Abstract
The kingdoms of life share many small molecule cofactors and coenzymes. Molybdenum cofactor (Moco) is synthesized by many archaea, bacteria, and eukaryotes, and is essential for human viability. The genome of the animal Caenorhabditis elegans contains all of the Moco biosynthesis genes, and surprisingly these genes are not essential if animals are fed a bacterial diet that synthesizes Moco. C. elegans lacking both endogenous Moco synthesis and dietary Moco from bacteria arrest development, demonstrating interkingdom Moco transfer. Our screen of E. coli mutants identified genes necessary for synthesis of bacterial Moco or transfer to C. elegans. Moco-deficient C. elegans developmental arrest is caused by loss of sulfite oxidase, a Moco-requiring enzyme, and is suppressed by mutations in either C. elegans cystathionine gamma-lyase or cysteine dioxygenase, blocking toxic sulfite production from cystathionine. Thus, we define the genetic pathways for an interkingdom dialogue focused on sulfur homeostasis.
Introduction:
Molybdenum cofactor (Moco) is a tricyclic pyranopterin with a coordinated molybdenum that is synthesized in multiple enzymatic steps from GTP (Fig. 1)1. Moco and the enzymes that synthesize Moco are conserved across archaea, bacteria, and eukarya2. The steps in Moco synthesis emerged from bacterial genetic analyses3. Human Moco deficiency causes seizures, progressive neurological damage, and neonatal death4,5. Moco is an essential cofactor for four animal enzymes; sulfite oxidase, xanthine oxidase, aldehyde oxidase, and the mitochondrial amidoxime reducing component4, but loss of sulfite oxidase activity is suspected to be the primary pathology in human Moco deficiency1. Because Moco is unstable, it has not been thought to be transferred between cells, tissues, or organisms.
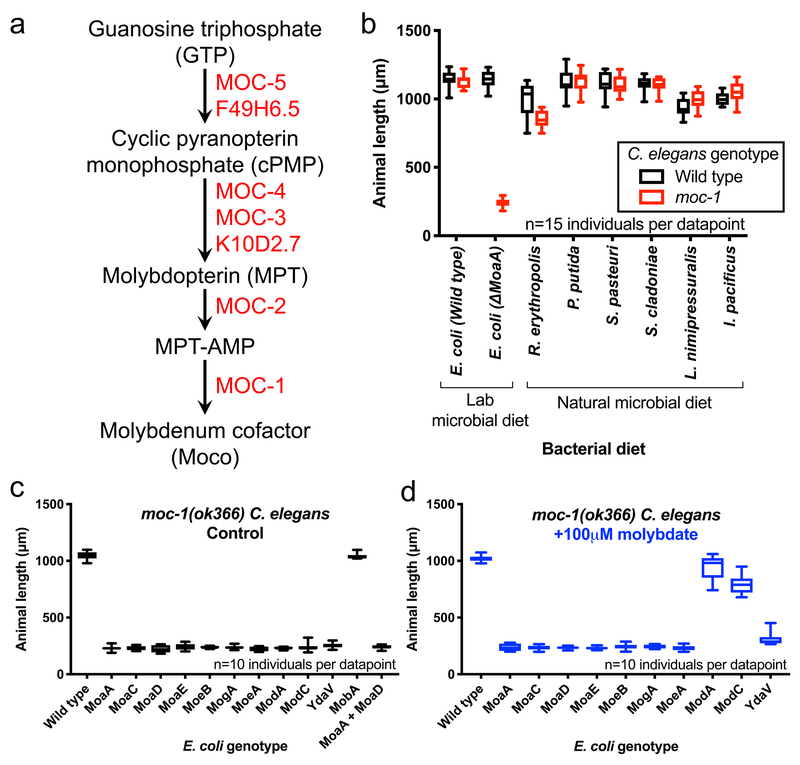
Figure 1: Moco acquisition and biosynthesis are redundantly required for life in C. elegans.
(A) By protein sequence homology to human and bacterial Moco biosynthetic enzymes, the inferred C. elegans Moco biosynthesis pathway is displayed. (B) The growth of wild-type and moc-1(ok366) animals on either wild-type or ΔMoaA E. coli (lab microbial diet) or 6 bacterial strains native to the Caenorhabditis ecosystem (natural microbial diet) was quantified after 72 hours of growth from the first larval stage. (C,D) The growth of moc-1(ok366) animals on either wild-type or mutant E. coli was quantified after 72 hours of growth from the first larval stage. Before being fed to C. elegans, the wild-type and mutant E. coli strains were initially cultured without (C, Control) or with 100μm sodium molybdate (D, +100μm molybdate) in LB. The mutant E. coli strains used here are nonpolar deletions of the indicated genes, except for the MobA mutant strain which still has the Kanamycin resistance cassette inserted into the MobA open reading frame12. Box plots display the median, upper, and lower quartiles while whiskers indicate minimum and maximum data points. Sample size (n) is displayed for each experiment.
Here we show that C. elegans can acquire mature Moco and a Moco precursor (cPMP, cyclic pyranopterin monophosphate) from its bacterial diet. We show C. elegans lacking endogenous Moco synthesis and dietary Moco arrest development due to an inability to detoxify sulfite. C. elegans cystathionine gamma lyase and cysteine dioxygenase, conserved components of the sulfur amino acid catabolism pathway, generate sulfite that is lethal when Moco is deficient6. In the absence of sulfur amino acid catabolism, Moco is no longer necessary. Thus, interkingdom transport of Moco from microbes to C. elegans acts in sulfite detoxification.
Results:
Moco synthesis and dietary acquisition in C. elegans:
We explored the function of Moco and its biosynthetic enzymes in the animal C. elegans. Genome sequence analysis using protein queries from E. coli Moco biosynthesis enzymes identified C. elegans homologs for each of the 7 core Moco-biosynthesis proteins (Fig. 1A, Supplementary Table 1)1. We characterized phenotypes of C. elegans with loss-of-function mutations in six predicted Moco biosynthesis enzymes: moc-1, moc-2, moc-3, moc-4 (T27A3.6), moc-5 (F49E2.1), and lin-467–9. Surprisingly, given that Moco biosynthesis is essential in humans, C. elegans strains carrying mutations in any of these genes were viable and grew normally on wild-type E. coli. We hypothesized that C. elegans acquires Moco from the E. coli, which is competent for Moco synthesis, on which it feeds. In the wild C. elegans consumes many different species of bacteria in rotting fruit. Because Moco biosynthesis is not universal to all bacteria, it is reasonable to expect C. elegans to import bacterial Moco as well as synthesize Moco de novo10. To test this hypothesis, we grew wild-type and Moco-biosynthesis mutant C. elegans strains on an E. coli strain with a deletion in MoaA, the first step in Moco biosynthesis. This ΔMoaA E. coli strain, which grows at a normal rate on LB or nematode growth media (NGM), cannot synthesize Moco or any of the Moco precursor metabolites. Wild-type C. elegans grows normally feeding on ΔMoaA E. coli, demonstrating that microbial Moco biosynthesis is not required for viability of wild-type C. elegans (Fig. 1B). C. elegans with a mutation in moc-1, moc-2, moc-3, moc-4, or moc-5 arrested or died at early larval stages when grown on ΔMoaA E. coli, in contrast to the healthy growth of these C. elegans mutants fed wild-type E. coli (Fig. 1B and Fig. 2A–F). Of the 6 putative C. elegans Moco-biosynthesis mutants tested, only the lin-46 mutant grew normally on ΔMoaA E. coli (Supplementary Fig. 1). LIN-46 and MOC-1 are paralogous, likely emerging from a MoeA duplication event. LIN-46 displays less sequence similarity to MoeA than MOC-1 (Supplementary Table 1), and thus may be functionally divergent. Mutations in lin-46 emerged from studies of miRNA biology, and were not characterized further here7. We also found evidence for maternal contribution of Moco: when moc-1, moc-2, moc-3, moc-4, and moc-5 mutant C. elegans mothers were fed ΔMoaA E. coli from the L4 stage of development, one stage prior to reproductive adulthood, progeny viability dropped dramatically (Supplementary Table 2). Embryo viability was not affected when wild-type mothers were fed wild-type or ΔMoaA E. coli. Thus, our data suggests that Moco is transferred from mother to progeny.
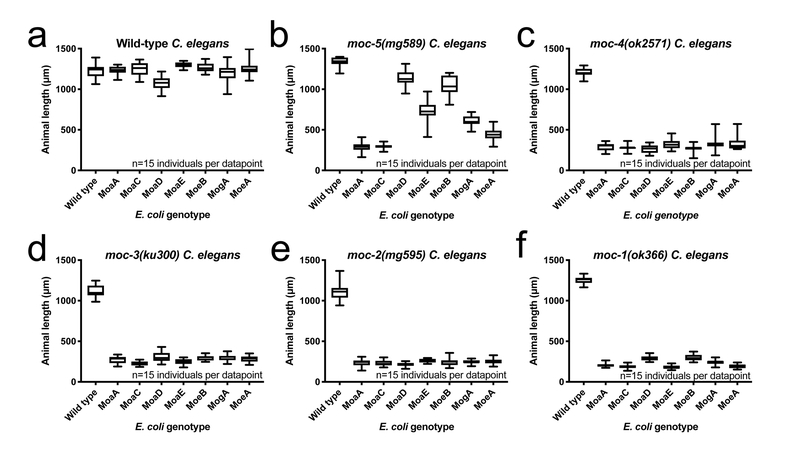
Figure 2: C. elegans acquire cPMP and Moco from dietary E. coli.
(A) Wild-type, (B) moc-5(mg589), (C) moc-4(ok2571), (D) moc-3(ku300), (E) moc-2(mg595), and (F) moc-1(ok366) animals were cultured from synchronized L1 larvae for 72 hours on wild-type and mutant E. coli disrupted at distinct steps in Moco biosynthesis. E. coli mutations used here are nonpolar and should not disrupt other genes in their operons. Animal lengths were determined for each condition. The moc-1(ok366) mutation is a 1.4kb deletion that is predicted to be a null allele. The moc-4(ok2571) mutation is a 1.8kb deletion that is predicted to be a null allele. moc-2(mg595) encodes a (Gly72Stop) followed by a frameshift that is likely a null allele. moc-3(ku300) is an established genetic reagents that represents a likely loss-of-function or null allele8. moc-5(mg589) is a missense mutation (G303R) substitution loss-of-function or null allele. Box plots display the median, upper, and lower quartiles while whiskers indicate minimum and maximum data points. Sample size (n) is displayed for each experiment.
We identify redundant pathways for maintaining sufficient Moco in C. elegans: endogenous Moco biosynthesis and acquisition from dietary E. coli. To test whether the ability of E. coli to synthesize Moco and transfer it to C. elegans is common in bacterial species associated with C. elegans in the wild, we examined the growth of moc-1 mutant C. elegans cultured on 6 bacterial strains isolated from the natural Caenorhabditis microbiome10,11. C. elegans moc-1 mutants grew and developed on all 6 natural bacterial isolates tested, representing 5 unique bacterial phyla (Fig. 1B, Supplementary Table 3). Thus diverse bacterial species from the natural Caenorhabditis habitat supply Moco to C. elegans, suggesting this is a conserved and robust host-microbe interaction. C. elegans may retain its Moco biosynthetic genes because not all bacterial species it naturally consumes synthesize Moco. Genome analyses showed that 325 out of 451 bacterial species carry Moco biosynthesis genes2. In eukaryotes, Moco biosynthetic genes are generally present, but particular fungal and protist species lack Moco biosynthesis.
C. elegans acquires microbial cPMP and Moco:
To define the comprehensive set of E. coli gene activities that are required for the synthesis or transfer of Moco to C. elegans, we tested growth of the moc-1 mutant animals on each of the approximately 4,000 mutant strains of the Keio E. coli deletion mutation collection12. We screened for E. coli mutant strains that permit growth of wild-type C. elegans, but not moc-1 mutant animals. This screen identified 12 E. coli genes that are specifically required for the viability of moc-1 mutant animals (Supplementary Fig. 2A). The genes identified include the 7 enzymes that synthesize Moco from GTP (MoaA/C/D/E, MoeA/B, and MogA). We also identified the E. coli ABC-type transport system for acquisition of environmental molybdenum for incorporation into Moco (ModA/B/C)13. Finally, we discovered 2 new E. coli genes that have no known function in Moco biosynthesis, YdaV and YehP.
Ten of the 12 E. coli mutations that disrupt the growth of moc-1 mutant C. elegans disrupt steps in Moco biosynthesis. These data strongly suggest that Moco or biochemical precursors in the E. coli Moco biosynthetic pathway are acquired and utilized by C. elegans. We used a matrix of E. coli and C. elegans Moco biosynthetic mutant combinations to determine which molecules are transferred from E. coli to C. elegans. Keio collection kanamycin resistant insertion/deletion mutations are likely polar, down regulating the expression of downstream genes in the operon and precluding disruption of single biochemical steps in Moco synthesis (Supplementary Fig. 2B,C). To precisely analyze E. coli single gene disruptions, the mutations were rendered nonpolar via Flp recombinase-mediated removal of each kanamycin cassette14. Of the 12 E. coli mutant strains identified in our screen, we generated nonpolar versions of all but the ModB mutation. Of the 11 nonpolar mutant strains generated, all but YehP recapitulated the larval lethality of moc-1 mutant C. elegans, consistent with the original polar Keio E. coli mutations. This result suggests that the downstream gene in the Yeh operon, YehQ, may also need to be inactive for E. coli to fail in Moco biosynthesis or transport (Supplementary Fig. 3A,B). We did not further analyze this operon. The 10 other nonpolar E. coli mutations were further analyzed for their ability to support the growth and development of C. elegans mutants that harbor defects at defined points in Moco biosynthesis.
We challenged wild-type, moc-1, moc-2, moc-3, moc-4, or moc-5 mutant C. elegans to grow on wild-type E. coli, or E. coli with nonpolar mutations in MoaA, MoaC, MoaD, MoaE, MoeA, MoeB, MogA, ModA, ModC, or YdaV (Fig. 2, Supplementary Fig. 3C–H). C. elegans moc-1, moc-2, moc-3, and moc-4 mutant animals were viable only on wild-type E. coli able to synthesize mature Moco (Fig. 2). This suggests that C. elegans acquire Moco from their diet: moc-1 mutant animals cannot utilize other Moco-precursors from bacteria to synthesize mature Moco, and the growth of moc-2, moc-3, and moc-4 mutants is not rescued when their respective downstream Moco-metabolites (MPT, MPT-AMP) are available from bacteria (Fig. 2C–E). These data suggest that moc-1, moc-2, moc-3, and moc-4 mutant animals grow and develop by acquiring mature Moco from E. coli.
In contrast, moc-5 mutant C. elegans displayed little or no growth on ΔMoaA or ΔMoaC E. coli, but near wild-type growth on E. coli lacking MoaD, MoaE or MoeB (Fig. 2A,B). MoaD, MoaE, and MoeB mutant E. coli accumulate cPMP (Supplementary Fig. 2B)15. moc-5 mutant C. elegans is unable to make its own cPMP, but possess all of the downstream machinery to utilize cPMP to synthesize mature Moco. Thus, these genetic data suggest that cPMP provided by the MoaD, MoaE or MoeB mutant E. coli is bioavailable and supports the growth of moc-5 mutant animals deficient in the production of cPMP. MogA and MoeA mutant E. coli also synthesize cPMP. Why do they not support the growth and development of moc-5 mutant worms as well as MoaD, MoaE, and MoeB mutant E. coli (Fig. 2B)? We speculate that cPMP does not accumulate in MogA and MoeA mutant E. coli because they possess the enzymes that convert cPMP to MPT or MPT-AMP, respectively.
What form of Moco is taken up by C. elegans from E. coli? While animals utilize the di-oxo form of Moco (referred to as ‘Moco’ in this manuscript), the majority of E. coli molybdenum-utilizing enzymes are in the DMSO reductase family that utilizes bis-MGD Moco. This dinucleotide Moco variant is not utilized by eukaryotes and requires the activity of the MobA enzyme for its synthesis in E. coli (Supplementary Fig. 2B)16. To test whether the bis-MGD form of Moco is transferred from E. coli to C. elegans, we tested the ability of moc-1 mutant C. elegans to grow on ΔMobA E. coli. moc-1 mutant animals grew normally on ΔMobA bacteria (Fig. 1C), supporting the model that di-oxo Moco and not bis-MGD Moco is transferred from E. coli to C. elegans.
To further test whether the mature Moco prosthetic group is transferred from E. coli to C. elegans, we utilized the E. coli Mod mutants identified in our screen for inviable moc-1 mutant C. elegans. The E. coli Mod operon encodes proteins necessary for molybdate import and thus biosynthesis of the molybdenum cofactor. Mod mutant E. coli fail to synthesize Moco when grown in standard culture media, but synthesize Moco when grown in media supplemented with 100μM molybdate17. Thus, Moco synthesis can be controlled in Mod mutant E. coli with molybdate supplementation. We cultured ModA and ModC mutant E. coli with and without 100μM supplemental molybdate and fed these bacteria to moc-1 mutant C. elegans. Only when grown with supplemental molybdate do ModA or ModC mutant E. coli support the growth of moc-1 mutant C. elegans (Fig. 1C,D). No other E. coli mutants displayed this molybdate suppression (Fig. 1D).
An alternate explanation for the ability of wild-type E. coli to support the growth of moc-1 mutant C. elegans is that the animal imports the gene products (mRNA or protein) of the bacterial Moco-biosynthesis enzymes. To exclude this possibility, we cultured moc-1 mutant C. elegans on a 1:1 mixture of nonpolar MoaA and MoaD deletion E. coli strains. Neither of these strains individually supports the growth of moc-1 mutant animals (Fig. 1C), yet the dietary combination of MoaA and MoaD bacterial strains possess all of the genes necessary for Moco biosynthesis. The MoaA + MoaD mixture caused larval arrest similar to either individual strain, suggesting that the complete set of E. coli Moco-biosynthetic mRNAs or enzymes present in the combination of the mutant bacterial strains are not transferred to complement C. elegans Moco deficiency (Fig. 1C). Rather, our data support a model that the Moco prosthetic group and cPMP produced by E. coli cells is transferred to C. elegans.
The C. elegans MOC-1 protein is homologous to bacterial and mammalian enzymes that insert molybdenum into MPT-AMP to synthesize mature Moco. We tested if the requirement for this enzyme could also be suppressed by high levels of molybdate. Indeed, moc-1 mutant animals grown on MoaA mutant E. coli supplemented with 5mM molybdate partially bypass the requirement for MOC-1 in the synthesis of Moco (Supplementary Fig. 4).
Moco-deficient phenotypes are promoted by CTH-2 and CDO-1:
To better understand why C. elegans lacking endogenous and dietary Moco are not viable, we sought genetic suppressors of this lethality. We EMS-mutagenized populations of moc-2, moc-3, and moc-4 C. elegans mutants and grew them on wild-type E. coli until the F2 generation. We then challenged the F2 generation animals to grow on ΔMoaA E. coli, which normally causes completely penetrant larval arrest (Fig. 2). We identified 49 independent mutant strains that grew robustly and reproduced in the absence of Moco (Supplementary Table 4). To determine the causative genetic lesions in these mutants, DNA from each strain was analyzed via whole genome sequencing18. Based on the presence of multiple independent alleles, we identified mutations in two different genes that allow the viability of Moco-deficient C. elegans cultured on Moco-deficient E. coli. A mutation in either of these 2 genes was present in all 49 of the strains isolated in our screen.
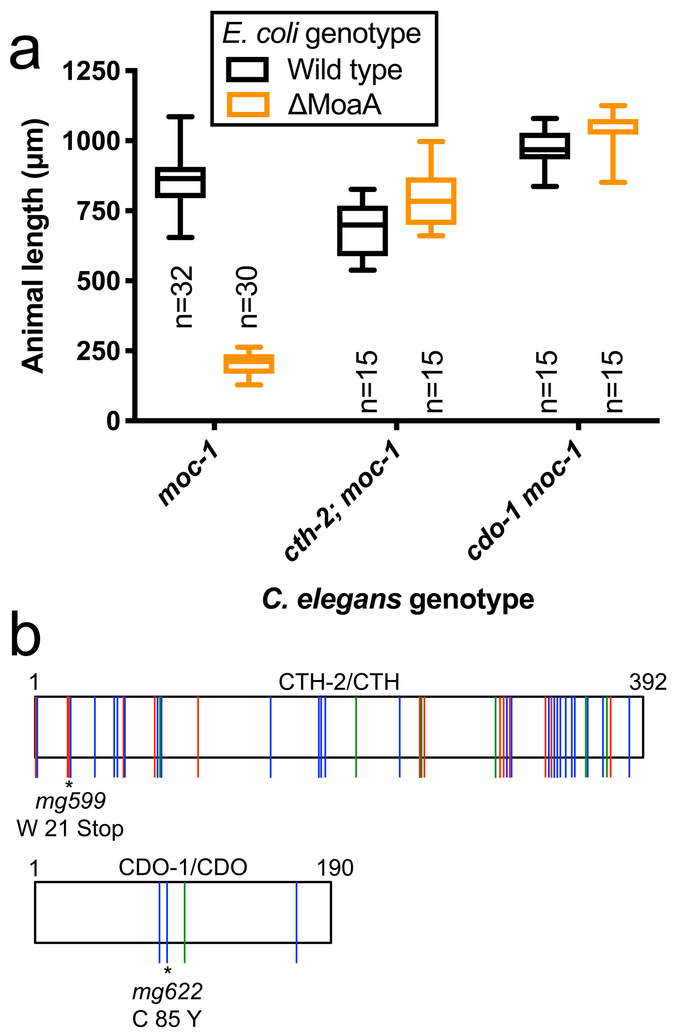
We identified 45 alleles of the C. elegans cystathionine gamma-lyase, cth-2/ZK1127.10. These 45 alleles affected 37 distinct amino acid positions and included 9 nonsense and 36 missense mutations, suggesting that cth-2 loss of function suppresses the growth arrest caused by C. elegans Moco deficiency (Fig. 3, Supplementary Table 4). Cystathionine gamma-lyase (CTH), an enzyme conserved across all kingdoms of life, catabolizes cystathionine into cysteine, ammonia, and α-ketobutyrate, on the main axis of sulfur amino acid metabolism (Fig. 4A)6. To demonstrate the requirement of cth-2 gene activity for the inviability of Moco-deficient C. elegans grown on Moco-deficient E. coli, we genetically transferred the cth-2(mg599) allele, predicted to cause a premature stop (W 21 Stop), into the moc-1 mutant background and assayed growth and development. Like wild type, cth-2 mutant animals grew well on both wild-type and ΔMoaA E. coli. moc-1 mutant animals grew well on wild-type E. coli, but displayed early larval arrest on ΔMoaA E. coli (Fig. 3A). cth-2; moc-1 double mutant animals grew well on both wild-type and ΔMoaA E. coli (Fig. 3A), demonstrating that the activity of cth-2 is required for larval arrest caused by C. elegans Moco deficiency. cth-2 is one of two cystathionine gamma-lyase-encoding genes in C. elegans. The CTH-1 protein, 86% identical to CTH-2, is not redundant with CTH-2 because only cth-2 alleles emerged from this screen.
Figure 3: cth-2 and cdo-1 are necessary for the growth arrest and death caused by Moco deficiency.
(A) moc-1(ok366), cth-2(mg599);moc-1(ok366), and cdo-1(mg622) moc-1(ok366) mutant animals were synchronized at the L1 stage and cultured on wild-type and ΔMoaA E. coli for approximately 48 hours. Animal lengths were determined for each condition. Box plots display the median, upper, and lower quartiles while whiskers indicate minimum and maximum data points. Sample size (n) is the number of individuals assayed and is displayed for each experiment. (B) Cartoons of CTH-2 (upper) and CDO-1 (lower) proteins. Colored vertical lines indicate amino acid substitutions identified in our screen as suppressors of (green) moc-2(mg595), (red) moc-3(ku300), or (blue) moc-4(ok2571) mutant C. elegans when cultured on ΔMoaA E. coli. *, reference alleles for each gene.
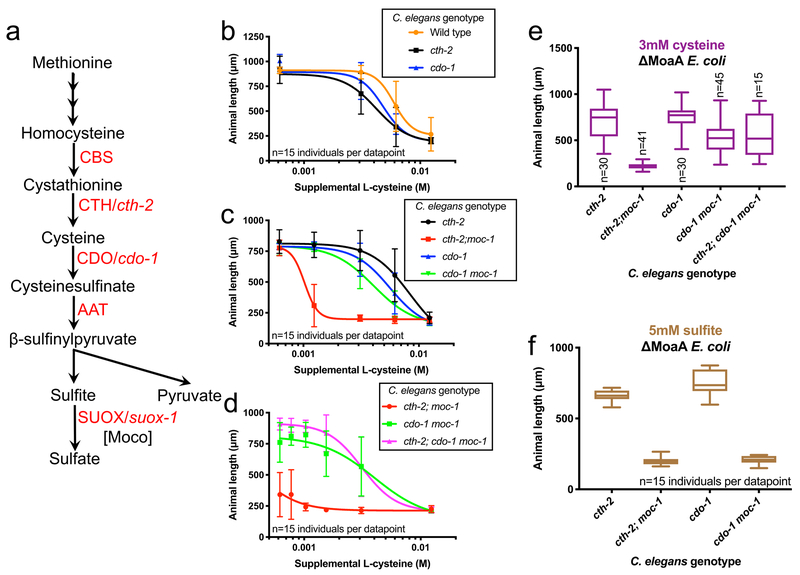
Figure 4: Endogenously produced sulfites inhibit growth and development during Moco deficiency.
(A) Simplified cartoon of sulfur amino acid catabolism beginning with methionine. Here we highlight the roles of CTH/cth-2, CDO/cdo-1, and the Moco requiring enzyme SUOX/suox-1. CBS is cystathionine beta synthase and AAT is aspartate aminotransferase. (B-F) Wild-type, cth-2(mg599), cdo-1(mg622), cth-2(mg599);moc-1(ok366), cdo-1(mg622) moc-1(ok366) and cth-2(mg599); cdo-1(mg622) moc-1(ok366) animals were cultured from the first larval stage on ΔMoaA E. coli supplemented with various concentrations of (B-E) cysteine or (F) 5mM sulfite. Animals were cultured for 48 hours and animal lengths were quantified. (B-D) Average values and standard deviation are displayed for each concentration of supplemental cysteine. (E,F) Growth of C. elegans mutants was scored on (E) 3mM supplemental cysteine or (F) 5mM supplemental sulfite. Box plots display the median, upper, and lower quartiles while whiskers indicate minimum and maximum data points. Sample size (n) is the number of individuals assayed and is displayed for each experiment. The data in Figure 4E summarizes the data in Figures 4B–D, highlighting the critical 3mM concentration of supplemental cysteine.
We also identified 4 alleles of C. elegans cysteine dioxygenase, cdo-1/F56F10.3. We identified 3 missense mutations affecting conserved amino acids in the cdo-1 active site and 1 nonsense mutation, suggesting that loss of function of cdo-1 suppresses the growth arrest caused by C. elegans Moco deficiency (Fig. 3, Supplementary Table 4). Cysteine dioxygenase (CDO) acts immediately downstream of CTH-2 in the sulfur amino acid metabolism pathway, catalyzing the reaction of cysteine with dioxygen to generate cysteine sulfinate (Fig. 4)6. To demonstrate the requirement of cdo-1 for the growth arrest of Moco-deficient C. elegans, we genetically transferred the cdo-1(mg622) allele (C85Y) into the moc-1 mutant background and assayed growth and development. cdo-1 moc-1 double mutant animals grew well on both wild-type and ΔMoaA E. coli, recapitulating the suppression identified in our screen (Fig. 3A). Additionally, cth-2 and cdo-1 mutations suppressed embryogenesis defects caused by maternal Moco deficiency (Supplementary Table 2).
Sulfites inhibit development during Moco deficiency:
We hypothesized that CTH-2 and CDO-1-dependent catabolism of sulfur amino acids generates a product that is toxic if Moco is deficient. To test this idea, we treated animals with cysteine, a product of CTH-2 and a substrate for CDO-1, and tested for reversion of the cth-2(lf) or cdo-1(lf) suppression of Moco-deficient larval arrest on ΔMoaA E. coli (Fig. 4A). Wild-type animals display half maximal growth rate (IC50) with 6.0mM cysteine. The cth-2 mutant animals display an IC50 of 8.4mM cysteine (Fig. 4B,C). In contrast, cth-2; moc-1 double mutant animals are sensitive to cysteine (IC50 1.0mM cysteine); at 3mM cysteine cth-2; moc-1 mutants arrest early in development (Fig. 4C,E). Thus cth-2; moc-1 double mutant animals are 8 times more sensitive to cysteine than the cth-2 single mutant, demonstrating that under Moco-deficient conditions, cysteine, a CTH-2 catabolic product, promotes larval arrest.
In contrast, cdo-1 and cdo-1 moc-1 mutant animals grew well on ΔMoaA E. coli at most cysteine concentrations, displaying normal IC50s of 5.6mM and 4.1mM cysteine respectively (Fig. 4C,E). These results demonstrate that the activity of CDO-1 is required for the larval arrest caused by supplemental cysteine during Moco deficiency. To further explore the relationship between cth-2, cdo-1, and cysteine, we analyzed the growth of cth-2; cdo-1 moc-1 triple mutant animals with cysteine. The cth-2; cdo-1 moc-1 triple mutant (IC50 of 3.1mM cysteine) displays growth on cysteine similar to cdo-1 moc-1 (IC50 of 4.0mM) double mutants and grows much better on cysteine than cth-2; moc-1 double mutants (IC50 of 0.4mM) (Fig. 4D,E). These data demonstrate that the function of cdo-1 is epistatic to cth-2 and cysteine supplementation and support the model that a cysteine catabolic product downstream of CDO-1 is responsible for the larval arrest caused by Moco deficiency.
Sulfites are produced via sulfur-amino acid catabolism downstream of both CTH-2 and CDO-1 (Fig. 4A). If cysteine-derived sulfites inhibit growth and development during Moco deficiency, supplemental sulfite should cause inviability independent of the activity of CTH-2 or CDO-1 during Moco deficiency. We cultured synchronized wild-type, cth-2, cdo-1, cth-2; moc-1, and cdo-1 moc-1 mutant animals on Moco-deficient E. coli supplemented with sulfite and analyzed growth. When supplemented with 5mM sulfite, wild-type, cth-2, and cdo-1 mutant animals display a slight developmental delay while cth-2; moc-1 and cdo-1 moc-1 double mutant animals arrest or die early in development (Fig. 4F). These data demonstrate that when animals are Moco deficient, sulfites cause developmental arrest independent both CTH-2 and CDO-1. We conclude that accumulation of sulfite generated by sulfur amino acid catabolism is toxic to C. elegans if animals are Moco deficient.
The previous experiments were all performed on animals fed ΔMoaA E. coli eliminating contribution of dietary Moco. We wondered if dietary Moco could support C. elegans growth and development when animals are challenged with supplemental cysteine or sulfite. We assayed the growth of moc-1 mutant C. elegans fed wild-type E. coli, competent for Moco synthesis, supplemented with 3mM cysteine or 5mM sulfite. Even when fed E. coli providing dietary Moco, moc-1 mutant animals were sensitive to supplemental cysteine and sulfite when compared to wild-type animals (Supplementary Fig. 5). Thus, sufficient Moco to respond to environmental challenges to sulfur homeostasis cannot be derived from dietary sources alone.
Moco-deficient defects explained by loss SUOX-1 activity:
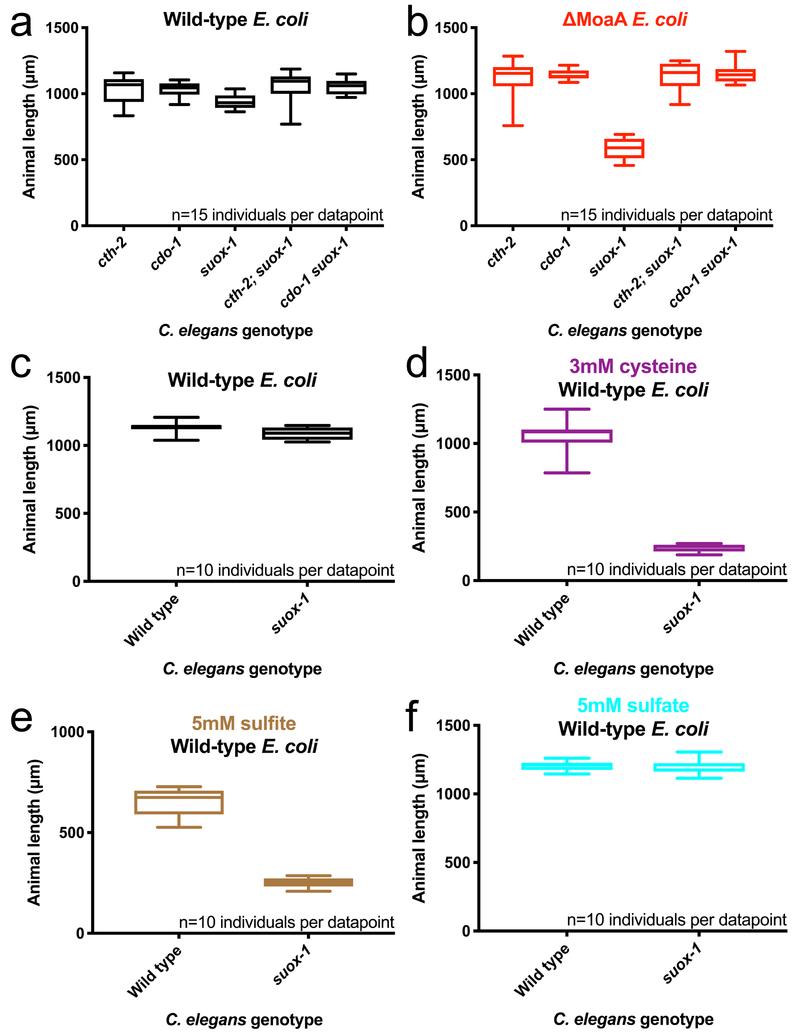
In animals, sulfite is oxidized to sulfate by the Moco-requiring enzyme, sulfite oxidase (suox-1/SUOX) (Fig. 4A). This reaction transfers electrons from sulfite to cytochrome C and the electron transport chain to generate sulfate, a proton gradient, and ATP19. Moco is also required for the function of 3 other animal enzymes; xanthine oxidoreductase, aldehyde oxidase, and mitochondrial amidoxime reducing component4. However, because the genetic suppressors of C. elegans Moco deficiency act in a pathway that produces sulfite, the pathology of C. elegans Moco deficiency is cleanly explained by a defect in sulfite oxidation. To test this model, we characterized suox-1 null and hypomorphic mutations. We found that suox-1 is an essential gene; suox-1 null mutant animals display completely penetrant larval lethality (0% of suox-1(mg663) null mutant animals are viable (n=16) when grown on wild-type E. coli). Furthermore, the larval lethality of suox-1 null mutant animals is suppressed by mutations in cth-2 (90% of cth-2(mg599); suox-1(mg663) animals were viable and fertile (n=19) when grown on wild-type E. coli). Thus, loss of suox-1 phenocopies Moco-deficient phenotypes in C. elegans.
We also characterized suox-1 missense mutations generated by the C. elegans Million Mutation Project (MMP) (Supplementary Table 5)20. One of the suox-1 alleles analyzed, suox-1(gk738847) (D391N), grew normally on wild-type E. coli, but displayed a severe developmental delay when cultured on ΔMoaA E. coli (Fig. 5A,B) and a reduced embryo viability when adult mothers were cultivated on Moco-deficient bacteria (Supplementary Table 2). Similar to Moco deficiency, these phenotypes were suppressed by loss of cth-2 or cdo-1 (Fig. 5B, Supplementary Table 2). Thus, the activities of cth-2 and cdo-1 are necessary for the developmental defects caused by reduced SUOX-1 activity. These data suggest that SUOX-1 detoxifies the sulfite produced by sulfur amino acid catabolism. Consistent with this model, the suox-1 hypomorphic mutant is hypersensitive to cysteine or sulfite, but not sulfate, (Fig. 5C–F). The phenotypic overlap between Moco deficiency and suox-1(lf) in C. elegans supports the model that the essential function of Moco is to maintain active sulfite oxidase protein and that Moco can either be synthesized in the animal cells or provided by the microbial diet.
Figure 5: suox-1 hypomorphic allele phenocopies Moco deficiency in C. elegans.
Analyses of the suox-1(gk738847) hypomorphic allele: cth-2(mg599), cdo-1(mg622), suox-1(gk738847), cth-2(mg599);suox-1(gk738847), and cdo-1(mg622) suox-1(gk738847) animals were cultured from the first larval stage on (A) wild-type or (B) ΔMoaA E. coli. (C-F) Wild-type or suox-1(gk738847) mutant animals were cultured from the first larval stage on (C) wild-type E. coli, or wild-type E. coli supplemented with (D) 3mM cysteine, (E) 5mM sulfite, or (F) 5mM sulfate. Animals were cultured for 72 hours and animal lengths were quantified. Box plots display the median, upper, and lower quartiles while whiskers indicate minimum and maximum data points. Sample size (n) is displayed for each experiment. Please note, the wild-type data points displayed in Figure 5C–F are also displayed in Supplementary Fig.5, as these assays were performed in parallel and utilize the same wild-type controls.
Tissue-specific rescue of suox-1, moc-1, and cth-2:
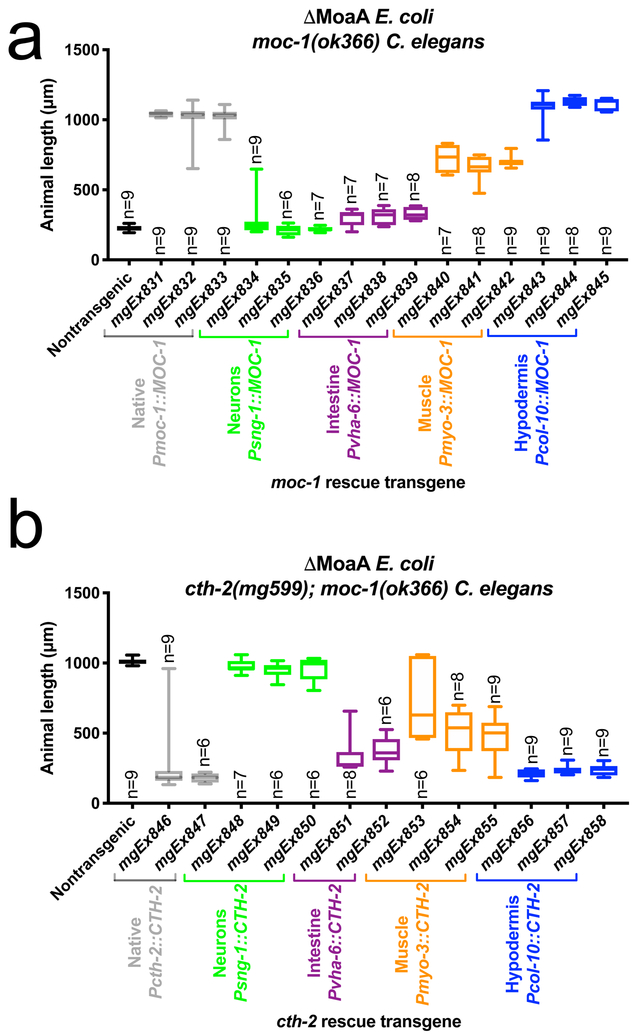
To determine in which tissues C. elegans SUOX-1 functions, we performed tissue-specific rescue experiments. The suox-1 genomic locus was expressed under the control of the rpl-28 (all tissues), sng-1 (neurons), vha-6 (intestine), myo-3 (muscle), or col-10 (hypodermis) tissue-specific promoters and tested for ability to rescue of the lethality of suox-1(mg663) null mutant animals. Transgenes expressing suox-1 in all tissues or only in the hypodermis rescued the lethality of suox-1(mg663) animals (3/3 independent transgenic lines were viable for each transgene). In contrast, expression of suox-1 in the neurons, muscle, or intestine failed to rescue the lethality of suox-1(mg663) (0/3 independent transgenic lines were viable for each of these transgenes). This suggests that the hypodermis is the primary site of C. elegans sulfite detoxification.
To determine the sites of MOC-1 activity, we performed similar tissue-specific rescue experiments. The moc-1 genomic locus was expressed under the control of its native promoter, or the sng-1 (neurons), vha-6 (intestine), myo-3 (muscle), or col-10 (hypodermis) tissue-specific promoters. Multiple transgenic strains expressing moc-1 under the control of these promoters were tested for their ability to rescue the larval arrest of moc-1(ok366) mutant animals grown on ΔMoaA E. coli. Transgenes expressing moc-1 under the control of its own promoter or the hypodermis-specific promoter rescued the larval arrest phenotype of moc-1 mutant animals when grown on ΔMoaA E. coli (Fig. 6A). The muscle-specific Pmyo-3::MOC-1 transgene caused consistent rescue, although not as complete as moc-1 driven by its own promoter or a hypodermal promoter (Fig. 6A). moc-1 expression in neurons or the intestine did not rescue the inviability of moc-1(ok366) animals grown on ΔMoaA E. coli (Fig. 6A). These data suggest that, similar to suox-1, the primary site of Moco biosynthesis is the C. elegans hypodermis. Distinct from suox-1, C. elegans muscle also appears to be a functional site of Moco synthesis via moc-1 activity.
Figure 6: Tissue specific rescue of suox-1, moc-1, and cth-2.
(A) Nontransgenic moc-1(ok366) and moc-1(ok366) transgenic animals expressing moc-1 under the control of the moc-1 (native, gray), sng-1 (neurons, green), vha-6 (intestine, purple), myo-3 (muscle, orange), or col-10 (hypodermis, blue) promoters were cultured for 72 hours from the first larval stage on MoaA mutant E. coli and animal lengths were quantified. Each rescue construct was tested with 3 independently isolated transgenes. (B) Nontransgenic cth-2(mg599); moc-1(ok366) and cth-2(mg599); moc-1(ok366) transgenic animals expressing cth-2 under the control of the cth-2 (native, gray), sng-1 (neurons, green), vha-6 (intestine, purple), myo-3 (muscle, orange), or col-10 (hypodermis, blue) promoters were cultured for 72 hours from the first larval stage on MoaA mutant E. coli and animal lengths were quantified. Each rescue construct was tested with 2–3 independently isolated transgenes. Box plots display the median, upper, and lower quartiles while whiskers indicate minimum and maximum data points. Sample size (n) is the number of individuals assayed and is displayed for each experiment.
To determine the functional tissues of CTH-2 activity, we similarly expressed the cth-2 genomic locus under the control of its native promoter or tissue-specific promoters. Multiple transgenic strains expressing cth-2 under the control of these promoters were tested for their ability to promote larval arrest of cth-2(mg599); moc-1(ok366) mutant animals grown on ΔMoaA E. coli. Transgenic cth-2 expression by the native cth-2, the hypodermal, or the intestinal promoters displayed strong rescue activity (Fig. 6B). cth-2 expressed in the muscle also displayed weak rescue activity while cth-2 expressed in neurons did not rescue (Fig. 6B). These transgenes did not affect cth-2; moc-1 double mutant worms when animals were cultured on wild-type E. coli. These data suggest that CTH-2-mediated sulfur amino acid catabolism occurs in the C. elegans hypodermis, intestine, and muscle.
Discussion
The ability to synthesize Moco is conserved across all kingdoms of life and organisms have previously been thought to rely solely upon de novo synthesis to maintain Moco homeostasis. Here show that C. elegans imports microbial Moco and the Moco precursor cPMP from the bacteria on which it feeds. The interkingdom transfer of these metabolites raises the possibility that transport of cPMP/Moco between cells, tissues, and organisms may be widespread. Ancient and nearly universal enzyme cofactors such as B vitamins, heme, and enzyme cofactors such as Moco, have probably been shared between organisms for billions of years. Organisms become reliant on exogenous sources of these cofactors, often via evolutionary loss of biosynthetic pathways. For example, C. elegans and some parasitic nematodes have lost heme biosynthetic enzymes, now acquiring heme solely from dietary sources21. The evolutionary maintenance or loss of cofactor biosynthesis and acquisition pathways is likely a function of bioavailability of the cofactor and the metabolic cost of producing the cofactor de novo. For C. elegans, it appears beneficial to have retained both the ability to synthesize Moco as well as to acquire it from its microbial diet.
In what state is Moco transferred from E. coli to C. elegans? There is no known pool of free or stored Moco in bacterial cells and the oxygen sensitivity of the mature cofactor make it unlikely that unbound Moco is the transferred molecule. It is possible that C. elegans ingests Moco bound to the bacterial Moco-utilizing proteins or the Moco-biosynthetic enzymes, and that this protein-cofactor interaction protects the cofactor during the uptake process. In this model, C. elegans would extract Moco from the E. coli proteins and incorporate it into its own Moco-requiring enzymes, such as SUOX-1. The mechanism of Moco transfer from bacteria to nematode remains to be discovered.
Could the inter-cellular transfer of cPMP and Moco be a conserved phenomenon? Cells from humans with Moco deficiency (MoCD) secrete the Moco-intermediate cPMP which can be acquired by cells in vitro22. This work laid the foundation for the idea of treating Moco deficiency with cPMP supplementation; 21 years later, a human MoCD patient was successfully treated with intravenous cPMP23. cPMP supplementation is only efficacious if patients possess the enzymes to convert cPMP into mature Moco. The successful treatment of human MoCD patients with cPMP parallels the ability of C. elegans to acquire cPMP from dietary E. coli, and suggest that this might be a conserved phenomenon. Furthermore, our demonstration of transfer of mature Moco from E. coli to C. elegans offers hope to deliver mature Moco, perhaps even from the microbiome, as a therapy to treat MoCD in patients that are refractory to cPMP treatment.
Why is Moco essential? In addition to its role as a cofactor, some Moco-biosynthetic enzymes have additional conserved activities such as tRNA thiolation (MOCS3) and promotion of glycine receptors clustering in neurons (Gephyrin)24–26. Our genetic suppressors of Moco-deficient larval arrest identified mutations in cystathionine gamma-lyase (cth-2) or cysteine dioxygenase (cdo-1). These enzymes catalyze sequential steps in the catabolism of methionine and cysteine. Our pharmacological studies demonstrate that sulfite produced by these enzymes is the key toxin that causes developmental arrest during Moco deficiency. While the exact mechanism by which sulfites are toxic to C. elegans is mysterious, sulfites are known to biochemically alter RNA, DNA, and proteins27–29. Sulfite also reacts with cystine to produce S-sulfocysteine (SSC), which is structurally similar to glutamate and a potent trigger of neural cell death30. Whether these modes of toxicity contribute to the inviability of Moco- and SUOX-1-deficient C. elegans is unknown. Humans are exposed to both endogenous sulfites, via catabolism of dietary methionine and cysteine6, and exogenous sulfites due to their widespread use as food preservatives27. These sulfites are detoxified by the Moco-requiring sulfite oxidase enzyme which oxidizes sulfite to sulfate to be excreted31,32. Our suppressor mutations in cth-2 or cdo-1 block the production of sulfites that accumulate during Moco deficiency, permitting growth and development. This is a conserved phenomenon: reducing sulfite production by limiting dietary intake of methionine and cysteine yields biochemical and clinical improvements in human patients with mild sulfite oxidase deficiency33,34. Thus, inhibition of cystathionine gamma-lyase or cysteine dioxygenase in human patients suffering from MoCD or sulfite oxidase deficiency might further alleviate the toxicity of endogenously produced sulfites.
Our tissue-specific rescue studies of suox-1 suggest that the hypodermis is the major site of SUOX-1-mediated sulfite detoxification. We also found that the hypodermis is the major site of Moco biosynthesis (Fig. 6A). These data suggest that Moco is both synthesized (via MOC-1) and utilized (via SUOX-1) primarily in the hypodermis. However, the muscle is also a site of MOC-1-mediated Moco biosynthesis, but is not a site of SUOX-1 activity. A possible explanation for this asymmetry of Moco synthesis and utilization is that Moco may be synthesized in muscle and transported to the hypodermis where it supports SUOX-1 activity. Intestine-specific expression of moc-1 does not rescue the moc-1 inviability when animals are grown on Moco-deficient E. coli, suggesting that the intestine is not a site of Moco biosynthesis. Given that dietary E. coli-derived Moco can support the growth and development of moc-1 mutant worms, these results suggest that dietary Moco is taken up through the intestine and transported to the hypodermis to support SUOX-1 activity. Therefore, the intestine may be specialized in uptake and distribution of dietary Moco while the muscle and hypodermis may be the primary sites of endogenous Moco synthesis. Identifying the mechanisms by which endogenous and dietary Moco are transported between animal tissues will be an exciting area of future research.
Online Methods:
Caenorhabditis elegans:
C. elegans strains were cultured at 20°C on nematode growth medium (NGM) seeded with E. coli OP50 unless otherwise noted35. The wild-type strain was Bristol N2. All C. elegans strains used in this work, their developmental stage at time of experiment, culture conditions, outcrossing status, and their sources are listed in Supplementary Table 3 or in the appropriate figure legend.
Bacteria:
The wild-type bacterial strains were E. coli (OP50) or E. coli (K-12) as indicated in the figure legends and Method Details section. All bacterial strains used in this work, their culture conditions, and their sources are listed in Supplementary Table 3 or the appropriate figure legends.
Plasmid construction and transgenesis:
All plasmids utilized in this manuscript are listed in Supplementary Table 3. All cloning was performed using isothermal/Gibson assembly in a pBluescript plasmid backbone36. Tissue-specific rescue plasmids were constructed by amplifying the suox-1, moc-1, and cth-2 genomic loci from wild-type C. elegans genomic DNA. Each locus was amplified from the start codon to the stop codon including all exons and introns and inserted into plasmids that contained the appropriate tissue specific promoter and C-terminal green fluorescent protein. For these plasmids, the stop codon was mutated to allow translation of C-terminal GFP. For Pmoc-1::MOC-1::GFP (pKW26) and Pcth-2::CTH-2::GFP (pKW23) the entire genomic locus including upstream promoter, 5’UTR, exons, and introns were amplified in a single polymerase chain reaction and inserted into plasmids that contained C-terminal GFP. The moc-1 promoter was the 874bp upstream of the moc-1 start codon. The cth-2 promoter was the 3,965bp of the cth-2 start codon. The tissue-specific promoters used in this work are rpl-28 (all tissues), sng-1 (neurons), vha-6 (intestine), myo-3 (muscle), and col-10 (hypodermis). The rpl-28 promoter was the 605bp upstream of the rpl-28 start codon. The sng-1 promoter used was the 2,080bp upstream of the sng-1 start codon. The vha-6 promoter used was the 934bp upstream of the vha-6 start codon. The myo-3 promoter was the 2,168bp upstream of the myo-3 start codon. The col-10 promoter was the 1,129bp upstream of the col-10 start codon. For information on the cloning of Pmoc-5::MOC-5 (pKW20) see “Identification of F49E2.1/moc-5 and W01A11.6/moc-2 as genes required for growth during dietary Moco deficiency.”
Transgenic animals containing extrachromosomal arrays were generated by injecting the gonad of worms with a plasmid of interest and a co-injection marker37. The co-injection marker was Pmyo-2::mCherry for all strains analyzed in this manuscript except for GR2258 which utilized a Pmyo-3::GFP co-injection marker38. All transgenic C. elegans strains are listed in Supplementary Table 3.
C. elegans growth assays:
C. elegans were synchronized at the first stage of larval development (L1). L1 animals were then cultured on NGM seeded with wild-type or mutant E. coli K-12. The strains of E. coli used are noted specifically for each assay and are listed in Supplementary Table 3.
For cysteine, sulfite, or sulfate sensitivity assays, NGM was seeded with wild-type or ΔMoaA E. coli K-12 and then supplemented with various concentrations of L-cysteine (0, 0.00062, 0.0012, 0.0031, 0.0062, and 0.12M), sodium metabisulfite (0.005M), or sodium sulfate (0.005M). Synchronized L1 animals were placed onto the various cysteine, sulfite, or sulfate culture conditions and were allowed to develop for 48 or 72 hours (specified in figure legends) at 20°C.
For all assays, after the growth period was completed live animals were imaged using an Axio Zoom.V16 microscope (Zeiss) equipped with an ORCA-Flash4.0 digital camera (Hamamatsu). Images were captured using ZEN software (Zeiss) and processed utilizing ImageJ. Animal length was measured from the tip of the head to the end of the tail. The concentration of supplemental cysteine that caused 50% of the wild-type growth (IC50) was calculated with nonlinear regression analyses using GraphPad Prism software.
Quantification of embryo hatching rate:
To determine the hatching rate of wild-type and mutant C. elegans when cultured under various growth conditions, we performed synchronized egg lays using young adults. Embryos were then scored for hatching 12–24 hours after being laid. To facilitate adult-specific Moco deficiency, in some experiments L4 animals were shifted onto either wild-type or ΔMoaA E. coli, allowed to develop to the young adult stage and their progeny were assayed for their ability to hatch. All experiments were performed at 20°C unless otherwise noted.
Screening for E. coli gene activities required for the growth and development of C. elegans lacking endogenous Moco biosynthesis:
To identify E. coli gene activities that are required for the growth of moc-1(ok366) animals, we utilized the Keio collection of deletion E. coli strains (NIG). Each of the approximately 4,000 deletion strains are derived from the E. coli K-12 strain BW2511312. Each strain was cultured overnight in LB with 50 μg/ml kanamycin. 0.1ml of this 2ml overnight culture was seeded on NGM and allowed to dry and grow overnight at room temperature (~22–23°C) creating a lawn of bacteria.
Concurrently, approximately 200,000 moc-1(ok366) animals grown on E. coli OP50, which is wild type for Moco biosynthetic pathway genes, were prepared with sodium hypochlorite to generate eggs and sterilize the E. coli from the parental feeding. These eggs were then allowed to hatch overnight without food and were synchronized at the L1 stage. 30–50 L1 animals were dispersed onto each unique deletion strain of E. coli. These animals were cultured for 2 days at room temperature (~22–23°C). Wells were then scored for growth. Here we report deletion strains of E. coli that caused early larval growth arrest of moc-1(ok366) animals.
To more cleanly analyze the E. coli strains identified in our screen, we used established methods to remove the kanamycin resistance cassette from each Keio strain of interest12. This process results in the deletion of the E. coli gene of interest and leaves behind a sequence that codes for a 34 amino acid peptide. Importantly, this peptide coding sequence is in frame, allowing downstream genes in that operon to be functional. Thus, these deletions are nonpolar and allowed genetic interpretation of the single gene knockout. Nonpolar mutant strains were utilized in Figure 1C,D, Figure 2, and Supplementary Figure 3. All other strains still contained the kanamycin resistance cassette and might be polar, thus affecting the entire operon of interest.
Identification of F49E2.1/moc-5 and W01A11.6/moc-2 as genes required for growth during dietary Moco deficiency:
To define C. elegans gene activities that were required for growth during dietary Moco deficiency, we carried out a chemical mutagenesis screen for animals that were unable to grow and develop on ΔMoaA E. coli. Wild-type C. elegans were mutagenized with ethyl methanesulfonate (EMS) and grown on wild-type E. coli for two generations. We then screened for mutant animals in the F2 generation that failed to develop when cultured on ΔMoaA E. coli but grew well on wild-type E. coli35. We isolated two mutant alleles in this screen, mg589 and mg639.
Using whole genome sequencing and linkage analyses we determined that mg589 was located on the X linkage group (LG) and identified 5 candidate lesions on LG X that might be the mg589 mutation. Candidate lesions were homozygous and were predicted to alter the protein sequence. One of these 5 candidates, F49E2.1, is homologous to E. coli MoaA and MoaC, genes with established roles in Moco biosynthesis. F49E2.1 has a long isoform F49E2.1b and a short isoform F49E2.1a, both of which are predicted to use the same start codon. The lesion in F49E2.1 changes a Gga glycine codon to an Aga encoding an arginine. This missense mutation results in a G303R change in the encoded peptide. Interestingly, glycine 303 is conserved in E. coli MoaA, C. elegans F49E2.1, and human MOCS1 suggesting its functional importance. Glycine 303 is present in both isoforms of F49E2.1.
Our linkage, sequencing, and homology data strongly suggested F49E2.1 was the gene affected by mg589. To independently demonstrate this, we rescued mg589 mutant animals with the F49E2.1 wild-type genomic locus. To generate the rescuing plasmid, we engineered pBluescript to contain the entire wild-type F49E2.1 locus from 1,502 base pairs upstream of the ATG start codon to 722 base pairs downstream of the F49E2.1b stop codon. Thus, the rescuing plasmid (pKW20, F49E2.1(+), also known as Pmoc-5::MOC-5) contained the endogenous promoter, exons, introns and untranslated regions of both F49E2.1a and F49E2.1b and no other genes.
Transgenic animals containing extrachromosomal arrays were generated by injecting the gonad of mg589 mutant worms with pKW20 and a Pmyo-3::GFP co-injection marker37,38. Pmyo-3::GFP is a plasmid with the myo-3 promoter driving the expression of green fluorescent protein (GFP) in the body wall muscles. Transgenic strains were selected and maintained based on the heritable transmission of GFP in the body wall muscle. We isolated 4 independently derived transgenic mg589 strains expressing F49E2.1(+).
To test the ability of F49E2.1 to rescue the mg589 phenotype, we cultured mg589 + F49E2.1(+) animals and their nontransgenic siblings on ΔMoaA E. coli. For all 4 transgenic strains isolated, the transgenic mg589 + F49E2.1(+) animals grew and developed well on ΔMoaA E. coli while the nontransgenic sibling mg589 animals did not. These results demonstrate that the F49E2.1 genetic locus rescued the mg589 growth defect during dietary Moco deficiency. Thus, based on linkage, sequencing, homology, and rescue data, we conclude that F49E2.1 is the gene affected by the mg589 mutation. We rename F49E2.1 as moc-5 for molybdenum cofactor biosynthesis.
Using whole genome sequencing and linkage analyses we determined that mg639 was located on LG V and identified 8 candidate lesions on LG V that might be the mg639 mutation. Candidate lesions were homozygous and were predicted to alter the protein sequence. One of these 8 candidates, moc-2, is homologous to E. coli MogA, a gene with an established role in Moco biosynthesis. The lesion in moc-2 changes a gCa alanine codon to a gTa encoding a valine. This missense mutation results in an A136V change in the encoded peptide.
Our linkage, sequencing, and homology data strongly suggested moc-2 was the gene affected by mg639. However, the sensitivity of animals carrying the mg639 mutation to growth on ΔMoaA E. coli was not as dramatic as other mutations in C. elegans Moco biosynthetic enzymes (Supplementary Fig. 6, Supplementary Table 2). Furthermore, A136V is likely to be a subtle loss-of-function as alanine and valine are very similar amino acids. Thus we speculated that mg639 might be a hypomorphic allele of moc-2. To test this hypothesis and independently demonstrate that moc-2 was the gene affected by mg639, we generated a presumptive null mutation in moc-2 using CRISPR/Cas9 technology9. This allele results in a Gly72Stop followed by a frameshift to completely alter the codons downstream of the new mutant stop codon. The newly isolated mutation was designated moc-2(mg595). moc-2(mg595) animals displayed complete larval arrest when cultured on ΔMoaA E. coli (Fig. 2E). This independently isolated mutation phenocopies moc-2(mg639) and displays a more severe growth defect on ΔMoaA E. coli. Thus, based on linkage, sequencing, homology, and phenocopy with an independent allele, we concluded that moc-2 is the gene affected by the mg639 mutation. Furthermore, we believe that mg639 is a hypomorphic allele of moc-2 while the newly generated mg595 is likely a null allele. mg595 is our reference allele for moc-2 and was used for all the work in this manuscript unless otherwise noted.
EMS-mutagenesis screens for suppressors of moc-2, moc-3, and moc-4 mutant growth arrest during Moco deficiency:
To define C. elegans gene activities that mediate the growth arrest of C. elegans moc mutants fed on Moco-deficient E. coli, we carried out a chemical mutagenesis screen for suppressors of C. elegans moc-2(mg595), moc-3(ku300), or moc-4(ok2571) strains grown on Moco mutant E. coli. moc-2(mg595), moc-3(ku300), and moc-4(ok2571) C. elegans were mutagenized with ethyl methanesulfonate (EMS) and grown for two generations on wild-type E. coli. F2 animals were then synchronized as L1s, and challenged to grow and develop on ΔMoaA E. coli. We screened >100,000 F1 genomes and isolated mutations that suppressed this larval arrest/lethality.
Identification of EMS-induced mutations via whole-genome sequencing and linkage analyses:
The methods we used to identify EMS-induced mutations via whole-genome sequencing have been previously described18. Briefly, whole-genome DNA was prepared from C. elegans utilizing the Gentra Puregene Tissue Kit (Qiagen) per the manufacturer’s instructions. Genomic DNA libraries were prepared utilizing the NEBNext genomic DNA library construction kit (New England Biolabs). DNA libraries were sequenced on an Illumina Hiseq instrument and deep sequencing reads were analyze using CloudMap39.
We performed linkage analyses by mating a mutant strain of interest with the unmutagenized parental strain. Linkage analysis of mg589 and mg639 was performed by mating wild-type males with mg589 or mg639 hermaphrodites. Linkage analysis was also performed for a number of the moc-3(ku300) suppressors and was performed by mating moc-3(ku300) males with moc-3(ku300);mg588 hermaphrodites (for example). The subset of moc-3(ku300) suppressors for which linkage analysis was performed is displayed in Supplementary Table 4. For each individual case, 20–50 F2 progeny of the linkage cross were collected that displayed the mutant phenotype. Each isolate represented a unique homozygous mutant strain that carried a unique set of recombination events. For each linkage cross, all F2 homozygous mutant animals were pooled and DNA was extracted. In downstream analyses, only EMS mutations that were tightly linked to the causative lesion remained homozygous, shrinking the candidate pool of causative lesions. The genes affected by mutations that were not analyzed using linkage analyses were identified because multiple alleles affected the same genes (i.e. cth-2 and cdo-1). We noticed that no cdo-1 mutations emerged from the screens to suppress moc-3(ku300) growth arrest on Moco-deficient E. coli. To ensure that cdo-1 mutations do not specifically suppress moc-2 and moc-4 mutant C. elegans, we constructed the moc-3(ku300); cdo-1(mg622) double mutant and attempted to culture the strain on Moco-deficient E. coli. As expected, cdo-1 loss of function also suppressed moc-3 mutant growth arrest on Moco-deficient E. coli (Supplementary Fig. 7).
Analysis of suox-1 null mutant:
The suox-1(mg663) allele is a 653 base pair deletion generated using CRISPR/Cas9 technology9. This mutation causes larval lethality and was balanced using the X balancer tmC2440.
Supplementary Material
Acknowledgements:
We thank the Caenorhabditis Genetics Center, Min Han, Victor Ambros, and Shohei Mitani for providing C. elegans strains. We thank Marie-Ann Félix for providing wild bacterial isolates and the National BioResource Project (NIG, Japan) for providing the Keio E. coli knockout collection. This work was funded by an NIH Grant (5R01GM044619–26) to G.R. and a Damon Runyon Fellowship (DRG-2293–16) to K.W.
Footnotes
Quantification and statistical analysis:
ImageJ was used for image analysis. GraphPad Prism software was used for calculations of IC50, median, upper, and lower quartiles. Sample size, dispersion, and precision measures are described in the figure legends. No data points were excluded from this study based off statistical tests.
Data Availability Statement:
Further information and reasonable requests for data, resources, sequences, and reagents should be directed to and will be fulfilled by the corresponding author.
Competing Interests:
The authors declare no competing interests.
References:
- 1.Schwarz G, Mendel RR & Ribbe MW Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y & Gladyshev VN Molybdoproteomes and Evolution of Molybdenum Utilization. Journal of Molecular Biology 379, 881–899 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacGregor CH Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J. Bacteriol 121, 1117–1121 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss J & Hahnewald R Molybdenum cofactor deficiency: Mutations in GPHN, MOCS1, and MOCS2. Hum. Mutat 32, 10–18 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Huijmans JGM et al. Molybdenum cofactor deficiency: Identification of a patient with homozygote mutation in the MOCS3gene. Am. J. Med. Genet 173, 1601–1606 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Stipanuk MH SULFUR AMINO ACID METABOLISM: Pathways for Production and Removal of Homocysteine and Cysteine. Annu. Rev. Nutr 24, 539–577 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Pepper ASR et al. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development 131, 2049–2059 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Kim S et al. Allele-specific suppressors of lin-1(R175Opal) identify functions of MOC-3 and DPH-3 in tRNA modification complexes in Caenorhabditis elegans. Genetics 185, 1235–1247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arribere JA et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuel BS, Rowedder H, Braendle C, Félix M-A & Ruvkun G Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. U.S.A 113, E3941–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirksen P et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14, 38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba T et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 1–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Self WT, Grunden AM, Hasona A & Shanmugam KT Molybdate transport. Res. Microbiol 152, 311–321 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuebbens MM & Rajagopalan KV Investigation of the Early Steps of Molybdopterin Biosynthesis in Escherichia coli through the Use of in Vivo Labeling Studies. J. Biol. Chem 270, 1082–1087 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Reschke S et al. Identification of a bis-molybdopterin intermediate in molybdenum cofactor biosynthesis in Escherichia coli. J. Biol. Chem 288, 29736–29745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosentel JK, Healy F, Maupin-Furlow JA, Lee JH & Shanmugam KT Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J. Bacteriol 177, 4857–4864 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehrbach NJ, Ji F & Sadreyev R Next-Generation Sequencing for Identification of EMS-Induced Mutations in Caenorhabditis elegans. Curr Protoc Mol Biol 117, 7.29.1–7.29.12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshino N & Chance B The properties of sulfite oxidation in perfused rat liver; interaction of sulfite oxidase with the mitochondrial respiratory chain. Archives of Biochemistry and Biophysics 170, 514–528 (1975). [DOI] [PubMed] [Google Scholar]
- 20.Thompson O et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Research 23, 1749–1762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao AU, Carta LK, Lesuisse E & Hamza I Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. U.S.A 102, 4270–4275 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JL, Wuebbens MM, Mandell R & Shih VE Molybdenum cofactor biosynthesis in humans. Identification of two complementation groups of cofactor-deficient patients and preliminary characterization of a diffusible molybdopterin precursor. J. Clin. Invest 83, 897–903 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veldman A et al. Successful Treatment of Molybdenum Cofactor Deficiency Type A With cPMP. PEDIATRICS 125, e1249–e1254 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Leimkühler S Shared function and moonlighting proteins in molybdenum cofactor biosynthesis. Biol. Chem 398, 1009–1026 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury MM, Dosche C, Löhmannsröben H-G & Leimkühler S Dual role of the molybdenum cofactor biosynthesis protein MOCS3 in tRNA thiolation and molybdenum cofactor biosynthesis in humans. J. Biol. Chem 287, 17297–17307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirsch J, Wolters I, Triller A & Betz H Gephyrin antisense oligonucleotides prevent glycine receptor clustering in spinal neurons. Nature 366, 745–748 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Gunnison AF Sulphite toxicity: a critical review of in vitro and in vivo data. Food Cosmet Toxicol 19, 667–682 (1981). [DOI] [PubMed] [Google Scholar]
- 28.Bailey JL & Cole RD Studies on the reaction of sulfite with proteins. J. Biol. Chem 234, 1733–1739 (1959). [PubMed] [Google Scholar]
- 29.Würfel M, Häberlein I & Follmann H Inactivation of thioredoxin by sulfite ions. FEBS Lett 268, 146–148 (1990). [DOI] [PubMed] [Google Scholar]
- 30.Kumar A et al. S-sulfocysteine/NMDA receptor-dependent signaling underlies neurodegeneration in molybdenum cofactor deficiency. J. Clin. Invest 127, 4365–4378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mårtensson J The effects of short-term fasting on the excretion of sulfur compounds in healthy subjects. Metab. Clin. Exp 31, 487–492 (1982). [DOI] [PubMed] [Google Scholar]
- 32.Wattiaux-de Coninck S & Wattiaux R Subcellular distribution of sulfite cytochrome c reductase in rat liver tissue. Eur. J. Biochem 19, 552–556 (1971). [DOI] [PubMed] [Google Scholar]
- 33.Touati G et al. Dietary therapy in two patients with a mild form of sulphite oxidase deficiency. Evidence for clinical and biological improvement. J. Inherit. Metab. Dis 23, 45–53 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Del Rizzo M et al. Metabolic stroke in a late-onset form of isolated sulfite oxidase deficiency. Mol. Genet. Metab 108, 263–266 (2013). [DOI] [PubMed] [Google Scholar]
Methods-only References:
- 35.Brenner S The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Mello CC, Kramer JM, Stinchcomb D & Ambros V Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frøkjær-Jensen C et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet 40, 1375–1383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minevich G, Park DS, Blankenberg D, Poole RJ & Hobert O CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dejima K et al. An Aneuploidy-Free and Structurally Defined Balancer Chromosome Toolkit for Caenorhabditis elegans. Cell Rep 22, 232–241 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.