REPLY
In their Letter to the Editor of mBio, written in response to our recent article on evolution of the global RNA virome (1), Holmes and Duchêne submit that the extreme sequence divergence between the RNA-dependent RNA polymerases (RdRps) makes it impossible to infer deep relationships between RNA viruses from any type of sequence analysis. We certainly agree with Holmes and Duchêne that extreme caution is due in the analysis and interpretation of deep phylogenies, and in particular, that alignment quality is central to our ability to resolve long-distance evolutionary relationships. If the alignment is largely wrong (i.e., does not align homologous protein sites) or noninformative (i.e., cannot be used to distinguish between alternative histories), it is of no utility for phylogenetic reconstruction. Moreover, even a correct and informative alignment does not guarantee correct phylogenetic reconstruction due to the technical limitations of the software, systematic biases of the available evolutionary models, and the fundamentally random nature of sequence divergence. Therefore, formal phylogenetic analysis should be accompanied by careful consideration of the associated biological data and examined in terms of the implications of the respective evolutionary scenarios.
Where exactly lies the boundary between an alignment that is suitable for phylogenetic reconstruction and one that is “highly unlikely to be accurate” is far from being an easy question. In the ideal situation (high sequence similarity, random homoplasy), one might need as little as O(log k) informative sites to resolve a tree of k sequences (2). With real-life data, it is critical that sequence similarity, even if extremely low between the most distant sequences, changes according to a pattern, consistent with the tree structure. Fortunately, the structure of proteins with their nearly invariant functional sites, strongly conserved structural core, variable bulk, and extremely fluid interface surfaces naturally provides such an essential pattern, with each class of sites in the scale of evolutionary rates allowing for good resolution at the appropriate range of distances. Furthermore, the very definition of a “random” site is not a trivial matter. An alignment site that contains all 20 amino acids might appear completely random, but in fact, its validity and utility greatly depend on the amino acid distribution pattern. An obvious hypothetical example is a site where 110 sequences have one amino acid, 110 sequences have another amino acid, and the remaining 18 sequences each have different amino acids. Such a site would contain a strong bipartition signal, and if the other positions of the discordant sequences show affinity with one of these two groups, it would be highly informative for tree reconstruction.
The alignment of 228 RNA-dependent RNA polymerases (RdRps) from RNA viruses and 10 reverse transcriptases (RTs) that was employed in our work (1) to construct the global tree of RNA viruses does indeed push the envelope of usable sequence similarity. As Holmes and Duchêne note, there are no invariant sites, no sites without gaps, more than 96% of the alignment columns contain more than 50% of gaps, and where sites are aligned, the similarity is low (the median distance between RTs and RdRps is 5.0 substitutions per site as estimated by PhyML).
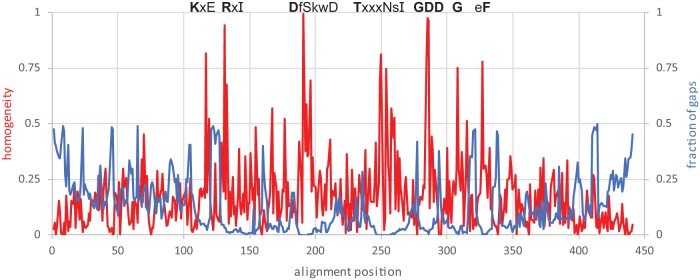
However, some of these metrics, although correctly calculated, do not give the full picture of the alignment properties. Although as indicated above, only 441 sites contain less than 50% of gaps in an alignment of the total length of 12,200, the median length of the RdRp core is 497 amino acids, so that actually, 89% of a typical sequence is part of a reasonable alignment. The plot of the conservation (alignment column homogeneity) and gap content shows multiple, sharp peaks of relatively high conservation and low gap content. Moreover, these regions correspond to well-known motifs that are conserved among the RdRps, across the evolutionary distance of more than five substitutions per site, on average (Fig. 1). Although this level of conservation might appear insufficient to capture the deepest relationships between the RNA viruses, one should keep in mind that, at the deepest level, there are few major clades to resolve (according to our analysis, the RT and five branches of RdRps). The alignment statistics rapidly improve at the shallower levels: even within each major branch, the clade-specific conservation is readily apparent (Table 1).
FIG 1.
Sequence conservation profile along the core alignment of the RdRps and RTs. The homogeneity metric is based on the BLOSUM62 scores between the consensus amino acid and the actual amino acids in the alignment column and are scaled from 1 (all residues are the same) to 0 (the score is not different from the random expectation). The fraction of gaps is computed using sequence weights (6). The amino acids conserved in five prominent motifs are shown. The conservation of a residue is indicated as follows: bold uppercase letter, homogeneity of ≥0.9; uppercase letter, homogeneity of ≥0.75; lowercase letter, homogeneity of ≥0.3; x, homogeneity of <0.3.
TABLE 1.
Branch-specific statistics of the alignment used to construct the RdRp phylogeny
| Alignment subseta |
Sites with homogeneity of: |
No. of sites with <50% gapsb |
|||
|---|---|---|---|---|---|
| ≥0.9 | ≥0.75 | ≥0.5 | ≥0.3 | ||
| Full | 4 | 8 | 20 | 55 | 441 |
| Br1 | 11 | 15 | 53 | 139 | 427 |
| Br2 | 8 | 14 | 37 | 128 | 443 |
| Br3 | 13 | 16 | 37 | 128 | 451 |
| Br4 | 8 | 13 | 24 | 72 | 435 |
| Br5 | 4 | 12 | 44 | 182 | 555 |
Br, branch.
Weighted fractions.
More generally, large and diverse sequence sets that, due to the hyperexponential growth of sequence databases, have become ubiquitous in today’s evolutionary studies, present an inherent conundrum for alignment construction and analysis. Random sequence-level events (mutations, deletions, and especially, insertions) affect the alignment metrics in a ratchet-like manner. Given enough sequences, apparent substitutions (rare real ones or sequencing errors) will be found in all sites, including the supposedly invariant ones. A deletion leaves a site in the “gapped” status, no matter how rare it is. A unique insertion (again, real or artefactual) leaves a trail of gaps in other sequences, bloating the alignment and complicating all types of analyses. Indeed, in the RdRp alignment discussed here, 6,527 of the 12,200 aligned sites contain nothing but gaps that are inherited from the larger original alignment of 4,627 sequences, and additional 2,054 sites harbor an effectively unique insertion. Although the case of the virus RdRp might be somewhat extreme, this type of alignment is by no means limited to virus proteins. In order to take advantage of the rapidly growing diversity of available sequences rather than being hampered by it, evolutionary biologists have to step up to the challenge and adopt appropriate approaches for the analysis of such “untidy” alignments, which is what we attempted to do in our study of the global RNA virome evolution.
Crucially, the conclusions derived from the RdRp tree are corroborated by additional information. In particular, the five major branches of RNA viruses and many clades within each branch possess additional signature genes that are present in the majority of the respective viruses and, in some cases at least, can be traced to the hypothetical ancestral viruses. These genes include a distinct serine protease of apparent bacterial origin in branch 2 (picornavirus-like and related viruses); the capping enzyme in branch 3 that consists of alpha-like and related viruses (albeit, most likely, convergently acquired by three large clades within this branch); a unique capsid protein in branch 4 (double-strand RNA viruses); capping enzyme and “cap-snatching” endonuclease, respectively, in two major clades within branch 5 (negative-sense RNA viruses). Furthermore, the monophyly of branches 2 and 3, and the main clades within each of these branches, is supported by clustering of the single jelly-roll capsid proteins, the second most common protein, after RdRp, in RNA viruses (1).
In summary, we strongly believe that, despite the extreme sequence divergence, the global evolutionary analysis of RNA viruses that is necessarily centered on the RdRp tree is informative and useful because it yields a unified framework for further study of virus diversity, evolution, and classification (3). In particular, the monophyly of the five major branches and the many clades within these branches is strongly supported. We have to emphasize, however, that the relationship between the five branches is a different matter. These deepest parts of the tree, in particular, the placement of the negative-sense RNA viruses (branch 5) within the dsRNA viruses (branch 4) have to be treated with utmost caution as we repeatedly point out in the original article. It should be noted that even this most unexpected aspect of the virus RdRp tree topology appears to be supported by analysis of the respective 3D structures, which demonstrates a pronounced structural similarity among the RdRps of negative-sense and double-stranded RNA viruses (4, 5).
Although we are reluctant to subscribe to the view of Holmes and Duchêne that the “very first moments” of RNA virus evolution are unknowable in principle, we concede that it might not be possible to reconstruct these stages with confidence. This, however, is no reason to give up on global analyses of virus evolution.
Footnotes
This is a response to a letter by Holmes and Duchêne https://doi.org/10.1128/mBio.00289-19.
Citation Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2019. Reply to Holmes and Duchêne, “Can sequence phylogenies safely infer the origin of the global virome?”: deep phylogenetic analysis of RNA viruses is highly challenging but not meaningless. mBio 10:e00542-19. https://doi.org/10.1128/mBio.00542-19.
REFERENCES
- 1.Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9:e02329-18. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erdos PL, Steel MA, Szekely LA, Warnow TJ. 1999. A few logs suffice to build (almost) all trees. Random Struct Alg 14:153–184. doi:. [DOI] [Google Scholar]
- 3.Kuhn JH, Wolf YI, Krupovic M, Zhang Y-Z, Maes P, Dolja VV, Koonin EV. 2019. Classify viruses - the gain is worth the pain. Nature 566:318–320. doi: 10.1038/d41586-019-00599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 5.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henikoff S, Henikoff JG. 1994. Position-based sequence weights. J Mol Biol 243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]