Abstract
Muscle wasting is a debilitating consequence of fasting, inactivity, cancer, and other systemic diseases that results primarily from accelerated protein degradation by the ubiquitin-proteasome pathway. To identify key factors in this process, we have used cDNA microarrays to compare normal and atrophying muscles and found a unique gene fragment that is induced more than ninefold in muscles of fasted mice. We cloned this gene, which is expressed specifically in striated muscles. Because this mRNA also markedly increases in muscles atrophying because of diabetes, cancer, and renal failure, we named it atrogin-1. It contains a functional F-box domain that binds to Skp1 and thereby to Roc1 and Cul1, the other components of SCF-type Ub-protein ligases (E3s), as well as a nuclear localization sequence and PDZ-binding domain. On fasting, atrogin-1 mRNA levels increase specifically in skeletal muscle and before atrophy occurs. Atrogin-1 is one of the few examples of an F-box protein or Ub-protein ligase (E3) expressed in a tissue-specific manner and appears to be a critical component in the enhanced proteolysis leading to muscle atrophy in diverse diseases.
In mammals, muscle protein serves as a primary reserve of amino acids that can be mobilized during fasting and disease to provide a source of amino acids for hepatic gluconeogenesis and energy production (1). An important physiological adaptation to fasting is an increase in the overall rate of breakdown of muscle proteins leading to a rapid loss of muscle mass and protein content. A similar rapid atrophy of muscle is a common debilitating feature of many systemic diseases including diabetes, cancer, sepsis, hyperthyroidism, and uremia (2, 3) and occurs in specific muscles upon disuse or nerve injury (4, 5). These different forms of muscle atrophy are characterized by a common set of biochemical changes (2, 3). Loss of muscle protein occurs primarily through enhanced protein breakdown because of activation of the ubiquitin (Ub)-proteasome pathway, as shown by inhibitor studies in intact muscles (6), increased content of Ub-protein conjugates, and by cell-free measurements of Ub-dependent proteolysis (7, 8). In addition, these various atrophying muscles show increases in mRNA for components of this pathway including polyubiquitin, E214k, and multiple proteasome subunits (2, 9, 10). However, the key ubiquitination enzymes active in degrading the bulk of muscle protein in normal or catabolic states remain unclear (11).
Proteins destined for degradation by the Ub-proteasome pathway are first covalently linked to a chain of Ub molecules, which marks them for rapid breakdown to short peptides by the 26S proteasome (12). The key enzyme responsible for attaching Ub to protein substrates is a Ub-protein ligase (E3) that catalyzes the transfer of an activated form of Ub from a specific Ub-carrier protein (E2) to a lysine residue on the substrate. Individual E3s ubiquitinate specific classes of proteins; hence, the identity of the proteins degraded by the proteasome is largely determined by the complement of E3s active in individual cells.
The present studies were undertaken to identify key factors that may be important in the acceleration of muscle proteolysis in catabolic states. To establish a comprehensive picture of the transcriptional adaptations that occur during various types of muscle atrophy, and that may be responsible for the activation of protein breakdown, we have used Incyte cDNA microarrays to compare mRNA levels in normal mouse muscles to those from atrophying muscles. Because much is known about the enhancement of proteolysis and other metabolic adaptations to fasting (13–15), we initially performed microarray experiments comparing poly(A)+ RNA from muscles of normal and food-deprived mice, and we have identified a group of genes whose transcripts increase markedly in the atrophying muscles. One expressed sequence tag was of particular interest because its level increased most dramatically on fasting. Therefore, we have cloned this protein and defined its properties. We demonstrate here that this protein has the properties of an E3 of the SCF class and is unusual in being expressed selectively in striated muscle. We have also studied further the expression of this gene on food deprivation and in several other models of human diseases in which there is a marked acceleration of muscle proteolysis. These studies demonstrate the existence of a unique ubiquitination enzyme that appears to increase when muscles undergo atrophy.
Methods
cDNA Library Production and Screening.
Total RNA was isolated from the gastrocnemius muscle of 2 d food-deprived mice by using TRIzol (Life Technologies, Grand Island, NY) reagent, and poly(A)+ RNA was purified by using the Oligotex mRNA isolation kit (Qiagen). A cDNA library was constructed from mouse RNA by using the Superscript Plasmid cDNA Synthesis system (Life Technologies). Analysis of the library revealed an average insert size of 2–3 kb. Hybridization screens were performed according to the procedures of Sambrook et al. (16) and Church and Gilbert (17). From the 30,000 recombinants screened, six positive clones were identified.
Creation of Atrogin-1 Mutants.
Atrogin-1 in BlueScriptII KS+ (Stratagene) served as the phagemid for the generation of single-strand DNA for site-directed mutagenesis of the F-box sequence. The plasmid was transformed into f+ Escherichia coli strain (RZ1032). A single colony was grown to mid-log phase, superinfected with M13KO7 helper phage, and grown overnight at 37°C. The bacteria were removed by centrifugation, and the phage was precipitated from the supernatant with NaCl-polyethylene glycol solution. Single-strand DNA was purified by phenol-chloroform extraction followed by ethanol precipitation and resuspension in water. An aliquot of the preparation was used as a template for oligonucleotide site-directed mutagenesis (16) to generate both the deletion of the F-box motif (ΔFb-atrogin-1; amino acids 228–284; primer 5′-GAAGTGGTACTGGCAGAGTCGATCGGTGATCGTGAGGCCTTTGAAG-3′) as well as for mutation of the first two residues of the F-box sequence (atrogin-13A; D-L-P to A-A-A; primer 5′-GTTGTAAGCACACAGCCGCGGCGGTGATCGTGAGG-3′). Oligonucleotide primers for the amplification of DNA fragments containing wild-type or mutant versions of the atrogin-1 gene were designed with flanking attB1 or attB2 sites for insertion into the GATEWAY donor vector pDONR201 (Life Technologies). Primers with the following sequences were synthesized by MWG Biotec (Ebersberg, Germany): AT1 (forward), 5′-attB1-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCTTGGGCAGGACTGGCGG-3′; and AT1 (reverse), 5′-attB2-GGGGACCACTTTGTACAAGAAAGCTGGGTTATTCAGAACTTGAACAAATTG-3′. The PCR products were cloned directly into pDONR201, and the resulting plasmids were used to transfer the atrogin-1 gene sequences into pDEST27 [glutathione S-transferase (GST) fusion vector] via homologous recombination.
Coprecipitation Experiments.
For expression of GST-, Myc5-, FLAG-, and hemagglutinin (HA)-tagged proteins, 293T cells were grown in DMEM (Life Technologies) supplemented with 10% FBS. GST-atrogin-1, GST-atrogin-13A, or GST-ΔFb-atrogin-1 in pDEST27 (5 μg) was cotransfected with 5 μg of Myc5-Skp1 (kindly provided by N. Ayad, Harvard Medical School) or HA-Skp1, FLAG-Cul1, and FLAG-Roc1 (kindly provided by K. Tanaka, Tokyo Metropolitan Institute of Medical Science) into 293T cells at 60–80% confluence by using Superfect reagent (Qiagen) according to the manufacturer's instructions. The cells were maintained in medium with 10% of FBS for 24 h before beginning the experiment. After harvest, the cells were resuspended in cell lysis buffer [20 mM Na-Hepes, pH 7.7/225 mM KCl/1% Triton X-100, supplemented with protease inhibitor mixture (Roche Biomedical)] and lysed by vortexing. The lysates were cleared by centrifugation at 10,000 × g for 20 min in an Eppendorf microcentrifuge. Coprecipitation was carried out by incubating 900 μl of cell lysate (containing 1.5–3.0 mg of protein) with 20 μl of glutathione-Sepharose beads (Amersham Pharmacia) for ≈16 h at 4°C with gentle rocking. The beads were then washed three times with 1 ml of cell lysis buffer containing 0.5% Triton X-100 and 225 mM KCl, resuspended in SDS loading dye, and resolved by SDS/PAGE. Proteins were transferred to PDVF membrane (Millipore) and probed with mouse monoclonal anti-HA, -FLAG, or -GST primary antibodies (Sigma) in 5% milk/PBS, at a dilution of 0.1 μg/ml, 0.1 μg/ml and 1:8000, respectively. Membranes were washed twice with PBS/0.1% Tween-20, once with PBS and incubated with an alkaline phosphatase-linked anti-mouse IgG fragment (Promega). Detection was by enhanced chemiluminescence (CDP-Star/Tropix, Bedford, MA).
Animal Models of Muscle Atrophy.
Six-week-old male C57BL6 mice were deprived of solid food for 1 or 2 d but given free access to water. During this period, the gastrocnemius lost 15–20% of its initial weight (R.T.J., S.H.L., M.D.G., and A.L.G., unpublished data). Both gastrocnemius muscles from 10 control or food-deprived mice were harvested and pooled to prepare total RNA for the gene microarray, cDNA library preparation, and northern analyses. Gastrocnemius muscles from uremic and diabetic rats and controls were kindly provided by S. R. Price, J. L. Bailey, and W. E. Mitch (Emory University, Atlanta). The uremic rats were prepared by 5/6 nephrectomy and demonstrate marked muscle atrophy and accelerated protein breakdown (18). Acute diabetes was induced by streptozotocin administration, and muscles were harvested 3 d later when proteolysis was accelerated and animals demonstrate marked muscle atrophy (19). Gastrocnemius muscles from rats implanted with Yoshida Ascites Hepatoma and controls were provided by V. Baracos (University of Alberta, Edmonton, AB, Canada). These muscles were harvested 6 d after tumor implantation and demonstrate marked muscle atrophy (20). A systematic analysis of the transcriptional changes in these different conditions will be presented elsewhere.
Results
Cloning and Structure of the Atrogin-1 Gene.
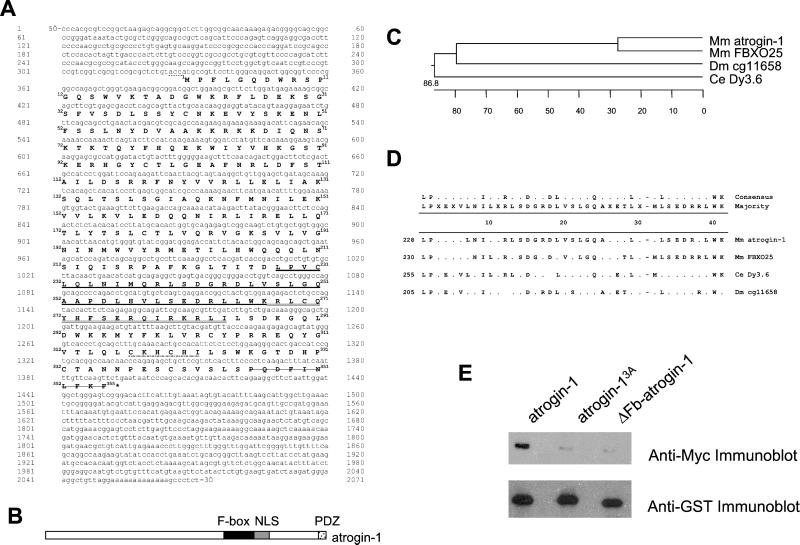
The gene transcript that was increased most dramatically (seven- to ninefold) on the Incyte microarray after food deprivation corresponded to an expressed sequence tag (GenBank accession no. AW051824) and was chosen for further study. [The mouse gene has subsequently also been identified by the RIKEN Mouse Gene Encyclopedia Project (NP_080622) (42)]. After conducting sequence analysis in available expressed sequence tag databases (http://www.labonweb.com), we identified a sequence extended at both 3′- and 5′- ends, which allowed us to clone a 1.1-kb fragment by PCR from a human skeletal muscle cDNA library. To identify the full-length cDNA, the PCR-amplified human gene fragment was used as a probe to screen a mouse cDNA library derived from the atrophying hindlimb muscles of mice deprived of food for 2 d. These tissues were chosen because of the high level of expression of this gene at this time point compared to levels in fed controls (see below). A positive clone containing a 2.1-kb insert was excised by complete SalI and NotI double digestion, subcloned into the BlueScriptII KS+, and sequenced in its entirety. The nucleotide and deduced amino acid sequences of the gene, which we term atrogin-1 (for atrophy gene-1), are shown in Fig. 1A.
Figure 1.
The atrogin-1 gene. (A) Sequences. The deduced amino acid sequence of the encoded polypeptide is shown in single-letter code below the nucleotide sequence in bold face type and is numbered beginning with the initiating methionine. Nucleotides are numbered in the 5′ to 3′ direction. The asterisk denotes the 3′-terminal stop codon. Residues constituting the F-Box motif, bipartite nuclear localization sequence, cytochrome c family heme-binding site signature, and PDZ domain are underlined, double underlined, dot dashed, and stricken-through respectively. A sequence matching the Kozak consensus that precedes the initiating methionine is dotted. (B) Schematic illustration of atrogin-1 protein. (C) Phylogenetic tree generated with Lasergene megalign software. C. elegans (Ce), D. melanogaster (Dm), and Mus musculus (Mm). (D) The family of atrogin-1 proteins. Multiple alignment of the amino acid sequences of the F-box motifs for representative members of the atrogin-1 superfamily. The consensus sequence at the top was based on conservation of a residue at any given position in >75% of sequences. Amino acids that differ from the consensus or majority sequence are denoted by (·). The GenBank accession no. for Mm atrogin-1 is NP_080622.1; for Mm FBX025, NP_080061; for Dy3.6, CAB09415; and for cg11658, AAF50023. (E) Association between atrogin-1 and Skp1. (Upper) The 293T cells were cotransfected with constructs encoding wild-type GST-atrogin-1 and Myc5-Skp1 (lane 1), GST-atrogin-13A and Myc5-Skp1 (lane 2), and ΔFb-atrogin-1 and Myc5-Skp1 (lane 3). Lysates were subjected to GST tag pull-down assay by using glutathione-Sepharose beads, and then the Myc5-Skp1 proteins were detected by using an anti-Myc antibody. (Lower) The membranes were also stripped and reprobed with anti-GST antibody to show that similar amounts of GST-tagged proteins were expressed.
Analysis of the mouse DNA sequence (nucleotides 1–2071) indicated the presence of a single 1,068-bp ORF with a predicted initiation codon (ATG) at nucleotide 328 and a termination codon at nucleotide 1,395. This ORF codes for a polypeptide of 355-aa residues with a molecular mass of 41,503 Da and a pI of 9.46. The initiation codon (nucleotide 328) is embedded in the sequence ACCATG, which matches perfectly with the eukaryotic translation initiation consensus sequence ACCATG (21). Because no other ATG was found upstream of this initiation codon, this region probably represents the complete coding sequence.
Comparison of the atrogin-1 cDNA sequence with the National Center for Biotechnology Information sequence database identified four other similar genes, including the human homolog, orthologs in both Caenorhabditis elegans (Dy3.6) and Drosophila melanogaster (cg11658), and a similar human protein, FBXO25. Human and mouse atrogin-1 are almost identical, differing in only 11 amino acids. Atrogin-1 shares slightly greater overall amino acid sequence identity with FBOX25 (60%) than with other members of the family. Atrogin-1 is 27% identical to C. elegans Dy3.6 and 26% identical to the cg11658 protein predicted from Drosophila genome sequence (Fig. 1C).
Atrogin-1 Is an F-Box Protein, a Component of an SCF E3.
A systematic search for functional domains within the atrogin-1 sequence by using the pfscan algorithm (http://www.isrec.isb-sib.ch) and cansite (http://www.cansite.bidmc.harvard.edu) indicated the presence of four conserved motifs (Fig. 1B). An F-box is present at amino acid 228–267 (Fig. 1D). This highly degenerate motif is found in a family of proteins, most of which function as one component of the SCF family of Ub-protein ligases (E3s) (22). The F-box protein functions as an adaptor that binds proteins to be ubiquitinated and, through the F-box, associates with the Skp1 protein (or a homolog) and thus with other components of the E3 complex (23). Atrogin-1 lacks the common motifs found in other F-box proteins that are known to interact with protein substrates (e.g., leucine-rich regions, WD40 repeats). However, at its extreme carboxy terminus (amino acids 346–355), atrogin-1 does contain a motif known to interact with proteins containing class II PDZ domains. PDZ domain-containing proteins bind to specific sequences at the extreme C-termini of target proteins (24, 25) and may represent a new type of substrate-binding region in F-box proteins. By using the convention proposed by Cenciarelli et al. (26) and Winston et al. (27), we categorize atrogin-1 as an FBXO class of F-box protein, X referring to its lack of a recognized substrate-binding region. Other features also present in the molecule include a potential bipartite nuclear localization sequence (amino acid 267–284) and a cytochrome c family heme-binding site signature (amino acid 317–322).
To demonstrate that the F-box domain in atrogin-1 is functional, we tested whether this protein associates with Skp1, the classical F-box-binding partner in SCF complexes. For these studies, we also created mutant forms of atrogin-1 in which the amino acids at positions 227–229 containing the first two highly conserved residues of the putative F-box were replaced by alanines (atrogin-13A) and a second mutant in which the putative F-box was deleted completely (ΔFb-atrogin-1). These wild-type and mutant forms of atrogin-1 were expressed as GST-fusions in 293T cells also expressing Myc5-Skp1, and the cell lysate was subjected to a GST pull-down assay. Atrogin-1 was precipitated together with Skp1, as detected by immunoblot analysis (Fig. 1E). However, both mutant forms that lacked the complete F-box sequence failed to associate with Skp1 (Fig. 1E, lanes 2 and 3). Thus, atrogin-1 can interact directly with Skp1 through the F-box domain. Further evidence for a direct interaction between atrogin-1 and Skp1 was our finding that the abundance of GST-atrogin-1 was increased when it was coexpressed with Myc5-Skp1 (data not shown). This in vivo stabilization depended on the F-box, because the abundance of mutant forms of atrogin-1 lacking the F-box was unaffected by Myc5-Skp1 coexpression (data not shown).
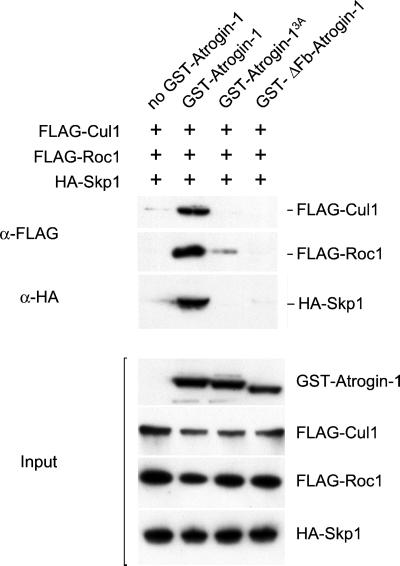
To learn whether atrogin-1 exists in cells as part of an SCF complex, we tested whether the other components, Cul1 and Roc1, were also associated with GST-atrogin-1 and Skp1. 293T cells were transiently transfected with constructs encoding FLAG- or HA-tagged versions of these various proteins, the cell lysed, and atrogin-1 was then precipitated. These pellets contained also Skp1 as well as Cul1 and Roc1 (Fig. 2), which were identified by immunoblot. In contrast, these other SCF subunits were not found coprecipitated with the versions of atrogin-1 containing a mutation or deletion in the F-box. These observations provide strong evidence that atrogin-1 is a component of an SCF E3. The functional significance of the other putative domains and its substrates are presently being studied.
Figure 2.
Formation in vivo of the SCFatrogin-1 complex. (Upper) The 293T cells were cotransfected with constructs encoding SCF components (HA-Skp1, FLAG-Cul1, and FLAG-Roc1) with and without different GST-tagged versions of atrogin-1 [no GST-atrogin-1 (lane 1), GST-atrogin-1 (lane 2), GST-atrogin-13A (lane 3), and GST-ΔFb-atrogin-1 (lane 4)]. Lysates were subjected to GST coprecipitation by using glutathione-Sepharose beads, and then the FLAG-Cul1 and FLAG-Roc1 proteins were detected by using an anti-FLAG antibody. The membrane was stripped and reprobed with an anti-HA antibody to detect HA-Skp1. (Lower) Western blots of total cell lysates to show that similar amounts of GST-atrogin-1, GST-atrogin-13A, GST-ΔFb-atrogin-1, FLAG-Cul1, FLAG-Roc1, and HA-Skp1 proteins were expressed.
Expression of Atrogin-1 in Atrophying Muscles.
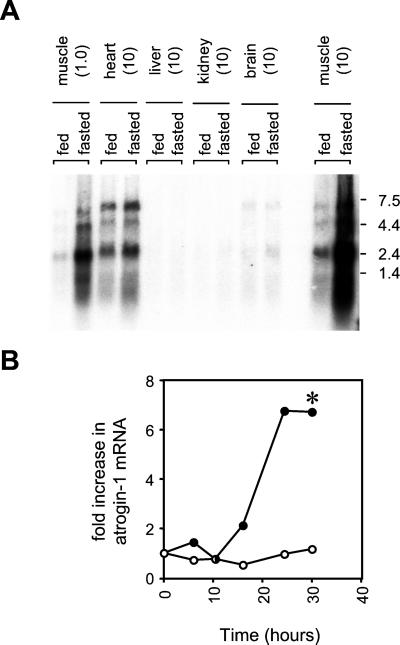
Northern blot analysis of total RNA from various tissues of control and food-deprived mice revealed three major transcripts (≈2.5, 5.0, and 7.3 kb) (Figs. 3A and 4). In fed controls, gastrocnemius muscles and to a much lesser extent heart were the only tissues expressing significant amount of these transcripts. Liver, kidney, brain, spleen, and testis (data not shown) contained only trace amounts. In addition to being restricted to striated muscle normally, Northern blot analysis showed that after fasting, the levels of all three transcripts increased at least sevenfold in skeletal muscles but not in heart or other tissues (Fig. 3A). The increase in atrogin-1 mRNA is among the largest we have identified for any gene in different types of atrophying muscle (see below). For example, polyubiquitin mRNA, whose expression has served as the benchmark for activation of Ub-mediated proteolysis in atrophying muscles, is increased typically 2- to 5-fold in muscles during fasting (28, 29).
Figure 3.
(A) Tissue distribution of atrogin-1. Total RNA was extracted from the pooled tissues of eight control or eight, 2 d food-deprived mice by using TRIzol reagent. RNA was electrophoresed on a 0.8% formaldehyde-agarose gel, transferred to a Zeta-Probe membrane (Bio-Rad), and UV crosslinked. The quantity of RNA loaded (in micrograms) for each tissue is in parentheses. A gel-purified KpnI/XbaI fragment of the atrogin-1-coding sequence (bp 1,500–2,368) was used as a probe for the atrogin-1 transcripts. The probe was labeled by random priming (Prime-It Kit, Stratagene). Hybridization was performed by the method of Church and Gilbert (17) at 65°C. Hybridized membranes were analyzed by using a Fuji Phosphorimager. Blots were stripped and rehybridizing with a mouse glyceraldehyde-3-phosphate dehydrogenase probe (GAPDH, Ambion, Austin, TX) to ensure equivalent gel loading. (B) Time course of atrogin-1 expression in gastrocnemius muscle. The 48 C57BL6 mice were split into two groups at time = 0: 24 animals were transferred to cages without solid food, whereas the remaining 24 animals were maintained with food. At indicated times, four animals from each group were killed, gastrocnemius muscles weighed, and total RNA extracted by using TRIzol reagent. Northern blots were performed as above by using 10 μg of total RNA/lane from the pooled control and fasted muscle at each time point. Atrogin-1 band intensities were quantitated by densitometry, and membranes reprobed with a mouse GAPDH probe to ensure equal RNA loading. The fold increase in atrogin-1 mRNA was derived by dividing the corrected atrogin-1 band intensity (atrogin-1/GAPDH) at each time point with the corrected band intensity at t = 0. ○ fed; ● fasted. *, Muscle weight loss was significantly greater (P < 0.05) in fasted compared to fed animals after 30 h but not earlier time points (see text).
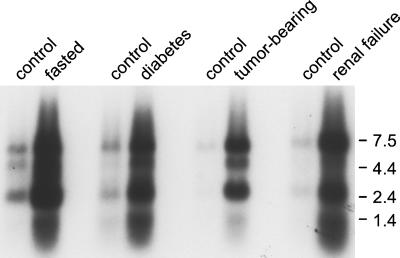
Figure 4.
Atrogin-1 expression in muscles atrophying because of different diseases. Total RNA was prepared from the gastrocnemius muscles of control and 2 d food deprived mice, control and 3-d streptozotocin-treated rats, control and rats bearing Yoshida ascites hepatoma for 6 d, and control and 5/6 nephrectomized rats. Previous studies have shown enhanced protein degradation in these animal models (18–20). Northern blots (10 μg of total RNA per lane) were preformed exactly as in Fig. 3.
To test whether the induction of atrogin-1 occurs before and may thus trigger the loss of muscle tissue mass, we examined the time course of its gene expression in the gastrocnemius muscle after food deprivation. The level of atrogin-1 mRNA was clearly increased as early as 16 h after food removal and reached a maximum level by 24 h, which was sustained at 30, 48, and 72 h (Fig. 3B and data not shown). In the same experiment, muscle weights (expressed as a proportion of initial body weight as an indirect measure of muscle weight loss) were compared between fasted and fed animals. In contrast to the rapid rise in the level of atrogin-1 mRNA, no significant decrease in the mass of the gastrocnemius muscle was detected in the fasted animals until 30 h after food removal. At 30 h, gastrocnemius weight (mg)/initial body weight (g) decreased by 8% (from 5.52 in the fed to 5.09 in fasted animals) corrected for body weight (P = 0.02, Mann–Whitney u test). Similarly, in previous experiments with larger groups of mice of similar age, significant muscle weight loss was always found 48 h after food removal but not at 24 h, whereas atrogin-1 mRNA was markedly increased at both these times (data not shown). Thus, the increase in expression of atrogin-1 precedes muscle weight loss and remains at a high level during the course of muscle atrophy. These findings suggest that activation of this gene is important in the development and progression of muscle protein loss in fasting.
To test whether this gene is induced generally in atrophying muscles, we analyzed the mRNA levels of atrogin-1 in muscles from a number of rat models of human diseases in which there is also enhanced proteolysis and marked muscle wasting. In atrophying muscles from rats with streptozotocin-induced diabetes, rats bearing a peritoneal Yoshida hepatoma, or rats with experimentally induced uremia, at times when proteolysis by the ATP-dependent cytosolic pathway and Ub conjugation are markedly accelerated (18–20), the mRNA levels of atrogin-1 were also increased at least 10-fold (Fig. 4). Thus, atrogin-1 is a muscle-specific gene, whose expression rises under diverse conditions when muscle wasting occurs.
Discussion
By using cDNA microarrays to define the transcriptional changes in atrophying muscle from food-deprived mice, we have discovered a new muscle-specific F-box protein termed atrogin-1. Because atrogin-1 is very strongly induced in many catabolic states, it is likely to play a key role in the generation of muscle atrophy. The strong induction of atrogin-1 expression at an early stage of muscle wasting, in fact before muscle weight loss was detectable, and the maintenance of its high expression during the period when overall proteolysis is accelerated, strongly suggest a role in both initiation and maintenance of the accelerated proteolysis.
The signals that trigger the loss of muscle mass in the varied pathological states modeled in this study differ. For instance, glucocorticoids and low insulin levels are essential for the enhanced proteolysis and muscle wasting in starvation and diabetes (28, 30, 31), whereas TNF-α, prostaglandins, and glucocorticoids play important roles in muscle wasting in experimental models of sepsis and cancer-induced cachexia, including that induced by the Yoshida hepatoma (32–35). Acidosis and glucocorticoids have been shown to be important in the enhancement of muscle proteolysis in uremic animals (36, 37). By themselves, glucocorticoids can stimulate protein breakdown in muscle (35), and in related studies, we have also found a marked up-regulation of atrogin-1 mRNA on treatment of animals or cultured muscle cells with glucocorticoids and also increased atrogin-1 mRNA in rat muscles undergoing atrophy because of unloading by hind-limb suspension (unpublished observations). Despite the different physiological signals, the induction of atrogin-1 in all these catabolic states is strong evidence for our suggestion that the muscle atrophy in these diseases (2, 3, 10, 35) proceeds through a common set of adaptations leading to enhanced proteolysis.
The changes seen here in mRNA for atrogin-1 are much larger than those reported by others and us for mRNAs encoding other components of this proteolytic pathway. For example, the expression of polyubiquitin increases in all forms of muscle atrophy; however this induction (two to five times) is much smaller than the increase in atrogin-1. mRNAs for Ub carrier protein (E2) genes, such as E214k, are increased two- to threefold in atrophying muscles (7), and similar increases may also occur with many other E2s (38) (unpublished observations). mRNA for E3α, the Ub protein ligase of the N-end rule pathway, is increased twofold in muscles from diabetic and septic animals (7, 39), and this ubiquitination pathway appears to contribute to the enhanced proteolysis in these catabolic states (7, 8). Because atrogin-1 shares no sequence homology with E3α, it is likely to act on different cell proteins (see below).
In SCF complexes, the F-box protein binds the substrate and links it to the other subunits (RING-H2, Cullin, and Skp1 proteins) and the E2 involved in ubiquitination. Accordingly, our coprecipitation experiments showed that the F-box in atrogin-1 is necessary for this protein to interact with the Skp1 protein because coprecipitation from cell lysates did not occur when critical residues in the F-box were mutated. Like certain other F-box proteins (40), atrogin-1 is stabilized in vivo by its interaction with Skp1. In fact, unless Skp1 was coexpressed with atrogin-1, only trace amounts of this protein could be found in the cultured cells. Moreover, atrogin-1, but not mutants in the F-box domain, assembled with Cul1, Skp1, and Roc1 to form the minimal SCF module (SCFAtrogin-1) characteristic of this family of E3s. Most F-box proteins bind to substrates through regions C-terminal to the F-box. Atrogin-1 may bind to substrates through the predicted PDZ-binding motif at its extreme C terminus or through an as yet undetermined region in its N-terminal portion. Other F-box proteins with similar C-terminal F-box domains are known (26, 27), although their substrate-binding domains have not been identified. In fact, in related studies, we have shown that these complexes can ligate 125I-ubiquitin to form high molecular weight conjugates (unpublished observation).
Also of appreciable interest is the presence in atrogin-1 of a putative nuclear localization signal. Many other F-box proteins also contain nuclear localization sequences (41). The presence of such a sequence in atrogin-1, along with its muscle-specific expression, suggests that it may function in ubiquitinating muscle-specific transcription factors or other nuclear proteins involved in muscle growth. However, atrogin-1 may also function to ubiquitinate cytosolic proteins such as components of the myofibril, which are rapidly degraded during muscle atrophy.
Previous studies have suggested that although diverse conditions can lead to rapid muscle atrophy, the loss of muscle tissue in each case results primarily from activation of a common biochemical program that stimulates muscle proteolysis (2, 3, 10, 35). The identification of a new E3 dramatically up-regulated in various types of atrophy further strengthens the argument for such a common “atrophy program” and for the existence of a group of atrophy-related genes. We will present further evidence to support this concept in subsequent papers based on transcriptional profile analysis of a variety of types of atrophying muscles. Identification of the transcription factors that regulate atrogin-1 expression will be an important step to understand the atrophy response. Also, defining the substrates that atrogin-1 ubiquinates will be essential to clarify its role in the atrophy process. Genetic manipulations or pharmacological agents that reduce atrogin-1 activity might even prove useful in counteracting the debilitating effects of muscle wasting.
Acknowledgments
We thank Sandy Ryeom for her help and advice; Vickie Baracos for providing muscles from tumor-bearing rats; James Bailey, Russ Price, and William E. Mitch for providing muscles from diabetic and uremic rats; and Dr. Edouard Vannier for providing the human muscle cDNA library. We are also grateful to Dr. Keiji Tanaka and Tomiki Chiba for providing cDNA plasmids and for advice on carrying out coprecipitation experiments. This study was supported by grants from the National Space Biomedical Research Institute and the Muscular Dystrophy Association (to A.L.G.) and from the National Institutes of Health (DK02707 to S.H.L.) During these studies, M.D.G. was a fellow of the Fundaçâo de Amparo à Pesquisa do Estado de Sâo Paulo. R.T.J. held a fellowship from US–UK Fulbright Commission and the Warren-Whitman-Richardson Fellowship.
Abbreviations
- Ub
ubiquitin
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Kettelhut I C, Wing S S, Goldberg A L. Diabetes Metab Rev. 1988;4:751–772. doi: 10.1002/dmr.5610040805. [DOI] [PubMed] [Google Scholar]
- 2.Lecker S H, Solomon V, Mitch W E, Goldberg A L. J Nutr. 1999;129:227S–237S. doi: 10.1093/jn/129.1.227S. [DOI] [PubMed] [Google Scholar]
- 3.Mitch W E, Goldberg A L. N Engl J Med. 1996;335:1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 4.Jaspers S R, Tischler M E. J Appl Physiol. 1984;57:1472–1479. doi: 10.1152/jappl.1984.57.5.1472. [DOI] [PubMed] [Google Scholar]
- 5.Tischler M E, Rosenberg S, Satarug S, Henriksen E J, Kirby C R, Tome M, Chase P. Metabolism. 1990;39:756–763. doi: 10.1016/0026-0495(90)90113-q. [DOI] [PubMed] [Google Scholar]
- 6.Tawa N E, Odessey R, Goldberg A L. J Clin Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecker S H, Solomon V, Price S R, Kwon Y T, Mitch W E, Goldberg A L. J Clin Invest. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solomon V, Baracos V, Sarraf P, Goldberg A L. Proc Natl Acad Sci USA. 1998;95:12602–12607. doi: 10.1073/pnas.95.21.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasselgren P O, Fischer J E. Ann Surg. 2001;233:9–17. doi: 10.1097/00000658-200101000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attaix D, Combaret L, Pouch M N, Taillandier D. Curr Opin Clin Nutr Metab Care. 2001;4:45–49. doi: 10.1097/00075197-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Jagoe R T, Goldberg A L. Curr Opin Clin Nutr Metab Care. 2001;4:183–190. doi: 10.1097/00075197-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 13.Li J B, Goldberg A L. Am J Physiol. 1976;231:441–448. doi: 10.1152/ajplegacy.1976.231.2.441. [DOI] [PubMed] [Google Scholar]
- 14.Wing S S, Haas A L, Goldberg A L. Biochem J. 1995;307:639–645. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina R, Wing S S, Haas A, Goldberg A L. Biomed Biochim Acta. 1991;50:347–356. [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey J L, Wang X, England B K, Price S R, Ding X, Mitch W E. J Clin Invest. 1996;97:1447–1453. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price S R, Bailey J L, Wang X, Jurkovitz C, England B K, Ding X, Phillips L S, Mitch W E. J Clin Invest. 1996;98:1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baracos V E, DeVivo C, Hoyle D H, Goldberg A L. Am J Physiol. 1995;268:E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 22.Kipreos E T, Pagano M. Genome Biol. 2000;1:1–7. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshaies R J. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 24.Ponting C P, Phillips C, Davies K E, Blake D J. BioEssays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 25.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 26.Cenciarelli C, Chiaur D S, Guardavaccaro D, Parks W, Vidal M, Pagano M. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 27.Winston J T, Koepp D M, Zhu C, Elledge S J, Harper J W. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 28.Wing S S, Goldberg A L. Am J Physiol. 1993;264:E668–E676. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- 29.Medina R, Wing S S, Goldberg A L. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg A L, Tischler M, DeMartino G, Griffin G. FASEB J. 1980;39:31–36. [PubMed] [Google Scholar]
- 31.Kettelhut I C, Pepato M T, Migliorini R H, Medina R, Goldberg A L. Braz J Med Biol Res. 1994;27:981–993. [PubMed] [Google Scholar]
- 32.Costelli P, Carbo N, Tessitore L, Bagby G J, Lopez-Soriano F J, Argiles J M, Baccino F M. J Clin Invest. 1993;92:2783–2789. doi: 10.1172/JCI116897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Martinez C, Llovera M, Agell N, Lopez-Soriano F J, Argiles J M. Biochem Biophys Res Commun. 1995;217:839–844. doi: 10.1006/bbrc.1995.2848. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Martinez C, Llovera M, Agell N, Lopez-Soriano F J, Argiles J M. Biochem Biophys Res Commun. 1994;201:682–686. doi: 10.1006/bbrc.1994.1754. [DOI] [PubMed] [Google Scholar]
- 35.Hasselgren P O. Curr Opin Clin Nutr Metab Care. 1999;2:201–205. doi: 10.1097/00075197-199905000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Price S R, Bailey J L, England B K. Miner Electrolyte Metab. 1996;22:72–75. [PubMed] [Google Scholar]
- 37.Mitch W E. Miner Electrolyte Metab. 1996;22:62–65. [PubMed] [Google Scholar]
- 38.Chrysis D, Underwood L E. Endocrinology. 1999;140:5635–5641. doi: 10.1210/endo.140.12.7217. [DOI] [PubMed] [Google Scholar]
- 39.Fischer D, Sun X, Gang G, Pritts T, Hasselgren P O. Biochem Biophys Res Commun. 2000;267:504–508. doi: 10.1006/bbrc.1999.1987. [DOI] [PubMed] [Google Scholar]
- 40.Mathias N, Johnson S, Byers B, Goebl M. Mol Cell Biol. 1999;19:1759–1767. doi: 10.1128/mcb.19.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blondel M, Galan J M, Chi Y, Lafourcade C, Longaretti C, Deshaies R J, Peter M. EMBO J. 2000;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Nature (London) 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]