Delineating the mechanisms that drive hepatic injury and hepatocellular carcinoma (HCC) progression is critical for development of novel treatments for recurrent and advanced HCC but also for the development of diagnostic and preventive strategies. Heat shock protein 70 (HSP70) acts in concert with several cochaperones and nucleotide exchange factors and plays an essential role in protein quality control that increases survival by protecting cells against environmental stressors.
KEYWORDS: DNA damage checkpoints, ERK negative feedback signaling, molecular chaperones, oxidative stress, p53 apoptosis
ABSTRACT
Delineating the mechanisms that drive hepatic injury and hepatocellular carcinoma (HCC) progression is critical for development of novel treatments for recurrent and advanced HCC but also for the development of diagnostic and preventive strategies. Heat shock protein 70 (HSP70) acts in concert with several cochaperones and nucleotide exchange factors and plays an essential role in protein quality control that increases survival by protecting cells against environmental stressors. Specifically, the HSP70-mediated response has been implicated in the pathogenesis of cancer, but the specific mechanisms by which HSP70 may support malignant cell transformation remains to be fully elucidated. Here, we show that genetic ablation of HSP70 markedly impairs HCC initiation and progression by distinct but overlapping pathways. This includes the potentiation of the carcinogen-induced DNA damage response, at the tumor initiation stage, to increase the p53-dependent surveillance response leading to the cell cycle exit or death of genomically damaged differentiated pericentral hepatocytes, and this may also prevent their conversion into more proliferating HCC progenitor cells. Subsequently, activation of a mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) negative feedback pathway diminishes oncogenic signals, thereby attenuating premalignant cell transformation and tumor progression. Modulation of HSP70 function may be a strategy for interfering with oncogenic signals driving liver cell transformation and tumor progression, thus providing an opportunity for human cancer control.
INTRODUCTION
The underlying general principles by which environmental factors and inherited predispositions regulate cancer initiation, growth, and metastasis are adequately defined (1–3). It has been, however, challenging to identify and validate the exact nature of initiating events that are strongly influenced by etiological background and determine the rates and spectrum of cancer-driving genetic alterations and unique molecular fingerprints in specific tumor types. In this context, primary liver cancer, and in particular hepatocellular carcinoma (HCC), which is an aggressive malignancy of increasing incidence and a leading cause of cancer-related deaths worldwide (4–6), continues to be a major focus of investigation for prevention and therapeutic purposes. Commonly, HCC develops after a long period of latency as a complication of chronic liver injury and inflammation, primarily induced by alcohol use, viral hepatitis, or nonalcoholic fatty liver disease. Hepatic cirrhosis represents another aspect of inflammatory and fibrotic chronic liver damage, but its exact tumor-promoting effect remains largely unexplored. Moreover, like for other cancer types, one of the hallmarks of HCC is the genetic complexity and heterogeneity uncovered by recent genome-wide deep-sequencing studies (7–10). Certainly, this genetic variation has a significant impact on the HCC treatment response and thus long-term survival. An interesting emerging concept suggests that the HCC formation is triggered by DNA adduct-forming agents, including reactive oxygen species (ROS), that increase in response to inflammation and environmental carcinogens. These events cause chronic liver damage and trigger the formation of initiating cancer cells, primarily through growth factor-driven conversion of differentiated pericentral hepatocytes into hepatic cell-like progenitor cells, as well as expansion of preexisting bipotential hepatic stem cells with a dominant role in regeneration of the injured liver (11–13). The propagation of pretransformed cancer progenitors into neoplastic and dysplastic foci seems to depend on multiple cycles of compensatory proliferation and death of hepatocytes induced by Jun N-terminal protein kinase (JNK) activation combined with loss of protective NF-κB and other proinflammatory signals as well as expression of antioxidant proteins (14, 15). Although this pathogenic mechanism of HCC development is supported from biochemical and genomic studies, the molecular mechanisms and mutational processes operating during the initiation and long evolution of HCC remain largely unknown.
In addition to genetic and epigenetic changes, equally important for survival of malignant cells and disease progression is the contribution of the chaperone-directed proteome homeostasis (proteostasis). This highly wired system coordinates protein synthesis, folding, trafficking, and degradation (16, 17) and is regulated by stress sensors, including heat shock proteins (HSPs), transcription factors such as HSF1, chromatin modifiers, and signaling pathways (18–20). By also affecting cellular metabolic and genomic stability, being an integral component of the DNA damage (DDR) response (21), the proteostasis network (PN) activity is vital to cellular fitness. Indeed, the programming of well-established core hallmarks of cancer, including oncogenic signaling, cellular energy metabolism, and genome integrity, relies on chaperone-mediated protein quality control (19, 20, 22–24). In line with its constitutive expression in cancer, accumulating evidence has demonstrated that the stress-inducible HSP70 (HSPA1A/HSPA1B) chaperone has a central role in tumor induction and progression (25–27). Indeed, HSP70 overexpression in the majority of human tumors, including HCC, is associated with disease progression, increased resistance to radiation and chemotherapy of tumor cells, and poor prognosis of cancer patients (22, 25). Furthermore, combined silencing of HSP70 and HSC70 (constitutively expressed) causes cell cycle arrest and tumor-specific apoptosis (28). Likewise, downregulation of HSP70 considerably enhances the tumor-inhibitory effects of pharmacologic HSP90 inhibitors, indicating a synergism of the HSP70/HSP90 chaperone machineries in cell survival and tumor promotion (29). Although targeting HSP70 function likely impacts a large array of cellular functions that are altered to sustain malignancy, including inhibition of cell proliferation by inducing cellular senescence and decreased tumor cell survival by promoting apoptosis (30, 31), the specific mechanisms by which HSP70 loss may affect growth of specific tumor types, including HCC, are not well understood. Moreover, the potential therapeutic utility of inhibiting HSP70 on DNA damage repair pathways to affect genomic stability and thus influencing the barrier for tumor initiation and progression is elusive (32, 33). This prompted us to investigate a possible causal relationship between protein homeostasis accomplished by the HSP70 molecular chaperone machinery and regulation of the carcinogen-induced DNA damage response (DDR) and growth factor signaling resulting in HCC initiation and progression.
RESULTS
Deletion of hsp70 suppresses chemical-induced liver tumorigenesis.
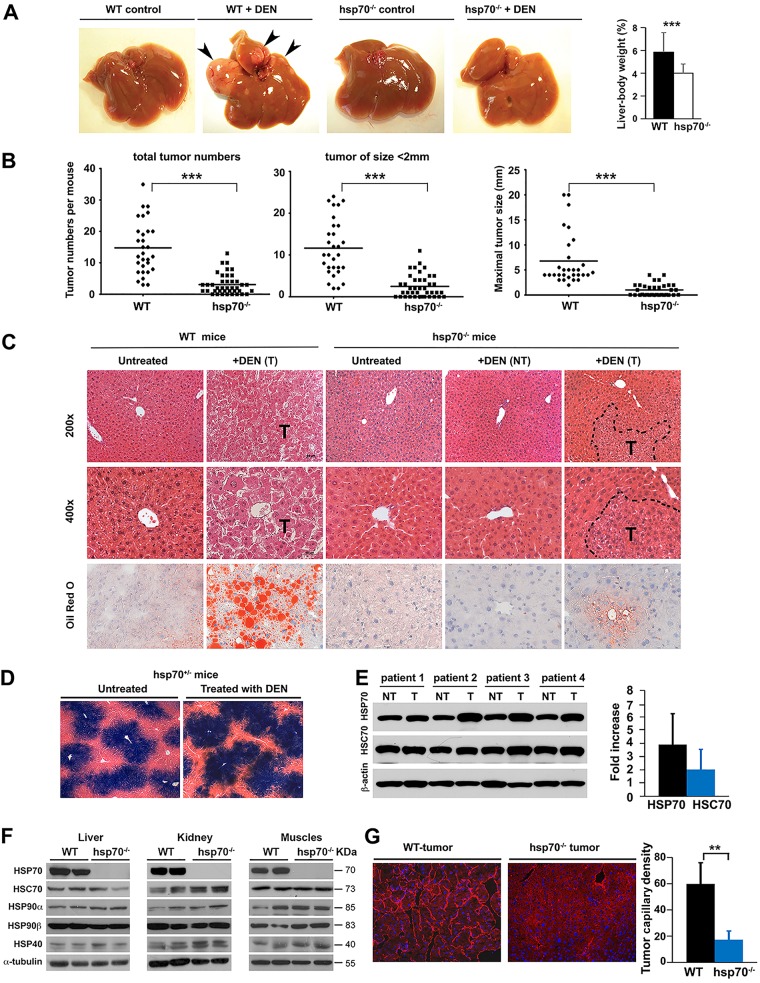
To investigate the specific role of stress-inducible HSP70 in cancer pathogenesis, we employed a diethylnitrosamine (DEN)-induced primary HCC mouse model that incorporates chronic liver injury, inflammation, and DNA damage, thus mimicking key pathological and histological features of human HCC (34, 35). DEN administration at postnatal day 14 (P14) is sufficient for HCC induction in 100% of male mice within 7 to 8 months. Particularly intriguing in this analysis was the markedly reduced tumor burden (numbers and size) in DEN-treated hsp70−/− mice compared to wild-type (WT) mice, indicating a potent inhibitory effect of HSP70 ablation on tumor development (Fig. 1A and B). Histological examination detected signs of liver pathology with severe steatosis in DEN-treated WT mice. In contrast, hsp70−/− mice displayed an almost-normal hepatic histology (Fig. 1C). Noticeably, the majority (around 80%) of HCC in WT mice expressed alpha-fetoprotein (α-FP), an established liver tumor marker (36), while the fewer small tumors that developed in a fraction of hsp70−/− mice were rarely positive for α-FP (around 20%) (data not shown). A strong basal transcriptional expression of hsp70 as revealed by reporter lacZ staining was also observed in the livers, but the expression patterns were indistinguishable between DEN-treated and untreated hsp70−/− mice (Fig. 1D). HSP70 deficiency had no significant effect on the expression of other members of the HSP family in different tissues (Fig. 1F). In addition, HSP70 was specifically upregulated in human HCCs (Fig. 1E), which is in agreement with previous reports implicating HSP70 as a marker for HCC development (37, 38). Another notable effect of hsp70 deletion was the inhibitory effect on tumor angiogenesis, which was verified by CD31 staining (Fig. 1G). This suggests that HSP70 may be required for tumor neovasculogenesis, although the possibility that the effect is simply a reflection of the different growth rates of tumors developed in mutant and WT mice cannot be ruled out. Taken together, these results indicate an important role for HSP70 in promoting hepatocarcinogenesis. Furthermore, the significant impact on tumor multiplicity strongly suggests that HSP70 inactivation may also interfere with HCC initiation and progression.
FIG 1.
HSP70 is needed for carcinogen-induced liver cancer development. (A to C) Analyses were performed 8 months after DEN injection at 14 days of age (P14). (A) Representative images of livers from WT and hsp70−/− mice at an advanced stage of disease. Tumor nodules are indicated (arrowheads). (B) Quantification of liver tumors in WT (n = 30) and hsp70−/− (n = 40) mice. (C) Histological analysis (H&E) of liver and tumor sections from WT and hsp70−/− mice. Top and middle panels, representative sections show large HCC and extensive pathology in DEN-treated WT mice. In contrast, hsp70−/− mice displayed an almost normal hepatic histology, with small tumors detected only in a fraction of DEN-treated mice. Broken lines depict the tumor (T) border. Original magnifications are indicated. Bottom panels, representative liver sections with staining for Oil Red O. Original magnification, ×400. NT, no tumors. (D) Histological β-gal staining images of liver sections from DEN-treated or untreated heterozygous hsp70+/−-lacZ reporter mice, visualizing hsp70 transcriptional expression. (E) Increased HSP70 expression protein level in human HCCs. A representative WB analysis of human HCC samples (paired normal and tumor tissues from the same patient) for expression of HSP70s is shown. Western blots for HSP70 or HSC70 were quantified by densitometry, and protein levels normalized to β-actin levels were expressed as relative fold increase to paired controls (tumor [T] to nontumor [NT] samples) (n = 10 patient samples per group). (F) HSP70 ablation does not affect the protein levels of other members of the HSP family in different tissues. A representative Western blot analysis of the indicated whole tissue extracts from 8- to 12-week-old WT and hsp70−/− mice using antibodies against HSP family members and α-tubulin as a loading control is shown (3 independent experiments). (G) Attenuation of tumor angiogenesis in DEN-treated hsp70−/− mice. Representative immunofluorescence of CD31 staining performed on liver sections, showing microvessel density (original magnification, ×400), is shown. Quantification of microvessels in WT (n = 3) and hsp70−/− (n = 4) HCCs per high-power field is indicated (P < 0.01 for WT versus hsp70−/−). For all panels, bars represent mean ± SD. Statistical significance is indicated (**, P < 0.01; ***, P < 0.001).
HSP70 inactivation sensitizes DEN-induced liver cell death but does not differentially affect compensatory proliferation of hepatocytes during HCC initiation and progression.
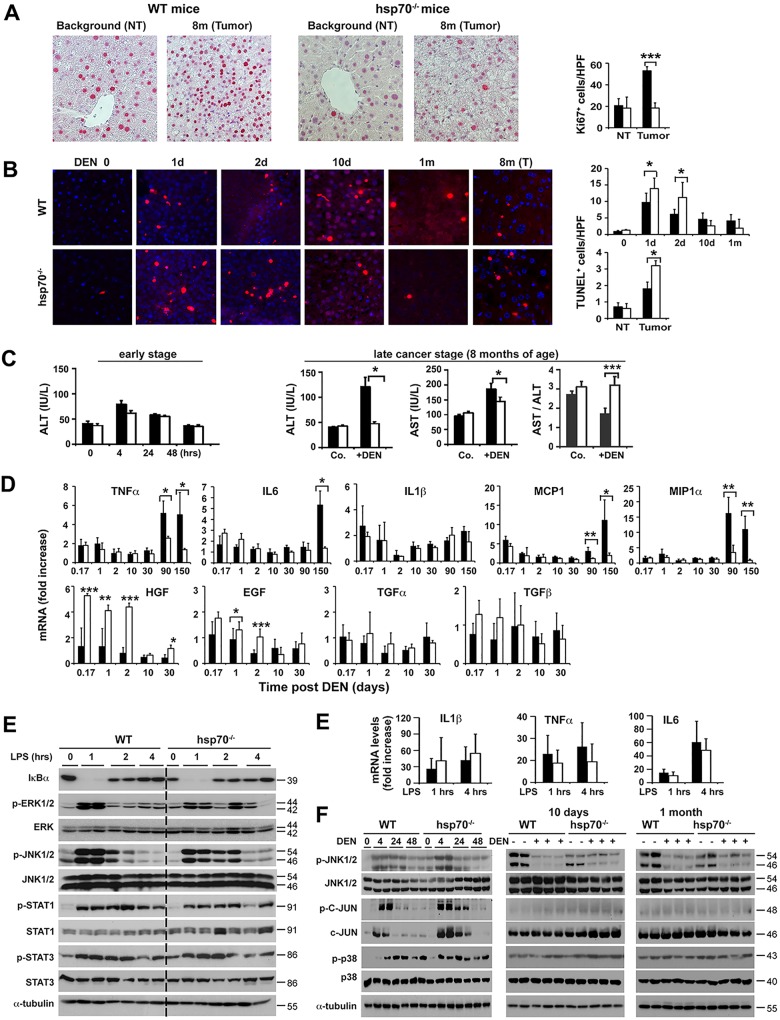
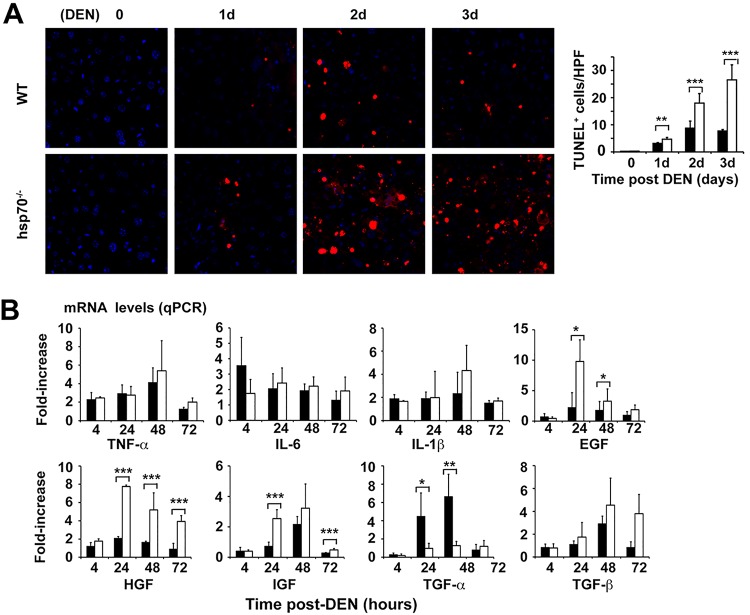
To investigate the specific mechanisms by which HSP70 loss induced resistance to HCC initiation, we compared early (short-term) and late effects of DEN injection on signal transduction and hepatic cell physiology. Short-term effects of DEN on liver integrity are marked by hepatocyte death, increased cytokine production, and compensatory hepatocyte proliferation and are proposed as good predictors of the outcome of the disease (35, 39). According to recent proposals, differentiation and conversion of DNA-damaged mature hepatocytes to HCC progenitor cells (HCC-PCs) seem to be a critical early step in HCC initiation, which takes place under conditions of intact p53-dependent signaling events (11, 12, 40). Staining for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) indicated a greater increase in apoptosis in hsp70−/− livers at 24 and 48 h post-DEN treatment (Fig. 2B). In support of this specific increased apoptotic activity, livers of hsp70−/− mice exhibited elevated poly(ADP-ribose) polymerase (PARP) cleavage in response to DEN treatment (see Fig. 6C). Importantly, as described below (see Fig. 7), the increase in hepatocyte death was not accompanied by enhanced compensatory cell proliferation in the pericentral (zone 3) region, which is composed of 7 or 8 concentric layers of hepatocytes around the central hepatic vein (41) and where DEN is preferentially metabolized and activated by Cyp2E enzymes. Interestingly, DEN administration led to a marked decrease in proliferating cells in the hsp70−/− livers. Of note, WT and hsp70−/− mice displayed increased but comparable serum alanine aminotransferase (ALT) levels at 4 to 48 h after DEN treatment (Fig. 2C), indicating that HSP70 loss might selectively induce the elimination of DNA-damaged liver cells by apoptosis without eliciting alterations consistent with an overly hepatic injury. As expected, DEN-induced tumors (late stage) in WT mice exhibited a sustained increase in cellular proliferation (Ki67+ cells) but also slightly increased apoptosis (and necrosis) (TUNEL+ cells) compared with nonmalignant liver tissue; high levels of serum ALT and aspartate transaminase (AST) and a decreased AST/ALT ratio, a useful parameter for nonalcoholic liver disease, were also detected in these mice, indicating enhanced liver injury, likely caused by the tumor progression (42). In contrast, DEN-treated hsp70−/− mice displayed unchanged number of proliferating cells between nontumor and small tumor areas but markedly increased numbers of apoptotic hepatocytes in tumor lesions (Fig. 2A and B). Enhanced cell apoptosis and an overall decrease in compensatory proliferation were also observed in DEN-exposed livers of hsp70−/− adult mice (8 to 12 weeks of age) (Fig. 3 and data not shown). Thus, the overall pattern of apoptosis and proliferation observed after initial genotoxic challenge suggests that the enhanced compensatory proliferation of pericentral hepatocytes, which correlates with augmented liver tumorigenesis, does not occur in the absence of HSP70. Therefore, HSP70 may be a factor that regulates a key step in HCC initiation.
FIG 2.
HSP70 deficiency increases apoptotic priming and potential elimination of DNA-damaged hepatocytes and exerts differential effects on inflammatory and growth factor responses induced by carcinogen challenge. (A and B) Ki67 and TUNEL staining of liver or tumor (T) sections from WT or hsp70−/− mice at the indicated time points after DEN administration at P14. Quantification of Ki67+ proliferating cells or TUNEL+ apoptotic hepatocytes per high-power field (HPF) is presented (right panels). Original magnifications, ×200 (Ki67) and ×400 (TUNEL). Bars show mean ± standard error of the mean (SEM) (n = 4 or 5 mice per group). (C) Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels. ALT and AST levels and ALT/AST ratio are shown for WT or hsp70−/− mice 8 months after DEN treatment (three right panels). Co., WT or hsp70−/− mice without DEN treatment served as controls. Bars show mean ± SEM (n = 5 mice per group). (D) Quantitative PCR of mRNA transcript levels of selected cytokines and growth factors in livers of DEN-treated mice. Bars represent mean ± SD (n = 4 or 5 mice). (E) Left, insignificant effect on hepatic LPS-induced signaling events in hsp70−/− mice. Representative WB analysis of IkBα degradation and levels of phosphorylated JNK1/2, STAT1, STAT3, and ERK in livers of WT and hsp70−/− mice are shown for the indicated time points after LPS injection (i.p. at 2.5 mg/kg of body weight) (from two experiments). Right, qPCR analysis of mRNA levels of inflammatory cytokines induced in response to LPS in the livers. Bars show mean ± SEM (n = 5 mice per group). (F) Effects of HSP70 ablation on DEN-induced MAPK signaling activation. Representative WB analyses of p-JNK1/2 (Y183/Y185), p-c-JUN (S63), and p-p38 (Y180/Y182) protein levels in the livers of WT and hsp70−/− mice at the indicated times post-DEN challenge are shown (from two experiments). For all panels, statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
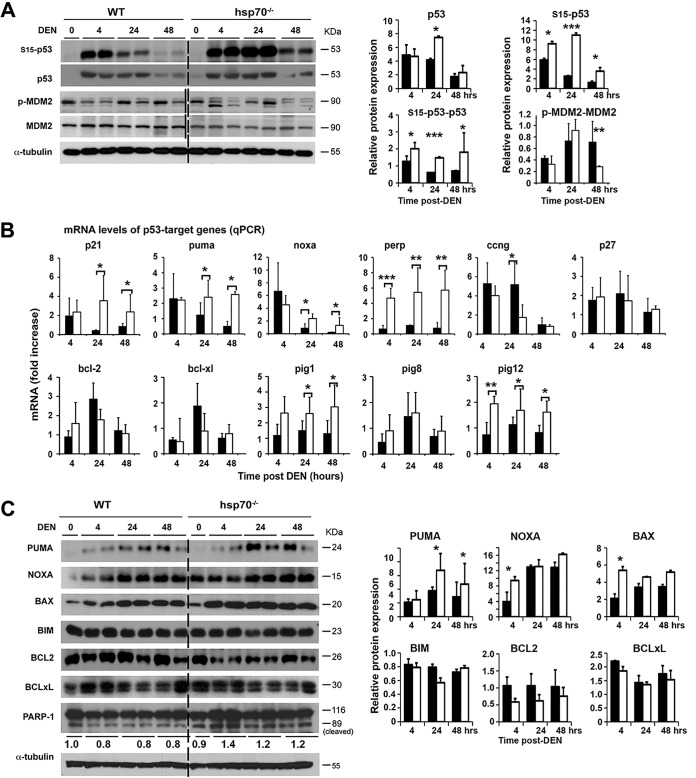
FIG 6.
HSP70 loss promotes p53-dependent cellular apoptosis that provides a barrier for liver cancer initiation and progression. (A) WB analysis to monitor levels of p53 and phosphorylated (p) forms of p53 and MDM2 activation following treatment of WT and hsp70−/− mice with DEN at P14. The level of p53 or p-p53 protein forms normalized to an α-tubulin loading control was expressed as relative fold increase compared to the control (WT without DEN treatment). The level of p-MDM2 normalized to total protein level was expressed as relative fold increase compared to the control (WT without DEN, indicated as 0) which was arbitrarily set at 1. (B) Expression of p53 target genes in the livers of DEN-treated mice determined by qPCR. Values are presented as relative mRNA expression. WT or hsp70−/− mice at P14 without DEN treatment served as a control. (C) WB analysis for expression of p53 target genes and pro- or antiapoptotic genes induced following DEN treatment of WT and hsp70−/− mice at P14. The protein level normalized to α-tubulin was expressed as relative fold increase compared to the control (WT without DEN treatment). The ratio of cleaved (89-kDa) to uncleaved (116-kDa) PARP1 expressed as relative fold increase compared to the control (WT without DEN, indicated as 0), which was arbitrarily set at 1, is shown below the panel. For all panels, bars represent mean ± SD (n = 5 or 6 mice per group); statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
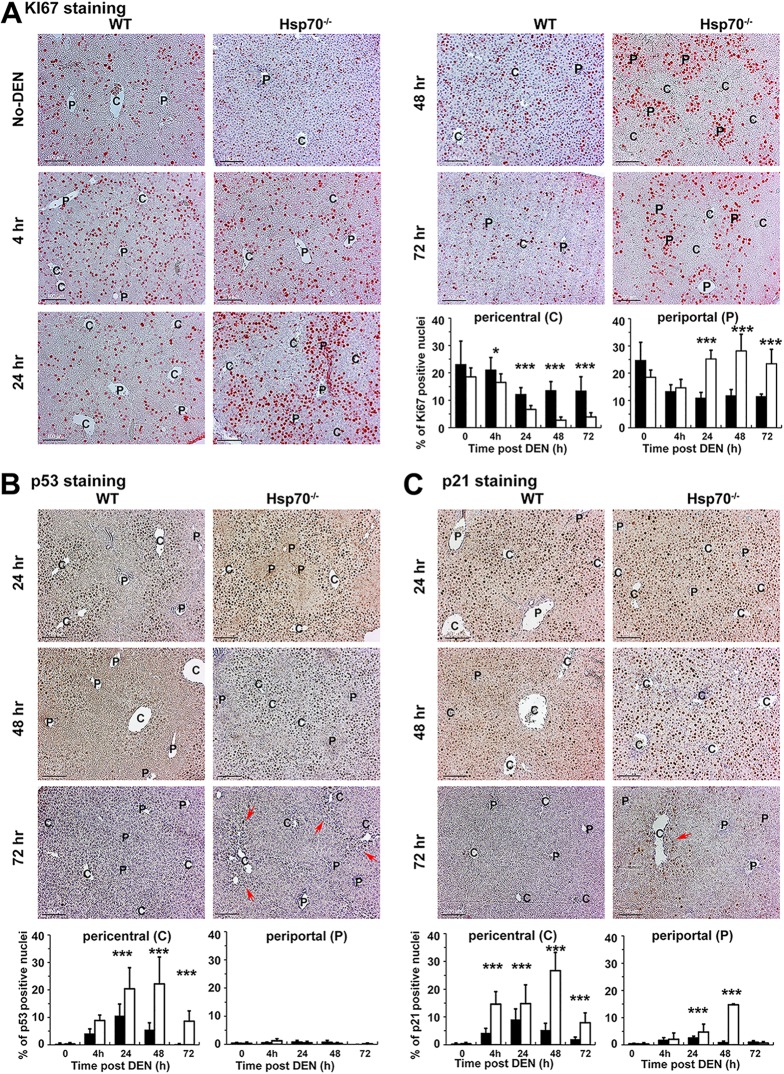
FIG 7.
HSP70 ablation suppresses HCC initiation by triggering an excessive p53 activation and p21 expression that leads to the cell cycle exit or death of pericentral DNA-damaged hepatocytes. Male mice of the indicated genotypes were exposed to DEN at P14, and nuclear expression of Ki67 (A) (indicated by pink) or p53 (B) or p21 (C) (indicated by brown and pink arrows, respectively) in the livers at the indicated time points was detected by IHC. The periportal (P) and pericentral (C) regions visualized by staining for GS are indicated. All scale bars = 200 μm. Original magnification, ×200. Quantification of Ki67, p53, and p21 staining data is presented. Bars show the percentage of positive hepatocytes in the pericentral region (n ≥ 20 fields from 3 or 4 mice per group; mean ± SD); statistical significance is indicated (*, P < 0.05; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
FIG 3.
Increased hepatocyte death associated with elevated growth factor production in livers of DEN-challenged adult hsp70−/− mice. (A) TUNEL staining of liver sections from WT or hsp70−/− mice at the indicated time points after DEN injection at 8 to 12 weeks of age. Quantification of TUNEL+ apoptotic hepatocytes per high-power field (HPF) is presented (right panels). Original magnifications, ×200 (Ki67) and ×400 (TUNEL). (B) Relative mRNA levels of cytokines and growth factors in livers of 8- to 12-week-old DEN-treated mice by quantitative reverse transcription-PCR (RT-PCR). Age-matched WT or hsp70−/− mice without DEN treatment served as a control. For all panels, bars represent mean ± SD (n = 5 mice per group); statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
Differential effects of HSP70 inactivation on DEN-induced inflammatory and growth factor responses.
Chronic inflammation is a critical factor in cancer development in different organs, including liver (43–45). The function of HSP70 in NF-κB activation and inflammatory responses, particularly, is less clear. HSP70, by interacting with regulatory components of NF-κB signaling pathways, may inhibit the inflammatory response (46, 47), but on the other hand, exogenous HSP70 has been shown to promote NF-κB activation and expression of proinflammatory cytokines and chemokines (48, 49). Analysis of inflammatory cytokines in the early tumor-initiating stage revealed rapid but similar levels of elevation of mRNA of interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), IL-1β, monocyte chemoattractant protein 1 (MCP1), and macrophage inflammatory protein 1α (MIP1α) in WT and hsp70−/− mice (Fig. 2D). This proinflammatory response persisted at a lower steady-state level in WT mice but dramatically increased during the progression stage (at 3 to 5 months), probably reflecting early events associated with tumor formation. In contrast, the initial inflammatory response in hsp70−/− mice was sustained at a lower steady-state level between 1 and 5 months of age. Consistent with the early-response results, the lipopolysaccharide (LPS)-induced degradation of IκBα (indicating NF-κB activation) was inhibited in the livers of both genotypes to similar extents (Fig. 2E). Moreover, LPS-induced phosphorylated extracellular signal-regulated kinase (p-ERK), p-JNK, p-p38, p-STAT1, or pY-STAT3 signals were stimulated at comparable levels (Fig. 2E). In contrast, we determined significantly increased mRNA expression of hepatocyte growth factor (HGF) and, to a lesser degree, of epidermal growth factor (EGF) in DEN-treated hsp70−/− mice compared to WT mice (Fig. 2D). We also confirmed a similar pattern in growth factor mRNA expression in DEN-treated adult mice (8 to 12 weeks of age) except that hsp70−/− mice displayed lower levels of tgfα and slightly but insignificantly elevated tgfβ compared to controls (Fig. 3B). Notably, increased transforming growth factor α (TGF-α), a ligand of EGF receptor signaling, and inhibition of TGF-β signaling have been shown to favor HCC formation (50). As the hepatic inflammatory response involves interactions between different cell types, including hepatocytes, stellate cells, and Kupffer cells, it is conceivable that HSP70 ablation might also affect cell-intrinsic cytokine-induced signaling in hepatocytes and thereby influence HCC development. However, this possibility is unlikely, because TNF-α or IL-6 stimulation of primary hepatocytes induced a similar effect on IκBα degradation or pY-STAT1, pY-STAT3, p-JNK, and p-AKT activation in both genotypes (data not shown). Thus, the absence of HSP70 significantly enhanced carcinogen-induced growth factor expression but had an insignificant impact on classical inflammatory cytokine responses during the tumor initiation phase.
Effects of HSP70 loss on DEN-induced p38 and JNK activation.
Given the specific requirement of the JNK1, c-JUN, and p38 pathways during liver cancer initiation (51, 52), we examined whether HSP70 inactivation may also affect the mitogen-activated protein kinase (MAPK)-mediated stress response. Consistently, DEN injection caused comparable levels of initial activation of p38, JNK, and its c-JUN substrate in the livers of mutant and WT control mice (Fig. 2F). Notably, while similar patterns of p-JNK1/2 activation were detected in mice with both genotypes, p-JNK1/2 activity was markedly repressed by day 10 and 1 month post-DEN treatment compared to that in untreated control mice. This negative effect was unexpected because previous studies indicated a negative role of HSP70 in regulating JNK activity (53). Thus, as discussed below, the effect of HSP70 deficiency on increasing cell death associated with reduced proliferation of DNA-damaged hepatocytes is unlikely to be a reflection of altered p38, JNK, or c-JUN activity.
Control of REF1 activity by HSP70 has a role in the activation of genotoxically induced DNA adduct formation and the repair pathway.
Alkylating agents such as DEN are metabolized by cytochrome P450 enzymes in the liver and generate a variety of covalent DNA adducts (54) that are repaired through multiple pathways, including direct DNA damage reversal, base excision repair (BER), and mismatch repair (MMR), of primary alkyl lesions or secondary lesions such as single- or double-strand DNA breaks (55). Defects in DNA damage repair function associated with incomplete DNA restoration are a relevant determinant of DNA mutation, genomic instability, and tumor progression and can sensitize tumor cells to DNA-damaging therapy (56, 57). Relevant to this, the characterization of the hepatitis C virus (HCV)-associated HCC genome identified more than 11,000 somatic substitutions with predominant T → C/A→ G transitions, indicating preferential DNA repair and high intratumoral heterogeneity (7). However, there is increasing appreciation in the literature for the existence of potent p53-dependent tumor-suppressive mechanisms, downstream of DNA damage, whose activation can efficiently coordinate the fate of genotoxically damaged hepatocytes to prevent accumulation of oncogenic mutations and inhibit tumor cell progression.
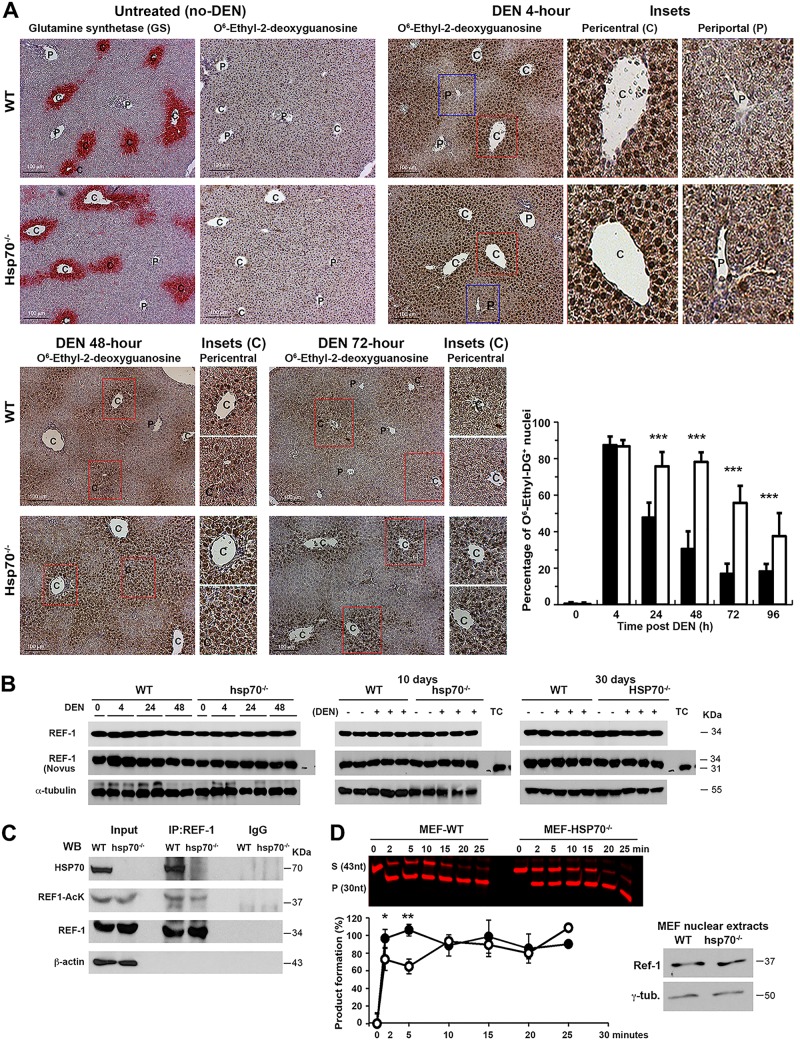
DEN injection into hsp70−/− or WT mice at P14 or 8 to 12 weeks of age (adult) induced comparable mRNA expression levels of DEN-catabolizing cytochrome P450 enzymes (e.g., CYP1A1, CYP1A2, and CYP2E1) (data not shown). This indicates that changes in DEN-induced activation and primary DNA adduct formation are unlikely to play a key role in HCC inhibition in hsp70−/− mice. To further examine the time course of DNA damage in situ, we performed immunohistochemistry (IHC) in P14 liver over 72 h post-DEN exposure. As expected, DEN administration caused centrilobular damage as indicated by the presence of mutagenic O6-ethyl-deoxyguanosine adducts (Fig. 4A). The positive cell staining was confined to the pericentral region, but no obvious difference in the maximum numbers of stained hepatocytes was observed between the genotypes at 4 h post-DEN treatment. However, at 72 h or 96 h post-DEN treatment, very little staining remained in WT liver, as opposed to significantly higher nuclear levels in the hsp70-deficient livers. This result led us to conclude that the initial formation of DNA adducts and intensity of genotoxically induced DNA damage were not affected by HSP70 loss but that its absence resulted in a prolonged recovery (repair) phase due to reduced rates of DNA adduct removal.
FIG 4.
A link between reduced REF1 activity in the absence of HSP70 and prolonged DNA damage and sustained levels of nuclear DNA adduct staining in DEN-exposed livers. (A) In situ detection and quantification of DNA adducts in livers of WT or hsp70−/− mice following DEN challenge at P14. Representative IHC photomicrographs for promutagenic O6-ethyl-deoxyguanosine DNA adducts at the indicated time points post-DEN administration are shown. All scale bars = 200 μm. Original magnification, ×200. The periportal (P) region and pericentral (C) region visualized by staining for glutamine synthetase (GS) are indicated. Quantification of O6-ethyl-deoxyguanosine staining data is presented. Bars show the percentage of positive hepatocytes in the pericentral region (n ≥ 20 fields from 3 mice per group; mean ± SD). (B) Western blot analysis for REF1 expression in livers of WT and hsp70−/− mice at the indicated time points after DEN injection at P14. Notably, probing protein levels of REF1 with two different antibodies raised against full-length protein revealed a slow-migrating band (about 34 kDa in liver extracts) and a fast-migrating band (about 31 kDa in control tumor cell lysates), which likely reflect the full-length (37-kDa) and N-terminal truncated (34-kDa) forms of REF1, respectively (124). Protein extracts from HEK293 were used as a control (TC) (from two experiments). (C) HSP70 interacts with REF1 in vivo and potentially regulates REF1 acetylation and activity. WT or hsp70−/− mice were DEN challenged at P14. Protein extracts from livers collected 10 days post-DEN treatment were subjected to immunoprecipitation using antibody to the endogenous REF1 protein. Proteins were subjected to WB analysis for HSP70, REF1, or total acetylated REF1 (from three experiments). (D) The absence of HSP70 negatively affects REF1 endonuclease activity. AP endonuclease activity of REF1 was measured in nuclear extracts (15 ng) from WT or hsp70−/− MEFs exposed to hydrogen peroxidase (200 μM) for 3 h. A representative image showing the conversion of the substrate (S) to product (P) at 0, 2, 5, 10, 15, and 20 min is shown. Western blot analysis confirmed the presence of similar amounts of REF1 protein in the nuclear extracts of the two cell lines (right panel). Quantification of the AP endonuclease activity of each reaction was calculated as (product/substrate at time zero) × 100% (lower panel). Bars show mean ± SD from three independent experiments. For all panels, statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
Since the vast majority of hepatocytes were proliferating in the early stage of tumor initiation, unresolved replication stress (single-strand DNA break accumulation and replication fork stalling) was considered a likely source of prolonged DNA damage in DEN-exposed livers of hsp70−/− mice (58). In support of this proposal, previous work suggested that HSP70 may improve DNA repair activity by binding and modulating the endonuclease activity of human apurinic/apyrimidinic (AP) endonuclease (HAPE1), a master regulator of the cellular response to oxidative stress and a key base excision repair enzyme (59). Western blot (WB) analysis did not reveal significant differences in the total protein level of APE1 (REF1) in livers of DEN-treated mice with the two genotypes (Fig. 4B). However, the present study provided strong evidence that HSP70 can be a potential activator of REF1 activity by demonstrating a direct protein interaction. Indeed, we detected immunoprecipitation (IP) of the endogenous REF1 with HSP70 in liver lysates from DEN-treated mice (Fig. 4C). Stimulation of the AP endonuclease activity of REF1 due to acetylation has also been suggested as a possible mechanism regulating the efficient repair of the AP site in mammalian cells (60). In comparison, IP lysates from hsp70−/− mice displayed slightly reduced levels of acetylated REF1 (Fig. 4C). This suggests that HSP70, through an unknown mechanism, may also impact posttranslational regulation of REF1 activity. Using an AP endonuclease assay to quantify REF1 activity induced by reactive oxygen species (ROS), we observed that the enzymatic activity was slightly but reproducibly reduced in hsp70−/− nuclear extracts from mouse embryonic fibroblasts (MEFs) (Fig. 4D). Notably, the REF1 nuclear protein levels did not differ between mice with the two genotypes. Collectively, these findings suggest a causative link between reduced REF1 activity in the absence of HSP70 and prolonged DNA damage and sustained amounts of nuclear DNA adduct staining.
Effects of HSP70 inactivation on the DEN-induced DDR and checkpoint activation.
One critical aspect of the DNA damage response (DDR) with respect to cellular protection from malignancy is the activation of checkpoint signaling pathways that block cell cycle progression, recruit DNA repair factors, and/or trigger senescence or programmed cell death (61). Enforcement of the DDR checkpoints relies on the activities of the upstream protein kinases ATM, ATR, and DNA-dependent protein kinase catalytic subunit (DNA-PKcs), the downstream transducer kinases CHK1 and CHK2, and targeting proteins such as histone H2AX and p53. ATM signaling is generally activated by double-strand DNA breaks, whereas ATR responds to a broader spectrum of DNA damage and guards the genome against destabilizing replication stress. Oxidative stress-induced DNA damage can also activate both ATM- and ATR-dependent signaling (62, 63). This information suggests that increased oxidative stress in hsp70−/− mice may potentiate DEN-induced DDR checkpoint signaling. Furthermore, ROS generated by intrinsic or extrinsic sources can influence p53-mediated cell fate decisions (64) and may disfavor HCC development in our model. Notably, hsp70 deletion has been shown to increase levels of spontaneous and radiation-induced chromosomal aberrations, suggesting that HSP70 may be involved in maintaining genomic stability and thus function as a tumor inhibitor (65). However, our results demonstrating that HSP70 inactivation suppresses HCC development are somewhat inconsistent with this idea. This prompted us to investigate the underlying mechanisms in more detail.
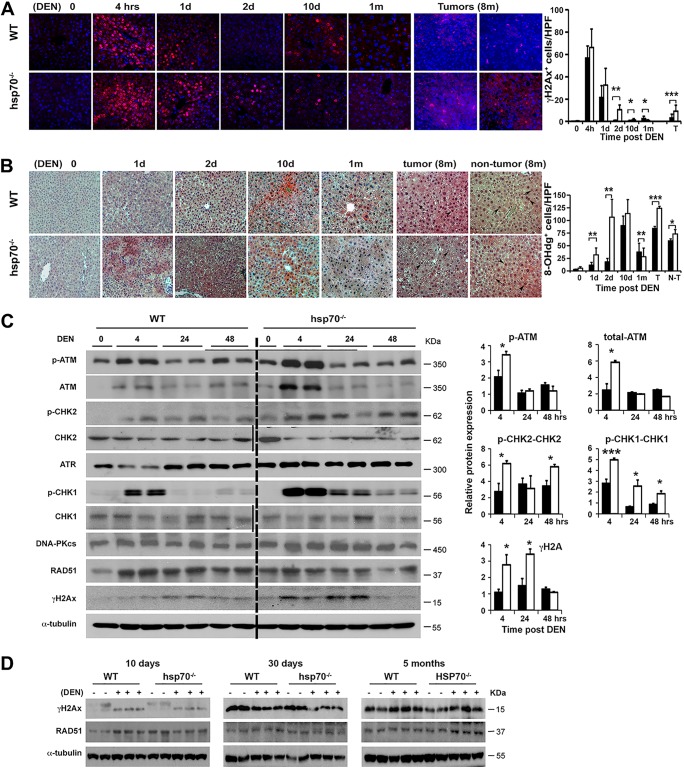
Next, the occurrence and extent of the DEN-induced DDR were determined by staining liver tissues for phosphorylated H2AX on Ser139 (γH2AX), which serves as a DDR sensor and helps maintain genome integrity (66). Untreated livers displayed extremely low levels of DDR and after DEN treatment showed an immediate induction of γH2AX-positive staining (Fig. 5A). The maximum number of γH2AX-positive hepatocytes was observed at 4 h, but quantification of the results did not reveal significant differences between the genotypes. However, in contrast to the case for WT mice, at 24 and 48 h post-DEN treatment, significantly more γH2AX+ hepatic cells remained in the hsp70−/− mice, thereby demonstrating prolonged DNA repair and/or checkpoint activation. By day 10, we observed substantially reduced γH2AX positivity, which persisted at low state-steady levels during the preneoplastic transformation phase (1 and 5 months post-DEN treatment). Remarkably, liver tumors at 8 months displayed a substantial number of γH2AX+ nuclei in hepatocytes but also in Kupffer cells and other accessory cells, with significantly higher numbers of positive cells detected in the mutant mice. By 24 and 48 h post-DEN treatment, we also observed a significantly greater accumulation of ROS-induced DNA damage lesions (revealed by staining for 8-hydroxy-2′-deoxyguanosine [8-OHdG]) in the livers of hsp70−/− mice than in those of WT mice (Fig. 5B). However, by day 10 post-DEN treatment both genotypes displayed equivalent numbers of 8-OHdG-positive cells that returned to steady-state levels by day 30. Consistent with enhanced oxidative stress, liver tumors of hsp70−/− mice at 8 months displayed increased 8-OHdG positivity, while a decreased signal was observed in aggressively grown WT tumors (Fig. 5B). Using WB analyses, we confirmed the differential expression pattern of γH2AX, ATM, p-ATM, p-CHK2, and ATR target p-CHK1 protein in the livers of hsp70−/− mice during the early stage of tumor initiation (Fig. 5C and D). No obvious differences were observed in the basal protein levels of DNA-PKcs and RAD51 among the genotypes. Consistent with the γH2AX immunostaining results, we also found an increased staining for p-ATM and p-CHK2 in the livers of hsp70−/− mice, which transiently persisted at significantly higher levels during the initiation stage of DEN-induced tumor development (data not shown). Furthermore, the increased damage signals detected by γH2AX and 8-OHdG staining during the tumor initiation stage in P14 mutant mice were confirmed in livers from adult mice exposed to DEN (data not shown).
FIG 5.
HSP70 loss enhances and prolongs the DNA damage response and checkpoint activation. (A) IF staining of γH2AX in liver sections from WT and hsp70−/− mice at the indicated time points after DEN injection at P14. Red labels, γH2AX-positive cells; blue labels, DAPI-labeled nuclei of the same field. Original magnification, ×200. The graph shows quantification of γH2AX+ cells per high-power field (HPF) (right panel). Bars show mean ± SEM (n = 3 mice per group). (B) The extent of ROS-induced DNA damage in liver sections from WT and hsp70−/− mice was analyzed by IHC staining for 8-OHdG (red labels). Representative images at the indicated times points after DEN injection at P14 are presented, and the graph shows quantification of 8-OHdG-positive cells per HPF (right panel). Data are shown as mean ± SEM (n = 5 mice per group). (C) Representative WB of DDR and checkpoint proteins in the livers of WT and hsp70−/− mice at the indicated time points after DEN injection at P14. The level of phosphorylated proteins normalized to the total level was expressed as relative fold increase with respect to the control (WT without DEN, indicated as 0), which was arbitrarily set at 1. Notably, no statistical difference in protein levels of ATR, DNA-PKcs, and RAD51 normalized to total protein (loading control) was detected between the genotypes. Bars show mean ± SEM (n = 5 mice per group). (D) WB analysis of DNA damage markers γH2AX and RAD51 in the livers of WT and hsp70−/− mice treated with DEN at P14 and assayed at the indicated times (from two experiments). For all panels, statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
Enhanced p53 activity by sustained genotoxically induced DDR in the absence of HSP70 provides a barrier for liver cancer initiation and progression.
Consistent with the strong DNA damage response (DDR) activation ascertained by elevation of γH2AX, p-ATM, and p-CHK2 in the livers of mutant mice, treatment with DEN induced a significantly higher level of p53 activity, as revealed by phosphorylation of p53 at Ser15 (Fig. 6A). Interestingly, the level of p-p53 at Ser15 persisted longer in the mutant mice. No significant changes in the induction and stabilization of total p53 were observed between the two genotypes. Consistent with this result, the level of p-MDM2 on Ser166, which increases its interaction with p300CBP, allowing MDM2-mediated ubiquitination and degradation of p53, was significantly reduced at 48 h post-DEN treatment below the untreated baseline levels in the mutant mice and compared to WT. To directly assess p53 activity, we then measured the induction of p53 target genes by quantitative real-time PCR (qPCR) after DEN injection at P14 or adult age (Fig. 6B and data not shown). As expected, a stronger induction of cycle-dependent kinase inhibitor p21 expression, which can lead to cellular senescence, but also much higher and sustained expression of several other p53 target genes that lead to cell death, including the Puma (Bbc3), Noxa (Pmaip1), and p53 apoptosis effector related to PMP-22 (Perp) genes, occurred in mutant mice. Interestingly, mRNA levels of p53-induced gene 1 (galectin-7) (Pig1) and p53-induced gene 12 (microsomal glutathione transferase homolog) (Pig12), which are the most highly expressed p53-dependent ROS-induced genes (67), were significantly higher in the livers of hsp70−/− mice than in those of WT mice. Of note, Pig1 is a member of the galactin PIG family that can stimulate superoxide production (68), and Pig12 is a microsomal glutathione transferase homolog and is involved in redox reactions (69). Because p53 directly induces production of ROS via transactivation of Pig genes (70), elevated expression of these genes in the livers of mutant mice may create a positive feedback loop for further increase in ROS levels, thereby contributing to death and elimination of hepatic cells with excessive DNA damage. Significant changes found at the mRNA level, including increased expression of PUMA, NOXA, and BAX, as well as slightly reduced or comparable levels of antiapoptotic protein B-cell lymphoma 2 (BCL2) or B-cell lymphoma-extra large (BCL-xL), respectively, in livers of mutant mice compared to the WT were confirmed at the protein level (Fig. 6C).
To further investigate what makes hsp70−/− mice resistant to HCC, we sought to determine the time course of the DNA damage response associated with p53 activation and compensatory cell proliferation by IHC analysis of livers from DEN-treated P14 mice. In mice without DEN exposure, the presence or absence of HSP70 had no apparent effect on the zonal distribution and the number of proliferating hepatocytes, as detected by Ki67 IHC (Fig. 7A). Metabolic DEN activation and DNA damage in the hsp70−/− livers led to a marked decrease in Ki67+ pericentral hepatocytes. Strikingly, however, this was accompanied by increased compensatory proliferation of hepatocytes in the periportal region (Fig. 7A). This effect, detected at 4 h post-DEN treatment, became more pronounced at 24 h, 48 h, and 72 h. In contrast, DEN administration caused a moderate and transient suppression of pericentral hepatocyte proliferation, which was, however, paralleled by a marked decrease in the number of proliferating periportal cells. Moreover, DEN administration led to p53 induction within 6 h in both WT and hsp70−/− livers (Fig. 7B). As expected, nuclear expression of p53 was confined to the pericentral region, and in comparison, hsp70−/− mice displayed slightly higher p53 induction, although this did not reach significance. However, p53 staining, which peaked at 24 h post-DEN treatment, rapidly declined to almost undetectable level at 72 h post-DEN treatment in WT liver, while it remained at a significantly higher level in hsp70−/− liver. p53 activation in DEN-treated liver was also accompanied by induction of p21, a critical p53 target that mediate its tumor suppression activity by inhibiting cell cycle progression (Fig. 7C). Similar to the p53 staining profile, DEN-exposed hsp70−/− liver displayed a significantly higher and sustained level of p21 staining than WT liver, in which DEN administration led to marginal and transient expression of p21. Notably, although the majority of positively p21-stained cells in the hsp70−/− liver were found in the pericentral region, in WT liver p21+ hepatocytes were distributed throughout the organ. This finding indicates that DEN-induced damage and p21 expression in the hsp70−/− mice may be regulated by additional signals other than p53. Taken together, these results highlight the key role of HSP70 in HCC initiation, and its ablation sensitizes pericentral hepatocytes to the genotoxic response but also triggers excessive p53 activation. This causes sustained p21 expression and cell cycle exit and blocks reprogramming of damaged mature hepatocytes to HCC-initiating precursors.
Loss of HSP70 inhibits DEN-induced HCC development by triggering a negative feedback signaling program that attenuates ERK and AKT signaling pathways.
Exome-wide mutational landscape analyses in neoplasms arising in the DEN mouse model revealed that HCCs carried a notably high burden of somatic mutations, with over 80% of tumors carrying mutations in the Hras, Braf, Egfr, and Apc genes, which dysregulate the RAS-MAPK and WNT/β-catenin signaling pathways (71, 72). Aberrant activation of the RAS pathway leads to a hypersensitivity to growth factor signaling, including receptor tyrosine kinase (RTK) ligand engagement and immortalization. Persistent growth factor signaling associated with RAS and mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) activation, however, may trigger cellular senescence through accumulation of p53, p16, and p19ARF, and this response can lead to tumor suppression (73, 74). Intriguingly, cellular senescence can also be induced through sustained activation of RAS and the downstream MAPK/ERK signaling cascade, but through a complex network of negative feedback mechanisms that limit the amplitude and duration of ERK and AKT signaling at several levels (75). In tumor cells this negative regulation is mediated by ERK-dependent phosphorylation and inhibition of upstream components of the pathway, including the EGF receptor, SOS, and RAF (76, 77), or expression of inhibitory proteins such as members of the sprouty (SPRY) and dual-specificity phosphatase (DUSP) families (78–80). Regardless of the potential contribution of RAS-induced senescence in early tumor inhibition, sustained activation of the MAPK/ERK pathway is a common feature of advanced HCC, underscored by the fact that RAF inhibitors improve survival of HCC patients (81). Furthermore, relief of the negative feedback inhibition of MAPK signaling in BRAF mutant melanomas by RAF inhibitors can attenuate their activity, increasing tumor resistance to drug treatment (82). Notably, HSP70 has been implicated in the downregulation of ERK activity, either by interfering with BAG1-mediated RAF-1 activity or by enhancing the activity of ERK-specific DUSPs such as DUSP1 (MKP1) or DUSP6 (MKP3) (30, 83, 84).
Consistent with increased growth factor production in the hsp70−/− mice during the early phase of tumor initiation (Fig. 2D), ERK and AKT activities as assessed by their phosphorylation status were markedly elevated and sustained longer in the livers of mutant mice than in those of WT controls (Fig. 8A). Surprisingly, however, in the mutant mice this initially stimulated ERK activity driven by a transient strong input stimulus dramatically decreased by days 10 and 30 to the nearly basal levels detected in control mice (without DEN treatment). In contrast, DEN-treated WT mice sustained high p-ERK levels for a long time. Similar to this finding, IHC of liver tissues confirmed this p-ERK1/2 suppression in hsp70−/− mice. Moreover, a complete rebound in ERK activity was not observed in the tumors with HSP70 loss, which displayed significantly lower numbers of p-ERK-positive cells than WT tumors (Fig. 8B). p-AKT at Ser473 protein levels also exhibited similar kinetics, except the magnitude of suppression was less. To dissect the molecular events underlying this p-ERK inhibition, we examined the effects on MAPK/ERK signaling regulators. As expected, levels of p-MEK1/2 were also reduced at days 10 and 30 post-DEN injection in the mutant mice, suggesting that a defect in sustaining activation of RTKs and RAS signaling or the downstream inhibitory loop likely was the cause of the suppression of ERK activity. Furthermore, despite interexperimental variation, we observed that ERK-dependent transcriptional output was markedly elevated and persisted longer in the livers of hsp70−/− mice and minimally involved SPRY (Spry2, Spry3, and Spry4) and DUSP (Dups2, Dusp4, Dusp5, and Dusp6) expression, which may contribute to suppression of RAS/ERK signaling pathways induced by different RTKs (Fig. 8C). Using Western blot analysis, we confirmed the significant changes that were found at the mRNA level for DUSP6 and other selected proteins (Fig. 8D). Similar patterns of enhanced p-ERK1/2, p-MEK1/2, and p-AKT activation as well as increased mRNA levels of several DUSP (Dusp2, Dusp5, Dusp6, and Dusp10) and SPRY (Spry1 through -4) genes during the tumor initiation stage were observed for DEN-treated adult mutant mice (data not shown). Thus, the initial enhanced ERK signaling activation in mice with HSP70 loss, likely triggered by growth factors and cell-intrinsic pathways, is accompanied by the induction of an adaptive ERK negative feedback loop that dramatically restricts ERK signaling, not only during the critical transition to the malignant conversion stage of HCC development but also at later stages of tumor progression.
FIG 8.
HSP70 ablation interferes with HCC progression through a DUSP6-mediated negative control of ERK and AKT signaling, which plays a prominent role in maintaining the pretransformed cancer progenitor population, expansion, and aggressiveness. (A) WB analysis of MAPK/ERK and AKT signaling proteins in the livers of WT and hsp70−/− mice at the indicated time points after DEN injection at P14. The level of phosphorylated proteins normalized to the total level is expressed as relative fold increase compared to the control (WT without DEN, indicated as 0). (B) IHC staining of liver sections of WT and hsp70−/− mice at the indicated time points after DEN injection. Red labels, p-ERK1/2-positive cells. Original magnification, ×200. (B1) Quantification of p-ERK1/2+ cells per high-power field (HPF). (C and D) mRNA transcript and protein levels of the indicated members of the Dusp and Spry families in livers of DEN-treated mice at the indicated time points. Values are presented as relative mRNA. Quantification of protein expression levels (normalized to WT without DEN) is shown below (from two experiments). (E) Basis of the inhibitory effect of HSP70 on the phosphorylation of ERK1 and ERK2. Attenuation of ERK activity involves the formation of HSP70-BAG3 complexes, which, by displacing BAG3 from its association with the DUSP6-ERK complex, negatively regulates the ERK kinase pathway. WT or hsp70−/− mice were DEN challenged at P14. Protein extracts from livers collected 10 days post-DEN treatment were immunoprecipitated with a DUSP6-specific antibody, and proteins were subjected to WB analysis for HSP70, DUSP6, p-ERK, and total ERK (from three experiments). Notably, quantification of the data by normalizing individual protein levels to IP DUSP6 levels revealed the following changes in the complex formation. In the absence of HSP70, the relative BAG3 and ERK1/2 levels were increased by ∼95% and ∼41% to 148%, respectively, over the WT level. Comparing WT with hsp70−/− mice, the estimated ratio of p-ERK to total ERK in the IP complexes was found to decrease by ∼44% to 48% in the mutant mice. For all panels bars represent mean ± SD (n = 5 mice per group); statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
We next sought to identify the molecular process targeted by this adaptive response. The fact that RTK and RAS input does not induce continuous MEK activity suggests that ERK negative feedback could in principle operate upstream of ERK or targeting members of the RAS/ERK signaling pathway. Because the DUSP6 (MKP3) protein profile best correlated with p-ERK suppression in hsp70−/− livers, we also reasoned that this negative feedback loop might directly target ERK, depending on a well-described class of negative regulators. Therefore, based on a previous report (85), we sought to test a model in which the negative loop on ERK activity in mutant mice that depends on the DUSP6 phosphatase activity could involve a second level of regulation with critical direct participation of BAG3, a cochaperone of HSP70 and potent regulator and enhancer of DUSP6-ERK1/2 interaction. Notably, a well-defined HSP70-BAG3 interaction network has been described to act as a broadly acting regulator of cancer cell signaling factors (86, 87). According to this finding, depletion of HSP70 not only is expected to increase ERK1/2-BAG3 interaction, which in turn suppresses p-ERK1/2 activity, but also could cause disruption of HSP70-BAG3 complexes, thus amplifying the antitumor effects. Using an antibody against DUSP6 for IP assays, we confirmed the predicted interaction of endogenous DUSP6 with BAG3, ERK1/2, and p-ERK1/2 in liver lysates from DEN-treated WT mice. Interestingly, a small amount of HSP70 was also part of this complex, which suggests a potential role for this protein in this process (Fig. 8E). Consistent with our model, IP assays performed on liver lysates of DEN-treated hsp70−/− mice revealed quantitative changes in the protein composition of DUSP6-ERK-BAG3 complexes. Compared to the WT control, the normalized relative amounts of BAG3 and ERK in the immunoprecipitated complexes prepared from liver lysates of hsp70−/− mice markedly increased, and the rate of phosphorylated ERK1/2 significantly decreased. These results suggest that in the absence of HSP70, elevated BAG3 levels in the complex result in a tighter interaction between ERK1/2 and DUSP6 phosphatase to reduce p-ERK1/2 activity rates. Because the chaperone activity of HSP70 plays a direct role in recovery of DUSP6 activity under heat stress conditions, we speculate that the HSP70 coimmunoprecipitated with DUSP6 in extracts of WT mice, in a physiological context, may function directly to stimulate DUSP6 phosphatase activity, providing a negative feedback loop for ensuring transient ERK signaling activation (84).
Loss of HSP70 limits growth of DEN-induced HCC through activation of pathways regulating the cellular senescence program.
Cellular senescence is prevalent in premalignant cells, and it is generally regarded as a mechanism that suppresses cell transformation by arresting the proliferation of cells harboring genomic damage (88). On the other hand, senescent cells can persist for long time and, through the secretion of inflammatory proteins that function in a cell-autonomous or paracrine manner, can promote cancer progression (89, 90). An association between HSP70/HSC70 and cell cycle regulators, cyclins (e.g., cyclin D [CCND]), or cyclin-dependent kinase (CDK) inhibitors (e.g., p16, p21, p27, and p57) has been proposed (91). Moreover, evidence indicates that HSP70 loss may exert inhibitory effects on tumor cell proliferation by regulating oncogene-dependent senescence that is critical for the early stages of malignant transformation (27, 28).
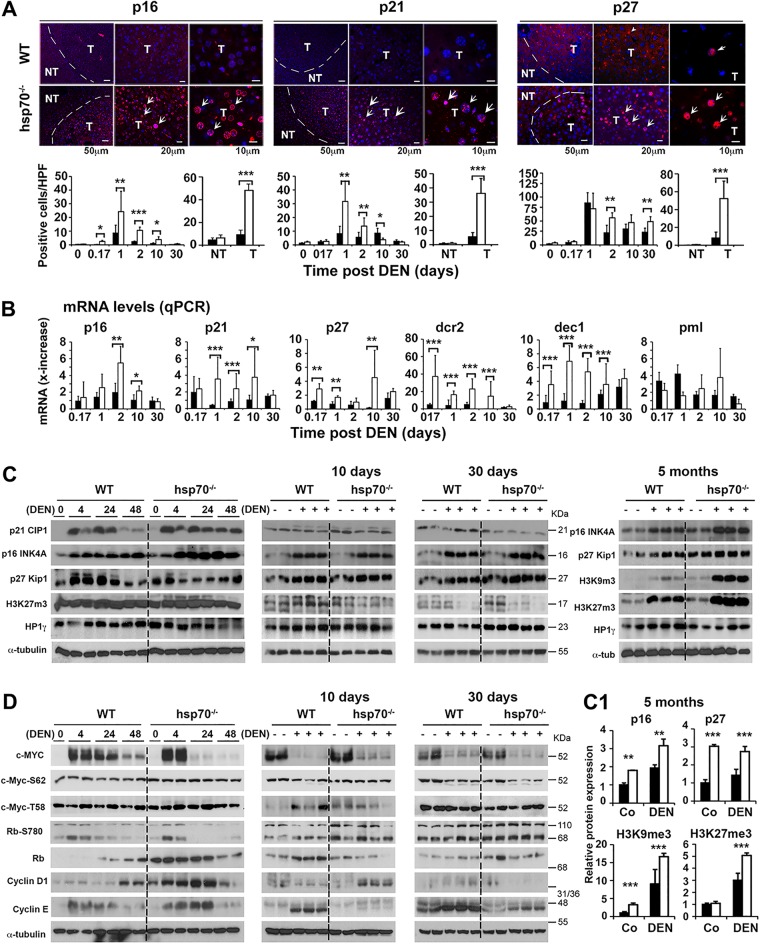
To investigate whether cellular senescence program may be modified by HSP70 loss during HCC development, we performed IHC of liver tissue sections from DEN-treated mice with a panel of antibodies specific to protein markers such as p16, p21, and p27, which have been causally linked to a senescence-like program (74). Notably, the presence of the β-galactosidase (β-gal) reporter gene in our targeting strategy used to generate the hsp70−/− mouse model prevented us from using SA-β-gal as marker for senescence. We observed that DEN treatment produced a robust cellular senescence-like response as shown by staining for p16, p21, or p27. Quantification of cells with nuclear expression of these markers revealed significantly higher numbers in the livers of DEN-treated hsp70−/− mice than in those of WT mice during the first 10 days after DEN injection (Fig. 9A). Although by 30 days after DEN treatment, only a few cells were found to express these markers, their numbers increased over time and were more abundantly detected in tumors developed in hsp70−/− mice than in WT mice. Time course studies confirmed that the mRNA expression levels of other senescence markers, including decoy receptor 2 (Dcr2), p15, delete in esophageal cancer 1 (Dec1), and promyelocytic leukemia protein (Pml), as detected by qPCR were also significantly higher in the whole livers of DEN-treated hsp70−/− mice than in WT controls during tumor initiation stages (Fig. 9B). Of note, the markers linked to senescence were not expressed in control livers without DEN treatment (data not shown). Thus, these results support the possibility that activation of the senescence program at advanced HCC stages partially accounts for the tumor growth inhibition in mutant mice.
FIG 9.
HSP70 ablation interferes with HCC growth through induction of a cellular senescence-like program. (A) Upper panels, representative immunofluorescence staining for expression of senescence signaling molecules in liver tumors (T) and nontumor areas (NT) of WT and hsp70−/− mice 8 months after DEN injection at P14. Lower panels, quantification of cells with nuclear staining for p16, p21, or p27 cells per high-power field (HPF) at the indicated time points, including T and NT areas. (B) qPCR of mRNA expression levels of key senescence-relevant signaling genes in livers of WT and hsp70−/− mice at the indicated times post-DEN treatment. Values are presented as relative mRNA expression. (C) WB analysis of senescence-relevant signaling proteins, including CDK inhibitors (p16, p21, and p27) and epigenetic modifiers in livers of WT and hsp70−/− mice at the indicated time points after DEN injection at P14. (C1) Quantification of protein expression levels (normalized to WT without DEN) from liver lysates collected 5 months post-DEN treatment (from two experiments). (D) WB analysis of senescence-related transcription factors (c-Myc and Rb) and cyclin kinases (cyclin D1 and cyclin E) in livers of WT and hsp70−/− mice at the indicated time points after DEN injection. Notably, c-MYC activity can be regulated by phosphorylation at Ser62, which stabilizes c-MYC, and Thr58, which promotes c-MYC degradation (125). In addition, the widely accepted model proposes that inactivation of Rb by hyperphosphorylation, including at Ser780, that is mediated by cyclin D/Cdk4/6 and cyclin E/Cdk2 complexes drives cell cycle progression by releasing E2F transcription factors (96). However, Rb may interact with E2F1, which is resistant to CDK phosphorylation; this interaction results in H3K27me3 at repetitive DNA elements. In our analysis, using three different Rb antibodies, we have been unable to detect the predicted full-length Rb protein (110 kDa); rather, we detected a fragment of about 68 kDa that probably reflects the cleavage Rb form. For all panels, bars represent mean ± SD (n = 3 to 5 mice per group); statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars indicate WT (filled) or hsp70−/− (open) mice.
To further determine the sequence of events that support a senescence program during DEN-induced liver cancer development, we profiled the time course expression of potential marker proteins, including cyclin kinases (cyclin D and cyclin E), CDK inhibitors (p16, p21, and p27), epigenetic regulators (H3K9me3, H3K27me3, and HP1γ), and relevant transcription factors (c-Myc and Rb) (Fig. 9C and D). As shown in Fig. 9C, a notably higher upregulation of p21 and p16 expression in the early phase of tumor initiation was confirmed in hsp70−/− mice. Most likely due to the absence of p53 stimulation at days 10 and 30 post-DEN treatment, p21 expression levels in both mouse genotypes were partially reduced to similar low steady-state basal levels. In contrast, after a transient slight reduction, p16 levels gradually increased again, with hsp70−/− mice displaying significantly higher levels at 5 months. Similarly, mutant mice expressed p27 at much higher levels at 5 months post-DEN treatment than WT mice. Moreover, the expression of the epigenetic modifiers proliferation-repressive H3K9me3 and activating H3K27me3 marks, which differentially regulate senescence programs (92–94), was dramatically induced in the livers of mutant mice at 5 months post-DEN treatment. Notably, epigenetic repression of repetitive DNA elements by H3K27me3, which requires p-RB-dependent recruitment of EZH2, helps to maintain stability and suppress neoplastic transformation (95, 96). Furthermore, beyond the potential link between epigenetic markers and senescence, the dramatic induced expression, especially of H3K9me modification, in the livers of mutant mice at 5 months post-DEN treatment suggests another interesting mechanism for tumor inhibition at the level of HCC progenitors. Indeed, methylation of H3K9me has been shown to progressively silence the fetal liver gene signature during liver maturation (e.g., AFP, insulin-like growth factor 2 [IGF2], GPC3, and H19), and this repressive function is likely to have a negative impact on the development of liver cancer progenitors and malignant cell transformation (97). The profile of expression of HP1γ, which has a role in transcription repression and propagation of heterochromatin structure (98), did not differ between the genotypes.
Expression of c-MYC, initially induced at 4 h and 24 h post-DEN treatment, rapidly declined to a much lower (nearly undetectable) level by day 10 or day 30 post-DEN treatment than in untreated control mice. The profiles of total c-MYC or its phosphorylated form did not significantly differ between the genotypes (Fig. 9D). In addition, no apparent changes in the expression profiles of Rb and p-Rb were detected. Interestingly the expression levels of cyclins (D and E), which were initially upregulated post-DEN treatment, gradually declined and were detected at significantly lower levels by day 30 post-DEN treatment in the hsp70−/− mice than in the WT controls. Collectively, our results suggest that activation of an adaptive senescence-like program by HSP70 loss may interfere with preneoplastic transformation and malignant cell progression.
HSP70 modulation by the small-molecule inhibitor PES attenuates carcinogen-induced HCC progression.
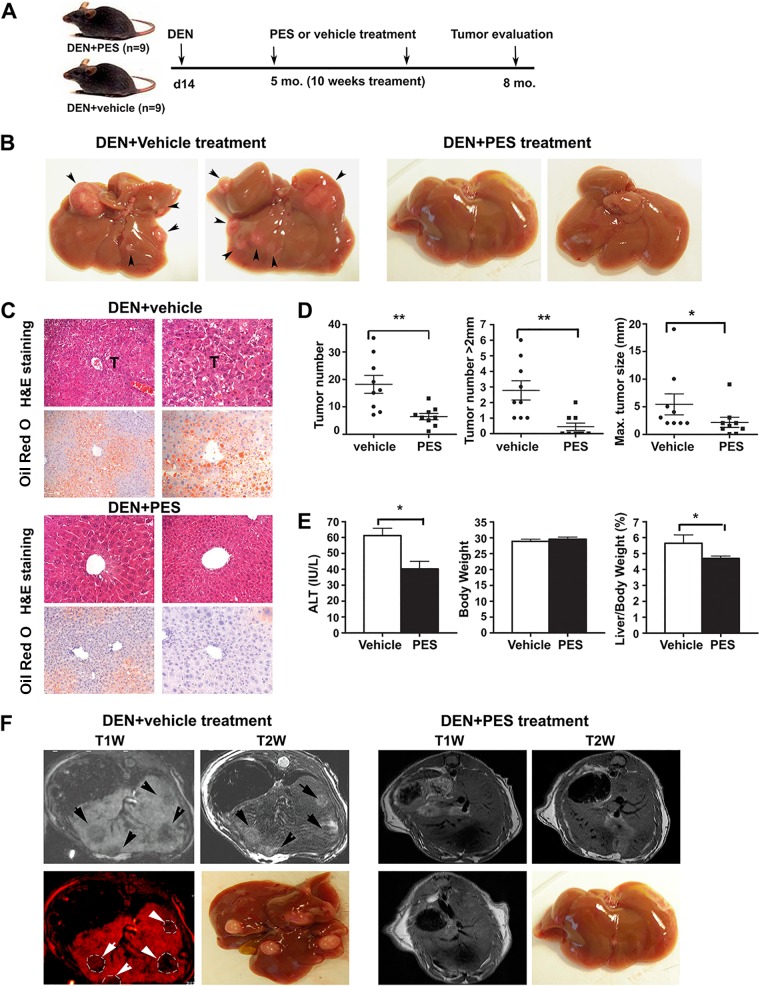
To test whether inhibition of HSP70 can reverse liver tumorigenesis in vivo without eliciting adverse physiological consequences, we used 2-phenylethynesulfonamide (PES), a small-molecule inhibitor for cytoplasmic HSP70s. PES was originally identified in a screen for molecules that would impair the mitochondrial localization of p53 (99), but recent studies suggest that it functions as antagonist of HSP70 and HSC70 substrate binding site activity and indirectly alters the activities of the cochaperone HSP90 machinery (100–102). Of note, the exact mechanism by which PES may modulate the HSP70 chaperone machinery remains to be fully elucidated, because recent biochemical and structural studies are inconclusive (103, 104). For these studies, WT mice were injected with DEN at day 14 after birth to initiate HCC. At tumor onset, at 5 months of age, the animals were treated with PES once per week for a period of 10 weeks. Mice were analyzed for liver tumor burden at 8 months of age. PES treatment markedly reduced liver tumorigenesis compared to that in mice treated with a vehicle control (Fig. 10). Liver tumor numbers per mouse and tumor burden were significantly decreased after PES treatment. Furthermore, tumor size distribution analysis showed that tumors detected in the PES-treated mice were relatively smaller than those in vehicle-treated controls, as revealed by magnetic resonance imaging (MRI) (Fig. 10F). It should also be noted that although a certain degree of HSP70/HSC70 inhibition by PES may be toxic to normal tissue, our study confirmed that PES treatment did not cause adverse effects on organ physiology, and we did not observe any signs of pathology in several tissues analyzed.
FIG 10.
Modulation of HSP70 function by PES attenuates carcinogen-induced HCC progression. (A) Schematics of the treatment protocol. A cohort of 18 WT male mice were injected with DEN at P14. Five months later, cohorts of mice were treated either with vehicle (DMSO) or with PES once a week for 10 weeks. At 8 months of age, the hepatic phenotype of the mice was examined by measuring tumor burdens using MRI and macroscopic inspection of livers, as well as by histological examination of liver sections. (B) Representative macroscopic photographs of livers from WT mice treated with PES or vehicle. Arrowheads indicate multiple tumor nodules in vehicle-treated mice, but there are no visible tumors in PES-treated animals. (C) Representative histological analysis (H&E) of livers, showing tumor (T) areas in vehicle-treated mice (upper panels). Oil Red O staining of representative sections shows extensive hepatic steatosis in vehicle-treated group but reduced or absent fat accumulation in the livers of PES-treated mice (lower panels). Original magnification, ×200. (D) Quantification of liver tumors in PES (n = 9 mice)- or vehicle (n = 9 mice)-treated mice. Bars represent mean ± SD. Statistical significance is indicated (*, P < 0.05; **, P < 0.01). (E) Tumor regression in PES-treated mice was associated with substantial amelioration of hepatic injury assessed by the serum ALT level and decreased ratio of liver to body weight. Bars are mean ± SD (n = 9 mice per group). (F) Representative MRI images, with liver tumors readily visible in vehicle-treated mice but significantly reduced tumor size or even an effective therapeutic response in PES-treated animals. Upper row, T1W and T2W images of liver, showing hypointense (T1W) and hyperintense (T2W) regions of tumors in vehicle-treated mice (left two panels) and no tumors in PES-treated mice (right panels). Lower row, T1W image, enhanced with pseudocolor, from a vehicle-treated mouse showing multiple tumors (left panel) and representative images from vehicle-treated and PES-treated animals; no tumors are visible in the PES-treated animal (compare second and fourth panels from left). Resolution of images: T1W, 200 by 200 by 500 μm; T2W, 100 by 100 by 500 μm. Tumor nodules are indicated (arrowheads).
DISCUSSION
Signature alterations in the proteome of malignant cells have been broadly recognized as a hallmark of cancer (105). The common notion that the metabolic reprogramming and reactivation of oncogenic signaling pathways supporting unlimited proliferation distinguish cancer cells from their normal counterparts and rely on protein homeostasis (nononcogene addiction) provides a rationale for targeting molecular chaperones, including HSP70, in cancer therapy (106). Considering the strong evidence indicating that HCC, experimentally induced by acute genotoxic challenge or persistent inflammatory liver injury, is initiated by regenerating mature hepatocytes that had acquired potential oncogenic mutations (11, 12, 40), a key challenge in deciphering the exact nature of initiating events is the identification of the regulators of the reprogramming process responsible for generation and maintenance of HCC-PCs. In this study, we have uncovered some key regulatory mechanisms by which HSP70 promotes adaptive mechanisms for HCC development. Our data suggest that during HCC initiation and progression, which are considered a multistep process tightly linked to progressive acquisition and accumulation of genetic and epigenetic alterations in a background of chronic liver injury induced by risk factors, HSP70 has a critical function as an information hub for sensing and channeling DNA damage signals. In this context, HSP70 plays a critical role in restricting the amplitude and duration of the DNA damage response. Consequently, in the presence of time-limiting downstream activation of DNA damage checkpoints and induction of tumor suppressor protein p53, a large fraction of genetically damaged liver cells become refractory to negative regulation by p53, whose sustained activation triggers apoptotic death, and this may lead to increased likelihood of mutagenesis and cancer progression. In the physiological context, our results provide another twist to prevailing notion that increased genomic instability provoked by constantly enhanced cell-intrinsic metabolic activity coupled with deficit in DNA repair signaling and elevated reactive oxidative burden is a main driver for transformation of tumor-initiating cells and has a supporting role in cancer progression (56). We show that HSP70 loss, despite creating a permissive genomic-instability environment, may prevent malignant transformation, presumably by increasing p53-dependent apoptotic priming of genomically damaged tumor-initiating cells, and it may also be detrimental for malignant cell growth and survival. In a model of spontaneous HCC linked to a chronic liver damage, in which tumor initiation proceeds very slowly, we envisage that HSP70 loss by causing a low degree of genomic instability may compromise cellular adaptability to adverse metabolic and oxidative stress triggered by genotoxic challenge and rapidly replicating cell proliferation and thus inhibits malignant cell transformation. Given the known ability of p53 to block reprogramming of fibroblasts into pluripotent stem cells (107), the induction of p53 and its targets, which is extended in the absence of HSP70, could also inhibit the potential for differentiation of DNA-damaged hepatocytes into HCC-PCs and sensitize their apoptotic death. Another surprising finding was the role of HSP70 in interfering with fail-safe MAPK/ERK negative signaling mechanisms that generally are activated to terminate oncogene- and growth factor-associated signaling during a premalignant condition and its role in preventing the induction of senescence-associated tumor-restraining mechanisms. These properties, which were found to be significantly induced in the absence of HSP70, may be a key step to prevent initiation and progression of HCC. Collectively, our results validate HSP70 as being an important mediator of HCC development by promoting tumor-initiating cells to escape self-limiting responses that restrain cell proliferation and transformation.
Progression models for many solid cancers, including HCC, revealed that malignant transformation and cancer dissemination driven by a succession of genomic alterations induced by DNA-damaging agents, including chemical carcinogens and ROS production, depend on successful adaptation of the cells to dramatic changes that occur in their genetic and biochemical makeup. Tumor initiation by carcinogens is associated with the formation of DNA lesions (DNA adducts), and if they persist, they may cause DNA damage-inducing mutagenesis and genomic instability and directly lead to cancer. Notably, malignant cells maintain a higher burden of oxidative stress than do normal cells, which drives cancer progression (108). Alternatively, optimal activation of the DDR and downstream checkpoints and relief of oxidative damage may lead to DNA repair and genomic restoration and thereby constrain the progression of preneoplastic lesions. Therefore, interfering with mechanisms that allow cancer cells to adapt to DNA damage events associated with oxidative stress is considered a potential therapeutic strategy for cancer. However, much remains to be elucidated about the mechanisms ensuring the survival of DNA-damaged cells with aberrant repair that contributes to susceptibility to liver cancer. Consistent with a dominant role of HSP70 in regulating the DDR, oxidative stress, and p53 activation, our findings suggest a progressive model of carcinogen-induced HCC (Fig. 11). We propose that HCC inhibition in mice with HSP70 loss results from the enforcement of a robust DEN-induced DDR and checkpoint signaling with a strong p53 induction. Notably, the functions of p53 extend beyond cellular surveillance, apoptosis, and metabolism to include regeneration and somatic and cancer stem cell reprogramming (109, 110). Its activation is also a hallmark of several DDR pathways. Although the precise mechanisms by which p53 exerts these biological functions remain to be fully elucidated, sensitivity of the cells to p53-mediated apoptosis or senescence seems to be dependent on several posttranslational modifications and extrinsic and intrinsic factors, such as the intensity and duration of the DDR and other stress signals and the complex interactions between components of the p53 pathway (111). Our results indicate that despite its limited extent, p53-mediated induction of proapoptotic genes (e.g., Puma, Noxa, and Perp genes) is not counterbalanced by the relatively high levels of p21 in the hsp70−/− mice and probably sensitizes cell killing rather than favoring p21-mediated growth arrest. This rewiring of the apoptotic machinery ensures the elimination of severely DNA-damaged cells and a reduced risk of accumulating mutations leading to cancer progression. In terms of the mechanisms by which HSP70 inactivation contributes to enhanced cell killing, it is logical to propose sustained checkpoint signaling due to impaired DNA repair, but additional mechanisms, including ROS or stress MAPK signaling (e.g., p38 and ERK), may directly contribute to stabilization and stronger phosphorylation of p53 and induction of its target genes (112, 113). This conclusion is further supported by our findings and previous reports showing that HSP70 inhibits ROS production (114, 115), and by binding and modulating the endonuclease activity of HAPE1 (59), it may improve DNA repair activity. Other mechanisms, such as enhanced ROS production in hsp70−/− cells that is positively regulated by both DDR-induced p53 target gene expression (PIG) and mitochondrial impairment, may augment the selective killing of DNA-damaged cells through p53-dependent or -independent mechanisms of action (116). However, the molecular basis of cooperation between HSP70 and carcinogen-induced DDR and repair complex formation awaits further investigation. Also, the mechanistic basis of the prevalence of the DEN-induced activation of the ROS and ERK pathways in the mutant mice and their effects on p53 activation and function will require further studies.
FIG 11.
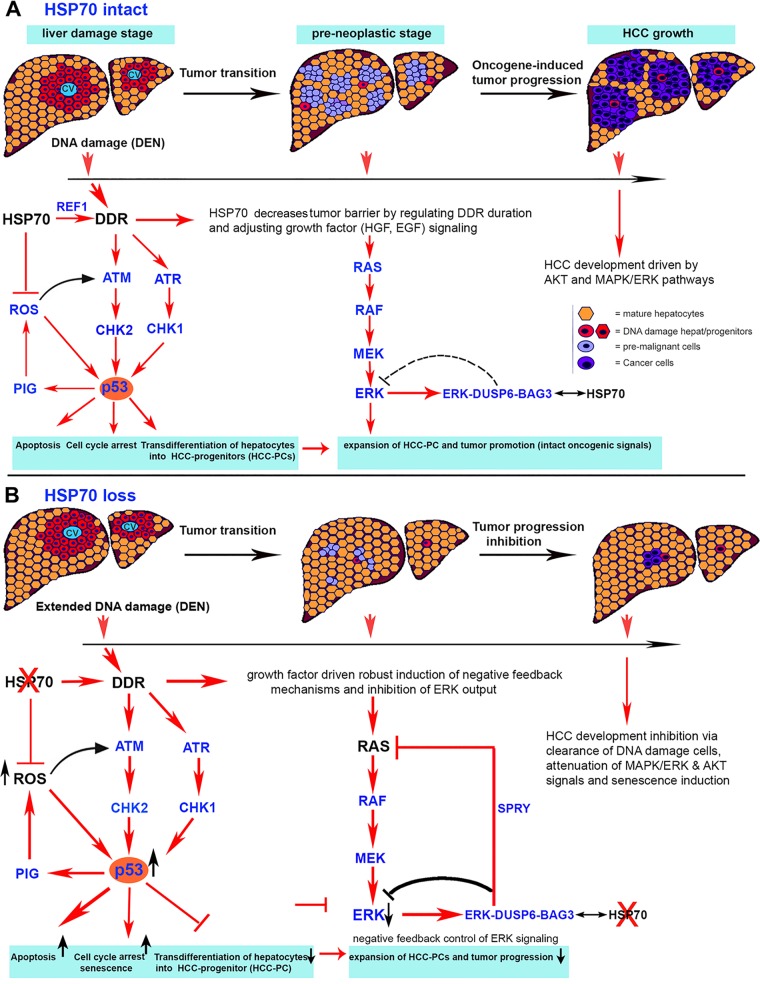
Schematic representation of the HSP70-REF1-ATM/ATR-p53 and HSP70-BAG3-DUSP6-ERK circuits, which control HCC formation and progression. (A) Mechanistically, our studies suggest a progressive model of carcinogen-induced HCC, in which HSP70, by promoting DNA repair (e.g., interacting with and regulating the activity of REF1/APE1, a key excision repair enzyme), limits the extent and duration of carcinogen-induced DNA damage. Consequently, a temporal stimulation of checkpoints that is mediated via ATM and ATR signals elicits a balanced adaptive p53 signaling that allows the survival and transdifferentiation of moderately DNA-damaged hepatocytes. In this context, HSP70-mediated inhibition of ROS production and oxidative stress may also contribute to this adaptation by downmodulating DDR checkpoint signaling pathways and p53 activation as well as limiting a direct ROS-mediated cell killing. Collectively, in the presence of HSP70, adaptive p53 activation and oxidative stress signaling may promote the transdifferentiation of initiated hepatocytes into proliferating HCC progenitors (HCC-PCs) bearing genomic alterations, thereby increasing the likelihood of mutagenesis and cancer progression. Another complementary mechanism by which HSP70 supports HCC development is based on its function to counteract negative feedback mechanisms that are activated to restrict RTK- and oncogene-mediated AKT and RAS/ERK signals. We propose that under HSP70 expression condition, BAG3 in the BAG3-DUSP6-ERK complex, which represses ERK activity, is displaced by BAG3-HSP70. Consequently, acting as a critical sensor and positive regulator of a sustained MAPK response and ERK signaling, HSP70 allows the proliferation of HCC-PCs and formation of preneoplastic cells harboring activated oncogenes. Furthermore, calibrating ERK, and perhaps AKT, signaling and reducing the high overall burden of oxidative stress present in neoplastic cells, HSP70 may prevent cellular senescence, which is a key to cancer progression. (B) In the absence of HSP70, robust carcinogen-induced DNA damage and checkpoint signaling associated with a strong and prolonged p53 activation may lead to eradication of cells with excessive DNA damage and prevent the transdifferentiation of initiated hepatocytes into cancer progenitors with high proliferative potential. Oxidative stress is a general hallmark of many cancers and has a crucial role in tumor progression. Therefore, the increased ROS tension observed in HSP70 deficiency predictably would promote cancer progression. However, we reason that the normally nonessential function of HSP70 protein for cell viability may be important for further sensitizing via ROS induction the killing not only of DNA-damaged cells in the earliest response to carcinogen treatment but also of malignant cells at advanced tumor stages. Furthermore, a robust induction of negative feedback mechanisms that is dependent on a negative regulator (DUSP6) leads to attenuation of ERK output, in the absence of HSP70, and may be sufficient to potently suppress RAS/MAPK signals that are essential for proliferation and acquisition of secondary mutations in initiating precursor cells. Collectively, we propose that the two levels of regulation in the absence of HSP70 might effectively antagonize HCC development.
Another central conclusion (Fig. 10) for this study is the powerful activation of ERK-dependent negative feedback mechanisms in the hsp70−/− mice, which may involve overexpression of SPRY and particularly DUSP6 phosphatase proteins. This mechanism probably provides an effective barrier to malignant progression by limiting or modulating growth factor-induced RTK signaling and ERK output. Positive feedback during the initial DEN-induced response potently causes a rapid onset of ERK activation in the mutant mice, which may arise from high levels of growth factor signaling combined with cell-intrinsic HSP70-mediated regulation of the RAS/ERK cascade (30, 83), The initial bursts of RAS/ERK response associated with a more upregulated expression of cell cycle regulators, including p16, p53, and p21, in the mutant mice, however, was not sustained and rapidly diminished upon activation of the negative feedback. Notably, in NF1-deficient cells with constitutive RAS-mediated mitogenic signaling, the initial burst of proliferation and subsequent downregulation of the RAS/ERK and PI3K/AKT pathways have been proposed to terminate the oncogenic signal and induce premature senescence (75). However, recent studies have demonstrated that oncogene-induced senescence programs depend on sustained expression and activity of ERK that promotes a selective protein degradation (117, 118). Intriguingly, in our study typical senescence markers (e.g., p16, p21, and p27) were highly expressed in slowly growing HCC in hsp70−/− mice. Moreover, a complete rebound in ERK activity was not observed in the tumors with HSP70 inactivation, which expressed much lower p-ERK levels than WT tumors (Fig. 5). Because physiological activation of ERK is self-limited in extent, one may ask how tumor cells in the mutant livers may have restored the senescence marker expression without a strong ERK output. We propose that during the initial and transition of premalignant stages in the mutant mice, a low level of oncogene-induced signals through acquisition of mutations cannot drive proliferation, because it is insufficient to substantially overcome the ERK-inactivating feedback mechanism. It is only during the advanced stages, when oncogenic signals in the WT tumors exceed critical thresholds, that these tumor suppressors (senescence markers) are substantially suppressed, for example, through acquisition of selective secondary mutations, a process that requires intense oncogene signals and cell proliferation. In contrast, partly restored but diminished ERK signaling in the mutant tumor cells may substantially lower the rates of adaptive secondary mutations but, with participation of intense DNA damage signals induced by oncogene and deregulated oxidative stress, may trigger a robust induction of cellular senescence with strong tumor cell-inhibitory effects. Finally, it must be kept in mind that a high ERK output is required to sustain the ERK negative feedback signaling, including the expression of SPRY and DUSP proteins. This implies that the observed loss of ERK activity in hsp70−/− tumors is likely to be multifactorial, with specific contributions from additional HSP70-dependent regulatory signals on the upstream components of the RAS/ERK signal transduction pathway. Further studies are required to elucidate the underlying mechanisms by which the HSP70 chaperone machinery might be involved in RAS/ERK attenuation in liver tumors.
Taken together, our results demonstrate that in the absence of HSP70, the prolonged DNA damage and sustained p53 induction in mature hepatocytes subjected to genotoxic challenge prevents acquisition and fixation of oncogenic mutations that are required for conversion to malignant initiating precursors. This may occur through p53-driven sustained p21 expression, cell cycle exit, and inhibition of the reprogramming of damaged mature hepatocytes to HCC-initiating precursors. Elimination of extensively DNA-damaged cells through p53-dependent apoptosis and oxidative stress-induced death may also be a potent tumor-suppressive mechanism during HCC initiation. For a cell population with moderate or low DNA damage that escapes elimination, mutations resulting in transformation are unlikely to be established unless the cells gain substantial survival advantages that largely depend on growth factor and ERK/MAPK signaling. Thus, attenuation of ERK activity at an early stage of the cell selection process reduces the risk for adaptive selective secondary mutations, which are key to cancer progression. At late stages, in a fraction of cells, partly rebound ERK signaling through acquisition of mutations in key oncogenes may fuel restricted proliferation and contribute to cell transformation, but oncogene and oxidative stress-induced senescence programs in the hsp70−/− tumor cells may provide a further barrier for rapid tumor progression. In addition, we provide a demonstration of potential HSP70 targeting for HCC prevention and perhaps therapy without adverse effects on the organ physiology or life span of the animals. A challenge in the future will be to determine how HSP70 members interact to promote tumor progression. In addition, how to practically manipulate the HSP70 machinery for clinical intervention is a future challenge (106). Although further studies are needed, increased expression of HSP70 in mouse models has been shown to have significant therapeutic potential for muscular dystrophy and cardioprotection and may delay progression of neurodegenerative diseases (119, 120), but its modulation also has desirable effects such as cancer protection.
MATERIALS AND METHODS
Animals.
hsp70i−/− (hsp70.1−/− and hsp70.3−/−) mice on a C57BL/6 genetic background were produced using a targeting strategy similar to that described for the generation of hsp70.1−/− or hsp70.3−/− mice (121): in the targeting vector, the entire coding sequences of the hsp70.1 and hsp70.3 genes, including the intergenic region of approximately 8 kb, were replaced with an in-frame lacZ gene sequence recreating a hsp70.1 transcription reporter mouse (unpublished data). The absence of HSP70 protein expression in tissues of hsp70i−/− mice was confirmed by WB analyses (Fig. 1D). Animal care and experiments were performed in accordance with the guidelines of Institutional Animal Care and Use Committee and NIH guidelines.
Liver tumor induction and analysis.
For HCC-induction, 14-day-old male mice were injected intraperitoneally (i.p.) with 25 mg/kg diethylnitrosamine (DEN) (Sigma), and animals were euthanized at the indicated ages. For tumor initiation in adult mice (8 to 12 weeks old) a higher dose of DEN (100 mg/kg) was used. For HCC analysis, mouse weight, liver weight, number of tumors per liver, maximal tumor size (millimeters), and liver/body weight percentage were determined.
Assays of liver enzymes and insulin in serum.
ALT and AST levels were detected using the liquid reagent kit according to the manufacturer’s instructions (Pointe Scientific Inc., MI).
Hepatocyte isolation.
Primary hepatocytes were isolated via two-step collagenase perfusion and isolation techniques (122) using liver perfusion medium and liver digest buffer as described in our previous report (123). Briefly, cells were allowed to attach for 2 to 3 h on collagen-coated plates in Dulbecco’s modified Eagle’s medium (DMEM) containing glucose (4.5 g/liter) and supplemented with 10% FBS, 2 mM sodium pyruvate, and 2% penicillin-streptomycin. After attachment, the medium was replaced with maintenance medium as described above, and the cells were incubated overnight before being used for treatment with growth factors.
Histology, IHC, and IF microscopy.
Livers (and tumors) were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline for at least 7 days and embedded in paraffin. Hematoxylin and eosin (H&E) staining was performed on paraffin sections (5 μm). For immunohistochemistry (IHC) staining, tissues were deparaffinized in xylene and rehydrated in a series of alcohol-water mixtures. Antigen retrieval was performed in 10 mM sodium citrate (pH 6.0) for 30 min or in Tris-EDTA (10 mM Tris base, 1 mM EDTA) buffer (pH 9.0) for 30 min. Tissue sections were blocked in 3% bovine serum albumin (BSA), incubated with primary antibody overnight at 4°C and biotin-conjugated secondary antibody for 1 h at room temperature (25°C) with extravidin-alkaline phosphatase substrate (30 min at 25°C), and developed with Fast Red (Roche). Sections were counterstained with hematoxylin. The ImmPRESS Excel amplified polymer staining kit (MP7601) was used for IHC, which required amplification of signal (p21, p53, and O6-deoxyguanosine). Antibody sources are provided in Table 1.
TABLE 1.
Antibodies
| Antigen | Host species and type | Use(s) | Source | Identifier |
|---|---|---|---|---|
| Ac-K | Rabbit polyclonal | WB | Cell Signaling | 9441 |
| ATM | Rabbit monoclonal (D2E2) | WB | Cell Signaling | 2873 |
| p-ATM (S1981) | Rabbit monoclonal (10H11.E12) | WB, IHC | Santa Cruz | 47739 |
| ATR | Rabbit (N19) polyclonal | WB | Santa Cruz | 1887 |
| AKT | Mouse monoclonal (F8) | WB | Santa Cruz | 271149 |
| p-AKT (S473) | Rabbit polyclonal | WB | Santa Cruz | 7985-R |
| β-Actin | Mouse monoclonal (C4) | WB | Santa Cruz | 47778 |
| BAG3 | Rabbit polyclonal (H-265) | WB, IP | Santa Cruz | 292154 |
| BAX | Rabbit polyclonal | WB | Cell Signaling | 2772 |
| BCL2 | Rabbit monoclonal (50E3) | WB | Cell Signaling | 2870 |
| BCL-xL | Rabbit polyclonal | WB | Cell Signaling | 2762 |
| BIM | Rabbit polyclonal | WB | Cell Signaling | 2819 |
| CHK1 | Mouse monoclonal (DCS-310) | WB | Santa Cruz | 56291 |
| p-CHK1 (S345) | Rabbit monoclonal (133D23) | WB | Cell Signaling | 2348 |
| CHK2 | Rabbit polyclonal | WB | Cell Signaling | 2662 |
| p-CHK2 (T68) | Rabbit polyclonal | WB, IHC | Novus Biologicals | NB100-92502 |
| CD31 | Rat monoclonal (MEC13.3) | IF (frozen) | BD | 550274 |
| Cyclin D1 | Rabbit monoclonal (92G2) | WB | Cell Signaling | 2978 |
| Cyclin E1 | Rabbit monoclonal (D7T3U) | WB | Cell Signaling | 20808 |
| DNA-PKcs | Rabbit (H163) | WB | Santa Cruz | 9051 |
| DUSP2 | Rabbit polyclonal | WB | Elabscience | E-AB-16397 |
| DUSP6 (MKP3) | Rabbit polyclonal | WB | Cell Signaling | 3058 |
| Rabbit polyclonal (H-130) | IP, WB | Santa Cruz | 28902 | |
| ERK1/2 | Rabbit polyclonal | WB, IHC | Santa Cruz | 94 |
| p-ERK1/2 (T202/Y204) | Rabbit monoclonal (D13.14.4E) | WB, IHC | Cell Signaling | 4370 |
| α-Fetoprotein | Goat polyclonal | WB | RD Systems | AF5369 |
| Glutamine synthetase | Mouse monoclonal (clone 6) | IHC | BD Transduction Lab | 610517 |
| HSP70 | Mouse monoclonal | WB | StressMarq Biosciences | SMC-100A/B |
| HSC70 | Rabbit polyclonal | WB | Abcam | 1427 |
| HSP90α | Rabbit polyclonal | WB | ENZO | ADI-SPS-771D |
| HSP90β | Rabbit monoclonal (D3F2) | WB | Cell Signaling | 7411 |
| HSP40 | Rabbit monoclonal (C64B4) | WB | Cell Signaling | 4871 |
| γH2AX (S139) | Rabbit polyclonal | WB, IHC | Cell Signaling | 2577 |
| IκBα | Rabbit polyclonal | WB | Cell Signaling | 4812 |
| H3K9me3 | Rabbit polyclonal | WB | EpiGentek | A-4036 |
| H3K27me3 | Rabbit polyclonal | WB | EpiGentek | A-4039 |
| HPIγ | Rabbit polyclonal | IHC | Bioss Inc. | 1421R |
| JNK1/2 | Rabbit monoclonal (56G8) | WB | Cell Signaling | 9258 |
| p-JNK1/2 (T183/Y185) | Rabbit monoclonal (81E11) | WB | Cell Signaling | 4668 |
| c-JUN | Rabbit monoclonal (60A8) | WB | Cell Signaling | 9165 |
| p-c-JUN (S63) | Rabbit monoclonal (54B3) | WB | Cell Signaling | 2361 |
| Ki67 | Rabbit monoclonal (SP6) | WB | Thermo Scientific | RM-9106 |
| MDM2 | Mouse monoclonal (SMP14) | WB | Santa Cruz | 965 |
| p-MDM2 (S166) | Rabbit polyclonal | WB | Cell Signaling | 3521 |
| MEK1/2 | Rabbit monoclonal (D1A5) | WB | Cell Signaling | 8727 |
| p-MEK1/2 (S217/221) | Rabbit monoclonal (41G9) | WB | Cell Signaling | 9154 |
| c-MYC | Rabbit monoclonal (Y69) | WB | Hoffmann-La Roche | 790-4628 |
| c-MYC (S62) | Rabbit polyclonal | WB | abm | Y408575 |
| c-MYC (T58) | Rabbit polyclonal | WB | abm | Y407783 |
| NOXA | Rabbit polyclonal (FL54) | WB | Santa Cruz | 30209 |
| 8-OH-dG | Rabbit polyclonal | IHC | Bioss Inc. | BS-1278R |
| O6-Ethyl-2-deoxyguanosine | Mouse monoclonal (EM21) | IHC | Squarix Biotechnology | SQX-SQM004 |
| TP53 | Mouse monoclonal (1C12) | WB | Cell Signaling | 2524 |
| p-p53 (S15) | Rabbit polyclonal | WB | Cell Signaling | 9284 |
| P38 | Rabbit polyclonal | WB | Cell Signaling | 9112 |
| p-p38 (Y180/Y182) | Rabbit monoclonal (3D7) | WB | Cell Signaling | 9215 |
| p16 | Mouse monoclonal (F12) | IF (paraffin) | Santa Cruz | 1661 |
| Rabbit (M156) | IF (paraffin) | Santa Cruz | 1207 | |
| Rabbit polyclonal | WB | Proteintech | 10883-1-IP | |
| p21 | Mouse monoclonal (SXM30) | WB, IF (paraffin) | BD | 556431 |
| Rabbit polyclonal | WB | Elabscience | E-AB-32448 | |
| Rabbit polyclonal (C-19) | WB | Santa Cruz | 397 | |
| Rabbit monoclonal (EPR18021) | IHC | Abcam | Ab188224 | |
| p27 | Rabbit polyclonal | WB | Cell Signaling | 2552 |
| Mouse monoclonal (F8) | IF (paraffin) | Santa Cruz | 1641 | |
| p53 | Rabbit polyclonal (CM5) | IHC, WB | Leica Biosystems | NCL-L-p53-CM5p |
| P27 | Rabbit polyclonal | WB | Elabscience | E-AB-32450 |
| PARP1 | Rabbit monoclonal (46D11) | WB | Cell Signaling | 9532 |
| PUMAα/β | Rabbit (H-136) | WB | Santa Cruz | 28226 |
| Rb | Rabbit polyclonal | WB | Elabscience | E-AB-32752 |
| Mouse monoclonal (IF8) | WB | Santa Cruz | 102 | |
| Rb (S780) | Rabbit monoclonal (C84F6) | WB | Cell Signaling | 3590 |
| REF1/APE1 | Mouse monoclonal (C-4) | WB, IP | Santa Cruz | 17774 |
| REF1 | Mouse monoclonal (13B8E5C2) | WB | Novus Biologicals | NB100-116 |
| RAD51 | Rabbit polyclonal (H92) | WB | Santa Cruz | 8349 |
| STAT1 | Rabbit polyclonal | WB | Cell Signaling | 9172 |
| p-STAT1 (S727) | Rabbit polyclonal | WB | Cell Signaling | 9177 |
| STAT3 | Rabbit monoclonal (D1A5) | WB | Cell Signaling | 8768 |
| p-STAT3 (Y705) | Rabbit monoclonal (D3A7) | WB | Cell Signaling | 9145 |
| SPRY1 | Rabbit monoclonal (D9V6I) | WB | Cell Signaling | 12993 |
| SPRY2 | Rabbit monoclonal (D3G1A) | WB | Cell Signaling | 14954 |
| SPRY3 | Mouse monoclonal (C-2) | WB | Santa Cruz | 374593 |
| SPRY3 | Goat polyclonal (N-17) | WB | Santa Cruz | 18603 |
| α-Tubulin | Rabbit monoclonal (EPR1568Y) | WB | Abcam | 68193 |
For immunofluorescence (IF) staining on paraffin-embedded tissues, after antigen retrieval the sections were blocked in 3% BSA, incubated with primary antibody overnight at 4°C and Cyt3-conjugated secondary antibody for 1 h at room temperature, and examined under an Axio10 imager fluorescence microscope (Carl Zeiss, Germany). Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) reagent. For liver, frozen tissues, air dried and fixed with 4% PFA sections, were blocked in 3% BSA for 30 min and incubated with primary antibody (CD31) overnight at 4°C and then with fluorescence-conjugated secondary antibody for 1 h at 25°C. The primary antibodies are listed in Table 1. An average of 10 randomly selected high-power fields (at a magnification of ×200 or ×400) were counted for each antibody and for each section.
TUNEL staining was performed with ApoptoTag Red in situ apoptosis detection (Chemicon/Millipore, Billerica, MA). Nuclei were counterstained with DAPI reagent. For Oil Red O staining, frozen tissue sections were fixed in 10% formalin for 10 min at 25°C. Slides were stained in 0.5% Oil Red O solution (Fisher Chemical Co.) for 8 min at 60°C. After rinsing with distilled water, slides were counterstained with Mayer's hematoxylin for 30 s.
qPCR.
Total RNA was isolated from liver tissue using TRIzol (Invitrogen, Carlsbad, CA) and reverse transcribed using the Improm-II reverse transcription system (Promega, Madison, WI). Quantitative real-time PCR (qPCR) was performed using iQ SYBR green Supermix (Bio-Rad) on a Stratagene Mx 3000 P qPCR system. Relative quantitative plots were constructed for quantity of RNA input for each gene of interest. qPCR was performed using gene-specific primers (primer sequences are presented in Table 2).
TABLE 2.
Primers for quantitative RT-PCR
| Gene name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| Bcl2 | TGGGATGCCTTTGTGGAACT | ACAGCCAGGAGAAATCAAACAG |
| Bcl-xl | GGCTGGGACACTTTTGTGGAT | GCGCTCCTGGCCTTTCC |
| Ccng1 | GCGAAGCATCTTGGGTGTGT | TCCTTTCCTCTTCAGTCGCTTT |
| Dusp1 | GGATATGAAGCGTTTTCGGCT | GGATTCTGCACTGTCAGGCA |
| Dusp2 | GTGCTCTTCTGAGGTGGCATA | TGGGAGCACAAGGCTTAAGCT |
| Dusp4 | CTACCTCGGCAGTGCCTATC | GACGGGGATGCACTTGTACT |
| Dusp5 | TGGATGTGAAGCCCACCTCA | CGCACTTGGATGCGTGGTAG |
| Dusp6 | CGACTGGAATGAGAACACTGGTGG | TCTAGATTGGTCTCGCAGTGCAGG |
| Dusp10 | CGCCTACTTGATGAAGCACA | AGGTTCGGGGAAATAATTGG |
| Dcr2 | AGCTAACCCAGCCCATAATCGTC | AGTTCCCTTCTGACAGGTACTGGC |
| Dec1 | GGAGACCTGTCAGGGATGGA | CCCGCCTGGACTTGTACACT |
| Egf | CCTTCACAGAGCACACCTCA | CAGGACATCCAGAGTCAGCA |
| Il-1β | CCCAAGCAATACCCAAAGAA | CATCAGAGGCAAGGAGGAAA |
| Il-6 | CTGCAAGAGACTTCCATCCAG | AGTGGTATAGACAGGTCTGTTGG |
| Inf-γ | AGG AAC TGG CAA AAG GAT GGT G | GTG CTG GCA GAA TTA TTC TTA TTG |
| Il-10 | GGACAACATACTGCTAACCGACTC | AAAATCACTCTTCACCTGCTCCAC |
| Igf1 | TGCAAAGGAGAAGGAAAGGA | TGTTTTGCAGGTTGCTCAAG |
| Hgf | AGGAACAGGGGCTTTACGTT | GTCAAATTCATGGCCAAACC |
| Mcp-1 | CTTCTGGGCCTGCTGTTCA | CCAGCCTACTCATTGGGATCA |
| Mip-1α | TTCTCTGTACCATGACACTCTGC | CGTGGAATCTTCCGGCTGTAG |
| Noxa | TCGCAAAAGAGCAGGATGAG | CACTTTGTCTCCAATCCTCCG |
| P21 | ACCCGGGTCCTTCTTGTGTTTC | CGTTTTCGGCCCTGAGATGTTC |
| Puma | GCGGCGGAGACAAGAAGA | AGTCCCATGAAGAGATTGTACATGAC |
| Perp | GCTGCAGCCACGCTTTTC | GGCGAAGAACGAGAGAATGAA |
| P16 | GCCGCACCGGAATCCT | TGCACCGTAGTTGAGCAGAAGA |
| P27 | GTGGACCAAATGCCTGACTC | TCTGTTCTGTTGGCCCTTTT |
| Pml | GCACCCGCTGCAAAGG | TGAGCTGTTCACACTCGAAACA |
| Pig1 | TGGACACTTCCGAGGTTGTCT | CCTCACGGCCCCATTTG |
| Pig8 | GGCATCTGTACCATCTCAAAGCT | TCGACGCTGTTCCTCTCTCTTC |
| Pig12 | GGCTTGGGATCCAGAGATGTC | GTGGCAGAGCTGCAGAAAGAA |
| Spry1 | ATGGATTCCCCAAGTCAGCAT | CCTGTCATAGTCTAACCTCTGCC |
| Spry2 | TCCAAGAGATGCCCTTACCCA | GCAGACCGTGGAGTCTTTCA |
| Spry3 | CCAGCATTACACCCTCTCCTT | TCAGAGCACCATCGACCTTTG |
| Spry4 | GCAGCGTCCCTGTGAATCC | GCAGCGTCCCTGTGAATCC |
| Tnf-α | GCCTCTTCTCATTCCTGCTT | CACTTGGTGGTTTGCTACGA |
| Tgf-α | CTGTGTGCTGATCCACTGCT | CAAGCAGTCCTTCCCTTCAG |
| Tgf-β | TTGCTTCAGCTCCACAGAGA | TGGTTGTAGAGGGCAAGGAC |
| 18S RNA | AGTCCCTGCCCTTTGTACACA | CGATCCGAGGGCCTCACTA |
WB analysis.
For Western blot (WB) analysis, liver tissue were homogenized in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 7.5]) with protease and phosphatase inhibitors (Roche Diagnostics, IN, and Thermo Scientific, IL). Samples were centrifuged at 10,000 × g for 20 min at 4°C. Thirty micrograms of protein was resolved by SDS-PAGE and immunoblotted as previously described (24). Membranes were probed with the antibodies listed in Table 1.
Co-IP.
For coimmunoprecipitation (co-IP), liver tissues were harvested from DEN-treated mice and lysed with RIPA buffer (20 mM Tris HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1% deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitors. After centrifugation, the supernatant (500 μg of protein) was incubated with a mouse antibody for REF1 (APE1) (C4; Santa Cruz) or rabbit polyclonal antibody for DUSP6 (MDK3) (H-130; Santa Cruz) at 4°C overnight, and immunocomplexes were precipitated using polyclonal antibody G-coupled Sepharose beads. The complexes were washed with RIPA buffer and were subjected to Western blotting using the indicated antibodies.
AP endonuclease activity assay.
A 43-mer 5.5-Cy-labeled oligonucleotide containing a single tetrahydrofuran (THF) (5′-GATCTGATTCCCCATCTCCTCAGTTTCACT(THF)CTGCACCGCATG-3′) and its reverse complementary oligonucleotide were synthesized by Midland Certified Company (Midland, TX). The duplex substrate DNA (2 μl from a 100 nM stock) was incubated with 15 ng of nuclear extracts of MEFs at 37°C for 3 to 20 min, during which the reaction rate in a 15-μl reaction mixture containing 50 mM Tris-HCl (pH 8.5), 50 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 100 μg/ml BSA was linear. The reaction was stopped with 10 μl 80% formamide–40 mM NaOH containing 0.05% xylene cyanol, followed by heating at 95°C for 5 min. The DNA was then separated by denaturing 20% polyacrylamide gel electrophoresis (8% urea, Tris-borate buffer) at 65°C. The substrate and cleaved products were analyzed with a gel imaging system (Odyssey Li-Cor CD7-1740). The AP endonuclease activity of each reaction was calculated as (product/substrate at time zero) × 100%.
Animal PES treatment and MRI studies.
PES (2-phenylethynesulfonamide or pifithrin-μ) was purchased from EMD Chemicals Inc. A cohort of wild-type (WT) C57BL/6 male mice were treated with DEN (25 mg/kg) at 14 days old. At 5 months after DEN treatment, mice were treated with either dimethyl sulfoxide (DMSO) or PES (40 mg/kg of body weight) once a week for 10 weeks. Before mice were euthanized at 8 months, liver tumors were monitored by abdominal magnetic resonance imaging (MRI). MR mouse liver T1W and T2W images were acquired using a 7T Bruker Biospec scanner (Georgia Cancer Center MRI Core). A three-dimensional modified driven-equilibrium Fourier transform (MDEFT) sequence was employed for exaggerated T1 weighting at higher field strength (TI/TR/TE = 1,100/4,000/3.5 ms; FOV = 2.56 by 2.56 by 2.30 cm3; matrix = 128 by 128 by 46). A two-dimensional rapid acquisition with refocused echoes (RARE) sequence was used to acquire T2W images with high in-plane resolution (∼100 by 100 μmol) (TR/TE = 2,900/46 ms; FOV = 2.56 by 2.56 cm2; matrix = 256 by 256; 20 slices; slice thickness = 0.5 mm; slice gap = 0.25 mm; 16 averages).
Statistics.
All experiments were performed with at least 3 to 20 male mice. Data are presented as mean ± standard deviation (SD). Statistical significance between experimental groups was assessed using Student's t test, and a P value of <0.05 was considered significant. For data sets with significant variant differences confirmed by Bartlett’s test for equality of variance, statistical significance was evaluated by Welch’s test.
ACKNOWLEDGMENTS
The research was supported by grants from the National Institutes of Health (CA121951 to D.M. and CA062130 and CA132640 to N.F.M.).
We declare that we have no conflict of interest.
W.C., X.J., J.P., and Y.W. performed the experiments and analyzed and participated in the interpretation of the data and generation of the figures. N.F.M. and D.M. designed and supervised the experiments, interpreted and analyzed the results, and wrote the manuscript.
REFERENCES
- 1.Cairns J. 1975. Mutation selection and the natural history of cancer. Nature 255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 2.Tomasetti C, Vogelstein B. 2015. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, Campbell PJ. 2017. Universal patterns of selection in cancer and somatic tissues. Cell 171:1029–1041. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spangenberg HC, Thimme R, Blum HE. 2009. Targeted therapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 6:423–432. doi: 10.1038/nrgastro.2009.86. [DOI] [PubMed] [Google Scholar]
- 5.Poon RT. 2011. Prevention of recurrence after resection of hepatocellular carcinoma: a daunting challenge. Hepatology 54:757–759. doi: 10.1002/hep.24569. [DOI] [PubMed] [Google Scholar]
- 6.El-Serag HB, Kanwal F. 2014. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatology 60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T, Sakamoto H, Wang L, Ojima H, Shimada K, Kosuge T, Okusaka T, Kato K, Kusuda J, Yoshida T, Aburatani H, Shibata T. 2011. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet 43:464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 8.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J. 2012. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulze K, Nault JC, Villanueva A. 2016. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol 65:1031–1042. doi: 10.1016/j.jhep.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 10.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S-I, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. 2016. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 11.He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K, Frazer KA, Harismendy O, Hatziapostolou M, Iliopoulos D, Suetsugu A, Hoffman RM, Tateishi R, Koike K, Karin M. 2013. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell 155:384–396. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mu X, Espanol-Suner R, Mederacke I, Affo S, Manco R, Sempoux C, Lemaigre FP, Adili A, Yuan D, Weber A, Unger K, Heikenwalder M, Leclercq IA, Schwabe RF. 2015. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J Clin Invest 125:3891–3903. doi: 10.1172/JCI77995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tummala KS, Brandt M, Teijeiro A, Grana O, Schwabe RF, Perna C, Djouder N. 2017. Hepatocellular carcinomas originate predominantly from hepatocytes and benign lesions from hepatic progenitor cells. Cell Rep 19:584–600. doi: 10.1016/j.celrep.2017.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. 2005. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. 2007. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell 11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 17.Hotamisligil GS. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akerfelt M, Morimoto RI, Sistonen L. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai C, Sampson SB. 2016. HSF1: guardian of proteostasis in cancer. Trends Cell Biol 26:17–28. doi: 10.1016/j.tcb.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Labbadia J, Morimoto RI. 2017. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol 27:895–905. doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumeni S, Evangelakou Z, Gorgoulis VG, Trougakos IP. 2017. Proteome stability as a key factor of genome integrity. Int J Mol Sci 18:E2036. doi: 10.3390/ijms18102036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosser DD, Morimoto RI. 2004. Molecular chaperones and the stress of oncogenesis. Oncogene 23:2907–2918. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- 23.Whitesell L, Lindquist SL. 2005. HSP90 and the chaperoning of cancer. Nat Rev Cancer 5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 24.Jin X, Moskophidis D, Mivechi NF. 2011. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab 14:91–103. doi: 10.1016/j.cmet.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciocca DR, Calderwood SK. 2005. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee S, Burns TF. 2017. Targeting heat shock proteins in cancer: a promising therapeutic approach. Int J Mol Sci 18:E1978. doi: 10.3390/ijms18091978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng L, Hunt C, Yaglom JA, Gabai VL, Sherman MY. 2011. Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene 30:2836–2845. doi: 10.1038/onc.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powers MV, Clarke PA, Workman P. 2008. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14:250–262. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Davenport EL, Zeisig A, Aronson LI, Moore HE, Hockley S, Gonzalez D, Smith EM, Powers MV, Sharp SY, Workman P, Morgan GJ, Davies FE. 2010. Targeting heat shock protein 72 enhances Hsp90 inhibitor-induced apoptosis in myeloma. Leukemia 24:1804–1807. doi: 10.1038/leu.2010.168. [DOI] [PubMed] [Google Scholar]
- 30.Gabai VL, Yaglom JA, Waldman T, Sherman MY. 2009. Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol Cell Biol 29:559–569. doi: 10.1128/MCB.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans CG, Chang L, Gestwicki JE. 2010. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem 53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sottile ML, Nadin SB. 2017. Heat shock proteins and DNA repair mechanisms: an updated overview. Cell Stress Chaperones doi: 10.1007/s12192-017-0843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Jin L, Shi T, Zhang B, Zhou Y, Zhou T, Bao W, Xiang H, Zuo Y, Li G, Wang C, Duan Y, Peng Z, Huang X, Zhang H, Xu T, Li Y, Pan X, Xia Y, Gong X, Chen W, Liu Y. 2017. Association between ambient particulate matter exposure and semen quality in Wuhan, China. Environ Int 98:219–228. doi: 10.1016/j.envint.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. 2004. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet 36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 35.Farazi PA, DePinho RA. 2006. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 36.Ariel I, Miao HQ, Ji XR, Schneider T, Roll D, de Groot N, Hochberg A, Ayesh S. 1998. Imprinted H19 oncofetal RNA is a candidate tumour marker for hepatocellular carcinoma. Mol Pathol 51:21–25. doi: 10.1136/mp.51.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, Hirohashi S. 2003. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology 37:198–207. doi: 10.1053/jhep.2003.50022. [DOI] [PubMed] [Google Scholar]
- 38.Shin E, Ryu HS, Kim SH, Jung H, Jang JJ, Lee K. 2011. The clinicopathological significance of heat shock protein 70 and glutamine synthetase expression in hepatocellular carcinoma. J Hepatobiliary Pancreat Sci 18:544–550. doi: 10.1007/s00534-010-0367-0. [DOI] [PubMed] [Google Scholar]
- 39.Karin M, Greten FR. 2005. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 40.Dhar D, Antonucci L, Nakagawa H, Kim JY, Glitzner E, Caruso S, Shalapour S, Yang L, Valasek MA, Lee S, Minnich K, Seki E, Tuckermann J, Sibilia M, Zucman-Rossi J, Karin M. 2018. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell 33:1061–1077. e1066. doi: 10.1016/j.ccell.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I, Itzkovitz S. 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorbi D, Boynton J, Lindor KD. 1999. The ratio of aspartate aminotransferase to alanine aminotransferase: potential value in differentiating nonalcoholic steatohepatitis from alcoholic liver disease. Am J Gastroenterol 94:1018–1022. doi: 10.1111/j.1572-0241.1999.01006.x. [DOI] [PubMed] [Google Scholar]
- 43.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. 2004. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 44.Grivennikov SI, Greten FR, Karin M. 2010. Immunity, inflammation, and cancer. Cell 140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luedde T, Schwabe RF. 2011. NF-kappaB in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senf SM, Dodd SL, McClung JM, Judge AR. 2008. Hsp70 overexpression inhibits NF-kappaB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB J 22:3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salminen A, Paimela T, Suuronen T, Kaarniranta K. 2008. Innate immunity meets with cellular stress at the IKK complex: regulation of the IKK complex by HSP70 and HSP90. Immunol Lett 117:9–15. doi: 10.1016/j.imlet.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. 2000. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med 6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 49.Anand PK, Anand E, Bleck CK, Anes E, Griffiths G. 2010. Exosomal Hsp70 induces a pro-inflammatory response to foreign particles including mycobacteria. PLoS One 5:e10136. doi: 10.1371/journal.pone.0010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baek JY, Morris SM, Campbell J, Fausto N, Yeh MM, Grady WM. 2009. TGF-beta inactivation and TGF-alpha overexpression cooperate in an in vivo mouse model to induce hepatocellular carcinoma that recapitulates molecular features of human liver cancer. Int J Cancer 127:1060–1071. doi: 10.1002/ijc.25127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakurai T, Maeda S, Chang L, Karin M. 2006. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci U S A 103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner EF, Nebreda AR. 2009. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 53.Park HS, Lee JS, Huh SH, Seo JS, Choi EJ. 2001. Hsp72 functions as a natural inhibitory protein of c-Jun N-terminal kinase. EMBO J 20:446–456. doi: 10.1093/emboj/20.3.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verna L, Whysner J, Williams GM. 1996. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol Ther 71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 55.Fu D, Calvo JA, Samson LD. 2012. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tubbs A, Nussenzweig A. 2017. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, Liu Y, Akbani R, Feng B, Donehower LA, Miller C, Shen Y, Karimi M, Chen H, Kim P, Jia P, Shinbrot E, Zhang S, Liu J, Hu H, Bailey MH, Yau C, Wolf D, Zhao Z, Weinstein JN, Li L, Ding L, Mills GB, Laird PW, Wheeler DA, Shmulevich I, Cancer Genome Atlas Research Network, Monnat RJ Jr, Xiao Y, Wang C. 2018. Genomic and molecular landscape of DNA damage repair deficiency across the Cancer Genome Atlas. Cell Rep 23:239–254. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 59.Kenny MK, Mendez F, Sandigursky M, Kureekattil RP, Goldman JD, Franklin WA, Bases R. 2001. Heat shock protein 70 binds to human apurinic/apyrimidinic endonuclease and stimulates endonuclease activity at abasic sites. J Biol Chem 276:9532–9536. doi: 10.1074/jbc.M009297200. [DOI] [PubMed] [Google Scholar]
- 60.Roychoudhury S, Nath S, Song H, Hegde ML, Bellot LJ, Mantha AK, Sengupta S, Ray S, Natarajan A, Bhakat KK. 2017. Human apurinic/apyrimidinic endonuclease (APE1) is acetylated at DNA damage sites in chromatin, and acetylation modulates its DNA repair activity. Mol Cell Biol 37:e00401-16. doi: 10.1128/MCB.00401-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ditch S, Paull TT. 2012. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci 37:15–22. doi: 10.1016/j.tibs.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willis J, Patel Y, Lentz BL, Yan S. 2013. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc Natl Acad Sci U S A 110:10592–10597. doi: 10.1073/pnas.1301445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA. 2003. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol Cell Biol 23:8576–8585. doi: 10.1128/MCB.23.23.8576-8585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. 2004. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol 24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371–383. doi: 10.1016/S0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. 1997. A model for p53-induced apoptosis. Nature 389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 68.Yamaoka A, Kuwabara I, Frigeri LG, Liu FT. 1995. A human lectin, galectin-3 (epsilon bp/Mac-2), stimulates superoxide production by neutrophils. J Immunol 154:3479–3487. [PubMed] [Google Scholar]
- 69.Lee SH, DeJong J. 1999. Microsomal GST-I: genomic organization, expression, and alternative splicing of the human gene. Biochim Biophys Acta 1446:389–396. doi: 10.1016/S0167-4781(99)00112-8. [DOI] [PubMed] [Google Scholar]
- 70.Abbas HA, Maccio DR, Coskun S, Jackson JG, Hazen AL, Sills TM, You MJ, Hirschi KK, Lozano G. 2010. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell 7:606–617. doi: 10.1016/j.stem.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aleksic K, Lackner C, Geigl JB, Schwarz M, Auer M, Ulz P, Fischer M, Trajanoski Z, Otte M, Speicher MR. 2011. Evolution of genomic instability in diethylnitrosamine-induced hepatocarcinogenesis in mice. Hepatology 53:895–904. doi: 10.1002/hep.24133. [DOI] [PubMed] [Google Scholar]
- 72.Connor F, Rayner TF, Aitken SJ, Feig C, Lukk M, Santoyo-Lopez J, Odom DT. 2018. Mutational landscape of a chemically-induced mouse model of liver cancer. J Hepatol 69:840–850. doi: 10.1016/j.jhep.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Downward J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 74.Collado M, Serrano M. 2010. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. 2006. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell 10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. 2005. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell 17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 77.Avraham R, Yarden Y. 2011. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 78.Hanafusa H, Torii S, Yasunaga T, Nishida E. 2002. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol 4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 79.Mason JM, Morrison DJ, Basson MA, Licht JD. 2006. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 80.Lee SA, Ladu S, Evert M, Dombrowski F, De Murtas V, Chen X, Calvisi DF. 2010. Synergistic role of Sprouty2 inactivation and c-Met up-regulation in mouse and human hepatocarcinogenesis. Hepatology 52:506–517. doi: 10.1002/hep.23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Min L, He B, Hui L. 2011. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin Cancer Biol 21:10–20. doi: 10.1016/j.semcancer.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 82.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, Huang A, Wong WL, Callahan MK, Merghoub T, Wolchok JD, de Stanchina E, Chandarlapaty S, Poulikakos PI, Fagin JA, Rosen N. 2012. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song J, Takeda M, Morimoto RI. 2001. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat Cell Biol 3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]
- 84.Yaglom J, O'Callaghan-Sunol C, Gabai V, Sherman MY. 2003. Inactivation of dual-specificity phosphatases is involved in the regulation of extracellular signal-regulated kinases by heat shock and hsp72. Mol Cell Biol 23:3813–3824. doi: 10.1128/MCB.23.11.3813-3824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falco A, Festa M, Basile A, Rosati A, Pascale M, Florenzano F, Nori SL, Nicolin V, Di Benedetto M, Vecchione ML, Arra C, Barbieri A, De Laurenzi V, Turco MC. 2012. BAG3 controls angiogenesis through regulation of ERK phosphorylation. Oncogene 31:5153–5161. doi: 10.1038/onc.2012.17. [DOI] [PubMed] [Google Scholar]
- 86.Colvin TA, Gabai VL, Gong J, Calderwood SK, Li H, Gummuluru S, Matchuk ON, Smirnova SG, Orlova NV, Zamulaeva IA, Garcia-Marcos M, Li X, Young ZT, Rauch JN, Gestwicki JE, Takayama S, Sherman MY. 2014. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res 74:4731–4740. doi: 10.1158/0008-5472.CAN-14-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yaglom JA, Wang Y, Li A, Li Z, Monti S, Alexandrov I, Lu X, Sherman MY. 2018. Cancer cell responses to Hsp70 inhibitor JG-98: Comparison with Hsp90 inhibitors and finding synergistic drug combinations. Sci Rep 8:3010. doi: 10.1038/s41598-017-14900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 89.Rodier F, Campisi J. 2011. Four faces of cellular senescence. J Cell Biol 192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pribluda A, Elyada E, Wiener Z, Hamza H, Goldstein RE, Biton M, Burstain I, Morgenstern Y, Brachya G, Billauer H, Biton S, Snir-Alkalay I, Vucic D, Schlereth K, Mernberger M, Stiewe T, Oren M, Alitalo K, Pikarsky E, Ben-Neriah Y. 2013. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell 24:242–256. doi: 10.1016/j.ccr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 91.Helmbrecht K, Zeise E, Rensing L. 2000. Chaperones in cell cycle regulation and mitogenic signal transduction: a review. Cell Prolif 33:341–365. doi: 10.1046/j.1365-2184.2000.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, Gregory BD, Adams PD, Berger SL. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sen P, Shah PP, Nativio R, Berger SL. 2016. Epigenetic mechanisms of longevity and aging. Cell 166:822–839. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ito T, Teo YV, Evans SA, Neretti N, Sedivy JM. 2018. Regulation of cellular senescence by Polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep 22:3480–3492. doi: 10.1016/j.celrep.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ishak CA, Marshall AE, Passos DT, White CR, Kim SJ, Cecchini MJ, Ferwati S, MacDonald WA, Howlett CJ, Welch ID, Rubin SM, Mann MRW, Dick FA. 2016. An RB-EZH2 complex mediates silencing of repetitive DNA sequences. Mol Cell 64:1074–1087. doi: 10.1016/j.molcel.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dick FA, Goodrich DW, Sage J, Dyson NJ. 2018. Non-canonical functions of the RB protein in cancer. Nat Rev Cancer 18:442–451. doi: 10.1038/s41568-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li G, Ji T, Chen J, Fu Y, Hou L, Feng Y, Zhang T, Song T, Zhao J, Endo Y, Lin H, Cai X, Cang Y. 2017. CRL4(DCAF8) ubiquitin ligase targets histone H3K79 and promotes H3K9 methylation in the liver. Cell Rep 18:1499–1511. doi: 10.1016/j.celrep.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 98.Bosch-Presegue L, Raurell-Vila H, Thackray JK, Gonzalez J, Casal C, Kane-Goldsmith N, Vizoso M, Brown JP, Gomez A, Ausio J, Zimmermann T, Esteller M, Schotta G, Singh PB, Serrano L, Vaquero A. 2017. Mammalian HP1 isoforms have specific roles in heterochromatin structure and organization. Cell Rep 21:2048–2057. doi: 10.1016/j.celrep.2017.10.092. [DOI] [PubMed] [Google Scholar]
- 99.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. 1999. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 100.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. 2009. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell 36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leu JI, Pimkina J, Pandey P, Murphy ME, George DL. 2011. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol Cancer Res 9:936–947. doi: 10.1158/1541-7786.MCR-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leu JI, Barnoud T, Zhang G, Tian T, Wei Z, Herlyn M, Murphy ME, George DL. 2017. Inhibition of stress-inducible HSP70 impairs mitochondrial proteostasis and function. Oncotarget 8:45656–45669. doi: 10.18632/oncotarget.17321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schlecht R, Scholz SR, Dahmen H, Wegener A, Sirrenberg C, Musil D, Bomke J, Eggenweiler HM, Mayer MP, Bukau B. 2013. Functional analysis of Hsp70 inhibitors. PLoS One 8:e78443. doi: 10.1371/journal.pone.0078443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Balaburski GM, Leu JI, Beeharry N, Hayik S, Andrake MD, Zhang G, Herlyn M, Villanueva J, Dunbrack RL Jr, Yen T, George DL, Murphy ME. 2013. A modified HSP70 inhibitor shows broad activity as an anticancer agent. Mol Cancer Res 11:219–229. doi: 10.1158/1541-7786.MCR-12-0547-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo J, Solimini NL, Elledge SJ. 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jego G, Hazoume A, Seigneuric R, Garrido C. 2013. Targeting heat shock proteins in cancer. Cancer Lett 332:275–285. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 107.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. 2009. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dizdaroglu M. 2012. Oxidatively induced DNA damage: mechanisms, repair and disease. Cancer Lett 327:26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 109.Vousden KH, Lane DP. 2007. p53 in health and disease. Nat Rev Mol Cell Biol 8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 110.Jain AK, Barton MC. 2018. p53: emerging roles in stem cells, development and beyond. Development 145:dev158360. doi: 10.1242/dev.158360. [DOI] [PubMed] [Google Scholar]
- 111.Dai C, Gu W. 2010. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med 16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. 2010. ATM activation by oxidative stress. Science 330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 113.Ray PD, Huang BW, Tsuji Y. 2012. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo S, Wharton W, Moseley P, Shi H. 2007. Heat shock protein 70 regulates cellular redox status by modulating glutathione-related enzyme activities. Cell Stress Chaperones 12:245–254. doi: 10.1379/CSC-265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dimitrovska M, Dervisevik M, Cipanovska N, Gerazova K, Dinevska-Kjovkarovska S, Miova B. 2018. Physiological and pharmacological inductors of HSP70 enhance the antioxidative defense mechanisms of the liver and pancreas in diabetic rats. Can J Physiol Pharmacol 96:158–164. doi: 10.1139/cjpp-2017-0394. [DOI] [PubMed] [Google Scholar]
- 116.Sinha K, Das J, Pal PB, Sil PC. 2013. Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch Toxicol 87:1157–1180. doi: 10.1007/s00204-013-1034-4. [DOI] [PubMed] [Google Scholar]
- 117.Shin J, Yang J, Lee J, Baek KH. 2013. Depletion of ERK2 but not ERK1 abrogates oncogenic Ras-induced senescence. Cell Signal doi: 10.1016/j.cellsig.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 118.Deschenes-Simard X, Gaumont-Leclerc MF, Bourdeau V, Lessard F, Moiseeva O, Forest V, Igelmann S, Mallette FA, Saba-El-Leil MK, Meloche S, Saad F, Mes-Masson AM, Ferbeyre G. 2013. Tumor suppressor activity of the ERK/MAPK pathway by promoting selective protein degradation. Genes Dev 27:900–915. doi: 10.1101/gad.203984.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nagai Y, Fujikake N, Popiel HA, Wada K. 2010. Induction of molecular chaperones as a therapeutic strategy for the polyglutamine diseases. Curr Pharm Biotechnol 11:188–197. doi: 10.2174/138920110790909650. [DOI] [PubMed] [Google Scholar]
- 120.Gehrig SM, van der Poel C, Sayer TA, Schertzer JD, Henstridge DC, Church JE, Lamon S, Russell AP, Davies KE, Febbraio MA, Lynch GS. 2012. Hsp72 preserves muscle function and slows progression of severe muscular dystrophy. Nature 484:394–398. doi: 10.1038/nature10980. [DOI] [PubMed] [Google Scholar]
- 121.Huang L, Mivechi NF, Moskophidis D. 2001. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol Cell Biol 21:8575–8591. doi: 10.1128/MCB.21.24.8575-8591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Seglen PO. 1976. Preparation of isolated rat liver cells. Methods Cell Biol 13:29–83. doi: 10.1016/S0091-679X(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 123.Qiao A, Jin X, Pang J, Moskophidis D, Mivechi NF. 2017. The transcriptional regulator of the chaperone response HSF1 controls hepatic bioenergetics and protein homeostasis. J Cell Biol 216:723–741. doi: 10.1083/jcb.201607091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhakat KK, Sengupta S, Adeniyi VF, Roychoudhury S, Nath S, Bellot LJ, Feng D, Mantha AK, Sinha M, Qiu S, Luxon BA. 2016. Regulation of limited N-terminal proteolysis of APE1 in tumor via acetylation and its role in cell proliferation. Oncotarget 7:22590–22604. doi: 10.18632/oncotarget.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]