Abstract
Hypobaric hypoxia (HH) is a major stress factor that is associated with physiological, biochemical, molecular and genomic alterations. Brain is the organ that reacts sensitively to oxygen deprivation, which leads to oxidative stress and cognitive function impairment. Our previous studies have reported that downregulation of brain derived neurotrophic factor (BDNF) leads to neurodegeneration and memory impairment. The aim of the present study was to investigate the effect of HH exposure on DNA methylation and its regulation in BDNF expression, neurodegeneration and spatial memory impairment. For this purpose, Sprague Dawley rats were exposed to HH at a simulated altitude of 25,000 feet for 14 days. Real-time polymerase chain reaction was used for transcriptional expression of DNA Methyltransferases (DNMTs) including DNMT1, DNMT3a and DNMT3b, and immunoblotting was used for the translational expression of DNMT1, DNMT3a, DNMT3b, Methyl CpG binding protein 2 (MeCP2), pMeCP2 and BDNF in rat hippocampus. Additionally, neuronal morphology alteration and neurodegeneration in CA1 region of hippocampus were investigated though Cresyl violet (CV) staining and Fluoro-Jade C staining respectively. Results obtained suggested that HH exposure increased the expression of DNMT1 DNMT3b at the mRNA as well as protein level, whereas no significant change was observed in the level of DNMT3a. Furthermore, the level of pMeCP2 and BDNF were significantly decreased; however, the expression level of MeCP2 was significantly increased. The CV and Fluoro-Jade C-positive cells were significantly enhanced in the CA1 region of hippocampus in the HH exposed group as compared to unexposed rats. Thus, the present study concluded that HH decreases neuronal activation by the upregulation of DNA methylation and MeCP2 and decreased the expression of pMeCP2, which result in the downregulation of BDNF. The decreased BDNF expression is associated with neuronal loss and spatial memory impairment. This study highlights that DNMT inhibition could be an important therapeutic target for neurodegenerative diseases.
Key Words: Hypobaric hypoxia, DNA methyltransferase, Methyl CpG binding protein 2, Brain-derived neurotrophic factor, Neurodegeneration
Introduction
Hypobaric hypoxia (HH) has been associated with alterations in neurophysiological functions. Exposure to HH leads to sleep disturbance [1], oxidative stress [2] and alteration in acetylcholine neurotransmitter [3]. HH exposure causes cognitive function impairment including learning and memory function, neurodegeneration in CA3 region of hippocampus, glutamate excitotoxicity and high influx of calcium-mediated apoptosis cascade [4]. It also decreases the neurotrophic factor brain derived neurotrophic factor (BDNF), which helps in neuroprotection [21]. It is still unclear how HH induces neurodegeneration and downregulates the BDNF expression that causes memory impairment. Epigenetic changes could be an important aspect in delineating the mechanism behind HH-induced memory impairment.
Recent data suggest that epigenetic modifications in brain play a principle role in the neuropathogenesis of psychiatric diseases [5], neurodevelopmental [6] and neurodegenerative diseases [7]. The mechanism of epigenetic regulation comprise of DNA methylation, histone modification, nucleosome remodelling and RNA-mediated targeting [8]. DNA methylation is the main epigenetic modification, that leads to environment-induced, stable and heritable changes in gene expression. DNA methylation is the covalent addition of methyl group catalysed by DNA Methyltransferase (DNMTs) at cytosine of specially CpG region of DNA that leads to transcriptional inactivation [9, 10]. There are different families of DNMTs that have been identified as DNMT 1, 3a and 3b. The functions of different DNMTs vary with different developmental stages [11]. DNA methylation leads to transcriptional repression by the binding of methyl CpG binding protein 2 (MeCP2) and methyl CpG binding domain proteins on the methylated DNA. Both the methyl CpG binding domain and MeCP2 proteins provide a binding site for histone deacetylase (HDAC) at the CpG dinucleotide and lead to the transcriptional repression of the target gene. However, histone acetylation and deacetylation, which are another type of epigenetic modification, catalysed through histone acetyl transferase and deacetyl transferase (HDAC) respectively [12, 13].
A previous study reported that reduced levels of methyl transferase in the brain protected it from ischemic injury [14]. Furthermore, another research concluded that DNA methylation plays an important role in the mechanism of memory formation and its storage [15, 16]; it has also suggested that DNMT inhibitor altered the methylation label on brain plasticity-related genes such as BDNF and reelin. Another study has reported that upregulation of DNA methylation led to contextual fear conditioning and inhibition of DNMT led to the blockage of contextual fear conditioning [17, 18]. In a study Kishi et al.[19] reported that MeCP2 is a transcription factor that repressed the expression of targeted genes involved in maturation and function of neurons [20].
Additionally, activity-dependent changes in gene expression occurs due to DNA methylation - for example, changes in BDNF expression due to the phosphorylation of the transcriptional repression complex including MeCP2, CpG and HDAC1 led to the detachment from the BDNF promoter region in addition to the demethylation of CpG of the BDNF gene result in transcriptional activation. BDNF is a main moderator of the activity-dependent process in brain and is involved in the regulation of neuronal development and plasticity. Another study has suggested that MeCP2 was found to bind at the promoter region of transcription variant III of BDNF and detached due to MeCP2 phosphorylation that result in transcriptional activation and increased expression [21]. There are several in vivo as well as in vitro studies that reported the increased phosphorylation of MeCP2, which was induced by the neuronal activity in neurons, although, the loss of the MeCP2 phosphorylation associated with the loss of a number of traits of brain functions [22, 23, 24, 25, 26, 27, 28]).
There are very few studies on epigenetic changes associated with learning and memory under HH. Thus, in the present study, we evaluated the mRNA as well protein levels of different DNMT1, DNMT3a and DNMT3b enzymes in the hippocampus of HH-exposed and -unexposed rats. Our study concluded that HH declines neuronal activation by the upregulation of DNA methylation and MeCP2 and decreased the expression of pMeCP2, which resulted in the downregulation of BDNF. The decreased BDNF expression is associated with neuronal loss and spatial memory impairment.
Material and Methods
Animal
Adult male Sprague Dawley (SD) rats weighing 220 ± 10 g were obtained from the animal house of the Institute. All animals were housed in the control environmental condition with a temperature of 23 ± 2°C and humidity 50 ± 10%. Rats were supplied food and water available ad libitum and the animal room was maintained at a 12 h/12 h light/dark cycle.
All experiments were performed according to the guidelines of the Committee for the purpose of control and supervision of experiments on animals, Government of India. All experimental protocols were approved by the Institutional Ethical Committee for Animal Care and Use.
HH Exposure
Animals were exposed to chronic HH at 25,000 ft (~7,600 m) in an animal decompression chamber by reducing the ambient barometric pressure. The temperature and humidity of the chamber were maintained at 25 ± 2°C and 55 ± 5% respectively. Rats were continuously exposed to HH for 14 days (14 DHH) with 10–15 min interval per day for the supply of water, food and changing of cage housing.
Morris Water Maze
Morris water maze (MWM) was used for the estimation of spatial memory impairment. MWM was performed according to the study published by Jain et al. [29], with some modification. MWM was composed of a circular pool with height 0.5 m and diameter 1.6 m filled with water. The temperature of water was maintained at 22–24°C. The circular pool was divided into four quadrants, designated as Zone 1, 2, 3, and 4. A hidden platform was placed in Zone 4 known as the platform zone and the platform remained 1 cm below water. The behaviour and performance of rats were monitored and recorded by a digital camera. The software of this camera was installed in the monitor with ANY Maze software. During the 7 acquisition training periods, rats were placed in water at midpoint of each quadrant. Rats were freely allowed to swim for 60 s or until they found and climbed on to a hidden platform. If rats were unable to find the hidden platform within 60 s, then it was guided to locate the platform and placed 10 s on platform. When the performance of all rats became equal, they were subjected for probe trial and memory test before HH exposure and after HH exposure for memory retention.
Gene Expression Studies
RNA Isolation
Total RNA were extracted from snap-freezed hippocampus of the unexposed and hypoxia-exposed rats by the TRIzol regent (Sigma) method. RNA quantification was done by Nano drop (Thermo-fisher) and integrity performed by the running of 2 µL RNA on 1% agarose gel. Single stranded cDNA synthesis was performed according to instructions given by the verso cDNA synthesis kit (AB-1453/A). The compatibility of cDNA synthesis was estimated by using the housekeeping gene as GAPDH.
Real-Time Polymerase Chain Reaction
Real time polymerase chain reaction (PCR) was used for the quantification of mRNA levels of DNMT1, DNMT3a and DNMT3b genes. The fold change of these genes was estimated by the use of SYBR green master mix (Thermofisher). The forward and reverse primer of specific genes such as DNMT1 F-3-CTGCGGACCCTGGATGTGTT-5, R-3-GGCTGCTGGTTCCCACATCT-5, DNMT3a F-3-CATCCGCCACCTCTTCGCTC-5, R-3-CTCTCCGTCCTCTCGTTCTTGG-5 and DNMT3b F-3-GTCATCCGCCACCTCTTCGCTC-5, R-3-CTTCTCTCCGTCCTCTCGTTCTTGG-5 was used for the quantification of cDNA expression in hippocampus. GAPDH gene quantification was used as internal control and normalization of the targeted genes expression. The relative quantification (fold change) of mRNA was estimated by the use of the ∆∆CT method.
Immunoblotting
Protein Extraction
Rats were anaesthetized with Xylazine (10 mL/kg body weight) and Ketamine (50 mL/kg body weight) by intraperitoneal (i.p.) injection and decapitated. Immediately whole brains were removed and the hippocampus regions were taken out. They were then snap freezed in liquid nitrogen and stored at −80°C until use. The hippocampus was homogenized in 1.5 mL micro tube containing the RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1% Sodium Dodecyl Sulfate (SDS)). A protease inhibitor cocktail (Sigma Aldrich) and a phosphatase inhibitor cocktail (Sigma Aldrich) were added before use. The sample was homogenized by a hand homogenizer followed by centrifugation and collection of supernatant. Protein concentrations of whole tissue homogenates were estimated by the Bradford method.
SDS Page for Immunoblotting
Protein levels in hippocampus were analysed through western blotting and analyses were performed according to previously described methods [30, 31]. An equal amount of protein extract was loaded on the SDS page gel that led to the separation of proteins according to their molecular weight. These separated proteins were transferred on to the nitrocellulose membrane followed by 2-h blocking in 3% BSA, which was prepared in 0.1% PBST (tween 20). After completion of blocking, the membranes were washed with 0.1% PBST and incubated with primary antibody anti DNMT1, DNMT3a and DNMT 3b, GAPDH MeCP2, BDNF and pMeCP2for overnight at 4°C. After the completion of primary antibody incubation, the membranes were washed thrice with 0.1% PBST followed by incubation in a horseradish peroxidase-linked secondary antibody (1: 5000, Millipore) for 2 h at room temperature. The immunoblots were developed with chemiluminescence reagents (Enhanced chemiluminescence (ECL); Sigma Aldrich) and image captured with MultiDoc-ItTM Imaging System. The band size of appropriated protein was identified by the use of prestained protein ladder (Thermo Scientific). The concentration of the identified protein was quantified through densitometry of immune blots by using Image J analysis software.
Histological Studies
Cresyl Violet Staining
Rats were anaesthsised with Xylazine and Ketamine and then perfused transcardially with chilled 1X PBS (pH = 7.4) and 4% para formaldehyde (pH = 7.4). After perfusion, the whole brain was stored in the same 4% para formaldehyde for 3–5 days. The brain was dehydrated in the series of 10, 20 and 30% sucrose dilution. Serial coronal sectioning of 10 and 30 μm was performed using cryostat. Coronal sections were taken on gelatin-coated slides and washed with distilled water followed by the incubation in Cresyl-Violet (CV) staining for 10 min. After the completion of incubation, slides were rinsed with water followed by dehydration in graded series of alcohol, that is, 50%, 70%, 90% and absolute alcohol. After dehydration, slides were incubated in Xylazine solution for 2 min and mounted on DPX mounting media.
Fluoro-Jade C Staining
Apoptotic neurons were estimated by the use of Fluoro-Jade C staining according to the previously described method [32] with some modifications. The procedure for cryo-sectioning was used similarly as described in Cresyl violet staining. Sections were immersed in 1% sodium hydroxide solution and then washed with 70% ethanol followed by distilled water washing. After washing, sections were incubated in 0.06% potassium permanganate solution for 20 min and rinsed twice with distilled water. Then sections were incubated with 0.0001% Fluoro-Jade C (Millipore, USA) staining for 20 min followed by 3 times distilled water rinsing. After washing, sections were dried on slides and incubated with Xylene for 2 min and mounted with DPX (Millipore, USA). The Fluoro-Jade C positive cells were manually counted in the CA1 region of HH exposed and control rats.
Statistical Analysis
Data was statistically assessed by the unpaired Student t test using graph Pad Prism software version 5. The value were represented as mean ± SEM of 4 animals for real-time PCR and western blotting and 6 animals per group for histological and behavioural study. The level of significance was set at p < 0.05.
Results
Effect of HH Exposure on Spatial Memory Impairment
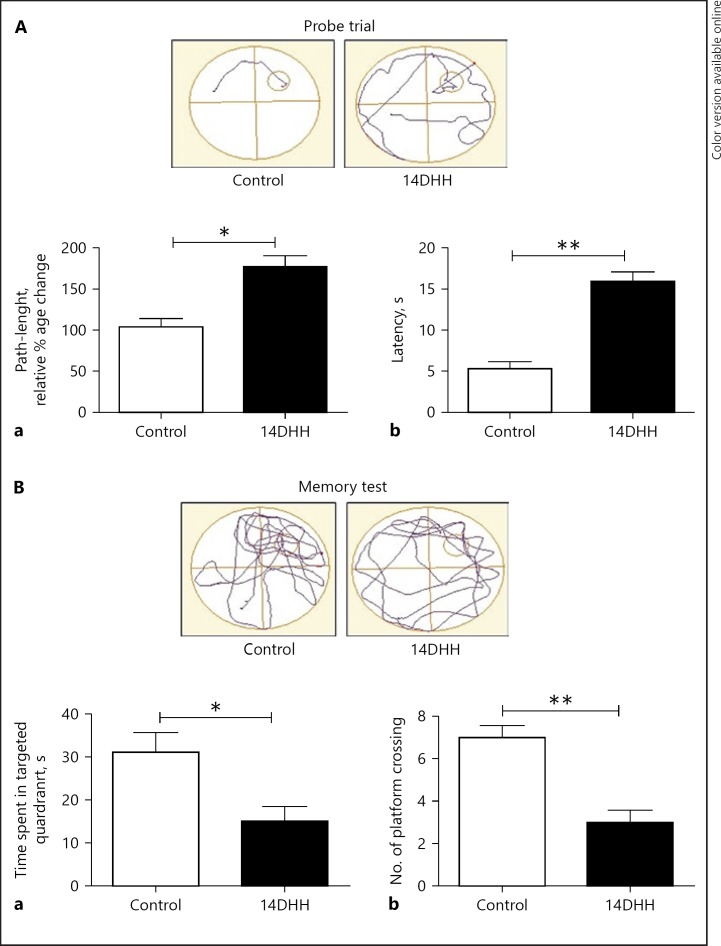
To explore the potential role of HH exposure on spatial memory, impairment was evaluated using MWM. After completion of 7 days training, probe trial and memory test were performed before and post HH exposure. A significant spatial memory impairment was observed in HH-exposed rats as compared to control (unexposed) rats. The probe trial showed that HH exposure resulted in a significant increase in path-length (Fig. 1Aa; p < 0.05) and latency (Fig. 1Ab; p < 0.01) as compared to unexposed rats. Furthermore, memory test after HH exposure showed that rats spent significantly less time in the targeted zone (Fig. 1Ba; p < 0.05) and number of platform entries (Fig. 1Bb; p < 0.01) as compared to control group.
Fig. 1.
Effect of HH exposure on spatial reference memory impairment. HH-exposed spatial memory impairment was estimated through Morris water maze (MWM). A, B Representative traces plot of MWM for probe trial and memory test respectively. The bar diagram (Aa, Ab) represent a significant increase in path-length (* p < 0.05) and latency (*** p < 0.01) in HH-exposed rats as compared to normoxia rats (control) respectively. The bar diagram of (Ba, Bb) represent a significant decrease in the time spent (* p < 0.05) and the number of crossing (** p < 0.01) in the targeted zone as compared to Normoxia rats (control) respectively. Data represents mean ± SEM of 4 independent experiments. Student t test was used to calculate significance.
Effect of HH Exposure on DNMT1, DNMT3a and DNMT3b mRNA Expression and Protein Level in Rat Hippocampus
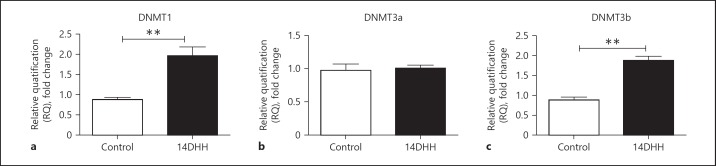
The mRNA expression of different DNMTs including DNMT1, DNMT3a and DNMT3b in rat's hippocampus was quantified through RT PCR. The bar diagrams in Figure 2a–c represent the fold change at the mRNA level of DNMT1, DNMT3a and DNMT3b in the hippocampus. HH exposure significantly increased the mRNA level of DNMT1 (Fig. 2a; p < 0.01) and DNMT3b (Fig. 2c; p < 0.01) as compared to control (unexposed) rats. However, there was no significant change observed in DNMT3a mRNA (Fig. 2b) in HH-exposed rats as compared to control (unexposed) rats.
Fig. 2.
The effect of HH on the mRNA level of different DNA Methyltransferases (DNMTs) in a rat's hippocampus. The fold change of different DNMTs as DNMT1, DNMT3a and DNMT3b was quantified through real-time PCR in both normoxia and HH-exposed rat's hippocampus. Fold changes of DNMT1, DNMT3a and DNMT3B have been shown through bar diagram as (a–c) respectively. Fold change in DNMT1 and DNMT3b was increased significantly (p < 0.01) in HH-exposed rats as compared to normoxia rats (Control). Data represents mean ± SEM of 4 independent experiments. Student t test was used to calculate significance; ** p value < 0.01.
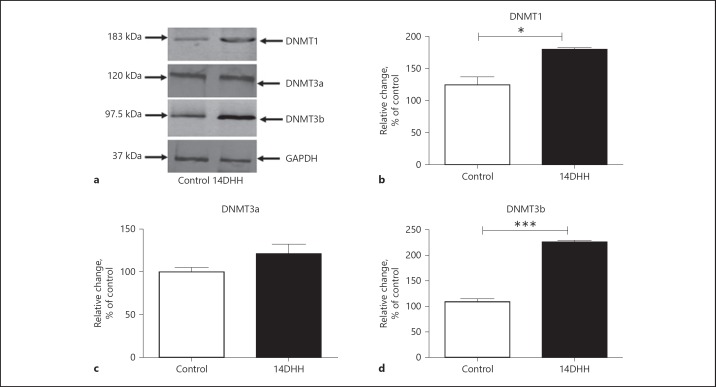
Furthermore, the protein levels of DNMTs (DNMT1, DNMT3a and DNMT3b) in hippocampus were quantified through western blotting using hippocampus lysates. The band of different DNMTs at appropriate molecular weight on nitrocellulose membrane is shown in Figure 3a. The expressions of DNMT1, DNMT3a and DNMT3b are represented in Figure 3a–c respectively. HH exposure resulted in a significant increase in levels of DNMT1 (Fig. 3b; p < 0.05) and DNMT3b (Fig. 3d; p < 0.001) as compared to control (unexposed) rats, although no significant changes were found in the level of DNMT3a (Fig. 3c) in HH-exposed rats as compared to control (unexposed) rats.
Fig. 3.
HH exposure increased the protein level of DNA Methyltransferases (DNMTs) in hippocampus of rats. The levels of DNMTs including DNMT1, DNMT3a and DNMT3b were estimated through western blotting shown in (a). The alterations in the level of DNMT1 (b) and DNMT3b (d) were significantly increased (* p < 0.05, *** p < 0.001) in HH-exposed rats as compared to normoxia rats (Control), although no significant change was observed in DNMT3a (c). Data represents mean ± SEM of 4 independent experiments. Student t test was used to calculate significance.
Effect of HH Exposure on MeCP2 and Phosphorylation of MeCP2 at Ser. 421 in Rats Hippocampus
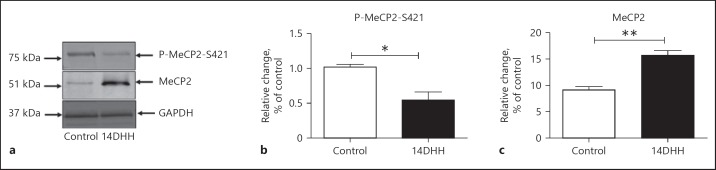
The expression level of total MeCP2 and specific phosphorylation of MeCP2 at Ser. 421 regulates the expression of BDNF, as we found that a band value of about 75 and 51 kDa (Fig. 4a) of pMeCP2 and MeCP2, respectively, was obtained on the nitrocellulose membrane. The densitometry quantification of pMeCP2 and MeCP2 showed significant alterations in the level of MeCP2 and pMeCP2 in HH exposed, and control rats were analysed through Student t test (unpaired test) using GraphPad Prism software. The levels of pMeCP2 and MeCP2 are shown in Figure 4b and c respectively. The level of MeCP2 was significantly increased (Fig. 4c; p < 0.01) in the HH exposed group as compared to control (unexposed) rats. Moreover, phosphorylation of MeCP2 was significantly decreased (Fig. 4b; p < 0.05) in the HH-exposed group when compared with control rats.
Fig. 4.
HH exposure alters the level of MeCP2 and MeCP2 phosphorylation in hippocampus. Alteration in the level of MeCP2 and MeCP2 phosphorylation at position Ser. 421 was analysed through western blotting in both HH-exposed and normal rats. An equal amount of proteins (in both control and HH exposed rats) was loaded for both MeCP2 and pMeCP2 (a). A significant decrease in MeCP2 phosphorylation (b; * p < 0.05) and increase in the level of MeCP2 (c; ** p < 0.01) were observed in the HH-exposed group as compared to control rats. Data represents mean ± SEM of 4 independent experiments. Student t test was used to calculate significance. MeCP2, methyl CpG binding protein 2.
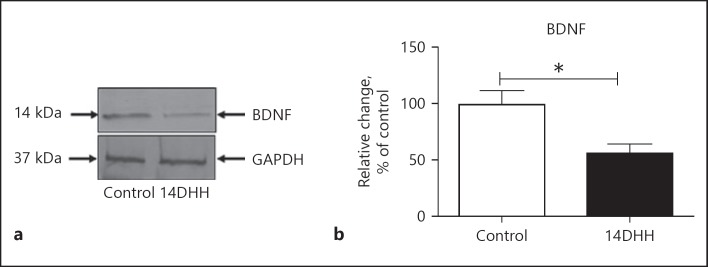
Effect of HH Exposure on the Expression of BDNF
The expression of BDNF protein was analysed through western blotting in hippocampus. Fig. 5a showed BDNF protein expression in respective groups. The densitometry analysis showed that HH exposure significantly decreased (Fig. 5b; p < 0.05) the level of BDNF as compared to unexposed rats.
Fig. 5.
HH exposure leads to alteration in the BDNF level in hippocampus. Change in the level of BDNF expression was quantified through western blotting in both normoxia (control) and HH-exposed rats. The immunoblotting of BDNF obtained on Nitrocellulose membrane (a) at corresponding molecular weight revealed a significant decrease in the level of BDNF expression in the HH-exposed group of rats (b; * p < 0.05) as compared to normoxia rats (Control). Data represents mean ± SEM of 4 independent experiments. Student t test was used to calculate significance. BDNF, brain derived neurotrophic factor.
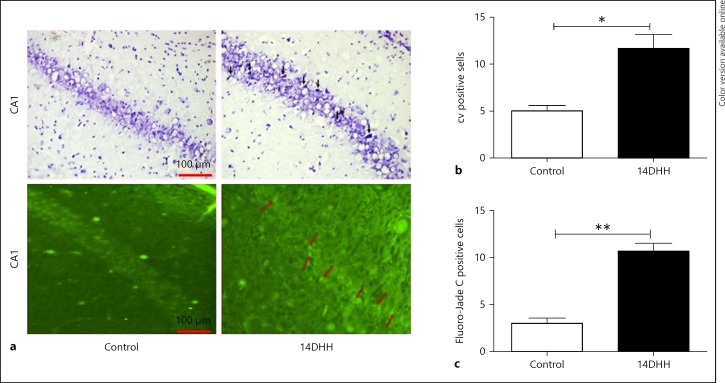
HH Induces Neuronal Morphology Alteration and Neurodegeneration in the CA1 Region of Hippocampus
Neuronal morphology alteration is the primary step of apoptotic neurons and loss of neurons associated with memory impairment. CV positive cells and Fluoro-Jade C positive cells in CA1 region had been shown in Figure 6a, as we found that HH exposure was associated with neuronal morphology alteration and neurodegeneration in CA1 region. The bar diagram shown in Figure 6b and c indicates that HH exposure significantly increased the CV-positive (p < 0.05) and Fluoro-Jade C-positive (p < 0.01) neurons, respectively, in the CA1 region of hippocampus as compared to control (unexposed) rats.
Fig. 6.
HH induced neuronal morphology and apoptosis in the CA1 region of hippocampus. Neuronal morphology alteration and apoptotic neurons were examined through CV staining and Fluoro-Jade C staining, respectively, as shown in (a). A significant increase in the number of CV positive neurons (b; * p < 0.05) and Fluoro-Jade C positive neurons (c; ** p < 0.01) were observed in the HH-exposed group as compared to Normoxia rats (Control). Data represents mean ± SEM of 6 independent experiments. Student t test was used to calculate significance. HH, hypobaric hypoxia; CV, crystal violet.
Discussions
The effect of HH on the expression of DNMTs and regulation of MeCP2 in BDNF expression is still an enigma. The present study investigated the possible impact of HH exposure on the level of different DNMTs (DNMT1, DNMT3a and DNMT3b) at the level of mRNA and protein and its involvement in alterations in spatial memory. Further, studies have been carried out to find the regulation of MeCP2 phosphorylation through neuronal activation, which regulate the expression of BDNF. BDNF is a neurotrophic factor that works as neuroprotective [33], regulate synaptic plasticity [34] and memory formation. There was an increase in the level of different DNMTs (DNMT1 and DNMT3b) at the mRNA as well as protein level in the hippocampus. HH decreases the neuronal activation that may have decreased the phosphorylation of MeCP2 at Ser. 421 (pMeCP2-S421). Phosphorylation of MeCP2 regulates the activity-dependent expression of BDNF, and decrease in the expression of BDNF could be associated with neurodegeneration and spatial memory impairment under HH exposure.
In our results, we have demonstrated that HH increases the levels of DNMT1 and DNMT3b at both mRNA and protein levels in the hippocampus, whereas there were no significance changes observed in DNMT3a. Our findings are supported by Zhang et al. [35] who reported that hypoxia induces the DNA methylation by increasing the expression of DNMTs in mice hippocampus both at the mRNA and protein levels. DNMT1 predominantly maintained DNA methylation after replication and favourably distinguished hemi methylated DNA [36]. Liu et al. [37] also reported that DNA methylation played an important role in DNA damage and repair under hypoxia/reperfusion-induced brain injuries. So, DNMT1 may play an important role in the methylation of newly incorporated cytosine after repairing [38]. The mechanism behind the upregulation of DNMT1 and DNMT3b under hypoxia is still not clear. A previous report demonstrated that the expression of DNMT1 and DNMT3b was upregulated by hypoxia-induced factor 1α (HIF1α) through the binding of HIF1α at hypoxia responsive element site present at the promoter region of DNMT1 and DNMT3b. The regulation of DNMT1 and DNMT3b through HIF1α provides adaptation to the cell against hypoxia. HH increases the expression of HIF1α [39], which may induce the expression of DNMT1 and DNMT3b. Changes in the expression of DNMT1 and DNMT3b are associated with the alteration of DNA methylation at the CpG region of the targeted genes. Increase in DNMTs activity leads to hypermethylation at the promoter of BDNF exon I and IV that result in the decrease of the BDNF mRNA level.
Additionally, we have found out the consequence of DNA methylation on spatial learning and memory in the rat model. During the probe trial, both the path-length and latency showed an increase after HH exposure. Furthermore, in memory recognition, the task time spent in the targeted zone and number of crossing in the targeted zone were decreased due to HH exposure. There may be a possible link between DNA hypermethylation and spatial memory impairment under HH. Our results are in agreement with those of Feng et al. [40] where they have also reported that DNMT1 and DNMT3a play an important role in synaptic plasticity and memory formation.
Next, we found out the possible mechanism that regulates the expression of BDNF through MeCP2 and pMeCP2 in hippocampus. MeCP2 is a main transcriptional repressor protein, unfavourably involved in the maturation of nervous system and also involved in the learning and memory formation [41]. MeCP2 controls the expression of genes through the regulation of chromatin structure and alters the interaction of protein at the promoter site of targeted gene i.e. BDNF etc. There was a significant increase in the level of MeCP2 in HH-exposed rats as compared to the unexposed group, which may be due to an increase in DNA methylation. Overexpression of MeCP2 has been associated with transcriptional inactivation and it plays an important role in neurodegeneration [42]. Additionally, our results revealed that HH-induced neuronal morphology alteration and neurodegeneration as quantified through CV and Fluoro-Jade C, respectively, in the CA1 region of hippocampus as compared to control rats. Similar findings are also reported by Dastidar et al. [43]. Furthermore, the study had been extended with these initial outcomes and it explored the possible role of MeCP2 and pMeCP2 in the neurodegeneration as well as regulation of BDNF expression. BDNF genes act as neuroprotective and involve in fear memory [44, 45] and spatial memory [46]. NMDA receptor antagonist MK801 leads to demethylation of BDNF exon III and IV promoter in hippocampus [47]. Next, we found the level of phosphorylation of MeCP2 at Ser. 421 (P-MeCP2-S-421) and BDNF in hippocampus of HH exposed and unexposed rats. Neuronal activation has been associated with the phosphorylation of MeCP2 at Ser. 421 (PMeCP2). The phosphorylation of MeCP2 at the promoter site of BDNF gene regulate different processes like neuronal morphology alteration, synapse formation and synaptic plasticity [20, 22, 23, 27]. Demethylation of promoter IV of BDNF resulted in an increased mRNA level of BDNF and phosphorylated MeCP2 [23, 48]. Interestingly, we found the decreased expression of P-MeCP2-S-421, and BDNF in HH-exposed rats as compared to control rats. Neuronal activity dependent increase in MeCP2 phosphorylation at ser 421 leads to removal of MeCP2 from BDNF promoter site which further may attribute to increased BDNF expression [21]. A previous reports suggested that the level of P-MeCP2-S-421 is directly correlated with the expression of BDNF [20, 49]. So, in the summary, HH induces DNA methylation that leads to the downregulation of BDNF expression, which may results in neurodegeneration and spatial memory impairment.
Conclusion
HH increases the expression of DNMT1 and DNMT3b in hippocampus at the mRNA and protein levels. An increase in the expression of DNMTs is associated with hypermethylation that leads to the transcriptional inactivation of the targeted genes by the binding of MePC2 at the hypermethylated site. Increase in DNA methylation is associated with neuronal damage that leads to neuronal morphology alteration and neurodegeneration in CA1 region of hippocampus. HH exposure decreased neuronal activation that is associated with a decrease in the phosphorylation of MeCP2 at Ser. 421 at the promotor of BDNF. This resulted in a decreased expression of BDNF, which worked as a neuroprotective and involved in long-term memory (LTM) formation. Therefore, a decrease in the expression of BDNF may lead to the neurodegeneration and spatial memory impairment under HH exposure. Consequently, MeCP2 and BDNF signaling pathway play an important role in the impairment of hippocampus functions [21].
Disclosure Statement
The authors have declared that there are no conflicts of interest to disclose.
Acknowledgements
This study was financially supported by the Defence Research and Development Organization (DRDO), Ministry of Defence, Govt. of India. RK thanks University Grant Commission of India for financial support in the form of JRF and SRF.
References
- 1.Saugy JJ, Schmitt L, Fallet S, Faiss R, Vesin JM, Bertschi M, Heinzer R, Millet GP. Sleep disordered breathing during live high-train low in normobaric versus hypobaric hypoxia. High Alt Med Biol. 2016;17:233–238. doi: 10.1089/ham.2016.0049. [DOI] [PubMed] [Google Scholar]
- 2.Jain V, Baitharu I, Barhwal K, Prasad D, Singh SB, Ilavazhagan G. Enriched environment prevents hypobaric hypoxia induced neurodegeneration and is independent of antioxidant signaling. Cell Mol Neurobiol. 2012;32:599–611. doi: 10.1007/s10571-012-9807-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muthuraju S, Maiti P, Pati S, Solanki P, Sharma AK, Singh SB, Prasad D, Ilavazhagan G. Role of cholinergic markers on memory function of rats exposed to hypobaric hypoxia. Eur J Pharmacol. 2011;672:96–105. doi: 10.1016/j.ejphar.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 4.Hota SK, Barhwal K, Ray K, Singh SB, Ilavazhagan G. Ceftriaxone rescues hippocampal neurons from excitotoxicity and enhances memory retrieval in chronic hypobaric hypoxia. Neurobiol Learn Mem. 2008;89:522–532. doi: 10.1016/j.nlm.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Gulchina Y, Xu SJ, Snyder MA, Elefant F, Gao WJ. Epigenetic mechanisms underlying NMDA receptor hypofunction in the prefrontal cortex of juvenile animals in the MAM model for schizophrenia. J Neurochem. 2017;143:320–333. doi: 10.1111/jnc.14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiers H, Hannon E, Schalkwyk LC, Bray NJ, Mill J. 5-hydroxymethylcytosine is highly dynamic across human fetal brain development. BMC Genomics. 2017;18:738. doi: 10.1186/s12864-017-4091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griñán-Ferré C, Corpas R, Puigoriol-Illamola D, Palomera-Ávalos V, Sanfeliu C, Pallàs M. Understanding epigenetics in the neurodegeneration of Alzheimer's disease: SAMP8 mouse model. J Alzheimers Dis. 2018;62:943–963. doi: 10.3233/JAD-170664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Baribault C, Ehrlich KC, Ponnaluri VKC, Pradhan S, Lacey M, Ehrlich M. Developmentally linked human DNA hypermethylation is associated with down-modulation, repression, and upregulation of transcription. Epigenetics. 2018:1–36. doi: 10.1080/15592294.2018.1445900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modrek AS, Golub D, Khan T, Bready D, Prado J, Bowman C, Deng J, Zhang G, Rocha PP, Raviram R, Lazaris C, Stafford JM, LeRoy G, Kader M, Dhaliwal J, Bayin NS, Frenster JD, Serrano J, Chiriboga L, Baitalmal R, Nanjangud G, Chi AS, Golfinos JG, Wang J, Karajannis MA, Bonneau RA, Reinberg D, Tsirigos A, Zagzag D, Snuderl M, Skok JA, Neubert TA, Placantonakis DG. Low-grade astrocytoma mutations in IDH1, P53, and ATRX cooperate to block differentiation of human neural stem cells via repression of SOX2. Cell Rep. 2017;21:1267–1280. doi: 10.1016/j.celrep.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina-Franco JL, Méndez-Lucio O, Due- ñas-González A, Yoo J. Discovery and development of DNA methyltransferase inhibitors using in silico approaches. Drug Discov Today. 2015;20:569–577. doi: 10.1016/j.drudis.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Schmalbach S, Petri S. Histone deacetylation and motor neuron degeneration. CNS Neurol Disord Drug Targets. 2010;9:279–284. doi: 10.2174/187152710791292684. Review 2010 9 279-284. [DOI] [PubMed] [Google Scholar]
- 13.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endres M, Meisel A, Biniszkiewicz D, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 17.Webb WM, Sanchez RG, Perez G, Butler AA, Hauser RM, Rich MC, O'Bierne AL, Jarome TJ, Lubin FD. Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol Learn Mem. 2017;142((Pt A)):66–78. doi: 10.1016/j.nlm.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Deng JH, Yan W, Han Y, Chen C, Meng SQ, Sun CY, Xu LZ, Xue YX, Gao XJ, Chen N, Zhang FL, Wang YM, Shi J, Lu L. Predictable chronic mild stress during adolescence promotes fear memory extinction in adulthood. Sci Rep. 201710;7:7857. doi: 10.1038/s41598-017-08017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X, Zhang X, Xue Z. Sevoflurane induces long-term memory impairment and increases MeCP2 phasphorylation in developing mice. Int J Clin Exp Med. 2017:10. [Google Scholar]
- 22.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent BDNF transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geranton SM, Morenilla-Palao C, Hunt SP. A role for transcriptional repressor methyl-CpG-binding protein 2 and plasticity-related gene serum- and glucocorticoid-inducible kinase 1 in the induction of inflammatory pain states. J Neurosci. 2007;27:6163–6173. doi: 10.1523/JNEUROSCI.1306-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geranton SM, Fratto V, Tochiki KK, Hunt SP. Descending serotonergic controls regulate inflammation-induced mechanical sensitivity and methyl-CpG-binding protein 2 phosphorylation in the rat superficial dorsal horn. Mol Pain. 2008;4:35. doi: 10.1186/1744-8069-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioural responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Zhong X, Chau KF, Williams EC, Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat Neurosci. 2011;14:1001–1008. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao LM, Horton E, Guo ML, Xue B, Jin DZ, Fibuch EE, Wang JQ. Cocaine increases phosphorylation of MeCP2 in the rat striatum in vivo: a differential role of NMDA receptors. Neurochem Int. 59:610–617. doi: 10.1016/j.neuint.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain V, Baitharu I, Prasad D, Ilavazhagan G. Enriched environment prevents hypobaric hypoxia induced memory impairment and neurodegeneration: role of BDNF/PI3K/GSK3β pathway coupled with CREB activation. PLoS One. 2013;8:e62235. doi: 10.1371/journal.pone.0062235. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Yang L, Mao L, Chen H, Catavsan M, Kozinn J, Arora A, Liu X, Wang JQ. A signaling mechanism from G alpha q-protein-coupled metabotropic glutamate receptors to gene expression: role of the c-Jun N-terminal kinase pathway. J Neurosci. 2006;26:971–980. doi: 10.1523/JNEUROSCI.4423-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang GC, Mao LM, Liu XY, Parelkar NK, Arora A, Yang L, Hains M, Fibuch EE, Wang JQ. In vivo regulation of Homer1a expression in the striatum by cocaine. Mol Pharmacol. 2007;71:1148–1158. doi: 10.1124/mol.106.028399. [DOI] [PubMed] [Google Scholar]
- 32.Xie Z, Enkhjargal B, Wu L, Zhou K, Sun C, Hu X, Gospodarev V, Tang J, You C, Zhang JH. Exendin-4 attenuates neuronal death via GLP-1R/PI3K/Akt pathway in early brain injury after subarachnoid hemorrhage in rats. Neuropharmacology. 2018;128:142–151. doi: 10.1016/j.neuropharm.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YC, Sanchez-Mendoza EH, Doeppner TR, Hermann DM. Post-acute delivery of memantine promotes post-ischemic neurological recovery, peri-infarct tissue remodelling, and contralesional brain plasticity. J Cereb Blood Flow Metab. 2017;37:980–993. doi: 10.1177/0271678X16648971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dandi E, Kalamari A, Touloumi O, Lagoudaki R, Nousiopoulou E, Simeonidou C, Spandou E, Tata DA. Beneficial effects of environmental enrichment on behavior, stress reactivity and synaptophysin/BDNF expression in hippocampus following early life stress. Int J Dev Neurosci. 2018 doi: 10.1016/j.ijdevneu.2018.03.003. pii: S0736-5748(17)30320-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhang S, Zhang Y, Jiang S, Liu Y, Huang L, Zhang T, Lu G, Gong K, Ji X, Shao G. The effect of hypoxia preconditioning on DNA methyltransferase and PP1γ in hippocampus of hypoxia preconditioned mice. High Alt Med Biol. 2014;15:483–490. doi: 10.1089/ham.2014.1042. [DOI] [PubMed] [Google Scholar]
- 36.Shao G, Zhang R, Zhang S, Jiang S, Liu Y, Zhang W, Zhang Y, Li J, Gong K, Hu XR, Jiang SW. Splice variants DNMT3B4 and DNMT3B7 overexpression inhibit cell proliferation in 293A cell line. In Vitro Cell Dev Biol Anim. 2013;49:386–394. doi: 10.1007/s11626-013-9619-z. [DOI] [PubMed] [Google Scholar]
- 37.Liu PK, Hsu CY, Dizdaroglu M, et al. Damage, repair, and mutagenesis in nuclear genes after mouse forebrain ischemia-reperfusion. J Neurosci. 1996;16:6795–6806. doi: 10.1523/JNEUROSCI.16-21-06795.1996. Daetstidar et al., (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin X, Dang H, Zhao X, Wang H. Effects of hypobaric hypoxia on rat retina and protective response of resveratrol to the stress. Int J Med Sci. 2017;14:943–950. doi: 10.7150/ijms.19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betz A, Jayatilaka S, Joshi J, Ramanan S, Debartolo D, Pylypiw H, Franke E. Chronic exposure to benzyl butyl phthalate (BBP) alters social interaction and fear conditioning in male adult rats: alterations in amygdalar MeCP2, ERK1/2 and ERα. Neuro Endocrinol Lett. 2013;34:347–358. [PubMed] [Google Scholar]
- 42.Subbanna S, Nagre NN, Shivakumar M, Umapathy NS, Psychoyos D, Basavarajappa BS. Ethanol induced acetylation of histone at G9a exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice. Neuroscience. 2014;258:422–432. doi: 10.1016/j.neuroscience.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dastidar SG, Bardai FH, Ma C, Price V, Rawat V, Verma P, Narayanan V, D'Mello SR. Isoform-specific toxicity of Mecp2 in postmitotic neurons: suppression of neurotoxicity by FoxG1. J Neurosci. 2012;32:2846–2855. doi: 10.1523/JNEUROSCI.5841-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal BDNF gene transcription after contextual fear conditioning. Genes Brain Behav. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuga GK, Muñoz VR, Gaspar RC, Nakandakari SCBR, da Silva A, SR, Botezelli JD, Leme JACA, Gomes RJ, de Moura LP, Cintra DE, Ropelle ER, Pauli JR. Impaired insulin signaling and spatial learning in middle-aged rats: The role of PTP1B. Exp Gerontol. 2018;104:66–71. doi: 10.1016/j.exger.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neeper SA, Gomez-Pinilla F, Choi J, Cotman C. Exercise and brain neurotrophins. Nature. 1995;373:109. doi: 10.1038/373109a0. [DOI] [PubMed] [Google Scholar]
- 49.Zhong X, Li H, Chang Q. MeCP2 phasphorylation is required for modulating synaptic scaling through mGluR5. J Neurosci. 2012;32:12841–1284. doi: 10.1523/JNEUROSCI.2784-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]