Abstract
Liquid-like droplets of biomacromolecules are emerging as a fundamental mechanism of cellular signaling, but designing synthetic mimics to form such membraneless organelles remains unexplored. Here we report the use of supramolecular assemblies of small peptides, as a mimic of biomacromolecular condensates, for intracellular sequestration of enzymes on endoplasmic reticulum (ER). Specifically, integrating a short peptide with naproxen (a nonsteroidal anti-inflammatory drug (NSAID) and a ligand of cyclooxygenase-2 (COX-2)) generates an enzymatic substrate that acts as a precursor for instructed-assembly. Slowly dephosphorylating the precursors by phosphatases forms the corresponding hydrogelators in cellular environment, which results in the supramolecular assemblies on ER. Consisting of the precursor and the hydrogelator molecules, the assemblies enable the sequestration of COX-2 and protein-tyrosine phosphatase 1B (PTP1B) on ER. Further structure-activity investigation reveals that the co-localization of COX-2 and PTP1B relies on the NSAID motif, the phosphotyrosine, and the enzymatic dephosphorylation of the precursor. This work, for the first time, illustrates the use of supramolecular processes for associating enzymes in cells, and may provide insights for understanding intracellular liquid condensates and a new strategy for modulating protein-protein interactions.
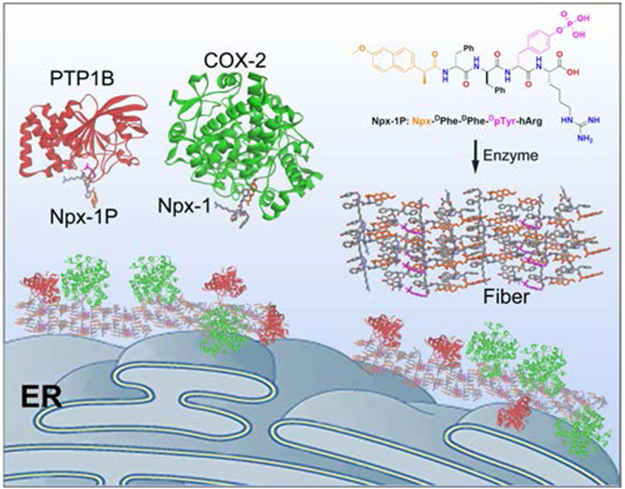
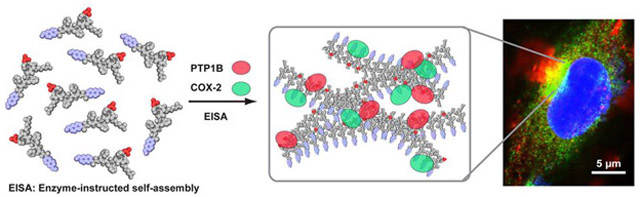
Graphical Abstract:
It is known that biomacromolecules are able to form liquid-phase condensates, such as Cajal bodies,1 U and P bodies,2 and nucleolus,3 inside cells via liquid-liquid phase separation. Recent studies have suggested that such a liquid phase condensation likely represents a ubiquitous cellular process for organizing intracellular space and for cellular signalings.4 For example, the binding of DNA to cyclic GMP–AMP synthase (cGAS) results in liquid droplets that concentrate an enzyme and reactants to activate innate immune signaling.5 Despite these exciting developments6,7 in biology, the use of synthetic molecules to mimic these membraneless organelles has yet to be explored. The key feature of liquid phase condensates is the non-covalent association of molecules at a high concentration to result in a liquid-liquid phase transition. This process coincides with sol-to-gel transition, a type of liquid-liquid phase transition, which is a usual consequence of enzyme-instructed self-assembly (EISA).8,9 Thus, we decide to use EISA to generate supramolecular assemblies of peptides in cellular environment to mimic the membraneless organelles for enzyme sequestration.
As a form of instructed-assembly,10 EISA refers to the formation of ordered superstructures of molecules as the consequence of enzymatic reactions. Like self-assembly, EISA relies on non-covalent interactions to form the assemblies from small, simple building blocks; unlike self-assembly, EISA includes an enzymatic reaction, a process that is away from equilibrium (ΔG < 0) and inherently irreversible.10 Recent studies have already demonstrated the promising applications of supramolecular assemblies in biomedicine,11–14 particularly the use of EISA of small molecules for selectively inhibiting cancer cells in vitro,15–19 for slowing tumor progression in animal models,20–23 and for molecular imaging applications.24–26 These promising developments imply that EISA generated supramolecular assemblies of small molecules should be able to mimic the biomacromolecular condensates (e.g., membraneless organelles) in cellular environment.
To demonstrate the concept of EISA for sequestration of enzymes, we use EISA to generate supramolecular assemblies to interact with multiple numbers of cyclooxygenase-2 (COX-2) and protein-tyrosine phosphatase 1B (PTP1B) simultaneously. Based on above concept, we designed EISA precursor Npx-1P, which consisted of a NSAID drug (naproxen), a self-assembling D-peptide backbone (D-Phe-D-Phe), an enzyme trigger (D-phosphotyrosine), and a positively charged homoarginine residue at the C-terminal. Such a design allows Npx-1P to interact selectively with COX-227 and to serve as a substrate for PTP1B28 simultaneously. Moreover, the use of homoarginine allows the supramolecular assemblies to form on ER via EISA as evidenced by our previous report that the fluorescent analogue of Npx-1P forms enzymatic assemblies on ER of cancer cells.29 As illustrated in Scheme 1, partially dephosphorylated by phosphatases, the precursor (Npx-1P) and its corresponding hydrogelator (Npx-1) co-assemble to form supramolecular assemblies that promote the association of COX-2 and PTP1B on endoplasmic reticulum (ER). Critical micelle concentration (CMC) measurement and transmission electron microscopy (TEM) confirm that enzymatic conversion of Npx-1P to Npx-1 results in increased self-assembling propensity and the phase transition (sol-gel). The formed supramolecular assemblies enrich COX-2 and PTP1B in a cell free condition, as evidenced by the TEM images and pull down analysis. Immunofluorescence staining indicates that the nanofibers, forming in cellular environment, interact with COX-2 and PTP1B and induce their association at ER of cells (Saos-230). Structure-activity relationship studies reveal that the COX-2 binding NSAID motif and the phosphatase substrate are essential for the association of the enzymes. As the first demonstration of EISA as a molecular process to enable intracellular sequestration of enzymes, this work illustrates enzymatic non-covalent synthesis of molecular condensates in cellular environment for conferring biological functions to supramolecular assemblies, a future direction of supramolecular chemistry.31
Scheme 1.
Illustration of instructed-assembly for intracellular sequestration of PTP1B and COX-2.
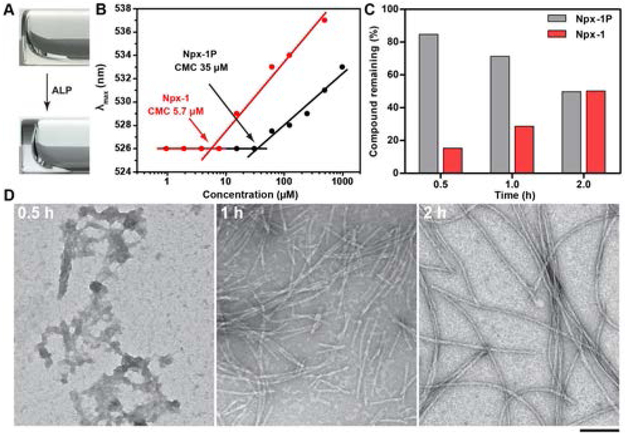
After synthesizing the precursor Npx-1P via solid-phase peptide synthesis,32 we first examined its liquid-liquid phase transition. Upon the addition of alkaline phosphatase (ALP), the Npx-1P undergoes a sol–gel transition when the concentration is about 400 µM (Figure 1A, Figure S9) in PBS buffer. We used liquid chromatography–mass spectrometry (LC-MS) and TEM to examine the enzymatic dephosphrylation of Npx-1P and the generation of supramolecular assemblies. Npx-1P hardly forms any ordered nanostructure at the concentration of 12.5 µM (Figure S10), agreeing with the CMC of Npx-1P (35 µM) (Figure 1B) in PBS buffer. The addition of ALP to the solution of Npx-1P in PBS for half an hour dephosphorylates 15% of Npx-1P to Npx-1 (Figure 1C) to form aggregates (Figure 1D). After 1 h incubation, the amount of dephosphorylated Npx-1P increases to 29%, which yields an Npx-1 concentration of 3.6 µM. While the concentration of Npx-1 in the above solution (3.6 µM) is lower than the CMC of Npx-1 (5.7 µM), short nanofibers form with a width of 7 ± 2 nm, suggesting the participation of Npx-1P at the mixture in the assembly process. The structural similarity between Npx-1P and Npx-1 favors their co-assemblies, as evidenced by the nanofibers with a uniform width of 7 nm in Figure 1D, since Npx-1P itself only forms uniform nanofibers with a width of 5 nm. (Figure S11) These results are similar to the previous reports of peptide coassembly.33,34 From 1 h to 2 h, the short nanofibers grow to long uniform nanofibers as 50% of Npx-1P being converted into Npx-1. Moreover, the dephosphorylation rate of Npx-1P by ALP decreased after 2 h incubation, (Figure S12) likely due to the co-assembly of Npx-1P and Npx-1, which reduces the amount of free Npx-1P. These results verify the phase/morphological transition resulted from EISA of Npx-1P and indicate the co-assembly of Npx-1P and Npx-1.
Figure 1.
(A) Sol-gel transition of Npx-1P (400 µM) upon adding ALP (1 U/mL). (B) CMCs of Npx-1P and Npx-1. (C) Dephosphorylation of Npx-1P (12.5 µM) after incubating with 0.1 U/mL ALP at different time. (D) TEM images of nanostructures formed before and after adding ALP (0.1 U/mL) to the solution of Npx-1P (12.5 µM)(scale bar = 100 nm). All the solutions were prepared in pH 7.4 PBS buffer.
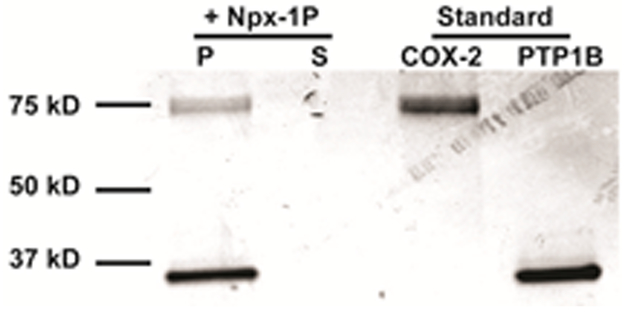
We then investigated the interactions between enzymatically formed assemblies and the enzymes (COX-2 and PTP1B). Being co-incubated with Npx-1P and after the addition of ALP, either PTP1B or COX-2 adheres to the nanofibers of Npx-1P and Npx-1 (Figure S13). The incubation of COX-2 and PTP1B with Npx-1P and ALP results in larger protein aggregates on the nanofibers. Some of the proteins appear to be away from the nanofibers in the TEM image, which are likely due to the dynamic nature of enzymatic assemblies. These observations suggest interaction and enrichment of COX-2 and PTP1B on the non-covalent assemblies formed by enzymatically dephosphorylation of Npx-1P. To further confirm the interactions between assemblies and proteins, we conducted the pull-down assay of PTP1B and COX-2 with the enzymatic assemblies (Figure 2), which reveals that the pellet, compared with the supernatant, of assemblies abundantly enrich both PTP1B and COX-2, further validating the enzyme sequestration by the supramolecular assemblies formed by EISA. In addition, after 1 h incubation, PTP1B (45 U/mL) converted 50%, 40% and 25% of Npx-1P to Npx-1 when the concentration of Npx-1P is 50 µM, 100 µM and 200 µM, respectively, but hardly hydrolyzed more Npx-1P from 1 h to 24 h (Figure S14). This observation likely results from the sequestration of PTP1B from solution to the assemblies, which reduces the amount of free enzymes.
Figure 2.
Pull down of COX-2 and PTP1B with enzymatic assemblies formed by Npx-1P in PBS. The image shows Coomassie staining of pellet (P) and supernatant (S) fractions.
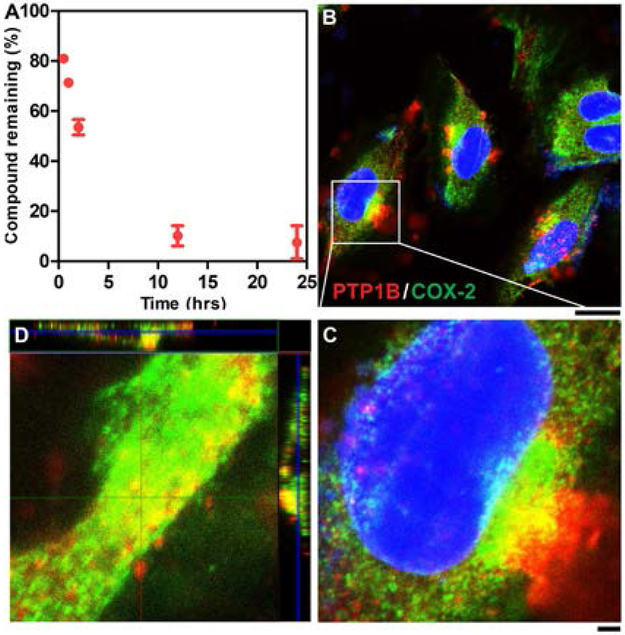
To verify the enzymatic transformation and self-assembly of Npx-1P in live cells, we quantified the conversion of Npx-1P after incubating it with Saos-2 cells at different time points. Figure 3A indicates that endogenous phosphatases convert 19%, 29%, and 46% of Npx-1P to Npx-1 after 0.5 h, 1 h, and 2 h incubation, respectively. This result implies the co-assembly of Npx-1P and Npx-1 in live cells, agreeing with the results from cell free condition (Figure 1). We next used immunofluorescence staining to verify whether the assemblies formed by co-assembly of Npx-1P and Npx-1 sequestrated the enzymes. Being incubated Saos-2 cells with Npx-1P (12.5 µM) after 0.5 h (Figure S15), both COX-2 and PTP1B form puncta and the green fluorescence from COX-2 start to overlap to red fluorescence from PTP1B at some regions. After 1 h incubation, more yellow fluorescence appears at the site of ER, indicating increased level of co-localization of the two enzymes (Figure 3B and C). Orthogonal Z-stack scanning (Figure 3D) also indicates the intracellular association of PTP1B and COX-2 upon incubation the cells with Npx-1P. We also examined the enzyme sequestration in HS-5 cells (as a control of Saos-2 cells since HS-5 express low level of ALPL35) and found that the addition of Npx-1P hardly induced the sequestration or co-localization of COX-2 and PTP1B in the cells (Figure S16). Together with our previous report29 that EISA precursors hardly form assemblies in HS-5 cells, this result indicates cell selective formation of molecular condensates for enzyme sequestration by EISA.
Figure 3.
(A) Dephosphorylation of Npx-1P (12.5 µM) after incubating with Saos-2 cells at different time. (B) CLSM images of Saos-2 cells treated with Npx-1P (12.5 µM) for 1 h and then stained with antibodies of PTP1B (red) and COX-2 (green). (C) The enlarged region of co-localization in (B) (scale bar = 2 μm). (D) Orthogonal Z-stack of co-localization of PTP1B and COX2.
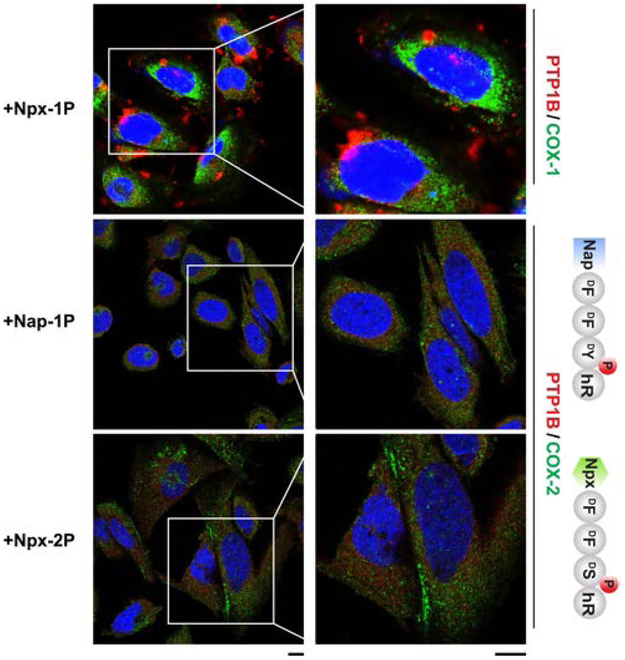
To evaluate the specificity of intracellular enzyme sequestration by the assemblies formed by EISA, we used immunofluorescence staining to examine the interactions of COX-1, an isozyme of COX-2. As shown in Figure 4, COX-1 hardly forms puncta inside Saos-2 cells, indicating that the supramolecular assemblies of Npx-1P and Npx-1 scarcely interact with COX-1 in Saos-2 cells. This results is likely due to the enhanced selectivity of Npx-1P towards COX-2,27 suggesting that the strong interaction between the naproxen-peptide conjugates and COX-2 is critical for the sequestration of COX-2. To further verify the molecular basis of Npx-1P for the sequestration of COX-2 and PTP1B, we designed two control molecules (Nap-1P, Npx-2P) by mutating fragments in Npx-1P (Scheme S1). Replacing the naproxen with naphthalene in Npx-1P yields Nap-1P, which hardly induces the puncta formation of COX-2 or PTP1B, further confirming the importance of the COX-2 binding moiety (i.e., naproxen). Having a D-phosphoserine to replace the D-phosphotyrosine (in Npx-1P), Npx-2P fails to cause the association of PTP1B or COX-2, agreeing with that phosphoserine significantly diminishes PTP1B-substrate interaction36,37 and self-assembling ability. These results, collectively, confirm the essential role of phosphotyrosine and naproxen for the sequestration of PTP1B and COX-2 by the assemblies of Npx-1P and Npx-1.
Figure 4.
CLSM images of Saos-2 cells stained with antibodies of PTP1B (red) and COX-1/COX-2 (green) after treating with Npx-1P, Nap-1P and Npx-2P for 1 h (scale bar = 10 μm).
In conclusion, this work illustrated a novel strategy to sequestrate intracellular enzymes selectively via instructed-assembly. Incorporating with specific protein interaction motifs, the precursors, via EISA process, form supramolecular assemblies that display the protein-binding motifs on the surface of the molecular assemblies due to the dynamic and adaptive properties of the peptide assemblies, thus interacting with multiple proteins in a manner to concentrate specific proteins (e.g., PTP1B and COX-2 in this work). This process is similar to formation of higher-order assemblies of proteins38 or protein phase transition,39 a ubiquitous phenomenon in biology for cell signaling. Moreover, employing synthetic small molecules to generate intracellular molecular assemblies (or condensates) represents a general way to create a multifunctional signal hub for modulating diverse protein-protein interactions,40 especially for controlling protein-protein interaction involving tertiary epitopes that are much more dynamic.41 Further exploration along this direction may lead to remarkable therapeutic benefits, as shown by the drugs that control protein-protein interactions.42,43
Supplementary Material
ACKNOWLEDGMENT
This work is partially supported by NIH (R01CA142746) and NSF (DMR-1420382). Z.F. is supported by NIH (F99CA234746).
Footnotes
Supporting Information
Materials, detailed experimental procedures, additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Gall JG; Bellini M; Wu Z; Murphy C Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 1999, 10, 4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Liu JL; Gall JG U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 11655–11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Boisvert FM; van Koningsbruggen S; Navascues J; Lamond AI The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol 2007, 8, 574–585. [DOI] [PubMed] [Google Scholar]
- (4).Shin Y; Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- (5).Du M; Chen ZJ DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Brangwynne CP; Eckmann CR; Courson DS; Rybarska A; Hoege C; Gharakhani J; Julicher F; Hyman AA Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science 2009, 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- (7).Li P; Banjade S; Cheng H-C; Kim S; Chen B; Guo L; Llaguno M; Hollingsworth JV; King DS; Banani SF; Russo PS; Jiang Q-X; Nixon BT; Rosen MK Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yang Z; Liang G; Xu B Enzymatic Hydrogelation of Small Molecules. Acc. Chem. Res 2008, 41, 315–326. [DOI] [PubMed] [Google Scholar]
- (9).Zhou J; Xu B Enzyme-instructed self-assembly: a multistep process for potential cancer therapy. Bioconjugate Chem. 2015, 26, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).He HJ; Xu B Instructed-Assembly (iA): A Molecular Process for Controlling Cell Fate. Bull. Chem. Soc. Jpn 2018, 91, 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lock LL; Reyes CD; Zhang P; Cui H Tuning Cellular Uptake of Molecular Probes by Rational Design of Their Assembly into Supramolecular Nanoprobes. J. Am. Chem. Soc 2016, 138, 3533–3540. [DOI] [PubMed] [Google Scholar]
- (12).Shamay Y; Shah J; Isik M; Mizrachi A; Leibold J; Tschaharganeh DF; Roxbury D; Budhathoki-Uprety J; Nawaly K; Sugarman JL; Baut E; Neiman MR; Dacek M; Ganesh KS; Johnson DC; Sridharan R; Chu KL; Rajasekhar VK; Lowe SW; Chodera JD; Heller DA Quantitative self-assembly prediction yields targeted nanomedicines. Nat. Mater 2018, 17, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sato K; Ji W; Palmer LC; Weber B; Barz M; Stupp SI Programmable Assembly of Peptide Amphiphile via Noncovalent-to-Covalent Bond Conversion. J. Am. Chem. Soc 2017, 139, 8995–9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ng KK; Takada M; Harmatys K; Chen J; Zheng G Chlorosome-Inspired Synthesis of Templated Metallochlorin-Lipid Nanoassemblies for Biomedical Applications. ACS Nano 2016, 10, 4092–4101. [DOI] [PubMed] [Google Scholar]
- (15).Wang H; Feng Z; Wang Y; Zhou R; Yang Z; Xu B Integrating Enzymatic Self-Assembly and Mitochondria Targeting for Selectively Killing Cancer Cells without Acquired Drug Resistance. J. Am. Chem. Soc 2016, 138, 16046–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang H; Feng Z; Wu D; Fritzsching KJ; Rigney M; Zhou J; Jiang Y; Schmidt-Rohr K; Xu B Enzyme-Regulated Supramolecular Assemblies of Cholesterol Conjugates against Drug-Resistant Ovarian Cancer Cells. J. Am. Chem. Soc 2016, 138, 10758–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Feng Z; Wang H; Zhou R; Li J; Xu B Enzyme-Instructed Assembly and Disassembly Processes for Targeting Downregulation in Cancer Cells. J. Am. Chem. Soc 2017, 139, 3950–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pires RA; Abul-Haija YM; Costa DS; Novoa-Carballal R; Reis RL; Ulijn RV; Pashkuleva I Controlling cancer cell fate using localized biocatalytic self-assembly of an aromatic carbohydrate amphiphile. J. Am. Chem. Soc 2015, 137, 576–579. [DOI] [PubMed] [Google Scholar]
- (19).Tanaka A; Fukuoka Y; Morimoto Y; Honjo T; Koda D; Goto M; Maruyama T Cancer cell death induced by the intracellular self-assembly of an enzyme-responsive supramolecular gelator. J. Am. Chem. Soc 2015, 137, 770–775. [DOI] [PubMed] [Google Scholar]
- (20).Du X; Zhou J; Wang H; Shi J; Kuang Y; Zeng W; Yang Z; Xu B In situ generated D-peptidic nanofibrils as multifaceted apoptotic inducers to target cancer cells. Cell Death Dis. 2017, 8, e2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liang C; Zheng D; Shi F; Xu T; Yang C; Liu J; Wang L; Yang Z Enzyme-assisted peptide folding, assembly and anti-cancer properties. Nanoscale 2017, 9, 11987–11993. [DOI] [PubMed] [Google Scholar]
- (22).Wang H; Wei J; Yang C; Zhao H; Li D; Yin Z; Yang Z The inhibition of tumor growth and metastasis by self-assembled nanofibers of taxol. Biomaterials 2012, 33, 5848–5853. [DOI] [PubMed] [Google Scholar]
- (23).Yuan Y; Wang L; Du W; Ding Z; Zhang J; Han T; An L; Zhang H; Liang G Intracellular Self-Assembly of Taxol Nanoparticles for Overcoming Multidrug Resistance. Angew. Chem., Int. Ed 2015, 54, 9700–9704. [DOI] [PubMed] [Google Scholar]
- (24).Ye DJ; Shuhendler AJ; Cui LN; Tong L; Tee SS; Tikhomirov G; Felsher DW; Rao JH Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat. Chem 2014, 6, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Yuan Y; Sun HB; Ge SC; Wang MJ; Zhao HX; Wang L; An LN; Zhang J; Zhang HF; Hu B; Wang JF; Liang GL Controlled Intracellular Self-Assembly and Disassembly of F-19 Nanoparticles for MR Imaging of Caspase 3/7 in Zebrafish. ACS Nano 2015, 9, 761–768. [DOI] [PubMed] [Google Scholar]
- (26).Zhan J; Cai YB; He SS; Wang L; Yang ZM Tandem Molecular Self-Assembly in Liver Cancer Cells. Angew. Chem., Int. Ed 2018, 57, 1813–1816. [DOI] [PubMed] [Google Scholar]
- (27).Li JY; Kuang Y; Gao Y; Du XW; Shi JF; Xu B D-Amino Acids Boost the Selectivity and Confer Supramolecular Hydrogels of a Nonsteroidal Anti-Inflammatory Drug (NSAID). J. Am. Chem. Soc 2013, 135, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gao Y; Shi JF; Yuan D; Xu B Imaging enzyme-triggered self-assembly of small molecules inside live cells. Nat. Commun 2012, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Feng Z; Wang H; Wang S; Zhang Q; Zhang X; Rodal AA; Xu B Enzymatic Assemblies Disrupt the Membrane and Target Endoplasmic Reticulum for Selective Cancer Cell Death. J. Am. Chem. Soc 2018, 140, 9566–9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Fogh J Human tumor cells in vitro; Plenum Press: New York, 1975. [Google Scholar]
- (31).Whitesides GM Complex Organic Synthesis: Structure, Properties, and/or Function? Isr. J. Chem 2018, 58, 142–150. [Google Scholar]
- (32).Merrifield RB SOLID PHASE PEPTIDE SYNTHESIS .1. SYNTHESIS OF A TETRAPEPTIDE. J. Am. Chem. Soc 1963, 85, 2149. [Google Scholar]
- (33).Hirst AR; Roy S; Arora M; Das AK; Hodson N; Murray P; Marshall S; Javid N; Sefcik J; Boekhoven J; van Esch JH; Santabarbara S; Hunt NT; Ulijn RV Biocatalytic induction of supramolecular order. Nat. Chem 2010, 2, 1089–1094. [DOI] [PubMed] [Google Scholar]
- (34).Ardoña HAM; Draper ER; Citossi F; Wallace M; Serpell LC; Adams DJ; Tovar JD Kinetically Controlled Coassembly of Multichromophoric Peptide Hydrogelators and the Impacts on Energy Transport. J. Am. Chem. Soc 2017, 139, 8685–8692. [DOI] [PubMed] [Google Scholar]
- (35).Zhou J; Du XW; Berciu C; He HJ; Shi JF; Nicastro D; Xu B Enzyme-Instructed Self-Assembly for Spatiotemporal Profiling of the Activities of Alkaline Phosphatases on Live Cells. Chem 2016, 1, 246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sarmiento M; Zhao Y; Gordon SJ; Zhang ZY Molecular basis for substrate specificity of protein-tyrosine phosphatase 1B. J. Biol. Chem 1998, 273, 26368–26374. [DOI] [PubMed] [Google Scholar]
- (37).Yudushkin IA; Schleifenbaum A; Kinkhabwala A; Neel BG; Schultz C; Bastiaens PIH Live-cell Imaging of enzyme-substrate interaction reveals spatial regulation of PTP1B. Science 2007, 315, 115–119. [DOI] [PubMed] [Google Scholar]
- (38).Wu H; Fuxreiter M The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell 2016, 165, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Boeynaems S; Alberti S; Fawzi NL; Mittag T; Polymenidou M; Rousseau F; Schymkowitz J; Shorter J; Wolozin B; Van den Bosch L; Tompa P; Fuxreiter M Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 2018, 28, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).van Dun S; Ottmann C; Milroy LG; Brunsveld L Supramolecular Chemistry Targeting Proteins. J. Am. Chem. Soc 2017, 139, 13960–13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Arkin MR; Tang Y; Wells JA Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing toward the Reality. Chem. Biol (Oxford, U. K.) 2014, 21, 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Souers AJ; Leverson JD; Boghaert ER; Ackler SL; Catron ND; Chen J; Dayton BD; Ding H; Enschede SH; Fairbrother WJ; Huang DCS; Hymowitz SG; Jin S; Khaw SL; Kovar PJ; Lam LT; Lee J; Maecker HL; Marsh KC; Mason KD; Mitten MJ; Nimmer PM; Oleksijew A; Park CH; Park CM; Phillips DC; Roberts AW; Sampath D; Seymour JF; Smith ML; Sullivan GM; Tahir SK; Tse C; Wendt MD; Xiao Y; Xue JC; Zhang HC; Humerickhouse RA; Rosenberg SH; Elmore SW ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med 2013, 19, 202–208. [DOI] [PubMed] [Google Scholar]
- (43).Schiff PB; Horwitz SB Taxol stabilizes microtubules in mouse fibroblast cells. Proc. Natl. Acad. Sci. U. S. A 1980, 77, 1561–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.