Abstract
LMO4 belongs to a family of LIM-only transcriptional regulators, the first two members of which are oncoproteins in acute T cell leukemia. We have explored a role for LMO4, initially described as a human breast tumor autoantigen, in developing mammary epithelium and breast oncogenesis. Lmo4 was expressed predominantly in the lobuloalveoli of the mammary gland during pregnancy. Consistent with a role in proliferation, forced expression of this gene inhibited differentiation of mammary epithelial cells. Overexpression of LMO4 mRNA was observed in 5 of 10 human breast cancer cell lines. Moreover, in situ hybridization analysis of 177 primary invasive breast carcinomas revealed overexpression of LMO4 in 56% of specimens. Immunohistochemistry confirmed overexpression in a high percentage (62%) of tumors. These studies imply a role for LMO4 in maintaining proliferation of mammary epithelium and suggest that deregulation of this gene may contribute to breast tumorigenesis.
The LIM domain defines a conserved cysteine-rich structure comprising two tandemly repeated zinc fingers and is found in a large group of diverse proteins (reviewed in refs. 1 and 2). This motif, originally identified in LIM-homeodomain transcription factors, may either occur alone (as one or more copies) or in association with heterologous domains such as a protein kinase or homeobox domain. Targeted gene disruption has established that LIM domain-containing proteins have critical functions in cell-fate specification and differentiation (3). The importance of LIM proteins in regulating normal growth and maturation is highlighted by the finding that inappropriate expression of proteins in the LMO (LIM-only) subclass can lead to leukemia (4).
The LMO family consists of four members (designated LMO1–LMO4), each of which comprises two tandem LIM domains. LMO1 and LMO2 were identified by virtue of their translocation in acute T cell leukemia (T-ALL) (5–7) and act as T cell oncogenes in transgenic mouse models (8–10). LMO2 is essential for embryonic hematopoiesis and is thought to function at the level of the pluripotent stem cell (11). Little is known about the function of LMO3, which was discovered on the basis of sequence homology. The most recently described member, LMO4, was isolated by virtue of its interaction with Ldb1/NL1/CLIM and in an expression screen with autologous serum (12–15). Ldb1 is a multifunctional adaptor protein that interacts with LMO proteins and other nuclear LIM-containing factors (16–19). The Lmo4 gene is widely expressed in both embryonic and adult tissues (12, 13, 15). Like the other members of this family, it is presumed that LMO4 is a transcriptional cofactor that primarily functions as a docking site for other factors. LMO4 may also contribute an activation or repression domain to influence transcriptional activity.
We sought to determine whether LMO4 plays a role in mammary development and in breast oncogenesis, given that it was initially deposited in the GenBank database as a breast tumor autoantigen (accession no. U24576). Here we report that the Lmo4 gene is developmentally regulated in mammary epithelium and that Lmo4 and its partner Ldb1 act as negative regulators of mammary epithelial differentiation. Overexpression of the LMO4 gene was found in more than 50% of primary breast cancers, suggesting that this protein contributes to the pathogenesis of breast cancer. Thus, deregulated expression of three members of this LMO family is associated with oncogenesis.
Materials and Methods
Cell Lines.
Most of the cell lines have been cited in previous studies (20). SCp2 mammary epithelial cells (21) were kindly provided by M. Bissell (University of California, Berkeley, CA).
In Situ Hybridization.
Mouse Lmo4 was isolated from mouse T cells by affinity chromatography using rabbit anti-Ldb1 antisera (19). The purification and cloning of Lmo4 will be described elsewhere (K.H., unpublished data). Full-length human LMO4 (ATCC no. T09407) or mouse Lmo4 cDNAs were cloned into pBluescript SK II(−) (Stratagene). Antisense and sense riboprobes were generated by using T3 or T7 RNA polymerase (Promega) with digoxigenin-UTP (Roche Diagnostics). Standard in situ hybridizations were performed as described (22). Breast cancer tissue arrays were either purchased from Clinomicslabs (Frederick, MD) or prepared from breast cancer samples at the Peter MacCallum Cancer Institute, Melbourne, Australia. All specimens evaluated were anonymous, archival tissue specimens. The Peter MacCallum Institutional Review Board approved the use of Peter MacCallum Cancer Institute specimens for tissue array analysis.
SCp2 Cell Differentiation Assay.
SCp2 mammary epithelial cells (21) were passaged in DMEM-F12 media containing DMEM-Ham's medium, 10% FCS, and insulin 5 μg/ml (Sigma). Full-length mouse cDNAs corresponding to Lmo4 or Ldb1 were cloned into both the pEF1α-puro and pEF1α-Flag-puro mammalian expression vectors (23). Protein expression was confirmed by transient transfection of 293T cells. Linearized expression vectors (10 μg) were introduced into SCp2 cells by using Superfect (Qiagen, Chatsworth, CA) and selected in puromycin for 8–10 days. Pools of stable transfectants expressing the appropriate gene or vector alone (control) were then used in the differentiation assay, essentially as described (21). Transfectants were induced to differentiate by the addition of DMEM-F12 containing insulin (5 μg/ml), hydrocortisone (1 μg/ml), and prolactin (5 μg/ml), which was generously provided by A. Parlow (National Hormone and Pituitary Program). After 96 h, cells were harvested directly for RNA extraction.
RNA Analysis and Reverse Transcription–PCR.
Northern blot analysis of poly(A)+ RNA was performed as described (24). Total RNA was isolated from SCp2 cells on extracellular matrix by using RNAzol (Tel-Test); cDNA synthesis and PCR were performed as described (25), with primers for β-casein, Wap, and Hprt (25). Sequences of the β-casein primers were: forward 5′-ATGAAGGTCTTCATCCTCGCCTGCC-3′ and reverse 5′-GCTGGACCAGAGACTGAGGAAGGTGC-3′. Sequences of the Wap primers were: forward 5′-TAGCAGCAGATTGAAAGCATTATG-3′ and reverse 5′-GACACCGGTACCATGCGTTG-3′. Samples were fractionated on agarose gels, ethidium bromide stained, blotted, then hybridized with specific internal oligonucleotides.
Immunoprecipitation and Western Blot Analysis.
Whole-cell lysates were generated from stably transfected SCp2 pools by lysing cells in KALB lysis buffer (26) containing protease inhibitors. Proteins were immunoprecipitated with anti-Flag M2 (Sigma) and protein G Sepharose (Amersham Pharmacia), and separated by SDS/PAGE (NOVEX, San Diego). After transfer, filters were blocked and incubated with rabbit antisera to either full-length mouse Lmo4 or a C-terminal Ldb1 polypeptide (19). Antibody binding was visualized with peroxidase-conjugated anti-mouse antibody by enhanced chemiluminescence (Amersham Pharmacia).
Immunohistochemistry.
For immunostaining, tissue arrays (Clinomicslabs) were incubated with supernatant containing rat anti-LMO4 monoclonal antibody, followed by incubation with biotinylated anti-rat IgG and HRP-Streptavidin (Dako), before detection by using diaminobenzidine (Dako), as described (27). Generation of the LMO4 monoclonal antibody will be described elsewhere. Other primary monoclonal antibodies used were: anti-ERα (1D5), anti-PgR (636), and anti-ErbB2 (all from Dako).
Results
Lmo4 Is Developmentally Regulated in the Mouse Mammary Gland.
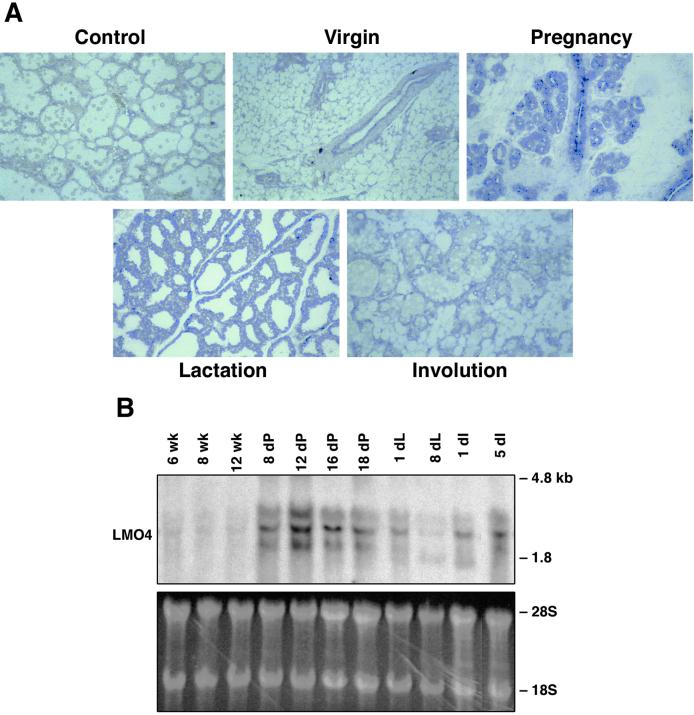
In situ hybridization analysis revealed that Lmo4 was prominently expressed in the lobuloalveolar units of the mammary gland during pregnancy (Fig. 1A). At this stage, a high rate of proliferation accompanies the formation and expansion of the lobuloalveoli. Lower levels of Lmo4 mRNA were present in the ductal epithelium of the virgin mammary gland, evident as a single layer, and in the early lactating and involuting mammary glands. Staining of the surrounding stroma was also apparent in the virgin mammary gland. The high level of Lmo4 in the pregnant mammary gland (Fig. 1A) was confirmed by Northern blot analysis, which revealed that levels of the two major transcripts (≈1.8 and 2.3 kb) peaked at midpregnancy (day 12) and remained high until late pregnancy (Fig. 1B). Although Lmo4 expression decreased during lactation, it seemed to be up-regulated during involution.
Figure 1.
Lmo4 is abundantly expressed in the lobuloalveolar units during pregnancy. (A) A single layer of ductal epithelium expressing Lmo4 transcripts is evident in the adult mammary gland. RNA expression was assessed by in situ hybridization by using sense (control) and antisense digoxigenin-labeled riboprobes corresponding to the full-length mouse Lmo4 sequence. Young adult virgin, 12-day pregnant, 2-day lactating, and 4-day involuting mammary glands were analyzed. The sense probe was hybridized to sections of 3-day involuting (shown), and to sections of lactating and pregnant mammary glands. (Original magnification ×50.) (B) Northern blot analysis of mammary glands from virgin, pregnant, lactating, and force-weaned (involuting) mice. Filters containing 20 μg of total RNA were hybridized with a mouse Lmo4 probe. The lower molecular weight bands may represent either an alternatively spliced variant of Lmo4 RNA or a cross-hybridizing species. The ethidium bromide-stained gel showing 28S and 18S ribosomal RNA provides a loading control.
Forced Expression of the Lmo4 and Ldb1 Genes Inhibits β-Casein Synthesis in Mammary Epithelial Cells.
Lmo4 binds with high affinity to Ldb1, a nuclear protein that serves as an adaptor for several LIM domain-containing proteins. To examine the role of Lmo4 and its partner protein Ldb1 in mammary differentiation, we introduced the genes into SCp2 mammary epithelial cells, which express moderate levels of Lmo4 and Ldb1 RNA (Fig. 3). The SCp2 cell line was originally isolated from the mammary gland of a midgestation mouse and mimics the essential features of mammary differentiation in the presence of extracellular matrix and a lactogenic stimulus (21). Differentiation of SCp2 cells is accompanied by the production of milk proteins, such as β-casein and whey acidic protein (Wap), which we have used here as molecular markers. Stable transfectants of SCp2 cells harboring Lmo4 or Ldb1 cDNAs (either epitope-tagged or untagged) together with a puromycin resistance marker, were generated and pools of cells assayed for their ability to undergo differentiation. For the latter assay, transfectants were plated on extracellular matrix in the presence or absence of a lactogenic stimulus (prolactin, insulin, and hydrocortisone).
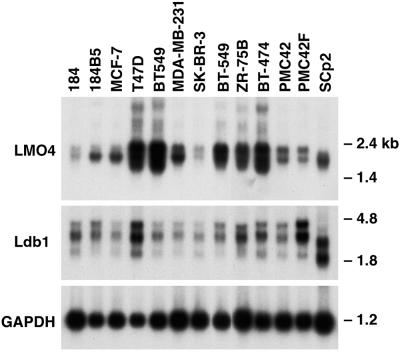
Figure 3.
LMO4 is overexpressed in several human breast cancer cell lines. Northern blot analysis of poly(A)+ RNA (3 μg) from human and mouse (SCp2) breast epithelial cell lines. Filters were hybridized sequentially with mouse Lmo4, Ldb1, and Gapdh cDNA probes. The lower molecular weight LDB1 transcript in human breast cancer cell lines may represent a cross-hybridizing species. Sizes of the Lmo4 and Ldb1 transcripts were lower in the mouse SCp2 cell line relative to those in human cell lines. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
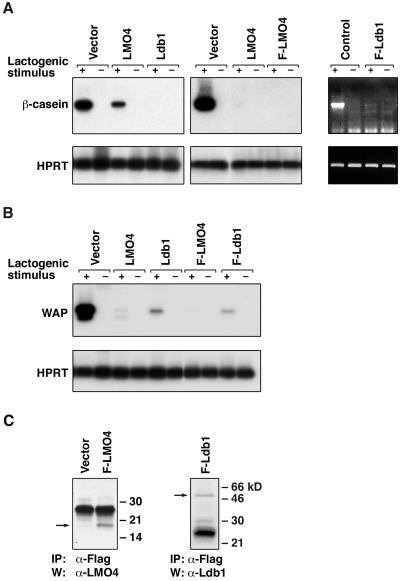
Overexpression of Lmo4 or Ldb1 markedly inhibited mRNA expression of β-casein and Wap on treatment with a lactogenic stimulus (Fig. 2). In contrast, transfectants expressing empty vector were indistinguishable from parental cells (Fig. 2 A and B). Expression of the Flag-tagged Lmo4 and Ldb1 proteins was readily detectable in several SCp2 transfectants, examples of which are shown in Fig. 2C, whereas expression of the untagged transgenes was verified by Northern blot analysis (data not shown). Forced expression of antisense Lmo4 RNA in SCp2 and HC11 mammary epithelial cells consistently augmented β-casein synthesis by severalfold, although Lmo4 protein levels were only slightly reduced by Western blot analysis (data not shown). Taken together, the data indicate that Lmo4 and its partner protein Ldb1 play a role in maintaining proliferation rather than differentiation of mammary epithelial cells.
Figure 2.
Lmo4 and Ldb1 inhibit β-casein and whey acidic protein (Wap) RNA synthesis in SCp2 mammary cells induced to differentiate. (A) Reverse transcription-PCR analysis was performed by using total RNA derived from stably transfected SCp2 pools that were stimulated (+) with prolactin, insulin, and hydrocortisone or unstimulated (−) for 96 h. β-casein and Hprt were used as markers of differentiation and loading, respectively. At least five independent transfections were performed. PCR products were fractionated by gel electrophoresis, blotted, and hybridized with internal oligonucleotide probes. The ethidium bromide- stained gel for Flag-Ldb1 is shown Right. (B) Reverse transcription-PCR analysis was performed on the same pools of transfectants as shown in A, using primers specific for Wap and Hprt. (C) Immunoprecipitation (IP) and Western blot analysis (W) confirmed expression of Flag-Lmo4 and Flag-Ldb1 in SCp2 transfectants. Lysates from cells expressing either gene or containing empty vector were subjected to immunoprecipitation by using mouse anti-Flag antibody and then blotted with rabbit anti-Lmo4 or rabbit anti-Ldb1 antibody. Arrows indicate relevant proteins; HPRT, hypoxanthine phosphoribosyltransferase.
Overexpression of LMO4 in Breast Cancer Cell Lines and Primary Invasive Cancers.
LMO4 RNA levels vary markedly between different human breast epithelial cancer cell lines (Fig. 3). High levels of LMO4 mRNA were detected in 5 of 10 breast cancer cell lines. Transcript levels were low in the immortalized cell line 184 but were substantially higher in a transformed variant of this line, 184B5 (20). LDB1 was expressed as two major transcripts (2.3 and 3.5 kb) among the panel of breast cancer cell lines and showed less variation in the extent of expression. The size of the Lmo4 and Ldb1 transcripts in SCp2 (Fig. 3), HC11, and EpH4 (not shown) mouse mammary epithelial cells was lower than that of the corresponding human RNAs, and reflects a species difference.
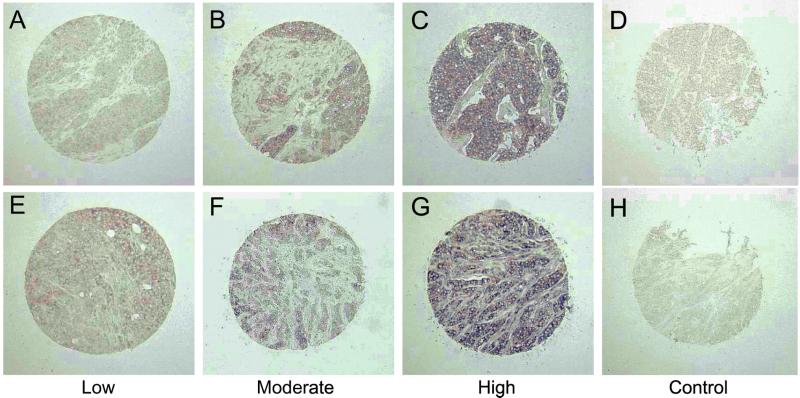
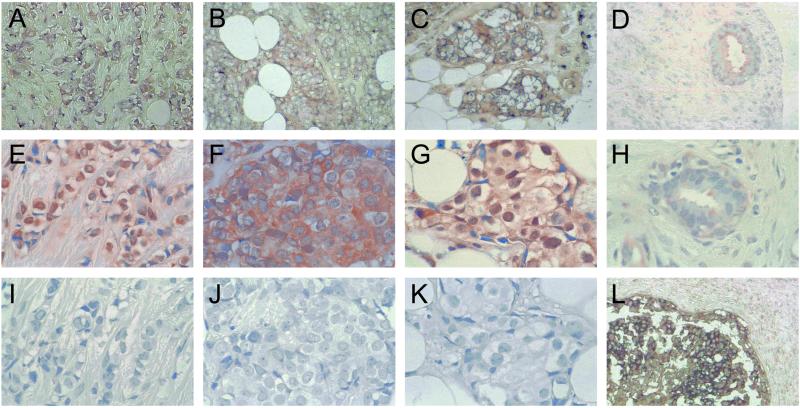
To assess whether LMO4 was up-regulated in primary breast cancers, we performed in situ hybridization on tissue arrays comprising 177 invasive breast cancers. Tumor specimens were scored as low/negative, moderate, or high for expression of LMO4 on the basis of their intensity of hybridization to the human LMO4 riboprobe. Two examples within each intensity group are given in Fig. 4 at low magnification. The control sense probe gave a negligible signal (Fig. 4 D and H). Strong to moderate staining of LMO4 RNA was evident in 99 of 177 (56%) cancers, most of which corresponded to infiltrating ductal carcinomas but also included invasive lobular and mixed ductal and lobular carcinomas. Fig. 5 depicts prominent expression of LMO4 in an invasive lobular carcinoma (Fig. 5A) and two infiltrating ductal carcinomas (Fig. 5 B and C). Low levels were detected in 7 of 20 (35%) benign breast fibroadenomas (Fig. 5D) or normal breast tissue samples. LMO4 RNA levels seemed to be elevated in ductal carcinoma in situ (Fig. 5L), in which 7 of 18 (38%) preinvasive tumors showed strong to moderate staining. A subset comprising 60 tumors was also analyzed by immunohistochemistry using a rat anti-LMO4 monoclonal antibody. Overexpression of LMO4 protein was observed in 62% of tumors. A comparison of in situ RNA and immunohistochemical staining for three tumors is shown in Fig. 5 (A and E, B and F, and C and G, respectively), revealing overexpression of LMO4 at both the RNA and protein levels. Benign breast tissue displayed either low (Fig. 5H) or undetectable levels of LMO4 protein. Anti-Ig antibody controls gave negligible staining for all tumor samples (Fig. 5 I–K). In this subset of 60 tumors, 35 (58%) scored high for RNA expression. Of these 35 tumors, 25 (71%) also expressed high levels of LMO4 protein, whereas 10 tumors displayed lower levels of protein. Conversely, 11 of 60 (17%) tumors that gave weak staining for LMO4 RNA scored high for protein expression. In summary, there was a strong correlation between RNA and protein overexpression in 71% of cases. The discordance observed for a minority of tumors is likely to reflect the numerous mechanisms that regulate the stability and turnover of RNA and protein, as well as the integrity of the primary cancer tissues collected by biopsy. For example, deregulation of a Cyclin D1 protein degradation pathway has been reported in breast cancer, such that Cyclin D1 protein levels are high despite low RNA expression (28).
Figure 4.
Overexpression of LMO4 RNA in primary breast cancer. In situ hybridization by using full-length human LMO4 sense and antisense riboprobes labeled with digoxigenin. Low magnification of tumor specimens displaying low (A and E), moderate (B and F), and high (C and G) levels of LMO4 mRNA. The LMO4 sense riboprobe gave negligible staining (D and H). Sense controls (D and H) correspond to the tumors shown in C and G, respectively.
Figure 5.
Overexpression of LMO4 RNA and LMO4 protein in primary breast cancer. In situ hybridization (with digoxigenin-labeled human LMO4 riboprobes) and immunohistochemistry (with a rat anti-LMO4 monoclonal antibody) were performed on tissue arrays containing archival breast specimens. High LMO4 RNA expression in an infiltrating lobular carcinoma (A), and two infiltrating ductal carcinomas (B and C) (antisense LMO4 probe); (D) benign fibroadenoma, displaying low expression of LMO4 RNA. Abundant LMO4 protein expression was detected in the corresponding infiltrating lobular and ductal cancer samples (E–G). Low levels of LMO4 protein were detected in the benign sample (H). Corresponding negative controls (Ig) for immunostaining are shown (I–K); (L) ductal carcinoma in situ, showing high expression of LMO4 RNA. Sense LMO4 probe gave no signal for any of the tumor samples. (Original magnification: A–D, ×100; E–K, ×200; L, ×50.)
Subcellular localization of LMO4 was variable. In some tumors, prominent nuclear staining was detected (Fig. 5 E and G), whereas in other tumors nuclear plus cytoplasmic staining was visualized (Fig. 5F). These findings are consistent with subcellular localization studies of LMO4 in transfected cells (12, 13). The same tumor set of 60 samples was evaluated for expression of the estrogen (ERα), progesterone, and ErbB2 receptors by immunohistochemistry, in which 32%, 40%, and 30% were positive for the respective receptors. No correlation was found between overexpression of LMO4 and expression of these markers.
Discussion
Here we describe a role for LMO4 as a negative regulator of mammary epithelial differentiation and present data that implicates this protein in breast cancer. Lmo4 was expressed predominantly in the alveolar epithelium of the mammary gland during pregnancy, which coincides with rapid proliferation of the lobuloalveolar units. Expression decreased during lactation but increased during involution, a stage entailing massive remodeling of the mammary gland. Compatible with a role for this gene in proliferating cells, overexpression of Lmo4 in mammary epithelial cells attenuated differentiation in β-casein-producing cells. Enforced expression of Ldb1, a multiadaptor protein that binds Lmo4 with high avidity, was also found to be a potent inhibitor of differentiation. These findings parallel the findings for LDB1 and LMO2 (another LMO family member) in a different cellular system, in which overexpression of either gene blocked erythroid cell maturation (19). Although LMO4 and LDB1 may not exert all their functions as a complex, the cumulative data suggest that one function of the LMO–LDB1 complex is to maintain the undifferentiated state.
Three members of the LMO family have now been implicated in oncogenesis. LMO1 and LMO2 are translocated in a subset of T-ALL resulting in deregulated expression of these genes at a stage when they would not normally be expressed (4). We have established that the LMO4 gene is overexpressed in ≈50% of breast cancer cell lines and primary breast cancers. Increased LMO4 expression was also observed in premalignant tissue relative to normal breast. Almost 40% of ductal carcinomas in situ expressed abundant LMO4 RNA, indicating that this gene may be an early marker in the progression of breast cancer. LMO4 was also isolated by screening breast cancer libraries with autologous serum from a patient with breast cancer (15, 29). This observation presumably reflects high levels of LMO4 in the breast cancer from this woman. Although LMO4 is often expressed in proliferating cells and organs, it is also present in postmitotic cells, and apparently is not a simple marker of cell growth (12, 13).
LMO genes are oncogenic in transgenic mice. Inappropriate expression of either LMO1 or LMO2 in the T cell lineage blocks T cell differentiation and leads to T cell tumors (4). These tumors arise with long latency suggesting that additional somatic mutations are required for overt tumorigenesis. The timing of transgene expression and the absolute level of expression are likely to be critical for perturbing the composition/stoichiometry of the LMO-containing multiprotein complexes, thus influencing the onset of tumorigenesis. It remains to be determined whether LMO4 is a potential oncoprotein in the mammary gland.
At least 20 genes, including those encoding MYC, Cyclin D1, EGFR, FGFR1, FGFR2, and AIB1, are overexpressed in breast cancer (30, 31). These genes are common targets for gene amplification, but they can also be overexpressed in the absence of gene amplification. The human LMO4 gene has been mapped to chromosome 1p22.3 (32), which has been reported to be deleted in some human cancers, including lymphoma, liver, brain, skin, and breast cancer (33–35). The 1p22.3 region also maps to regions of amplification in many different human cancers (36). High-resolution mapping can now reveal copy number gains and losses within a relatively small region (37, 38), and further, has been used to identify high-level amplification in a chromosomal region reported to undergo allelic loss (37). It remains to be established whether LMO4 is amplified or deregulated by other means in breast cancer.
The molecular mechanism(s) by which LMO4 participates in mammary gland development have not been elucidated. There is no evidence that LMO4 or other LMO members can bind DNA. Rather, LMO4 functions as a protein–protein interaction surface, associating with at least two other proteins, LDB1 (16–19) and the transcription factor, deformed epidermal autoregulatory factor 1 (13). Identification of target genes for LMO4-containing transcriptional complexes in breast epithelium should provide insight into how LMO4 may contribute to the pathogenesis of breast cancer.
Acknowledgments
We are grateful to Hania Lada and Nadeen Jonas for expert technical assistance, Steven Mihajlovic for histology, and Dennis Advani and Brigitte Mesiti for assistance with graphics. This work was supported by the Victorian Breast Cancer Research Consortium, The National Breast Cancer Foundation (Australia), and the Rotary Bone Marrow Research Laboratories, Melbourne, Australia.
Abbreviations
- LMO
LIM-only protein
- LDB1
LIM domain-binding protein 1
References
- 1.Sanchez-Garcia I, Rabbitts T H. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 2.Dawid I B, Toyama R, Taira M. C R Acad Sci Ser III. 1995;318:295–306. [PubMed] [Google Scholar]
- 3.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 4.Rabbitts T H. Genes Dev. 1998;12:2651–2657. doi: 10.1101/gad.12.17.2651. [DOI] [PubMed] [Google Scholar]
- 5.Boehm T, Baer R, Lavenir I, Forster A, Waters J J, Nacheva E, Rabbitts T H. EMBO J. 1988;7:385–394. doi: 10.1002/j.1460-2075.1988.tb02825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehm T, Foroni L, Kaneko Y, Perutz M F, Rabbitts T H. Proc Natl Acad Sci USA. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royer-Pokora B, Loos U, Ludwig W D. Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 8.McGuire E A, Rintoul C E, Sclar G M, Korsmeyer S J. Mol Cell Biol. 1992;12:4186–4196. doi: 10.1128/mcb.12.9.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, Rabbitts T H. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- 10.Neale G A, Rehg J E, Goorha R M. Leukemia. 1997;11, Suppl. 3:289–290. [PubMed] [Google Scholar]
- 11.Warren A J, Colledge W H, Carlton M B, Evans M J, Smith A J, Rabbitts T H. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 12.Kenny D A, Jurata L W, Saga Y, Gill G N. Proc Natl Acad Sci USA. 1998;95:11257–11262. doi: 10.1073/pnas.95.19.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugihara T M, Bach I, Kioussi C, Rosenfeld M G, Andersen B. Proc Natl Acad Sci USA. 1998;95:15418–15423. doi: 10.1073/pnas.95.26.15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grutz G, Forster A, Rabbitts T H. Oncogene. 1998;17:2799–2803. doi: 10.1038/sj.onc.1202502. [DOI] [PubMed] [Google Scholar]
- 15.Racevskis J, Dill A, Sparano J A, Ruan H. Biochim Biophys Acta. 1999;1445:148–153. doi: 10.1016/s0167-4781(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 16.Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Nature (London) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 17.Jurata L W, Kenny D A, Gill G N. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach I, Carriere C, Ostendorff H P, Andersen B, Rosenfeld M G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 19.Visvader J E, Mao X, Fujiwara Y, Hahm K, Orkin S H. Proc Natl Acad Sci USA. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas A M, Grant S L, Goss G A, Clouston D R, Sutherland R L, Begley C G. Int J Cancer. 1998;75:64–73. doi: 10.1002/(sici)1097-0215(19980105)75:1<64::aid-ijc11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 21.Desprez P-Y, Roskelley C, Campisi J, Bissell M J. Mol Cell Differ. 1993;1:99–110. [Google Scholar]
- 22.Wilkinson D. In Situ Hybridisation. New York: IRL; 1992. [Google Scholar]
- 23.Huang D C, Cory S, Strasser A. Oncogene. 1997;14:405–414. doi: 10.1038/sj.onc.1200848. [DOI] [PubMed] [Google Scholar]
- 24.Visvader J, Begley C G, Adams J M. Oncogene. 1991;6:187–194. [PubMed] [Google Scholar]
- 25.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson S E, Willson T A, Farley A, Starr R, Zhang J G, Baca M, Alexander W S, Metcalf D, Hilton D J, Nicola N A. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armes J E, Trute L, White D, Southey M C, Hammet F, Tesoriero A, Hutchins A M, Dite G S, McCredie M R, Giles G G, Hopper J L, Venter D J. Cancer Res. 1999;59:2011–2017. [PubMed] [Google Scholar]
- 28.Russell A, Thompson M A, Hendley J, Trute L, Armes J, Germain D. Oncogene. 1999;18:1983–1991. doi: 10.1038/sj.onc.1202511. [DOI] [PubMed] [Google Scholar]
- 29.Racevskis J, Dill A, Stockert R, Fineberg S A. Cell Growth Differ. 1996;7:271–280. [PubMed] [Google Scholar]
- 30.Kallioniemi A, Kallioniemi O P, Piper J, Tanner M, Stokke T, Chen L, Smith H S, Pinkel D, Gray J W, Waldman F M. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuny M, Kramar A, Courjal F, Johannsdottir V, Iacopetta B, Fontaine H, Grenier J, Culine S, Theillet C. Cancer Res. 2000;60:1077–1083. [PubMed] [Google Scholar]
- 32.Tse E, Grutz G, Garner A A, Ramsey Y, Carter N P, Copeland N, Gilbert D J, Jenkins N A, Agulnick A, Forster A, Rabbitts T H. Mamm Genome. 1999;10:1089–1094. doi: 10.1007/s003359901167. [DOI] [PubMed] [Google Scholar]
- 33.Hoggard N, Brintnell B, Howell A, Weissenbach J, Varley J. Genes Chromosomes Cancer. 1995;12:24–31. doi: 10.1002/gcc.2870120105. [DOI] [PubMed] [Google Scholar]
- 34.Emi M, Matsumoto S, Iida A, Tsukamoto K, Nakata T, Yokota T, Akiyama F, Sakamoto G, Yoshimoto M, Kasumi F, Nakamura Y. Breast Cancer Res Treat. 1997;4:243–246. doi: 10.1007/BF02966514. [DOI] [PubMed] [Google Scholar]
- 35.Tsukamoto K, Ito N, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, Nakamura Y, Emi M. Cancer. 1998;82:317–322. [PubMed] [Google Scholar]
- 36.Knuutila S, Bjorkqvist A M, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy M L, Tapper J, Pere H, et al. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 37.Bärlund M, Tirkkonen M, Forozan F, Tanner M M, Kallioniemi O, Kallioniemi A. Genes Chromosomes Cancer. 1997;20:372–376. doi: 10.1002/(sici)1098-2264(199712)20:4<372::aid-gcc8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 38.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W L, Chen C, Zhai Y, et al. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]