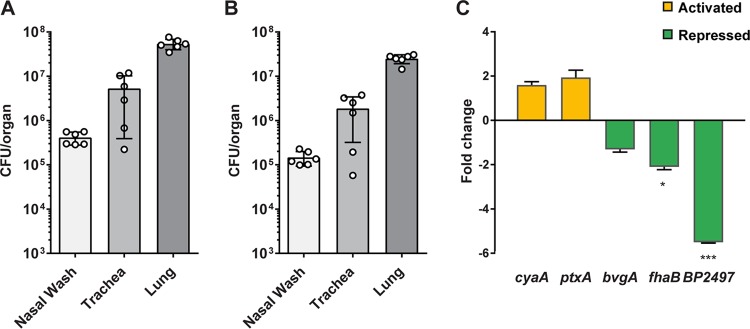
FIG 2.
Isolation of B. pertussis and qRT-PCR analysis from infected NSG mouse lungs. NSG mice were infected by intranasal administration of B. pertussis strain UT25. At 3 days postinfection, the nasal wash, trachea, and lung were collected, and the numbers of input and output bacteria (postfiltration) were quantified. (A) Bacterial load in each tissue after homogenization. (B) Number of bacteria recovered from each tissue after implementing the filtration strategy. (C) qRT-PCR analysis was performed on B. pertussis strain UT25 grown in SSM broth and compared to RNA from the B. pertussis isolated from NSG lungs by the filtration protocol. Fold change in gene expression levels between in vivo and in vitro growth conditions is shown. Data are from three biological replicates with three technical replicates each. Standard deviations were calculated from variations between biological replicates and compared using a t test (*, P < 0.05; ***, P < 0.001).