1E, cotransfection with dsRNA-GFP had no effect on β-gal activity of cell extracts, but dsRNA-LacZ inhibited the activity by ≈60% for both F9 and P19 cells.
The dose dependence of the dsRNA effect was investigated in P19 cells, using GFP as the reporter. The inhibitory effect on the number and mean fluorescence of GFP-positive cells was proportional to the amount of dsRNA-GFP (Fig. 1F). Replacement of dsRNA-GFP with a GFP-specific ssRNA of either sense or antisense orientation resulted in no effect (sense RNA) or only 25% inhibition (antisense RNA) (Fig. 1F).
dsRNA Inhibition of Endogenous Genes.
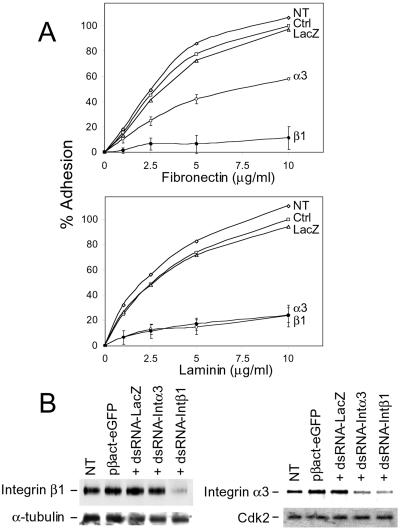
To obtain further evidence for the specificity of the dsRNA effect and to demonstrate that dsRNA can target mRNAs transcribed from endogenous genes, we performed RNAi experiments directed toward mRNAs encoding two subunits of cell surface receptor proteins, integrins α3 and β1. These proteins turn over rapidly (35), and their presence at the cell surface can be monitored by simple adhesion assay. The integrin β1-subunit heterodimerizes with many α-subunits, leading to receptors with varying ligand specificities, e.g., integrin α5β1 is a receptor for fibronectin, whereas integrin α3β1 binds only weakly to fibronectin but strongly to laminin (36–38). Hence, inactivation of integrin β1 should affect cell adhesion to both fibronectin and laminin, whereas inactivation of integrin α3 primarily should inhibit adhesion to laminin. Using Abs against mouse integrins α3 and β1, we verified by indirect immunofluorescence that both proteins indeed are present on the surface of F9 cells. We also found that preincubation of F9 cells with antiintegrin β1 Abs, but not control Abs, inhibits their adhesion to laminin by 80–90% (data not shown).
dsRNAs of ≈700 bp, containing integrin α3 or β1 mRNA sequences, were used to measure the RNAi effect, with dsRNA-LacZ as a control. Fluorescence of GFP, expressed from the cotransfected plasmid pβact-eGFP, was used to separate transfected from nontransfected cells by FACS. As already shown in Fig. 1A, cotransfection of pβact-eGFP with either dsRNA-α3 or -β1 had no inhibitory effect on GFP expression in F9 cells. The sorted GFP-positive cells were tested for their capacity to bind to laminin and fibronectin. Transfection of dsRNA-β1 inhibited binding of cells to both laminin and fibronectin by 80–90% (Fig. 2A). Cotransfection with dsRNA-α3 inhibited binding to laminin by 80%, whereas adhesion to fibronectin was reduced by only 45%, consistent with the limited contribution of integrin α3/β1 to binding to fibronectin (37). Transfection with the pβact-eGFP plasmid alone, or cotransfected with pβact-eGFP and dsRNA-LacZ, had no effect on the binding of cells to either laminin or fibronectin (Fig. 2A).
Figure 2.
dsRNA-mediated inhibition of integrin β1 and α3 expression in F9 cells. (A) Effect of dsRNA-β1 (β1) and dsRNA-α3 (α3) on adhesion to fibronectin (Upper) and laminin (Lower) of the FACS-sorted, GFP-positive cells. Aliquots of 103 cells were spotted in the plate wells precoated with indicated concentrations of either fibronectin or laminin. Values represent means of four determinations originating from two independent transfection and sorting experiments. SD values are shown only for cells treated with dsRNA-α3 and dsRNA-β1. Other curves represent nontransfected cells (NT) and cells transfected with pβact-eGFP alone (Ctrl) or in the presence of dsRNA-LacZ (LacZ). One hundred percent values correspond to the number of control (Ctrl) cells (60) attached at 10-μg/ml ligand concentration. (B) Western blot analysis of protein extracts (10 μg) from indicated transfected cells (described in A) with Abs specific for integrins β1 and α3. Blots were reprobed with anti-α-tubulin or α-Cdk2 Abs, as indicated.
The specificity of the dsRNA effect was assessed further by Westerns, using extracts from transfected and sorted F9 cells and Abs against integrins β1 and α3. There was a strong decrease in the level of integrin β1 in cells transfected with dsRNA-β1 but not dsRNA-α3 or dsRNA-LacZ and an ≈2.5- to 3-fold decrease of integrin α3 in cells treated with either dsRNA-α3 or dsRNA-β1, with control dsRNA-LacZ having no effect (Fig. 2B). The effect of dsRNA-β1 on integrin α3 accumulation is not surprising, because integrin α3 functions exclusively as a heterodimer with integrin β1 and elimination of the partner most probably would lead to its destabilization (38).
Inhibition of GFP Expression in ES Cells.
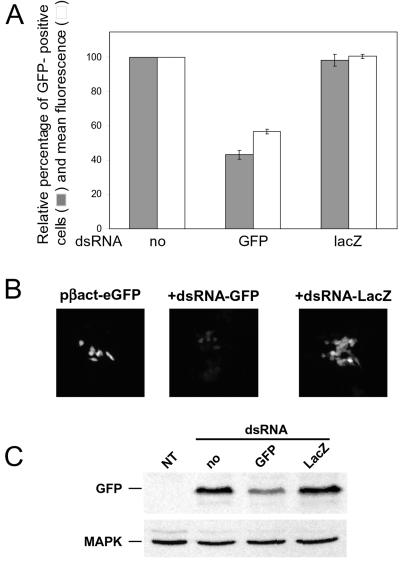
The effect of dsRNA on GFP expression also was assessed in mouse ES cells. ES cells were cotransfected with plasmid pβact-eGFP in the presence of either dsRNA-GFP or dsRNA-LacZ, and the GFP-positive cells were analyzed by FACS. About 30% of pβact-eGFP-transfected cells expressed GFP (data not shown). Whereas cotransfection of dsRNA-LacZ did not inhibit GFP expression, cotransfection with dsRNA-GFP reduced the number of fluorescent cells by about 60%; in addition, the mean fluorescence of the positive cells was reduced by 40% (Fig. 3A). The inhibitory effect of dsRNA-GFP, and not dsRNA-LacZ, on GFP expression in ES cells was confirmed by in vivo fluorescence microscopy performed on living cells (Fig. 3B) and by Western blots (Fig. 3C).
Figure 3.
Inhibition of GFP expression in mouse ES cells. (A) Effect of dsRNA on the number and fluorescence intensity of ES cells transfected with either pβact-eGFP alone or in the presence of dsRNA-GFP or dsRNA-LacZ. Values were calculated relative to the eGFP-positive cells transfected with only pβact-eGFP. They are averages of two independent experiments, with the range indicated. (B) A representative example of the in vivo imaging of ES cells transfected with pβact-eGFP alone or cotransfected with dsRNA-GFP or dsRNA-LacZ, as indicated. The fluorescence intensity scale obtained from pβact-eGFP-transfected cells was applied to cells cotransfected with pβact-eGFP and dsRNA-eGFP or dsRNA-LacZ. Cells were grown on coverslips and imaged 48 h posttransfection. (C) Western blot analysis of GFP levels in extracts of ES cells transfected with either pβact-eGFP alone or cotransfected with either dsRNA-GFP or dsRNA-LacZ. NT, nontransfected cells. GFP was detected with α-GFP Ab. (Lower) Reprobing with α-mitogen-associated protein kinase Ab.
EC Cell Extracts Process dsRNA to ≈23-nt Fragments.
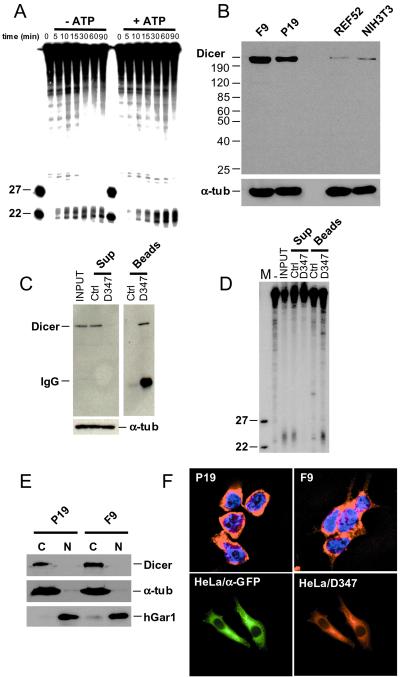
In plants, nematodes, Drosophila, and also in Drosophila cell-free extracts, dsRNA is processed to short, 21- to 23-nt siRNAs (8–11), acting as effectors of RNAi by directing the nucleolytic activity to the target mRNA (6, 9). An enzyme implicated in the generation of siRNAs is the multidomain protein Dicer, originally identified in Drosophila (12). Immunoprecipitates containing the overexpressed human Dicer were shown previously to process dsRNA to ≈22-nt RNAs (12), but formation of siRNAs in mammalian cell extracts has not been demonstrated.
We prepared cytoplasmic extract from P19 cells and tested it for dsRNA-processing activity. A fraction of the extract was preincubated with hexokinase and glucose to remove any endogenous ATP. As shown in Fig. 4A, the 32P-labeled, 727-bp-long dsRNA is cleaved to ≈23-nt fragments in the extract, and addition of ATP and the ATP-regenerating system stimulates processing approximately 2-fold. Extensive dialysis of the extract, combined with the hexokinase and glucose treatment, did not increase further the ATP effect. Incubation with the 32P-labeled ssRNA did not generate ≈23-nt fragments (data not shown).
Figure 4.
In vitro cleavage of dsRNA and cellular localization of Dicer. (A) Processing of dsRNA to ≈22-nt RNAs in cytoplasmic extract of F9 cells. Reactions were incubated for the indicated time, in the presence or absence of ATP, and analyzed by PAGE-Urea. Extracts used for reactions without ATP were preincubated with hexokinase and glucose. Size markers are 22- and 27-nt oligoribonucleotides. (B) Western blot analysis of total cell extracts prepared from F9, P19, NIH 3T3, and REF52 cells. Proteins (≈75 μg) were separated in an 8% SDS/PAGE, and the blot was probed sequentially with α-Dicer D347 and anti-α-tubulin Abs. (C) Immunodepletion of Dicer from F9 cytoplasmic extract by incubation with D347 Ab-coated Protein A-Sepharose beads. As control (Ctrl), the extract was incubated with noncoated Protein A-Sepharose beads. Supernatant (Sup) and beads fractions were analyzed by Western blots, using D347 and anti-α-tubulin Abs. (D) Analysis of the dsRNA-processing activity present in the immunoprecipitation fractions described in C. dsRNA (123 bp) was used as a substrate. (E) Western blot analysis of cytoplasmic (C) and nuclear (N) fractions of EC cells using D347 Ab. Reprobing of the blot with Abs against the nuclear protein hGAR1 and the cytoplasmic α-tubulin indicated no major cross-contamination of fractions. (F Upper) Indirect immunolocalization of endogenous Dicer in F9 and P19 cells, using D347 Ab and Texas red-labeled α-rabbit secondary Ab. Nuclei are stained with 4′,6-diamidino-2-phenylindole. (Lower) Localization of the CFP-Dicer fusion protein in HeLa cells transfected with pCFP-Dicer, using α-GFP and α-Dicer D347 Abs.
Dicer Is Involved in dsRNA Cleavage in EC Cell Extracts.
To demonstrate the presence of Dicer in EC cells and extracts derived from them, polyclonal Abs were raised against the polypeptide derived from the RNase III domain of the protein. When tested by Western blots with whole-cell extracts, the affinity-purified Ab D347 visualized a protein band migrating at ≈220 kDa. A band of identical mobility also was recognized by two other Abs raised against different Dicer peptides (not shown). Interestingly, extracts from EC cells contained much higher levels of Dicer than extracts prepared from cultures of differentiated cells such as rat REF52 and mouse NIH 3T3 (Fig. 4B) or human HeLa cells (not shown).
To directly show that Dicer is responsible for the generation of ≈23-nt RNAs in P19 cell extracts, the protein was immunodepleted by using D347 Ab coupled to Protein A-Sepharose beads (Fig. 4C). Supernatants and pellets of immunoprecipitation and control reactions were tested for dsRNA-processing activity. The immunodepleted extract did not cleave 123-bp-long dsRNA to ≈23-nt RNAs, but the activity was present in the immunoprecipitation pellet (Fig. 4D). Similar results were obtained with extracts originating from F9 cells (data not shown).
Dicer Is Localized in the Cytoplasm.
Fractionation of EC cell extracts revealed that Dicer is present in the cytoplasmic and not nuclear compartment (Fig. 4E). Indirect immunofluorescence experiments, performed with EC cells and Ab D347, likewise demonstrated a cytoplasmic localization, with no significant staining in the nucleus (Fig. 4F). Finally, we determined the localization of Dicer fused to CFP expressed in transfected HeLa cells. Probing with α-GFP and D347 Abs revealed an identical pattern of cytoplasmic localization with no signal in the nucleus (Fig. 4F). Control transfections, with the plasmid expressing CFP alone, indicated both nuclear and cytoplasmic localization (data not shown).
Discussion
Elicitation of RNAi in response to long dsRNA in cultured mammalian cells generally has been unsuccessful, most likely because of the known, nonspecific effects of dsRNA on gene expression and cell growth, mediated by the dsRNA-dependent protein kinase PKR and RNase L, both acting as effectors of the IFN response. These inhibitory effects have impeded studies of RNAi in mammals, in particular, the initial steps of dsRNA cleavage (see Introduction). Here, we showed that RNAi can be induced by dsRNA in nondifferentiated EC or ES mouse cells grown in culture. We exploited the properties of EC cells to gain some insight into the RNAi machinery in mammals. We found that extracts prepared from EC cells catalyze processing of dsRNA into ≈23-nt siRNAs and that the RNaseIII-like enzyme catalyzing this reaction, Dicer, localizes to the cytoplasm of P19, F9, and HeLa cells.
The observation that the RNAi response to dsRNA can be triggered effectively in undifferentiated embryonic cells is in line with previous findings that these cells are deficient in some of the dsRNA- and IFN-activated enzymes and that induction of IFN genes by dsRNA or viral infection in these cells is impaired (19, 21–25). Our findings are also consistent with reports showing that RNAi can be induced in mouse oocytes and early embryos by microinjection of dsRNA and that it persists until 6.5 days postimplantation (13, 14). It will be interesting to investigate how the RNAi response to dsRNA is modulated upon differentiation of embryonic cells in vitro, a process accompanied by induction of the IFN response. Knockout mice and cell lines deficient for PKR and RNase L (ref. 39 and references therein) could be used to measure directly the effects of these enzymes on RNAi.
Transfection of siRNA (15, 16) or dsRNA (ref. 17; this work) into mammalian cells allows for only transient inhibition of gene expression. Demonstration that dsRNA has no apparent unspecific effects in cultured embryonal cells suggests that longer-term inhibition could be achieved in these cells by the stable expression of dsRNA hairpins targeting genes of interest. The inhibition of gene activity described in this work was between 60% and 90%, and functional assays generally correlated well with the determination at the protein or mRNA level. However, inhibition of integrin α3 adhesion to laminin appeared to be stronger than would have been expected from observed partial depletion. The ligand-binding activity of integrins is regulated dynamically through cellular-signaling mechanisms, and multiple integrin molecules cooperate to induce adhesion (40). Thus, integrin α3β1 on F9 cells conceivably binds laminin with low affinity, and a partial decrease in cell surface levels may strongly affect cooperative interactions and, thus, ligand binding.
Analysis of RNAi in Drosophila extracts provided compelling evidence that siRNAs, generated by the cleavage of dsRNA, are effector molecules guiding a multicomponent nuclease, RISC, to the target mRNA (6, 8, 9, 12). Formation of 21- to 23-nt-long RNAs also has been documented for plants and C. elegans (10, 11). In Drosophila extracts and immunoprecipitates from Drosophila cells overexpressing Dicer, siRNA generation was shown to be ATP-dependent (8, 12), with RNAs produced in the presence of ATP having slightly higher mobility (8). We found that cytoplasmic extracts prepared from EC cells actively process dsRNA to ≈23-nt fragments. However, addition of ATP stimulated this reaction only modestly, ≈2-fold, and the RNAs generated had lower rather then higher mobility. The function of ATP in RNAi remains to be established.
Dicer is an evolutionarily conserved protein whose involvement in RNAi has been established for Drosophila and C. elegans (4, 12, 41). Immunoprecipitates containing the overexpressed tagged human Dicer were shown previously to process dsRNA to ≈22-nt fragments (12). By immunodepleting the protein with specific antibodies, we demonstrated that Dicer indeed is involved in the generation of siRNAs in mammalian cell extracts. Although RNAi and PTGS have been suggested to occur in the cytoplasm (1–3), none of the proteins directly involved in these processes had been localized. Our cell-fractionation and immunolocalization experiments showed that Dicer resides in the cytoplasm of mammalian cells, consistent with the absence in Dicer proteins of conserved sequences resembling nuclear localization signals. CAF, one of the three Dicer-like proteins in Arabidopsis, contains an N-terminal extension bearing two nuclear localization signals (42). Small, ≈23-nt RNAs derived from viroids, rod-shaped RNA pathogens replicating in the nucleus, can be detected in plants (43). Thus, in plants, Dicer-like proteins also may function in the nucleus. In addition to dsRNA processing, Dicer is involved in the biogenesis of regulatory stRNAs in C. elegans and HeLa cells (4, 5). The localization studies suggest that processing of stRNA precursors takes place in the cytoplasm. It will be interesting to determine whether the remarkably high levels of Dicer in EC cells are related to its function in stRNA synthesis and whether this is a more general property of embryonic and/or undifferentiated cells.
Acknowledgments
We thank P. Kopp for help and advice with ES cell experiments, S. Massa for FACS analyses, V. Jäggin and D. Roman for performing cell sorting, H. Brinkhaus, A. Littlewood-Evans, and M. Leu for assistance with fluorescent microscopy, and F. Asselbergs, F. Meins, and G. Thomas for discussions or comments on the manuscript. Friedrich Miescher Institut is a part of the Novartis Research Foundation.
Abbreviations
- FACS
fluorescence-activated cell sorting
- β-gal
β-galactosidase
- ds
double-stranded
- EC
embryonal carcinoma
- ES
embryonic stem
- GFP
green fluorescent protein
- PTGS
posttranscriptional gene silencing
- RNAi
RNA interference
- siRNA
small interfering RNA
- ss
single stranded
References
- 1.Hammond S M, Caudy A A, Hannon G J. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 2.Matzke M A, Matzke A J, Pruss G J, Vance V B. Curr Opin Genet Dev. 2001;11:221–227. doi: 10.1016/s0959-437x(00)00183-0. [DOI] [PubMed] [Google Scholar]
- 3.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A, Pasquinelli A E, Conte D, Li N, Parrish S, Ha I, Baillie D L, Fire A, Ruvkun G, Mello C C. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, McLachlan J, Pasquinelli A E, Balint E, Tuschl T, Zamore P D. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 6.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature (London) 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 7.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton A J, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 11.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 13.Svoboda P, Stein P, Hayashi H, Schultz R M. Development (Cambridge, UK) 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 14.Wianny F, Zernicka-Goetz M. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 15.Caplen N J, Parrish S, Imani F, Fire A, Morgan R A. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. . (First Published July 31, 2001; 10.1073/pnas.171251798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 17.Ui-Tei K, Zenno S, Miyata Y, Saigo K. FEBS Lett. 2000;479:79–82. doi: 10.1016/s0014-5793(00)01883-4. [DOI] [PubMed] [Google Scholar]
- 18.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 19.Silverman R H. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan J F, editors. New York: Academic; 1997. pp. 515–551. [Google Scholar]
- 20.Clemens M J, Elia A. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 21.Burke D C, Graham C F, Lehman J M. Cell. 1978;13:243–248. doi: 10.1016/0092-8674(78)90193-9. [DOI] [PubMed] [Google Scholar]
- 22.Barlow D P, Randle B J, Burke D C. Differentiation. 1984;27:229–235. doi: 10.1111/j.1432-0436.1984.tb01433.x. [DOI] [PubMed] [Google Scholar]
- 23.Francis M K, Lehman J M. Mol Cell Biol. 1989;9:3553–3556. doi: 10.1128/mcb.9.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 25.Krause D, Silverman R H, Jacobsen H, Leisy S A, Dieffenbach C W, Friedman R M. Eur J Biochem. 1985;146:611–618. doi: 10.1111/j.1432-1033.1985.tb08695.x. [DOI] [PubMed] [Google Scholar]
- 26.Kalvakolanu D V, Sen G C. Proc Natl Acad Sci USA. 1993;90:3167–3171. doi: 10.1073/pnas.90.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludin B, Doll T, Meili R, Kaech S, Matus A. Gene. 1996;173:107–111. doi: 10.1016/0378-1119(95)00899-3. [DOI] [PubMed] [Google Scholar]
- 28.Hogan B, Beddington R, Costantini F, Lavy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 29.Pogacic V, Dragon F, Filipowicz W. Mol Cell Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer M, Kaech S, Knutti D, Matus A. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 31.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 32.Verdel A, Curtet S, Brocard M P, Rousseaux S, Lemercier C, Yoshida M, Khochbin S. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 33.Muller U, Bossy B, Venstrom K, Reichardt L F. Mol Biol Cell. 1995;6:433–448. doi: 10.1091/mbc.6.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny J C, Muller U. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 35.Vekeman S, Jaspers M, Cassiman J J. FEBS Lett. 1993;327:207–212. doi: 10.1016/0014-5793(93)80171-p. [DOI] [PubMed] [Google Scholar]
- 36.Sonnenberg A, de Melker A A, Martinez de Velasco A M, Janssen H, Calafat J, Niessen C M. J Cell Sci. 1993;106:1083–1102. doi: 10.1242/jcs.106.4.1083. [DOI] [PubMed] [Google Scholar]
- 37.DiPersio C M, Shah S, Hynes R O. J Cell Sci. 1995;108:2321–2336. doi: 10.1242/jcs.108.6.2321. [DOI] [PubMed] [Google Scholar]
- 38.Hemler M E. In: Guidebook to the Extracellular Matrix, Anchor and Adhesion Proteins. Kreis T, Vale R, editors. London: Oxford Univ. Press; 1999. pp. 196–212. [Google Scholar]
- 39.Zhou A, Paranjape J M, Der S D, Williams B R, Silverman R H. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz M A, Schaller M D, Ginsberg M H. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 41.Knight S W, Bass B L. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobsen S E, Running M P, Meyerowitz E M. Development (Cambridge, UK) 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 43.Papaefthimiou I, Hamilton A, Denti M, Baulcombe D, Tsagris M, Tabler M. Nucleic Acids Res. 2001;29:2395–2400. doi: 10.1093/nar/29.11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]