The Gram-negative bacterium Pseudomonas aeruginosa is an important opportunistic pathogen that causes infections in cystic fibrosis and hospitalized patients. Therapeutic treatments are limited due to the emergence and spread of new antibiotic-resistant strains. In this context, the development of a vaccine is a priority. Here, we used an attenuated strain of Salmonella enterica serovar Typhimurium as a vehicle to express and deliver the Pseudomonas antigen PcrV. This vaccine induced the generation of specific antibodies in mice and protected them from lethal infections with P. aeruginosa. This is an important step toward the development of an effective vaccine for the prevention of infections caused by P. aeruginosa in humans.
KEYWORDS: PcrV, Pseudomonas aeruginosa, Salmonella-delivered vaccines, SseJ, type III secretion
ABSTRACT
Pseudomonas aeruginosa is a common Gram-negative opportunistic pathogen that is intrinsically resistant to a wide range of antibiotics. The development of a broadly protective vaccine against P. aeruginosa remains a major challenge. Here, we used an attenuated strain of Salmonella enterica serovar Typhimurium as a vehicle to express P. aeruginosa antigens. A fusion between the S. enterica type III secretion effector protein SseJ and the P. aeruginosa antigen PcrV expressed under the control of the sseA promoter was translocated by Salmonella into host cells in vitro and elicited the generation of specific antibodies in mice. Mice immunized with attenuated Salmonella expressing this fusion had reduced bacterial loads in the spleens and lungs and lower serum levels of proinflammatory cytokines than control mice after P. aeruginosa infection. Importantly, immunized mice also showed significantly enhanced survival in this model. These results suggest that type III secretion effectors of S. enterica are appropriate carriers in the design of a live vaccine to prevent infections caused by P. aeruginosa.
IMPORTANCE The Gram-negative bacterium Pseudomonas aeruginosa is an important opportunistic pathogen that causes infections in cystic fibrosis and hospitalized patients. Therapeutic treatments are limited due to the emergence and spread of new antibiotic-resistant strains. In this context, the development of a vaccine is a priority. Here, we used an attenuated strain of Salmonella enterica serovar Typhimurium as a vehicle to express and deliver the Pseudomonas antigen PcrV. This vaccine induced the generation of specific antibodies in mice and protected them from lethal infections with P. aeruginosa. This is an important step toward the development of an effective vaccine for the prevention of infections caused by P. aeruginosa in humans.
INTRODUCTION
Pseudomonas aeruginosa is an environmentally ubiquitous, Gram-negative, opportunistic bacterial pathogen. It is one of the more commonly reported nosocomial pathogens (1). P. aeruginosa forms biofilms in the upper airways of cystic fibrosis patients and repeatedly colonizes the lower airways, leading to chronic lung infection (2). It is also a common pathogen linked to burn wound infections (3), ventilator-associated pneumonia in intensive care unit patients (4), and urinary infections in patients with catheters in the upper urinary tract (5). In addition, P. aeruginosa is a leading cause of life-threatening infections in immunocompromised hosts with underlying diseases such as cancer or AIDS (6).
P. aeruginosa is intrinsically resistant to a wide range of antibiotics (7) and possesses adaptation strategies that facilitate its persistence in the environment, such as biofilm formation (8). In addition, the increasing selection of additional acquired resistance mechanisms, via mutations or horizontal gene transfer, has led to the emergence of multidrug-resistant strains (9).
In this context, the development of vaccines that limit the spread of P. aeruginosa infections is a major challenge. This has been the focus of research efforts for almost half a century, and over the last 25 years, multiple P. aeruginosa vaccines have been assessed in clinical trials (10). However, with the recent failure of the IC43 vaccine in a phase II clinical trial (11), there are currently no approved vaccines against P. aeruginosa or vaccines in advanced stages of clinical development (12). Renewal of the P. aeruginosa vaccine pipeline is thus a high priority.
Many antigens and delivery protocols have been tested as vaccine candidates, but to increase the efficacy of vaccination, novel approaches are clearly needed. Such approaches may combine previously tested P. aeruginosa antigens with delivery methods that were successful for other antigens.
The protective efficacy of outer membrane proteins OprF and OprI have been shown in animal models and clinical trials (13, 14). Another promising candidate is the P. aeruginosa V antigen (PcrV), the tip protein of the type III secretion system (T3SS), which is critical for its assembly and regulation. These secretion systems are present in many Gram-negative pathogens and symbionts and inject effector proteins into host cells to interfere with host cellular processes (15). Blockade of PcrV by specific antibodies inhibits the translocation of type III effector proteins, and immunization with recombinant PcrV or administration of anti-PcrV antibodies can protect animals from lethal P. aeruginosa infections (16, 17). Killed whole-cell and live attenuated P. aeruginosa vaccines present multiple antigens to the immune system but may exhibit some toxicity or residual virulence, whereas the use of recombinant proteins is safer but may induce a weaker immune response (18).
The use of live attenuated bacterial or viral pathogens is an interesting alternative for delivering heterologous antigens (19). Salmonella is among the first bacteria used for this purpose and has well-established protocols for genetic manipulation. Additional advantages of Salmonella-based vaccines are the low cost of production, absence of animal products, safety, and elicitation of efficient humoral and cellular immune responses via stimulation of innate and adaptive immunity (20). Salmonella enterica is a facultative intracellular pathogen that, once inside the host cell, resides in a modified phagosome known as the Salmonella-containing vacuole (SCV) (21). S. enterica possesses two T3SSs, T3SS1 and T3SS2, encoded in Salmonella pathogenicity islands 1 (SPI1) and 2 (SPI2), respectively (22–24). T3SS1 translocates effector proteins through the host plasma membrane and is required for invasion of nonphagocytic cells. T3SS2 is necessary for intracellular survival and secretes effectors from inside the SCV. Previous studies have shown that T3SS-mediated translocation can be used for efficient delivery of heterologous antigens that are fused to effector proteins to the cytosol of antigen-presenting cells (25–27). In fact, live S. enterica vaccines based on these delivery systems have been shown to produce protective immunity against other bacterial and viral pathogens (28–31) and are also being used in the field of tumor immunotherapy (32–36).
The aim of this study was to develop a vaccine against P. aeruginosa using S. enterica T3SS effectors as carriers for relevant antigens. A strong protective immune response against the PcrV antigen was obtained using the SseJ effector as a carrier.
RESULTS
Construction and evaluation of Salmonella vectors for delivery of P. aeruginosa antigens.
A previous study showed that the effector SseJ was an appropriate carrier for inducing a robust immune response against the model antigens ovalbumin and listeriolysin in vitro and in vivo (25). We used the promoter of the Salmonella SPI2 gene sseA (PsseA) to direct the expression of fusion proteins containing the Salmonella effector SseJ as well as Pseudomonas antigens OprF/I and PcrV (Fig. 1A and B and Materials and Methods). SseJ is secreted through the T3SS2 from inside the SCV (37), and PsseA responds to intravacuolar signals and is induced several hours after invasion (38, 39). The constructs were prepared using the low-copy-number plasmid pWSK29 as the vector. S. enterica serovar Typhimurium strain 14028 containing the plasmids with the hybrid genes was grown in low-magnesium minimal medium (LPM), a medium that imitates intravacuolar conditions, and the expression of fusion proteins was monitored by Western blotting against the FLAG tag that was also added. The results shown in Fig. 1C indicate that both plasmids yielded significant levels of protein production. Strains containing these plasmids were tested for their capacity to translocate the fusion proteins into RAW264.7 macrophages. Translocation was detected for both fusions, SseJ-OprF/I-FLAG and SseJ-PcrV-FLAG, by Western blotting using an anti-FLAG antibody (Fig. 1D). Translocation was T3SS2 dependent, since it was not observed in an ssaV mutant background (SsaV is an essential component of T3SS2).
FIG 1.
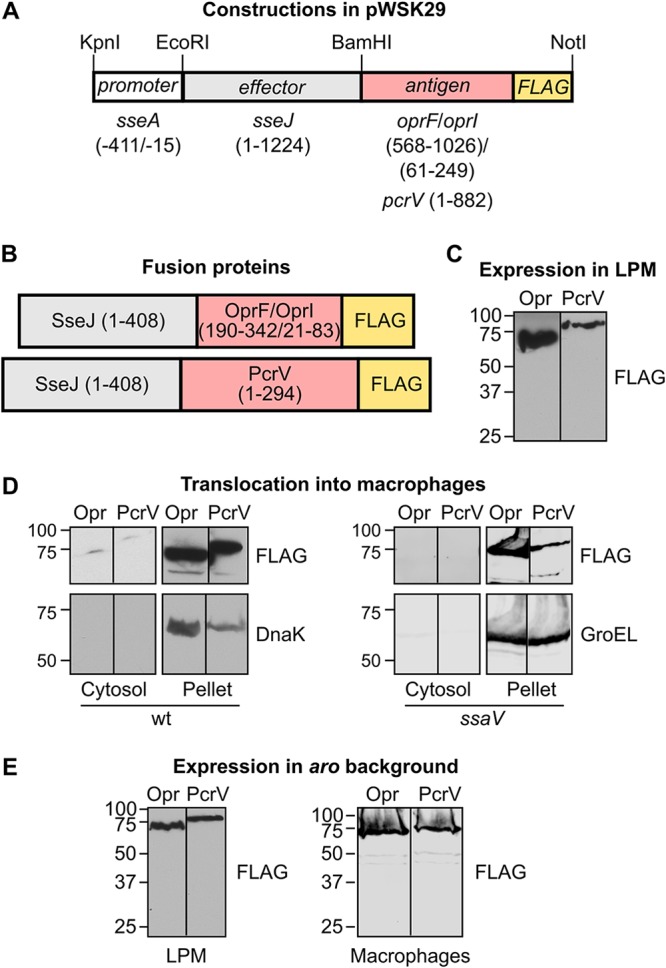
Generation of protein fusions for delivery of Pseudomonas aeruginosa antigens. (A) Vector pWSK29 was used for the generation of two plasmids in three steps. In the first step, part of the coding region of antigens OprF/OprI (see Materials and Methods for the generation of the hybrid gene oprF-oprI) or the complete coding region (without the stop codon) for the antigen PcrV from P. aeruginosa was amplified by PCR. DNA encoding the FLAG epitope and recognition sites for BamHI and NotI endonucleases was added with the primers used for amplification. These amplicons were cleaved with BamHI and NotI and ligated into pWSK29 previously cleaved with the same restriction enzymes. In the second step, the coding region of sseJ without the stop codon was amplified by PCR and ligated to the plasmids obtained in the first step previously cleaved with EcoRI and BamHI restriction enzymes. Finally, the promoter region of sseA (nucleotides −411 to −15 relative to the translation start site) was amplified by PCR and added to the plasmids obtained in the second step previously cleaved with endonucleases KpnI and EcoRI. Numbers in parentheses indicate base pairs relative to the start of the coding region. (B) Fusion proteins that are expected to be produced under the appropriate conditions after the introduction of the plasmids described in panel A in S. Typhimurium. Numbers in parentheses refer to amino acids included in the fusions. Both fusions include the complete amino acid sequence of SseJ and a C-terminal FLAG epitope to facilitate identification of the fusions by Western blotting. The expected molecular masses are 70.18 kDa for SseJ-OprF/I-FLAG and 79.23 kDa for SseJ-PcrV-FLAG. (C) Derivatives of S. Typhimurium strain 14028 carrying plasmids encoding OprF/I (Opr) or PcrV fusion proteins were grown in LPM. A FLAG tag was used for detection of the proteins by immunoblotting. Molecular weights in kDa are indicated on the left. (D) Salmonella strains that were derivatives of the wild-type strain (wt) or the ssaV mutant (carrying a nonfunctional T3SS2) expressing SseJ in fusion with Pseudomonas antigens OprF/I (Opr) or PcrV with a FLAG tag were grown under noninvasive conditions. These bacteria were used to infect RAW264.7 cells for 8 h. Cells were lysed with 1% Triton X-100 in PBS and centrifuged 15 min at 15,000 × g. The pellets and the filtered concentrated supernatants (cytosol) were analyzed by immunoblotting with anti-FLAG antibodies to detect translocation of fusion proteins into the host cytosols. Incubations with anti-DnaK or anti-GroEL antibodies were used as controls of contamination of the cytosol fraction with nonsecreted bacterial proteins. (E) Expression of the fusion proteins SseJ-OprF/I (Opr) and SseJ-PcrV (PcrV) was analyzed in an aroA aroB (aro) background. Bacteria were grown in LPM (left) or were grown for 24 h at 37°C with shaking in LB medium and used to infect RAW264.7 macrophages (right). Protein extracts were analyzed by immunoblotting with an anti-FLAG antibody.
Antibody responses induced by candidate vaccines against P. aeruginosa.
The plasmids generated as described above were introduced into an attenuated strain of S. Typhimurium with mutations in aroA and aroB. Expression of the fusion proteins was detected again in this background, both in LPM (imitating intravacuolar conditions) and during infection of macrophages (Fig. 1E), and C57BL/6 mice were immunized by intraperitoneal injections with the attenuated strains carrying the plasmids. Mice immunized with Salmonella aroA aroB without pWSK29 derivatives were used as the controls. Indirect enzyme-linked immunosorbent assays (ELISAs) were performed using sera collected 21 days after immunization. As shown in Fig. 2, immunization with Salmonella expressing SseJ-PcrV elicited detectable levels of antigen-specific total IgG in all mice, whereas Salmonella expressing SseJ-OprF/I was not able to induce a specific response. As expected, control mice had no detectable antigen-specific IgG.
FIG 2.
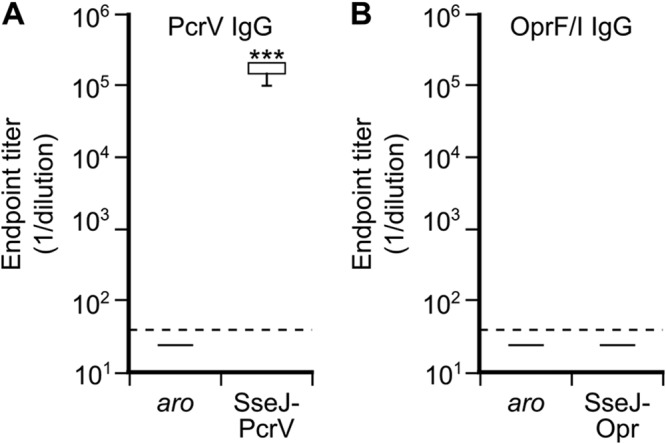
Antibody responses to immunization with Salmonella strains expressing Pseudomonas antigens. Mice were inoculated intraperitoneally with plasmid-less S. Typhimurium aroA aroB (aro) or with the same Salmonella strain carrying pWSK29 derivatives expressing PcrV or Opr fusion proteins as indicated. Serum samples were collected from vaccinated and control mice 21 days after immunization and levels of PcrV (A) or OprF/I (B) specific total IgG were measured by ELISA. Box and whisker plots represent the interquartile ranges and ranges, respectively, and horizontal lines represent median values. ***, P < 0.001 compared to levels in control mice using the Mann-Whitney U test.
Protective responses induced by candidate vaccines.
Vaccine efficacy was tested by infecting immunized and control mice with P. aeruginosa strain PAO1 by intraperitoneal injections, and survival was monitored over 7 days. As shown in Fig. 3, most of the mice vaccinated with Salmonella expressing the SseJ-PcrV construct survived the challenge, whereas mice vaccinated with Salmonella expressing SseJ-OprF/I or control Salmonella were not significantly protected. Statistical analysis of these survival data showed significant protection for the group vaccinated with Salmonella expressing SseJ-PcrV, whereas there was no significant difference between the control group and the group vaccinated with Salmonella expressing SseJ-OprF/I.
FIG 3.
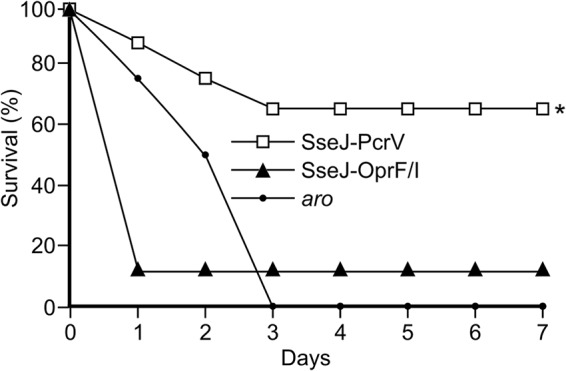
Survival curve of challenged mice. Groups of 8 C57BL/6 mice were intraperitoneally vaccinated with an aroA aroB mutant of S. Typhimurium (aro) or with the same strain carrying the plasmids pIZ2160 or pIZ2267 for expression of the fusion proteins SseJ-OprF/I-FLAG (from the PsseA promoter) or SseJ-PcrV-FLAG (from the PsseA promoter), respectively. Twenty-one days after the immunization, mice were infected with P. aeruginosa strain PAO1 by intraperitoneal injection, and the survival of mice was monitored for 7 days. Statistical significance was determined by log rank analysis. *, P < 0.05 compared to levels in control mice inoculated with Salmonella aroA aroB.
Effect of vaccination on tissue bacterial loads, postinfection serum cytokine levels, and survival.
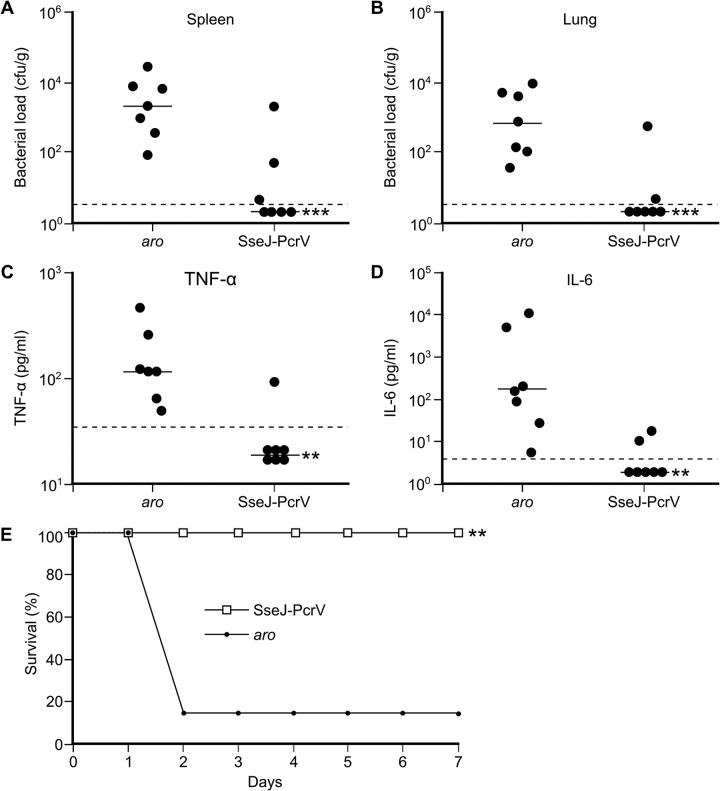
A new immunization experiment was carried out using the protective vaccine (attenuated Salmonella expressing SseJ-PcrV) or attenuated Salmonella without a pWSK29 derivative as a control. Immunized and control mice were challenged intraperitoneally with P. aeruginosa PAO1 21 days postimmunization. Twelve hours after infection, spleen and lung bacterial loads were determined (Fig. 4A and B). Vaccination with Salmonella expressing the SseJ-PcrV fusion dramatically reduced the number of P. aeruginosa in spleens and lungs. To characterize the effect of immunization on cytokine levels, sera were also collected 12 h after the infection with P. aeruginosa, and the levels of tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) were determined (Fig. 4C and D). Levels of both cytokines were significantly lower in vaccinated mice than in control mice. Finally, the survival of vaccinated mice was monitored for 7 days after challenge with P. aeruginosa (Fig. 4E). All mice vaccinated with Salmonella expressing SseJ-PcrV were protected from challenge, whereas 6 of 7 mice inoculated with control Salmonella died in less than 48 h.
FIG 4.
Effect of vaccination on bacterial loads, postinfection proinflammatory cytokine levels, and survival. C57BL/6 mice were intraperitoneally vaccinated with an aroA aroB mutant of S. Typhimurium (aro) or with the same strain carrying the plasmid pIZ2267 for expression of the fusion protein SseJ-PcrV-FLAG from the PsseA promoter. Twenty-one days after the immunization, mice were infected with P. aeruginosa strain PAO1 by intraperitoneal injection. Twelve hours postinfection, groups of 7 mice were euthanized to enumerate the spleen (A) and lung (B) bacterial loads and to determine serum levels of TNF-α (C) and IL-6 (D). Data points represent bacterial loads or cytokine levels from individual mice, and horizontal lines represent median values from groups of mice. (E) Survival of other groups of infected mice was monitored for 7 days. **, P < 0.01; ***, P < 0.001 compared to levels in control mice inoculated with Salmonella aroA aroB. Differences in bacterial loads and cytokine levels were tested using the Mann-Whitney U test, and differences in survival were analyzed by the log rank test.
DISCUSSION
This study shows that the T3SSs of Salmonella can be used as vectors for efficient delivery of P. aeruginosa antigens and elicitation of protective immunity against this opportunistic pathogen. In particular, the expression of a fusion between the Salmonella effector SseJ and the Pseudomonas antigen PcrV under the control of PsseA yielded successful results in the preclinical study shown here. Since PsseA responds to intravacuolar signals and SseJ is a specific substrate of the T3SS2, the fusion protein SseJ-PcrV is expected to be expressed by Salmonella inside the vacuole of the antigen-presenting cell and translocated from the vacuole into the host cell cytosol. Previous findings suggested that the translocation of fusion proteins results in major histocompatibility complex (MHC) class I- and MHC class II-restricted presentation and that a fusion of the model antigen listeriolysin to SseJ induced T cell responses after vaccination in mice (25). Here, we show that mice immunized with Salmonella expressing a fusion of SseJ to PcrV, a relevant P. aeruginosa antigen, produced high levels of specific anti-PcrV IgG and were able to survive an otherwise lethal infection with Pseudomonas. Protection was observed in two independent experiments shown in Fig. 3 and Fig. 4E. The differences in the results between these experiments could be due to multiple factors that influence this kind of experiment (differences in the lots of mice that were used, the exact state of the Salmonella cultures used for the vaccinations, different Pseudomonas cultures used for challenge, reactions of individual mice to intraperitoneal injections, etc.), but importantly, both experiments confirmed that this vaccine conferred statistically significant protection against Pseudomonas. The protective effect of this vaccination was confirmed because immunized mice displayed lower bacterial loads in different organs and lower serum levels of proinflammatory cytokines. PcrV was shown previously to induce protective immunity when delivered as a purified recombinant protein antigen, alone or in combination with other antigens (16, 40–43). In a recent report, mice were immunized intraperitoneally with recombinant PcrV formulated with one of three different adjuvants (43). These vaccines were effective in inducing specific immunity against PcrV and in conferring protection against a lethal dose of P. aeruginosa.
Surprisingly, a similar construct with the OprF/I antigen was unable to elicit the production of specific antibodies or to provide protection against Pseudomonas. The reasons for the superior immunogenicity of PcrV compared to that of OprF/I in our system are unclear. The recombinant hybrid protein OprF(190-342)/OprI(21-83) was shown to be immunogenic and protective in previous studies (13, 44). Glutathione transferase (GST)-OprF/I elicited specific antibodies against P. aeruginosa and OprF peptides and protected immunodeficient mice against 975-fold the 50% lethal dose of P. aeruginosa, whereas GST alone had no effect (45). A vaccine based on this hybrid protein, IC43, was evaluated in a phase II clinical trial in mechanically ventilated intensive care unit patients, although it did not produce significant differences in infection rates (11).
PcrV is an essential component of the T3SS. This system and its effectors are the major virulence determinants of P. aeruginosa (46). Thus, the T3SS is an appealing target for new therapies (47). PcrV, as part of the translocon, the most accessible part of the T3SS, has been selected for targeting by antibodies developed for clinical use (48). However, few vaccines have been described based on this antigen, presumably because of solubility problems during purification of the protein (41). The methodology employed here circumvents these problems, since the antigen is produced by Salmonella and directly translocated through the T3SS2 into the antigen-presenting cells. The use of an in vivo inducible promoter, such as PsseA, instead of a constitutive promoter is advantageous, because it enhances stable expression and immunogenicity of foreign antigens expressed by Salmonella (49). The pWSK29 vector carries the pSC101 replicon that produces 6 to 8 copies per cell (50) and is sufficiently stable for in vivo applications (25).
The aim of this study was to characterize the ability of this vaccination approach to produce an antibody response and to protect against infection in a preclinical model. Although this is an important step toward the development of an effective vaccine for the prevention of infections with P. aeruginosa, additional aspects could be optimized in future experiments carried out in this model. (i) Tolerability and toxicity studies would be necessary in order to advance this vaccine candidate. (ii) Other effectors could be tested as carriers for the PcrV antigen, taking into account that the fusion with the highest level of expression in vitro is not necessarily the most immunogenic in vivo (25). (iii) Since Salmonella is also a pathogen, strains used to develop a live vaccine should be attenuated. However, they should also be able to reach, multiply, and persist temporarily in lymphoid organs to stimulate protective immune responses. Thus, a balance between attenuation and immunogenicity is essential for the success of a live vaccine. The aro mutants of S. Typhimurium used here are auxotrophic for aromatic amino acids and several essential vitamins. These mutants are safe and immunogenic in mouse models (51), where this serovar can reach systemic organs, and have previously been used as carriers of heterologous antigens (52). Since S. Typhimurium causes only self-limited intestinal disease in immunocompetent humans and does not reach systemic sites in this host, the human-restricted S. enterica serovar Typhi is also being tested as a live vector for humans (53). A double aro mutant of S. Typhi is highly immunogenic, but additional attenuation could be desirable in humans; therefore, other means of enhancing vaccine efficacy and safety have been proposed (54). In particular, the safety of live attenuated vaccines needs to be determined in immunocompromised individuals, since they represent a relevant target for vaccination against Pseudomonas. In fact, an aro mutant may be virulent in immunodeficient mice (55), although moderate immunosuppression, causing increased susceptibility to wild-type Salmonella, does not affect resistance to an aroA Salmonella vaccine strain (56). Other attenuating mutations or combinations of mutations could be used to increase safety (55). (iv) Another aspect to be considered for future development of this vaccine is the route of immunization. The intraperitoneal injection is employed in many preclinical studies but is not typically used in the clinical setting as a route of immunization. However, mice can also be inoculated with Salmonella via the intravenous, subcutaneous, oral, intranasal, and rectal routes (57, 58), and some of these protocols should be tested in animal models before progressing into the clinical stages. It is also worth noting that the cell-mediated immune response was not evaluated in this study, an aspect that could be further characterized in future studies.
In conclusion, this study shows that the SPI2-related T3SS of Salmonella is an appropriate vehicle to deliver the Pseudomonas antigen PcrV and to generate a protective live vaccine against this opportunistic pathogen.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and strain construction.
Bacterial strains used in this study are described in Table 1. S. Typhimurium strains were derived from the wild-type strain ATCC 14028. Transductional crosses using phage P22 HT 105/1 int201 (59) were used for strain construction (60).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 67 |
| BL21(DE3) | F−- ompT gal dcm lon hsdSB(r− m−; E. coli B strain), with DE3, a λ prophage carrying the T7 RNA pol gene | Stratagene |
| Pseudomonas aeruginosa | ||
| PAO1 | Reference strain | 68 |
| Salmonella enterica serovar Typhimuriuma | ||
| 14028 | Wild type | ATCC |
| SV4338 | aroA551::Tn10 | Laboratory stock |
| SV5136 | ssaV::Cm | Laboratory stock |
| SV8462 | ΔaroB::Km | Laboratory stock |
| SV9699 | aroA551::Tn10 ΔaroB::Km | This work |
| Plasmids | ||
| pET15b | 6×His fusion vector, Apr | Novagen |
| pET15b-OprF/I | OprF/I cloned with NdeI and BamHI | This work |
| pGEX-4T-2 | GST fusion vector, Apr | GE Healthcare |
| pIZ2160 | pWSK29-PsseA-SseJ-OprF/I-FLAG | This work |
| pIZ2267 | pWSK29-PsseA-SseJ-PcrV-FLAG | This work |
| pIZ2338 | pGEX-4T-2-PcrV | This work |
| pWSK29 | Low-copy-number vector, Apr | 50 |
Derivatives of these strains were used as indicated in the text.
Bacterial culture.
The standard culture medium for all bacteria was Luria-Bertani (LB) broth. For SPI2-inducing conditions, bacteria were inoculated in LPM at pH 5.8 (61) and incubated overnight at 37°C with shaking. The following supplements were added to LPM to allow growth of aro strains: Phe, 40 μg/ml; Trp, 40 μg/ml; Tyr, 40 μg/ml; p-aminobenzoate, 10 μg/ml; 2,3-dihydroxybenzoate, 10 μg/ml. Solid medium contained 1.5% agar. Antibiotics were used at previously described concentrations (62).
DNA amplification by PCR and sequencing.
Amplification reactions were carried out in a T100 thermal cycler (Bio-Rad) using KAPA HiFi DNA polymerase (Kapa Biosystems) or MyTaq Red DNA polymerase (Bioline) according to the instructions of the suppliers. The oligonucleotides are described in Table 2. Constructs were sequenced with an automated DNA sequencer (Stab Vida, Oeiras, Portugal).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence 5′→3′ |
|---|---|
| Amplification of pcrV with FLAG tag for cloning in pWSK29 | |
| pcrVfwBam | ATCGGGATCCGAAGTCAGAAACCTTAATGC |
| pcrVFLAGrevNot | GCATGCGGCCGCCTATTTATCGTCGTCATCTTTGTAGTCGATCGCGCTGAGAATGTCGC |
| Fusion of oprF and oprI with FLAG tag | |
| oprFfwBam | ATCGGGATCCGCTCCGGCTCCGGAACCGGTTGCCGAC |
| oprFrev | TTCAACGCGACGGTTGATAGCGCG |
| oprF/oprIfw | GAAGGCCGCGCTATCAACCGTCGCGTTGAAAGCAGCCACTCCAAAGAAAC CGAAGCT |
| oprIFLAGrevNot | GCATGCGGCCGCCTATTTATCGTCGTCATCTTTGTAGTCCTTGCGGCTGGCTTTTTCCAG |
| Amplification of sseJ | |
| sseJfwEco | CCTAGAATTCGTAAGGAGGACACTATGCC |
| sseJrevBam | ACGTGGATCCTTCAGTGGAATAATGATGAG |
| Amplification of sseA promoter | |
| PsseAfwKpn | GCTAGGTACCAGAAGAGAACAACGGCAAG |
| PsseArevEco | CACTGAATTCACGATAGATAATTAACGTGC |
| Amplification of pcrV for cloning in pGEX-4T-2 | |
| pcrVfwBam | ATCGGGATCCGAAGTCAGAAACCTTAATGC |
| pcrVrevEco | ATCGGAATTCCTAGATCGCGCTGAGAATGT |
Plasmids.
The plasmids used in this study are listed in Table 1. The construction of derivatives of pWSK29 for vaccine assays was carried out in three steps (Fig. 1): (i) cloning of P. aeruginosa DNA fragments encoding antigens PcrV and OprF(190-342)/OprI(21-83) in fusion with FLAG using BamHI and NotI restriction sites; (ii) addition of S. Typhimurium DNA encoding effector SseJ using EcoRI and BamHI restriction sites; (iii) addition of PsseA using KpnI and EcoRI. The hybrid gene oprF-oprI was generated using the gene splicing by overlap extension approach (63) as previously described (45) using oligonucleotides shown in Table 2. To generate plasmid pIZ2338, the coding region of pcrV was amplified by colony PCR using lysed P. aeruginosa PAO1 as the template and cloned into pGEX-4T-2 using BamHI and EcoRI restriction sites. To generate plasmid pET15b-OprF/I, oprF-oprI was first synthetized in vitro (GenScript) and then transferred from the original vector (pUC57) into pET15b using NdeI and BamHI restriction enzymes.
Expression and purification of recombinant proteins in E. coli.
Plasmids pET15b-OprF/I, expressing 6×His-OprF/I, or pIZ2338, expressing GST-PcrV, were transformed into E. coli BL21(DE3). Bacteria were grown in LB with ampicillin at 37°C. At mid-exponential-growth phase, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce expression, and incubation was pursued for 4 h. The cells were harvested by centrifugation, and the cell pellet was resuspended in NP-40 lysis buffer (64). A commercial mixture of protease inhibitors (Sigma-Aldrich) was added to the suspension, and lysis was performed by sonication. The cell lysate was centrifuged at 15,000 × g for 30 min at 4°C, and the fusion proteins were isolated from the supernatants by affinity chromatography with Ni-nitrilotriacetic acid (NTA) agarose beads (Qiagen) or glutathione agarose beads (Sigma-Aldrich) according to the manufacturers’ protocols.
Mammalian cell culture.
RAW264.7 cells (murine macrophages, ECACC number 91062702) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and 2 mM l-glutamine. Penicillin (60 μg/ml) and streptomycin (100 μg/ml) were included in the culture medium (except for bacterial infection experiments). Cells were maintained in a 5% CO2 humidified atmosphere at 37°C.
Bacterial infection of cultured cells and protein translocation assays.
RAW264.7 cells were plated 24 h before infection in 6-well plates at 8 × 105 cells per well and incubated 24 h at 37°C with 5% CO2 in medium without antibiotics. Bacteria carrying plasmids expressing FLAG fusions were grown in LB medium with ampicillin for 24 h at 37°C with shaking, were added to the cell monolayers at a multiplicity of infection of 250 bacteria/cell, and then were incubated at 37°C with 5% CO2. The cell culture was washed twice with phosphate-buffered saline (PBS) 1 h postinfection, overlaid with DMEM containing 100 μg/ml gentamicin and ampicillin, and incubated for 1 h. The culture was then washed twice with PBS, covered with DMEM with gentamicin (16 μg/ml) and ampicillin (100 μg/ml), and incubated for 6 h. To study the translocation of FLAG fusions, infected mammalian cells were lysed with 1% Triton X-100. Cell lysates were processed for electrophoresis and Western blotting as previously described (65). Anti-DnaK (1:5,000; Assay Designs) and anti-GroEL (1:20,000; Sigma-Aldrich) antibodies were used as loading or contamination controls.
Analysis of expression of fusion proteins.
Salmonella strains carrying plasmids expressing FLAG fusions were grown overnight in LB medium at 37°C with shaking. Bacteria were then washed, diluted 1:50 in LPM, and incubated at 37°C with shaking for 8 h. The bacteria were then pelleted by centrifugation and lysed by boiling in Laemmli sample buffer. Proteins were separated by SDS-PAGE and analyzed by immunoblotting with a monoclonal antibody directed to the FLAG epitope (1:10,000; Sigma-Aldrich). Goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody (1:5,000; Bio-Rad) was used as the secondary antibody.
Mouse immunization and infection with P. aeruginosa.
Mice were maintained in the IBiS facility, and their care was in accordance with institutional guidelines. Attenuated S. Typhimurium strain SV9699 (aroA551::Tn10 ΔaroB::Km) (Table 1) with appropriate plasmids was grown overnight at 37°C with shaking in LB with ampicillin, diluted in fresh medium (1:100), and grown to an optical density at 600 nm (OD600) of 0.3 to 0.6. Vaccination was carried out in 6- to 8-week-old female C57BL/6 mice (Charles River Laboratories) by a single intraperitoneal injection with 0.2 ml of PBS containing 2 × 105 CFU of Salmonella. Mice were infected with P. aeruginosa strain PAO1 on day 21 postimmunization by intraperitoneal injection with 9 × 106 bacteria (3.6× the 50% lethal dose [LD50]) in 0.2 ml of PBS, and survival was monitored for 7 days. Mice that received the same infection were housed together with up to 5 mice per cage. Mice were monitored twice daily and culled using thiopental at the end of the experiments. The procedures (intraperitoneal injections) made the use of analgesics unnecessary. To minimize animal suffering, euthanasia using thiopental was carried out immediately when detecting severe clinical signs: hunching, labored breathing, severe weight loss, inactivity, or lethargy. Experiments involving the use of animals were approved by the Committee on Ethics and Experimentation of the Consejería de Agricultura, Pesca y Desarrollo Rural (Junta de Andalucía, Spain) (permit number 18-01-16-005) and were in accordance with the EU Directive 2010/63/EU for animal experiments.
Spleen and lung bacterial loads and serum cytokine levels.
Postinfection bacterial loads were determined in vaccinated and control mice 12 h after infection. Mice were euthanized with thiopental, and after the collection of blood samples from the retro-orbital sinus, spleens and lungs were aseptically removed, weighed, and homogenized in 2 ml of physiological saline. Serial log dilutions were plated on agar plates for bacterial quantification. Serum levels of tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) were determined in mice at 12 h postinfection using DuoSet ELISA kits (R&D Systems).
Enzyme-linked immunosorbent assays.
For indirect ELISAs, 96-well plates were coated with 100 μl per well of 1-μg/ml solutions of purified 6×His-OprF/I or GST-PcrV by incubating at 4°C overnight. ELISAs were performed using sera collected on day 21 as described previously (66). Antibody titers were measured against the OprF/I or PcrV antigens and were defined as the dilution in which spectrophotometric readings were at least 0.1 units greater than that in the background wells (wells containing no serum).
Statistical analysis.
Antibody titers, bacterial loads, and cytokine levels were compared using the Mann-Whitney U test. Survival data were compared using the log rank test. Statistics were performed using SPSS version 24.0 software (SPSS Inc.). P values of 0.05 or less were considered significant.
ACKNOWLEDGMENTS
This work was supported by Spanish Ministerio de Economía, Industria y Competitividad and the European Regional Development Fund (grant numbers SAF2013-46229-R and SAF2016-75365-R to F.R.-M.). The funding source was not involved in the research or the preparation of the article.
M.J.M. is a founder and scientific advisor for the biotechnology company Vaxdyn, S.L.
REFERENCES
- 1.National Nosocomial Infections Surveillance System. 2004. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control 32:470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzopardi EA, Azzopardi E, Camilleri L, Villapalos J, Boyce DE, Dziewulski P, Dickson WA, Whitaker IS. 2014. Gram negative wound infection in hospitalised adult burn patients-systematic review and metanalysis. PLoS One 9:e95042. doi: 10.1371/journal.pone.0095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramírez-Estrada S, Borgatta B, Rello J. 2016. Pseudomonas aeruginosa ventilator-associated pneumonia management. Infect Drug Resist 9:7–18. doi: 10.2147/IDR.S50669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lara-Isla A, Medina-Polo J, Alonso-Isa M, Benítez-Sala R, Sopeña-Sutil R, Justo-Quintas J, Gil-Moradillo J, González-Padilla DA, García-Rojo E, Passas-Martínez JB, Tejido-Sánchez Á. 2017. Urinary infections in patients with catheters in the upper urinary tract: microbiological study. Urol Int 98:442–448. doi: 10.1159/000467398. [DOI] [PubMed] [Google Scholar]
- 6.Rolston K. 2017. Infections in cancer patients with solid tumors: a review. Infect Dis Ther 6:69–83. doi: 10.1007/s40121-017-0146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moradali MF, Ghods S, Rehm B. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K, Yoon SS. 2017. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J Microbiol Biotechnol 27:1053–1064. doi: 10.4014/jmb.1611.11056. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigo-Troyano A, Sibila O. 2017. The respiratory threat posed by multidrug resistant Gram-negative bacteria. Respirology 22:1288–1299. doi: 10.1111/resp.13115. [DOI] [PubMed] [Google Scholar]
- 10.Grimwood K, Kyd JM, Owen SJ, Massa HM, Cripps AW. 2015. Vaccination against respiratory Pseudomonas aeruginosa infection. Hum Vaccin Immunother 11:14–20. doi: 10.4161/hv.34296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rello J, Krenn C-G, Locker G, Pilger E, Madl C, Balica L, Dugernier T, Laterre P-F, Spapen H, Depuydt P, Vincent J-L, Bogár L, Szabó Z, Völgyes B, Máñez R, Cakar N, Ramazanoglu A, Topeli A, Mastruzzo MA, Jasovich A, Remolif CG, Del Carmen Soria L, Andresen Hernandez MA, Ruiz Balart C, Krémer I, Molnár Z, von Sonnenburg F, Lyons A, Joannidis M, Burgmann H, Welte T, Klingler A, Hochreiter R, Westritschnig K. 2017. A randomized placebo-controlled phase II study of a Pseudomonas vaccine in ventilated ICU patients. Crit Care 21:22. doi: 10.1186/s13054-017-1601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Moigne V, Gaillard J-L, Herrmann J-L. 2016. Vaccine strategies against bacterial pathogens in cystic fibrosis patients. Med Mal Infect 46:4–9. doi: 10.1016/j.medmal.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Baumann U, Mansouri E, von Specht B-U. 2004. Recombinant OprF-OprI as a vaccine against Pseudomonas aeruginosa infections. Vaccine 22:840–847. doi: 10.1016/j.vaccine.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Lee NG, Jung SB, Ahn BY, Kim YH, Kim JJ, Kim DK, Kim IS, Yoon SM, Nam SW, Kim HS, Park WJ. 2000. Immunization of burn-patients with a Pseudomonas aeruginosa outer membrane protein vaccine elicits antibodies with protective efficacy. Vaccine 18:1952–1961. doi: 10.1016/S0264-410X(99)00479-X. [DOI] [PubMed] [Google Scholar]
- 15.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 16.Moriyama K, Wiener-Kronish JP, Sawa T. 2009. Protective effects of affinity-purified antibody and truncated vaccines against Pseudomonas aeruginosa V-antigen in neutropenic mice. Microbiol Immunol 53:587–594. doi: 10.1111/j.1348-0421.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- 17.Sawa T, Yahr TL, Ohara M, Kurahashi K, Gropper MA, Wiener-Kronish JP, Frank DW. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med 5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 18.Sharma A, Krause A, Worgall S. 2011. Recent developments for Pseudomonas vaccines. Hum Vaccin 7:999–1011. doi: 10.4161/hv.7.10.16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin IYC, Van TTH, Smooker PM. 2015. Live-attenuated bacterial vectors: tools for vaccine and therapeutic agent delivery. Vaccines 3:940–972. doi: 10.3390/vaccines3040940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheminay C, Hensel M. 2008. Rational design of Salmonella recombinant vaccines. Int J Med Microbiol 298:87–98. doi: 10.1016/j.ijmm.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol 11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella Typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galán JE, Curtiss R. 1989. Cloning and molecular characterization of genes whose products allow Salmonella Typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegazy WAH, Xu X, Metelitsa L, Hensel M. 2012. Evaluation of Salmonella enterica type III secretion system effector proteins as carriers for heterologous vaccine antigens. Infect Immun 80:1193–1202. doi: 10.1128/IAI.06056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panthel K, Meinel KM, Sevil Domènech VE, Trülzsch K, Rüssmann H. 2008. Salmonella type III-mediated heterologous antigen delivery: a versatile oral vaccination strategy to induce cellular immunity against infectious agents and tumors. Int J Med Microbiol 298:99–103. doi: 10.1016/j.ijmm.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Ramos-Morales F. 2012. Impact of Salmonella enterica type III secretion system effectors on the eukaryotic host cell. ISRN Cell Biol 2012:1–36. doi: 10.5402/2012/787934. [DOI] [Google Scholar]
- 28.Chen L-M, Briones G, Donis RO, Galán JE. 2006. Optimization of the delivery of heterologous proteins by the Salmonella enterica serovar Typhimurium type III secretion system for vaccine development. Infect Immun 74:5826–5833. doi: 10.1128/IAI.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panthel K, Meinel KM, Sevil Domènech VE, Geginat G, Linkemann K, Busch DH, Rüssmann H. 2006. Prophylactic anti-tumor immunity against a murine fibrosarcoma triggered by the Salmonella type III secretion system. Microbes Infect 8:2539–2546. doi: 10.1016/j.micinf.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Rüssmann H, Shams H, Poblete F, Fu Y, Galán JE, Donis RO. 1998. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science 281:565–568. [DOI] [PubMed] [Google Scholar]
- 31.Rüssmann H, Igwe EI, Sauer J, Hardt WD, Bubert A, Geginat G. 2001. Protection against murine listeriosis by oral vaccination with recombinant Salmonella expressing hybrid Yersinia type III proteins. J Immunol 167:357–365. doi: 10.4049/jimmunol.167.1.357. [DOI] [PubMed] [Google Scholar]
- 32.Jellbauer S, Panthel K, Hetrodt JH, Rüssmann H. 2012. CD8 T-cell induction against vascular endothelial growth factor receptor 2 by Salmonella for vaccination purposes against a murine melanoma. PLoS One 7:e34214. doi: 10.1371/journal.pone.0034214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X, Zhou P, Cai J, Yang G, Liang S, Ren D. 2010. Tumor antigen delivered by Salmonella III secretion protein fused with heat shock protein 70 induces protection and eradication against murine melanoma. Cancer Sci 101:2621–2628. doi: 10.1111/j.1349-7006.2010.01722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong G, Husseiny MI, Song L, Erdreich-Epstein A, Shackleford GM, Seeger RC, Jäckel D, Hensel M, Metelitsa LS. 2010. Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int J Cancer 126:2622–2634. doi: 10.1002/ijc.24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y-J, Hou Y, Huang H, Liu G-R, White AP, Liu S-L. 2008. Two oral HBx vaccines delivered by live attenuated Salmonella: both eliciting effective anti-tumor immunity. Cancer Lett 263:67–76. doi: 10.1016/j.canlet.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa H, Sato E, Briones G, Chen L-M, Matsuo M, Nagata Y, Ritter G, Jäger E, Nomura H, Kondo S, Tawara I, Kato T, Shiku H, Old LJ, Galán JE, Gnjatic S. 2006. In vivo antigen delivery by a Salmonella Typhimurium type III secretion system for therapeutic cancer vaccines. J Clin Invest 116:1946–1954. doi: 10.1172/JCI28045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao EA, Miller SI. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella Typhimurium. Proc Natl Acad Sci U S A 97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jantsch J, Cheminay C, Chakravortty D, Lindig T, Hein J, Hensel M. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell Microbiol 5:933–945. doi: 10.1046/j.1462-5822.2003.00334.x. [DOI] [PubMed] [Google Scholar]
- 39.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 40.Jiang M, Yao J, Feng G. 2014. Protective effect of DNA vaccine encoding Pseudomonas exotoxin A and PcrV against acute pulmonary P. aeruginosa infection. PLoS One 9:e96609. doi: 10.1371/journal.pone.0096609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, Gu J, Yang L, Gao C, Jing H, Wang Y, Zeng H, Zou Q, Lv F, Zhang J. 2017. Protective efficacy of the trivalent Pseudomonas aeruginosa vaccine candidate PcrV-OprI-Hcp1 in murine pneumonia and burn models. Sci Rep 7:3957. doi: 10.1038/s41598-017-04029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holder IA, Neely AN, Frank DW. 2001. PcrV immunization enhances survival of burned Pseudomonas aeruginosa-infected mice. Infect Immun 69:5908–5910. doi: 10.1128/IAI.69.9.5908-5910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamaoka S, Naito Y, Katoh H, Shimizu M, Kinoshita M, Akiyama K, Kainuma A, Moriyama K, Ishii KJ, Sawa T. 2017. Efficacy comparison of adjuvants in PcrV vaccine against Pseudomonas aeruginosa pneumonia. Microbiol Immunol 61:64–74. doi: 10.1111/1348-0421.12467. [DOI] [PubMed] [Google Scholar]
- 44.Priebe GP, Goldberg JB. 2014. Vaccines for Pseudomonas aeruginosa: a long and winding road. Expert Rev Vaccines 13:507–519. doi: 10.1586/14760584.2014.890053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Specht BU, Knapp B, Muth G, Bröker M, Hungerer KD, Diehl KD, Massarrat K, Seemann A, Domdey H. 1995. Protection of immunocompromised mice against lethal infection with Pseudomonas aeruginosa by active or passive immunization with recombinant P. aeruginosa outer membrane protein F and outer membrane protein I fusion proteins. Infect Immun 63:1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galle M, Carpentier I, Beyaert R. 2012. Structure and function of the type III secretion system of Pseudomonas aeruginosa. Curr Protein Pept Sci 13:831–842. doi: 10.2174/138920312804871210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anantharajah A, Mingeot-Leclercq M-P, Van Bambeke F. 2016. Targeting the type three secretion system in Pseudomonas aeruginosa. Trends Pharmacol Sci 37:734–749. doi: 10.1016/j.tips.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Warrener P, Varkey R, Bonnell JC, DiGiandomenico A, Camara M, Cook K, Peng L, Zha J, Chowdury P, Sellman B, Stover CK. 2014. A novel anti-PcrV antibody providing enhanced protection against Pseudomonas aeruginosa in multiple animal infection models. Antimicrob Agents Chemother 58:4384–4391. doi: 10.1128/AAC.02643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunstan SJ, Simmons CP, Strugnell RA. 2003. In vitro and in vivo stability of recombinant plasmids in a vaccine strain of Salmonella enterica var. Typhimurium. FEMS Immunol Med Microbiol 37:111–119. doi: 10.1016/S0928-8244(03)00065-8. [DOI] [PubMed] [Google Scholar]
- 50.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 51.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 52.Bachtiar EW, Sheng K-C, Fifis T, Gamvrellis A, Plebanski M, Coloe PJ, Smooker PM. 2003. Delivery of a heterologous antigen by a registered Salmonella vaccine (STM1). FEMS Microbiol Lett 227:211–217. doi: 10.1016/S0378-1097(03)00683-9. [DOI] [PubMed] [Google Scholar]
- 53.Galen JE, Pasetti MF, Tennant S, Ruiz-Olvera P, Sztein MB, Levine MM. 2009. Salmonella enterica serovar Typhi live vector vaccines finally come of age. Immunol Cell Biol 87:400–412. doi: 10.1038/icb.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Kong Q, Curtiss R. 2013. New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb Pathog 58:17–28. doi: 10.1016/j.micpath.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanCott JL, Chatfield SN, Roberts M, Hone DM, Hohmann EL, Pascual DW, Yamamoto M, Kiyono H, McGhee JR. 1998. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med 4:1247–1252. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- 56.Izhar M, DeSilva L, Joysey HS, Hormaeche CE. 1990. Moderate immunodeficiency does not increase susceptibility to Salmonella Typhimurium aroA live vaccines in mice. Infect Immun 58:2258–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon R, Tennant SM, Galen JE, Levine MM. 2011. Mouse models to assess the efficacy of non-typhoidal Salmonella vaccines: revisiting the role of host innate susceptibility and routes of challenge. Vaccine 29:5094–5106. doi: 10.1016/j.vaccine.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spreng S, Dietrich G, Weidinger G. 2006. Rational design of Salmonella-based vaccination strategies. Methods 38:133–143. doi: 10.1016/j.ymeth.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 59.Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet 119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 60.Maloy SR. 1990. Experimental techniques in bacterial genetics. Jones and Bartlett, Boston, MA. [Google Scholar]
- 61.Baisón-Olmo F, Galindo-Moreno M, Ramos-Morales F. 2015. Host cell type-dependent translocation and PhoP-mediated positive regulation of the effector SseK1 of Salmonella enterica. Front Microbiol 6:396. doi: 10.3389/fmicb.2015.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cordero-Alba M, Ramos-Morales F. 2014. Patterns of expression and translocation of the ubiquitin ligase SlrP in Salmonella enterica serovar Typhimurium. J Bacteriol 196:3912–3922. doi: 10.1128/JB.02158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 64.Bernal-Bayard J, Cardenal-Muñoz E, Ramos-Morales F. 2010. The Salmonella type III secretion effector, Salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J Biol Chem 285:16360–16368. doi: 10.1074/jbc.M110.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardenal-Muñoz E, Ramos-Morales F. 2011. Analysis of the expression, secretion and translocation of the Salmonella enterica type III secretion system effector SteA. PLoS One 6:e26930. doi: 10.1371/journal.pone.0026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McConnell MJ, Hanna PC, Imperiale MJ. 2006. Cytokine response and survival of mice immunized with an adenovirus expressing Bacillus anthracis protective antigen domain 4. Infect Immun 74:1009–1015. doi: 10.1128/IAI.74.2.1009-1015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 68.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]