SUMMARY
The ability to direct targeted intercellular interactions has the potential to enable and expand the use of cell-based therapies for regenerative medicine, tissue engineering, and immunotherapy. While genetic engineering approaches have proven effective, these techniques are not amenable to all cell types and often yield permanent modifications with potentially long-lasting adverse effects, restricting their application. To circumvent these limitations, there is intense interest in developing non-genetic methods to modify cell membranes with functional groups that will enable the recognition of target cells. While many such techniques have been developed, relatively few have been applied to directing specific cell-cell interactions. This review details these non-genetic membrane engineering approaches – namely, hydrophobic membrane insertion, chemical modification, liposome fusion, metabolic engineering, and enzymatic remodeling – and summarizes their major applications. Based on this analysis, perspective is provided on the ideal features of these systems with an emphasis on the potential for clinical translation.
INTRODUCTION
Cell therapy has rapidly emerged as an invaluable tool in translational medicine and has led to significant advances in several diverse fields, including tissue engineering, regenerative medicine, and immunotherapy. Stem cells – in particular mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs), and, more recently, induced pluripotent stem cells (iPSCs) – have emerged as both a cornerstone of regenerative medicine and a versatile therapy for immune disorders. Meanwhile, T cells have been at the forefront of cancer immunotherapy for over a decade. However, the efficacy of these cellular therapies is ultimately contingent upon the ability to appropriately control the fate and function of the therapeutic cells. Specifically, cells must be successfully directed to engage in the cell-cell interactions required for a productive outcome. For example, systemic infusion of MSCs following a myocardial infarction results in less than 1% accumulation of cells in the ischemic myocardium (Barbash et al., 2003). In contrast, MSCs induced to upregulate chemotactic receptors prior to infusion exhibit more than a 2-fold increase in ischemic tissue homing (Cheng et al., 2008). For this reason, cellular engineering has become a crucial area of interest in cell therapy research.
Initial efforts in cellular engineering involved either preconditioning cells ex vivo via exposure to various stimuli (such as pharmacological agents, soluble cytokines, or stimulatory ligands) or the simultaneous administration of supportive adjuvant therapies. The goals of these approaches were to enhance the in vivo function of the infused cells, generate longer cell lifetimes, and promote self-renewal mechanisms to combat the inherent variability of cell biodistribution. While these strategies increased overall in vivo cell retention, they did little to directly influence the desired cell-cell interactions.
Over the past decade, genetic engineering has emerged as the most utilized and clinically efficacious cellular engineering approach. Indeed, genetically-engineered chimeric antigen receptor (CAR) T cells were recently approved by the United States Food and Drug Administration (FDA) for certain B cell malignancies (Kuehn, 2017). In this approach, exogenous genetic material is incorporated into the desired cell’s genome where it encodes an artificial cell surface receptor that targets an antigen of interest (Curran et al., 2012; Sadelain et al., 2017). While genetic engineering is a robust strategy, it is associated with a number of significant drawbacks. For instance, the process is time consuming and produces results with variable and often unpredictable efficiency. Furthermore, not all cell types are amenable to such genetic alteration without deleterious effects – stem cells in particular. Finally, the genetic modification is typically permanent and irreversible, yielding significant adverse events in patients and raising long-term safety concerns for clinical applications (Bonifant et al., 2016).
An alternative to genetic engineering is the use of bispecific ligands (e.g., bispecific antibodies, bispecific T cell engagers, etc.) designed to interface between two antigen expressing cells (Huehls et al., 2014). This approach has also demonstrated clinical efficacy, particularly in the context of T cell tumor targeting (Mullard, 2014). However, these ligands are not tethered to the cell surface, and because they generally rely on monovalent interactions with known membrane antigens, their cell-directing abilities are both dynamic and transient; coupled with their rapid clearance, this necessitates constant exposure to free bispecific ligand and makes them cumbersome to administer to patients (Garber, 2014).
In an attempt to circumvent the limitations associated with genetic modifications and small bispecific ligands, effort has been made to develop non-genetic strategies to engineer cell surfaces with targeting elements capable of directing specific cell-cell interactions. Typically, these approaches are more transient (or reversible) and applicable to numerous cell types, including stem cells. Furthermore, non-genetic approaches have been developed around lipid-, glycan-, and protein-based modifications, which is in contrast to genetic engineering methodologies focusing primarily on protein expression.
This review summarizes the non-genetic membrane engineering approaches used in directing specific cell-cell interactions and discusses their current applications, both in vitro and in vivo. The most ideal features of these systems are described with emphasis on how these modifications can be applied to achieve the greatest translational benefit. Finally, the most promising strategies to date are outlined, and perspective is provided on future trends in this rapidly growing field.
NON-GENETIC APPROACHES TO MEMBRANE ENGINEERING
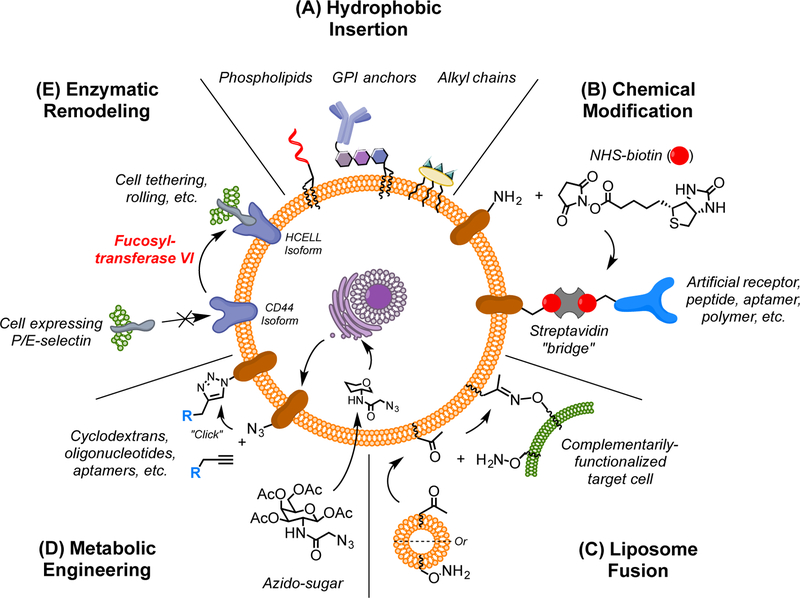
Several diverse, non-genetic approaches to cell membrane engineering have been developed. Typically, they fall into one of five broad categories: (1) the insertion of hydrophobic moieties into the lipid bilayer; (2) the direct chemical modification of cell surface constituents; (3) the membranous fusion of functionalized liposomes; (4) the use of functionalized sugar analogs to metabolically label surface glycoproteins; and (5) the enzymatic remodeling of membrane proteins (Figure 1).
Figure 1. Non-Genetic Membrane Engineering Approaches to Direct Intercellular Interactions.
Several non-genetic methods for cell membrane modification have been used to direct cell-cell interactions. (A) Hydrophobic groups will spontaneously insert into the lipid bilayer, tethering any conjugated cargo or ligands to the cell surface. (B) Functional groups naturally present on the cell surface can be reacted with chemical moieties to covalently tether a variety of species; in many cases, biotin is installed and streptavidin is used as a “bridge” to link other ligands to the cell surface. (C) Liposomes bearing various functional groups can be spontaneously fused with the cell membrane, thereby decorating the surface with the respective modifications. (D) Cells can be grown in media supplemented with functionalized sugar analogs, enabling the incorporation of bioorthogonal groups into surface glycoproteins; these groups can then be conjugated to exogenous ligands. (E) Enzymes can be used to modify naturally-existing proteins and glycans on the cell surface, creating divergent isoforms or tethering synthetic targeting elements.
Hydrophobic Insertion
Integral cell surface proteins are anchored into the membrane through hydrophobic transmembrane domains that interact with the hydrophobic interior of the lipid bilayer (White and von Heijne, 2005). The hydrophobic effect dictates both the incorporation and orientation of integrated proteins and locks them on the cell surface, barring cell membrane internalization (Simons and Sampaio, 2011). These same principles have been utilized for cellular engineering, where the desired ligand or chemical moiety is conjugated to a hydrophobic anchor and then incubated with the to-be-labeled cell population. As a thermodynamically favored process (Fernandez-Vidal et al., 2011; Gorfe et al., 2008; Jeong et al., 2013), the hydrophobic anchor will spontaneously insert into the lipid bilayer and effectively tether the attached species to the membrane (Figure 1A).
Such membrane insertion has been accomplished with a variety of hydrophobic tags, including alkyl chains (Jeong et al., 2013), lipid moieties (Kean et al., 2012), and glycophosphatidylinositol (GPI) anchors (Hamdy et al., 2005). Multiple methods for attaching these tags to the desired cargo have also been developed. Perhaps the most common approach is the chemical ligation of the tag by peptide couplings with activated esters (Ko et al., 2009), sulfhydryl-based Michael additions (Won et al., 2014), or click ligations between azide groups and various alkynes (Lu et al., 2015). In contrast, others have devised methods to produce hydrophobically-tagged ligands recombinantly, thereby omitting the additional conjugation step (de Kruif et al., 2000; Hamdy et al., 2005).
The relative ease with which cell surfaces can be “painted” with these hydrophobically-tagged species has made it a popular approach for the attachment of a wide variety of cargo. Targeting elements – including aptamers (Altman et al., 2013; Xiong et al., 2013), antibody-derived single chain variable fragments (scFvs) (de Kruif et al., 2000), and peptides (Kean et al., 2012; Lim et al., 2017) – are commonly installed on the cell membrane in this fashion. By first anchoring proteins A and G, several laboratories have functionalized cells with full antibodies (Kim and Peacock, 1993; Ko et al., 2009) and Fc-tagged fusion proteins (Lo et al., 2013). Oligonucleotides have been installed as well, enabling the clustering of cells functionalized with complimentary sequences (Selden et al., 2012; Todhunter et al., 2015). Finally, some have attached larger cell-targeting scaffolds, including nanoparticles (Csizmar et al., 2018; Gabrielse et al., 2014; Jeong et al., 2013) and polymers (Takeo et al., 2015). Notably, these methods were further developed into synthetic targeting systems used to direct artificial cell-cell interactions in vitro.
Chemical Modification
A very straightforward membrane engineering strategy is the direct chemical conjugation of small molecules and tethered cargo to functional groups that preexist on the cell surface (Figure 1B). These natural functional groups (such as free amines present on lysine side chains) are attractive targets for the covalent conjugation of various ligands since they require no preconditioning of the cell, chemical or otherwise. Furthermore, this approach can be used to modify essentially any cell membrane component – including proteins, lipids, carbohydrates, and glycans – albeit in a largely non-specific manner.
The use of N-hydroxysuccinimide (NHS)-activated esters to modify lysine residues and other surface amines is by far the most common strategy for chemical modification. This approach has long been used to non-specifically biotinylate cell surfaces for a host of applications, and several groups have since applied this scheme to the induction of intercellular interactions. In a typical workflow, cells are first biotinylated with an NHS-biotin conjugate before incubating them with tetravalent streptavidin. The streptavidin binds to the biotin moieties on the cell surface where it serves as a “bridge” to connect biotinylated ligands to the cell (Figure 1B). This streptavidin bridging approach has been used to functionalize cell surfaces with P-selectin binding aptamers (Zhao et al., 2011), sialyl LewisX (sLeX) motifs (Sarkar et al., 2011), and antibodies recognizing epithelial growth factor receptor (EGFR) (Koyfman et al., 2009). These membrane-engineered cells were then capable of interacting with selectin-expressing endothelial cells (in vitro (Zhao et al., 2011) and in vivo (Sarkar et al., 2011)) and with EGFR-expressing HEK 293T and HeLa cells (Koyfman et al., 2009), respectively. Finally, streptavidin has been used to bridge directly between biotinylated cells to generate cell clusters and layers (Gong et al., 2013).
Of course, NHS-esters can be used to conjugate species other than biotin to the cell surface, including various bioorthogonal functional groups. This approach was used to install reactive maleimide groups on the surface of MSCs which, through the Michael addition of a sulfhydryl group, were subsequently labeled with thiolated peptides targeting E-selectin (Cheng et al., 2012a).
Few groups have used chemistries other than NHS-activated esters to functionalize cell surfaces for the purpose of directing cell-cell interactions. Early work demonstrated that erythrocytes could be labeled with anti-CD90.2 (Thy-1.2) antibodies via chromic chloride coupling (Chiarantini et al., 1992). More recently, polymers composed of N-vinylpyrrolidone and 3-(acrylamide)phenylboronic acid, the latter of which binds cell membrane glycoproteins through covalent boronate ester bonds, were able to cross-link cells to form non-specific aggregates in culture media (Amaral and Pasparakis, 2015).
Liposome Fusion
Lipid membrane fusion is a critical process in normal cell biology and has been the subject of intense investigation. Not surprisingly, membrane fusion has been readily adapted for several applications, including membrane engineering. When incubated with cells, functional group-containing liposomes spontaneously fuse with the cell membrane to present the respective functional groups on the cell surface (Figure 1C). Such liposomes can be readily formed by dissolving the desired lipid species in a volatile solvent (e.g., chloroform), concentrating/drying them under high vacuum, resuspending them in the desired aqueous buffer, and finally applying an agitating stimulus (commonly sonication) to drive the formation of unilamellar vesicles (Dutta et al., 2011a; Sarkar et al., 2010).
This approach was used to from vesicles composed of the biotinylated lipid 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (Bio-PE), which were subsequently fused with MSC membranes to functionalize the cell surface with biotin (Sarkar et al., 2010). Streptavidin was then used as a bridge (see above) to tether biotinylated sLeX motifs to the surface.
The Yousaf group has extensively advanced liposome fusion technology, and their work provides the most robust examples of forming targeted intercellular interactions via this approach. In their system, liposomes composed of POPC (palmitoyl-oleoyl phosphatidylcholine), DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), and either dodecanone or O-dodecyloxyamine are spontaneously fused to cell membranes to present ketone or oxyamine groups, respectively, on the cell surface (Dutta et al., 2011a). Cells bearing the complementary ketone and oxyamine moieties were then linked to one another via a covalent oxime ligation. When applied to multiple cell types, the Yousaf group has been able to form multicellular sheets (Dutta et al., 2011b), spheroids (O’Brien et al., 2015), and microtissues (Rogozhnikov et al., 2016).
Metabolic Modification
The technique of metabolically introducing novel functional groups to the plasma membrane was first pioneered in 1992 (Kayser et al., 1992) and capitalized upon the promiscuity of natural carbohydrate biosynthesis pathways to metabolically incorporate non-physiologic amino sugar analogues into membrane glycoconjugates (Figure 1D).
Expanding upon this approach, Jurkat cells were metabolically labeled with the azide-functionalized mannose derivative, N-azideacetylmannosamine (ManNAz), affording the incorporation of azido sialic acid (SiaNAz) residues on the surface (Gartner and Bertozzi, 2009). The SiaNAz residues were then reacted with phosphine-conjugated ssDNA oligonucleotides via Staudinger Ligation or with difluorinated cyclooctyne (DIFO)-conjugated ssDNA oligonucleotides via copper-free click chemistry. Cells bearing complementary oligonucleotide sequences were able to assemble into aggregates.
A similar strategy was used to metabolically label cells with peracetylated N-azidoacetylgalactosamine (Ac4GalNAz), introducing azido groups into cell surface glycoconjugates (Shi et al., 2016). Copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) was then used to install a pegylated β-cyclodextrin (β-CD). The β-CD was then used to bind homobifunctional azobenzene species capable of bridging between β-CD-functionalized cells. Similarly, azobenzene-conjugated aptamers targeting mucin 1 (MUC1) were installed on the metabolically-engineered cells, enabling the recognition of MUC1+ targets.
In addition to azido sugar moieties, a synthetic methacryloyl-modified analog of N-acetyl mannosamine (ManNAc), termed ManM, was used to install methacryloyl groups in surface sialic acid residues (Sugimoto et al., 2015). The ManM groups were then coupled to thiol-terminated aptamers targeting protein tyrosine kinase-7 (PTK7) via a light-assisted (505 nm) thiol-ene reaction utilizing eosin Y as a photosensitizer. Ultimately, aptamer-functionalized macrophages were able to recognize and bind to PTK7+ human T lymphoblasts in vitro.
Enzymatic Remodeling
A less common approach to cell-surface engineering involves the use of enzymes to modify naturally-present proteins and glycans (Figure 1E) (Capicciotti et al., 2017). One method takes advantage of the popular “sortagging” technique. Here, the Staphylococcus aureus enzyme sortase A is used to conjugate appropriately tagged ligands to the cell surface. Notably, this approach has been used to conjugate antibodies onto naturally exposed N-terminal glycine residues on T cells (Swee et al., 2015).
Additionally, a process known as glycosyltransferase-programmed stereosubstitution (GPS) has been employed to convert CD44 on the surface of neural stem cells (NSCs) into an isoform capable of binding to E-selectin (Merzaban et al., 2015). Specifically, the α(1,3)-linkage specific fucosyltransferase VI was used to install a fucose group onto terminal 2-lactosamine units. When those loactosamines were then capped with an α(2,3)-linked sialic acid, sLex epitopes capable of being recognized by P- and E-selectin residues were created. NSCs modified in this manner were then able to interact with E-selectin expressing endothelial cells and demonstrated increased transendothelial extravasation in vitro.
APPLICATIONS FOR ENGINEERED CELL-CELL INTERACTIONS
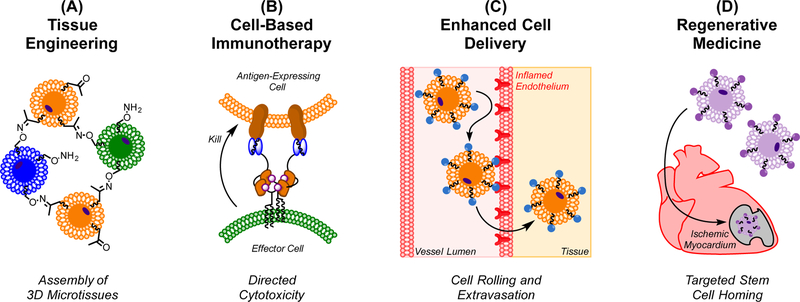
Though membrane-engineered cells have been explored in numerous contexts (Wang et al., 2015), their use to direct cell-cell interactions has been most frequently applied to four fields – tissue engineering, cell-based immunotherapy, targeted cell adhesion, and regenerative medicine (Figure 2).
Figure 2. Applications of Engineered Cell-Cell Interactions.
Non-genetically engineered cell-cell interactions are broadly applicable to a variety of fields. (A) The linking of multiple tissue types in vitro can be applied to tissue engineering. (B) Cytotoxic T cells can be directed to – and subsequently kill – antigen expressing cells for immunotherapy. Alternatively, engineered antigen presenting cells can be used to prime T cells against a particular target. (C) Therapeutic cells can be modified to more efficiently tether, roll, and extravasate through the vasculature, improving their delivery to target tissues. (D) Regenerative stem and progenitor cells can be functionalized with targeting ligands that enhance their accumulation within sites of injury.
Tissue Engineering
The ability to generate fully-functional tissues and organs in vitro has been a long-standing goal of biomedical science. Requisite to this aim is the capacity to direct the constituent cell types to interact with one another in a manner that recapitulates the parent tissue’s usual in vivo development. To this end, several groups have used membrane engineered cells to direct the formation of ordered microtissues composed of different cell types. For example, Yousaf and coworkers have long used a liposome fusion approach to functionalize cell surfaces with bioorthogonal ketone and oxyamine groups. Taking advantage of the oxime ligation, they first formed directed cell clusters of complimentarily-labeled cells, including murine fibroblasts (Dutta et al., 2011a), which could be further assembled into ordered cellular spheroids and sheets (Dutta et al., 2011b). Additional control over the order and cellular hierarchy of these microtissues was achieved using a microfluidic chamber to regulate the addition and ligation of ketone and oxyamine-labeled cells (O’Brien et al., 2015). Ultimately, they were able to use this methodology to assemble fibroblasts, cardiomyocytes, and HUVECs into a functional three-dimensional (3D) cardiac tissue that was able to beat spontaneously and synchronously. Importantly, neighboring cells in this tissue both developed physiologic intercellular junctions – including the gap-junction protein connexin 43 – and produced extracellular matrix, suggesting a more accurate recapitulation of the normal cardiac environment in vivo (Rogozhnikov et al., 2016). Furthermore, the induced cell-cell interactions could be reversed by UV irradiation when a photocleavable group was incorporated into the oxyamine component (Luo et al., 2014). This approach was then applied to fibroblasts, adipocytes, and hMSCs to generate 3D microtissues in vitro (Luo et al., 2014). Similarly, when hMSCs and fibroblasts were modified with hydroquinone and oxyamine groups, respectively, the formation of cell clusters could be induced by chemical or electrochemical oxidation of the hydroquinone to a reactive quinone moiety, which then coupled to oxyamine-labeled cells. The cell clustering could then be reversed via reduction of the quinone back to a hydroxyquinone (Pulsipher et al., 2014). Unfortunately, neither of these reversal mechanisms – photocleavage or electrochemical reduction – are currently applicable to in vivo applications.
Rather than covalently couple cell types together, the liposome-fusion approach can also be used to functionalize cell surfaces with targeting elements. Fibroblasts were first modified with a hydrazone-coupled ligand, and then that ligand was exchanged in situ for an oxyamine-conjugated RGD peptide. These RGD-functionalized fibroblasts were then able to recognize and adhere to other integrin-expressing fibroblasts, enabling the formation of tissue microlayers in vitro (Luo et al., 2015).
Another approach to assembling cells for tissue engineering involves cross-linking biotinylated cells with a tetravalent streptavidin moiety. In one example, cell surfaces are non-specifically labeled at primary amines with a reactive NHS-biotin; labeled cells are then clustered together by the addition of streptavidin, whose multiple biotin binding domains enable the formation of a “bridge” between labeled cells. The Jiang group used this technique to assemble layers of Jurkat cells on top of HUVEC cells and, separately, HUVEC cells on top of fibroblasts in vitro. Using fabricated two-dimensional (2D) stress-induced rolling membranes (SIRMs), they were able to pattern HUVECs on top of smooth muscle cells, and then roll the 2D substrate into a 3D tube resembling a blood vessel (Gong et al., 2013). Similarly, a hexagonal microarray assembled from biotinylated ssDNA oligonucleotides was able to attach to biotinylated Jurkat cells via streptavidin bridges (Koyfman et al., 2009). When biotinylated anti-EGFR antibodies were attached to the microarray, EGFR-expressing HEK 293T and HeLa cells could be captured. In this manner, the microarrays were also able to form intercellular bridges between EGFR-expressing cells, enabling the formation of cell pairs and clusters (Koyfman et al., 2009).
Gartner and Bertozzi have used oligonucleotides to assemble metabolically-engineered cells. SiaNAz-functionalized Jurkat cells were labeled with ssDNA oligonucleotides, and cells bearing complementary sequences were assembled into aggregates (Gartner and Bertozzi, 2009). The order of cells in these aggregates could be somewhat controlled stoichiometrically. Furthermore, these cellular interactions could be reversed by two strategies: (1) melting of the hybridized ssDNA sequences at 37 ºC, and (2) degradation by DNase. This DNA-programmed assembly of cells (DPAC) has also been used to form specific 3D arrangements of cells and microtissues (Todhunter et al., 2015). In this approach, phospholipid-conjugated ssDNA oligonucleotides are hydrophobically inserted into cell membranes. Cells labeled with complimentary ssDNA sequences could then patterned into microtissues in a Matrigel matrix.
Cell-Based Immunotherapy
The therapeutic potential of cell-directed immunotherapies is underscored by the clinical successes of CAR T cells (Kuehn, 2017). However, the limitations of the CAR platform have driven many groups to seek non-genetic methods to introduce targeting elements to the T cell surface. For instance, early work by the Al-Katib group sought to deliver cytotoxic payloads directly to CD20-expressing tumor cells through targeted erythrocytes. They developed and expressed a glycosylphosphatidylinositol (GPI)-fused anti-CD20 single chain variable fragment (scFv) that was hydrophobically inserted into red blood cell (RBC) membranes. These anti-CD20 RBCs were then able to recognize and bind to B cells in vitro, forming visible rosettes (Hamdy et al., 2005).
Using metabolic engineering, Qu and coworkers sought to target peripheral blood mononuclear cells (PBMCs) to breast adenocarcinoma cells. PBMCs were labeled with Ac4GalNAz to yield cell surface azide groups that were subsequently conjugated to a β-CD. Azobenzene-conjugated anti-MUC1 aptamers were then installed on the PBMCs, enabling them to target the MUC 1+ breast adenocarcinoma MCF-7 cell line in vitro (Shi et al., 2016).
The Wagner group has utilized a chemically self-assembled nanoring (CSAN) to non-covalently introduce targeting elements to T cell surfaces. CSANs incorporating anti-EpCAM scFvs and phospholipids were able to hydrophobically insert into T cell membranes, enabling the T cells to selectively recognize and eradicate EpCAM-expressing MCF-7 cells in co-culture with EpCAM-negative U87 cells (Gabrielse et al., 2014).
Using the “sortagging” approach, Swee and coworkers installed anti-mouse class-II MHC antibodies onto activated CD8+ T cells. These T cells were then able to selectively recognize and kill antigen-expressing splenocytes in vitro (Swee et al., 2015).
Aside from T cells, macrophages have also been explored as immunotherapeutic mediators. Logtenberg and coworkers sought to enhance the anti-tumor response of cancer vaccines by targeting irradiated tumor cells to antigen presenting macrophages. They expressed and purified lipid-fusions of scFvs targeting CD14, CD32, and CD64. When these lipid-scFv fusions were hydrophobically inserted into the membrane of Jurkat cells, the cells targeted to and were phagocytosed by macrophages (de Kruif et al., 2000). Finally, metabolically functionalized murine macrophage-like cells (RAW264.7) have been labeled with aptamers targeting the circulating cancer cell marker PTK7. These aptamer-targeted macrophages were able to recognize and phagocytose PTK7+ CCRF-CEM (T lymphoblast) cells in vitro (Sugimoto et al., 2015).
Unlike most approaches that drive immune cells to eradicate a particular target, Ko et al. took advantage of the immunosuppressive capabilities of MSCs to design a targeted cellular therapy for inflammatory bowel disease (IBD). PPG-painted MSCs were armed with antibodies to target them to the inflamed endothelium within the colon and draining mesenteric lymph nodes. Following intravenous infusion in a murine model of IBD, the functionalized MSCs exhibited increased localization within the inflamed tissues, reduced inflammation (assessed by colon length and histology), an increase in the percentage of regulatory T cells, and improved overall survival (Ko et al., 2010).
Enhanced Cell Delivery
A major barrier to the targeted delivery of stem cells is the inefficiency with which they extravasate from the vasculature and accumulate within the target tissue (Allen et al., 2017; Cheng et al., 2012b). To address this, many groups have sought to surface engineer stem cells with targeting elements designed to promote their adhesion to, rolling along, and extravasation through the vascular endothelium. The Karp laboratory has had a longstanding interest in modifying MSCs to bind to selectin-expressing surfaces. In one approach, MSCs were non-specifically biotinylated with NHS-biotin; then, streptavidin was used to tether biotinylated sLeX motifs to the MSCs. These sLeX-labeled MSCs then exhibited a robust rolling response on inflamed endothelium in vivo and homed to inflamed tissue with higher efficiency than native MSCs (Sarkar et al., 2011). Rather than tether sLeX motifs to biotinylated MSCs, the group has also tethered biotinylated selectin-binding aptamers to the MSCs through the streptavidin bridge. Similar to before, these aptamer-modified MSCs were able to recognized selectin-expressing HUVECs and neutrophils (Zhao et al., 2011).
The Dennis group has also shown interest in this area, having labeled protein G with NHS-conjugated palmitic acids and hydrophobically inserting them into MSC membranes. The palmitated protein G (PPG) modified MSCs were further labeled with antibodies targeting intercellular adhesion molecule 1 (ICAM-1), enabling their recognition of HUVEC cells in vitro, even under physiologic flow conditions (Ko et al., 2009). Neelamegham and coworkers similarly inserted (PPG) into MSC membranes, but rather than use it to bind a pre-existing antibody, they bound a custom fusion protein consisting of the first 19 amino acids of PSGL-1 (the pan-selectin binder “P-selectin glycoprotein ligand-1”) fused to a human IgG tail. A core-2 sLeX motif is also engineered at the N-terminus to confer “leukocyte-like” tethering and rolling properties. Ultimately, MSCs functionalized with protein G and this fusion protein interacted with P- and E-selectin expressing HUVECs and exhibited tethering and rolling properties similar to leukocytes in vitro (Lo et al., 2013).
Beyond selectins, some groups have used lipid-anchored glycan mimetics to modulate the bulk properties of a cell’s native glycocalyx. The insertion of long glycoprotein mimetics into the cell membrane drove the reorganization of cell surface receptors and, specifically, the clustering of integrins (Paszek et al., 2014). This enhanced the adhesion of mammary epithelial cells (MECs) to the extracellular matrix, and identified a potential mechanism for promoting metastasis of malignant cells. Likewise, cells adorned with longer glycoprotein mimetics exhibited enhanced metastatic potential in vivo and increased cell proliferation within the metastatic niche (Woods et al., 2017). Thus, these techniques can be used as tools to elucidate fundamental cell biology and explore how cell interact with their environments through both cellular contacts and membrane milieus.
Regenerative Medicine
Targeting stem cells to sites of tissue injury is an attractive approach to regenerative medicine. Few groups have applied non-genetic cell surface engineering approaches to this problem. Dennis and coworkers used phage display to identify peptides that selectively recognize ischemic myocardium in a mouse model of myocardial infarction (MI). They then synthesized these MI-targeting peptides with a palmitic acid tail, enabling their hydrophobic insertion into MSC membranes. In a mouse model of MI, targeted MSCs injected into the tail vein homed to and accumulated within ischemic myocardium to a greater extent than non-targeted MSCs (Kean et al., 2012).
The Sackstein group used enzymatic glycoengineering to generate neural stem cells (NSCs) capable of binding E-selectins. These engineered NSCs were shown to interact with E-selectin expressing HUVECs in vitro, and in a mouse model of experimental autoimmune encephalitis, were able to undergo transendothelial extravasation to accumulate in the brain, spleen, and liver parenchyma (Merzaban et al., 2015).
PERSPECTIVE
Given the diverse applications of surface-engineered cells – and the multitude of approaches to generating such cells – it is unlikely that a single engineering system will prove sufficient for all applications. Still, results from both the bench and clinic offer insights into what features of the membrane-engineering system may be beneficial for a given application. For example, while genetically-engineered CAR T cells have proven efficacious for the treatment of certain hematologic malignancies, their success in treating solid tumors has been hampered by both “on-target off-tumor” toxicities and an inability to provide functional replacements for the damaged tissues (e.g., B cell aplasia can be partly overcome by regular infusions of intravenous immunoglobulins) (Bonifant et al., 2016; Jackson et al., 2016; Sadelain et al., 2017). This highlights the notion that, while permanent, genetically-encoded modifications may be beneficial in some instances, a temporary or reversible cell-targeting modification may prove superior under other circumstances.
Based upon such examples and the current literature at large, we propose a short list of “ideal” features for non-genetic membrane engineering approaches as they relate to the formation of targeted cell-cell interactions. We postulate that fulfillment of most of these criteria will facilitate both preclinical success and clinical adoption of these technologies. Specifically, such systems should be: (1) stable under physiologic conditions; (2) universally applicable to all cell types; (3) phenotypically innocuous to the modified cell; and (4) reversible upon administration of a controlled stimulus.
Stability
In vivo stability is a potential limiting factor for certain approaches, particularly lipid- and oligonucleotide-based modifications. For instance, the half-life of lipid-tagged scFv fragments on cell surfaces in culture media at 37 ºC was reported to range from mere minutes to only several hours (de Kruif et al., 2000). Similarly, targeted peptides synthesized with palmitic acid tails exhibited a cell surface half-life of less than one hour in culture media at 37 ºC (Kean et al., 2012). The reason for this is likely multifaceted. Many lipid-anchored constructs can diffuse out of the plasma membrane and partition between the cell surface and the aqueous phase (Palte and Raines, 2012). The extent of this partitioning can be modulated by varying the hydrophobicity of the anchor, where an increase in net hydrophobicity serves to more stably anchor ligands to the membrane (Weber et al., 2014). Additionally, some lipid-conjugated agents undergo rapid endocytosis (Capicciotti et al., 2017; Rabuka et al., 2008). Once internalized, the species may be degraded, become trapped within endosomes, or incorporated into intracellular membranes, making them inaccessible for mediating cell-cell interactions (Dutta et al., 2011a; Woods et al., 2015).
In contrast, approaches using more than one lipid anchor have demonstrated prolonged stability on the cell surface. Protein G, when non-specifically coupled to a palmitic acid moiety, remained ≥25% incorporated into to the cell membrane at 48 h (Lo et al., 2013). Similarly, when eight diacyl phospholipids were incorporated into a self-assembling nanoring, the construct remained stably bound (≥20%) to the cell surface for over 72 h (Csizmar et al., 2018; Gabrielse et al., 2014). On these longer time-scales, cell division serves to dilute the concentration of the surface modification as well. Furthermore, physiologic membrane recycling can return certain hydrophobic ligands to the cell surface, prolonging their overall display and propagating the modification to daughter cells (Woods et al., 2015). These results indicate that hydrophobically inserted modifications can persist on cell surfaces for meaningful periods of time, provided appropriate anchoring or recycling mechanisms.
Aside from the stability of the membrane-anchoring component of the system, the cell-targeting component is also subject to scrutiny. Certain aptamers used in this context will denature and lose their binding efficacy at 37 ºC, limiting their use in vivo (Altman et al., 2013). Similarly, complementary oligonucleotides have been used to form targeted cell associations (Gartner and Bertozzi, 2009), but these early double-stranded DNA (dsDNA) constructs melted at 37 ºC, dissociating the cells and restricting this approach to ex vitro applications. Thermal stability could be increased using chemically modified oligonucleotides or by increasing the GC content of the strands. While this could enable in vivo use, enzymatic degradation and toll-like receptor (TLR) activation remain potential barriers.
Universality
Another important aspect of membrane engineering approaches is the degree to which they can be generalized to different cell types. This is especially true for tissue engineering applications where a key goal is to drive interactions between the numerous cell lineages comprising a functional tissue or organ. Fortunately, many of the non-genetic approaches reviewed here have each been applied to a wide array of cell types – including lymphocytes (Gabrielse et al., 2014), MSCs (Kean et al., 2012), cardiomyocytes (Rogozhnikov et al., 2016), and vascular endothelial cells (Gong et al., 2013) – suggesting that non-genetic alterations relying solely upon membrane modification may be uniquely flexible and thus find use in a variety of applications.
Nevertheless, some approaches have been shown to have a more limited scope or variable degrees of efficacy. For example, the enzymatic glycoengineering approach described by (Merzaban et al., 2015) requires that the target cell express the protein substrate (CD44) of the modifying enzyme (fucosyltransferase VI). Meanwhile, the hydrophobic insertion of fluorescently labeled phospholipid conjugates sometimes has variable efficacy amongst different cell types (Lim et al., 2017), suggesting that the insertion kinetics may differ based upon cell size, morphology, or the local cell membrane constituency.
Innocuity
In order for a membrane modification to be universal it must also be innocuous. This is especially important for stem cells, as the modification should neither drive the differentiation of the cell nor reduce its multipotency. Genetically encoded modifications have met little success in the arena of stem cells, as the genetic manipulation typically causes a decrease in pluripotency (Rombouts and Ploemacher, 2003). However, non-genetic approaches – including hydrophobic insertion (Dennis et al., 2004; Ko et al., 2009), chemical modification (Cheng et al., 2012a), liposome fusion (Pulsipher et al., 2014), metabolic engineering (Du and Yarema, 2010), and enzymatic remodeling (Merzaban et al., 2015) – have been successfully applied to stem and progenitor cell populations without altering their multi-lineage differentiation capabilities. Consequently, these non-genetic approaches, which don’t typically rely upon the engagement of a physiologic receptor on the cell membrane, may be uniquely suited to directed stem cell therapies. Still, innocuity is not guaranteed for these techniques, as the hydrophobic insertion of bulky glycopeptide mimetics has profound effects on the ECM adherence and metastatic potential of cells (Paszek et al., 2014; Woods et al., 2017). Thus, the degree to which these surface modifications alter the cell’s phenotype likely need to be defined on an individual basis and in a context-dependent manner.
Reversibility
Temporospatial control over the cell surface modification is highly desirable, as it has the potential to reduce the incidence and severity of adverse clinical events and thereby increase the scope of amenable applications. For example, the inability to remove CARs from T cells leads to both persistent B cell aplasia and immense difficulty translating the CAR platform to solid tumors. In this context, a reversible cell surface modification might enable the rapid removal of targeting capability from engineered lymphocytes, enabling the recovery of healthy B and plasma cells or reducing off-tumor toxicities.
While the short half-life of some methods can be viewed as an inherent reversibility, the inability to specifically trigger the removal of such ligands from the cell membrane is a disadvantage. To this end, many groups have incorporated temporospatial control mechanisms into their surface modification approaches. For example, the introduction of photoisomerizable (Shi et al., 2016) and photocleavable (Luo et al., 2014) groups into the modifications has allowing their removal upon UV irradiation. Electrochemical reversibility has also been demonstrated, with the formation and cleavage of oxime linkages reliant upon oxidation and reduction, respectively (Pulsipher et al., 2014). Additionally, several groups have utilized temperature as a stimulus via thermally-reversible polymers (Amaral and Pasparakis, 2015), oligonucleotides (Gartner and Bertozzi, 2009), and aptamers (Altman et al., 2013). Enzymatic reversibility has been shown, with exposure to DNase triggering the degradation of oligonucleotides responsible for cell-cell adhesion (Gartner and Bertozzi, 2009; Xiong et al., 2013). Aside from the systemic administration of human DNase to degrade DNA-based interactions, however, few of these reversal mechanisms are directly applicable to in vivo applications.
Thus far, the Wagner group has demonstrated the most readily translatable mechanism for reversal. In their approach, phospholipid-functionalized nanorings incorporating an EpCAM-targeting scFv were hydrophobically inserted into cell membranes. The nanorings could be disassembled in the presence of clinically-relevant concentrations of the orally bioavailable and FDA-approved antibiotic, trimethoprim, driving the dissociation of the targeting construct and providing a pharmacologic trigger for reversing the cell-cell interactions (Csizmar et al., 2018; Gabrielse et al., 2014).
Clinical Translatability
While many of these non-genetic membrane modifications have demonstrated preclinical success at directing cell-cell interactions, none have been successfully translated into the clinic to date. While the barriers to clinical translation are both steep and somewhat application specific, incorporating the “ideal” features previously discussed may help overcome these difficulties. For example, tissue engineering modifications should be both applicable to multiple cell types to facilitate organ construction and readily removable to reduce the potential immunogenicity of the implanted tissue. Immunotherapy modifications ought to be rapidly and specifically reversible, providing an “off switch” should cellular toxicities arise. Cell delivery modifications should be stable enough to persist on the membrane throughout extended circulation, vessel adhesion/rolling, and extravasation through the endothelium. Finally, regenerative medicine modifications intended to enhance stem cell homing should be innocuous and not alter the stem cell’s pluripotency prematurely.
CONCLUSIONS
Non-genetic membrane engineering is a powerful tool for directing targeted cell-cell interactions. A variety of approaches have been developed for this purpose, including hydrophobic insertion, chemical modification, liposome fusion, metabolic engineering, and enzymatic remodeling. Collectively, these techniques have demonstrated significant preclinical results in the fields of tissue engineering, cell-based immunotherapy, targeted cell delivery, and regenerative medicine. Still, significant barriers to clinical translation still exist for these methodologies. To that end, work is underway to make these systems more stable, universal, innocuous, and reversible – features which may help overcome the adverse events noted with other cell surface engineering modalities. Ultimately, these non-genetic membrane engineering systems are poised to broadly enable and expand the use of cell-based therapies across multiple disciplines.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from the National Institutes of Health R21 CA185627 (CRW), F30 CA210345 (CMC), T32 GM008244 (CMC), and the University of Minnesota.
REFERENCES
- Allen TA, Gracieux D, Talib M, Tokarz DA, Hensley MT, Cores J, Vandergriff A, Tang J, de Andrade JB, Dinh PU, et al. (2017). Angiopellosis as an Alternative Mechanism of Cell Extravasation. Stem Cells 35, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman MO, Chang YM, Xiong X, and Tan W (2013). Modifying cellular properties using artificial aptamer-lipid receptors. Sci Rep 3, 3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AJ, and Pasparakis G (2015). Macromolecular cell surface engineering for accelerated and reversible cellular aggregation. Chem Commun (Camb) 51, 17556–17559. [DOI] [PubMed] [Google Scholar]
- Barbash IM, Chouraqui P, Baron J, Feinberg MS, Etzion S, Tessone A, Miller L, Guetta E, Zipori D, Kedes LH, et al. (2003). Systemic delivery of bone marrow-derived mesenchymal stem cells to the infarcted myocardium: feasibility, cell migration, and body distribution. Circulation 108, 863–868. [DOI] [PubMed] [Google Scholar]
- Bonifant CL, Jackson HJ, Brentjens RJ, and Curran KJ (2016). Toxicity and management in CAR T-cell therapy. Mol Ther Oncolytics 3, 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capicciotti CJ, Zong C, Sheikh MO, Sun T, Wells L, and Boons G-J (2017). Cell-Surface Glyco-Engineering by Exogenous Enzymatic Transfer Using a Bifunctional CMP-Neu5Ac Derivative. Journal of the American Chemical Society 139, 13342–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Byrska-Bishop M, Zhang CT, Kastrup CJ, Hwang NS, Tai AK, Lee WW, Xu X, Nahrendorf M, Langer R, et al. (2012a). Stem cell membrane engineering for cell rolling using peptide conjugation and tuning of cell-selectin interaction kinetics. Biomaterials 33, 5004–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Shen D, Xie Y, Cingolani E, Malliaras K, and Marbán E (2012b). Brief report: Mechanism of extravasation of infused stem cells. Stem Cells 30, 2835–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ou L, Zhou X, Li F, Jia X, Zhang Y, Liu X, Li Y, Ward CA, Melo LG, et al. (2008). Targeted migration of mesenchymal stem cells modified with CXCR4 gene to infarcted myocardium improves cardiac performance. Mol Ther 16, 571–579. [DOI] [PubMed] [Google Scholar]
- Chiarantini L, Droleskey R, Magnani M, and DeLoach JR (1992). In vitro targeting of erythrocytes to cytotoxic T-cells by coupling of Thy-1.2 monoclonal antibody. Biotechnol Appl Biochem 15, 171–184. [PubMed] [Google Scholar]
- Csizmar CM, Petersburg JR, Hendricks A, Stern LA, Hackel BJ, and Wagner CR (2018). Engineering Reversible Cell-Cell Interactions with Lipid Anchored Prosthetic Receptors. Bioconjug Chem 29, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran KJ, Pegram HJ, and Brentjens RJ (2012). Chimeric antigen receptors for T cell immunotherapy: current understanding and future directions. J Gene Med 14, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kruif J, Tijmensen M, Goldsein J, and Logtenberg T (2000). Recombinant lipid-tagged antibody fragments as functional cell-surface receptors. Nat Med 6, 223–227. [DOI] [PubMed] [Google Scholar]
- Dennis JE, Cohen N, Goldberg VM, and Caplan AI (2004). Targeted delivery of progenitor cells for cartilage repair. J Orthop Res 22, 735–741. [DOI] [PubMed] [Google Scholar]
- Du J, and Yarema KJ (2010). Carbohydrate engineered cells for regenerative medicine. Adv Drug Deliv Rev 62, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Pulsipher A, Luo W, Mak H, and Yousaf MN (2011a). Engineering cell surfaces via liposome fusion. Bioconjug Chem 22, 2423–2433. [DOI] [PubMed] [Google Scholar]
- Dutta D, Pulsipher A, Luo W, and Yousaf MN (2011b). Synthetic chemoselective rewiring of cell surfaces: generation of three-dimensional tissue structures. J Am Chem Soc 133, 8704–8713. [DOI] [PubMed] [Google Scholar]
- Fernandez-Vidal M, White SH, and Ladokhin AS (2011). Membrane partitioning: “classical” and “nonclassical” hydrophobic effects. J Membr Biol 239, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielse K, Gangar A, Kumar N, Lee JC, Fegan A, Shen JJ, Li Q, Vallera D, and Wagner CR (2014). Reversible re-programing of cell-cell interactions. Angew Chem Int Ed Engl 53, 5112–5116. [DOI] [PubMed] [Google Scholar]
- Garber K (2014). Bispecific antibodies rise again. Nat Rev Drug Discov 13, 799–801. [DOI] [PubMed] [Google Scholar]
- Gartner ZJ, and Bertozzi CR (2009). Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc Natl Acad Sci U S A 106, 4606–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Zheng W, Huang Z, Zhang W, Xiao D, and Jiang X (2013). A Strategy for the Construction of Controlled, Three-Dimensional, Multilayered, Tissue-Like Structures. Advanced Functional Materials 23, 42–46. [Google Scholar]
- Gorfe AA, Baron R, and McCammon JA (2008). Water-membrane partition thermodynamics of an amphiphilic lipopeptide: an enthalpy-driven hydrophobic effect. Biophys J 95, 3269–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy N, Goustin AS, Desaulniers JP, Li M, Chow CS, and Al-Katib A (2005). Sheep red blood cells armed with anti-CD20 single-chain variable fragments (scFvs) fused to a glycosylphosphatidylinositol (GPI) anchor: a strategy to target CD20-positive tumor cells. J Immunol Methods 297, 109–124. [DOI] [PubMed] [Google Scholar]
- Huehls AM, Coupet TA, and Sentman CL (2014). Bispecific T-cell engagers for cancer immunotherapy. Immunology And Cell Biology 93, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson HJ, Rafiq S, and Brentjens RJ (2016). Driving CAR T-cells forward. Nat Rev Clin Oncol 13, 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JH, Schmidt JJ, Kohman RE, Zill AT, DeVolder RJ, Smith CE, Lai MH, Shkumatov A, Jensen TW, Schook LG, et al. (2013). Leukocyte-mimicking stem cell delivery via in situ coating of cells with a bioactive hyperbranched polyglycerol. J Am Chem Soc 135, 8770–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser H, Geilen CC, Paul C, Zeitler R, and Reutter W (1992). Incorporation of N-acyl-2-amino-2-deoxy-hexoses into glycosphingolipids of the pheochromocytoma cell line PC 12. FEBS Lett 301, 137–140. [DOI] [PubMed] [Google Scholar]
- Kean TJ, Duesler L, Young RG, Dadabayev A, Olenyik A, Penn M, Wagner J, Fink DJ, Caplan AI, and Dennis JE (2012). Development of a peptide-targeted, myocardial ischemia-homing, mesenchymal stem cell. J Drug Target 20, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, and Peacock JS (1993). The use of palmitate-conjugated protein A for coating cells with artificial receptors which facilitate intercellular interactions. J Immunol Methods 158, 57–65. [DOI] [PubMed] [Google Scholar]
- Ko IK, Kean TJ, and Dennis JE (2009). Targeting mesenchymal stem cells to activated endothelial cells. Biomaterials 30, 3702–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko IK, Kim BG, Awadallah A, Mikulan J, Lin P, Letterio JJ, and Dennis JE (2010). Targeting improves MSC treatment of inflammatory bowel disease. Mol Ther 18, 1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyfman AY, Braun GB, and Reich NO (2009). Cell-targeted self-assembled DNA nanostructures. J Am Chem Soc 131, 14237–14239. [DOI] [PubMed] [Google Scholar]
- Kuehn BM (2017). The Promise and Challenges of CAR-T Gene Therapy. In Jama (United States), pp. 2167–2169. [DOI] [PubMed]
- Lim KS, Lee DY, Valencia GM, Won YW, and Bull DA (2017). Cell surface-engineering to embed targeting ligands or tracking agents on the cell membrane. Biochem Biophys Res Commun 482, 1042–1047. [DOI] [PubMed] [Google Scholar]
- Lo CY, Antonopoulos A, Dell A, Haslam SM, Lee T, and Neelamegham S (2013). The use of surface immobilization of P-selectin glycoprotein ligand-1 on mesenchymal stem cells to facilitate selectin mediated cell tethering and rolling. Biomaterials 34, 8213–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Gao J, and Guo Z (2015). Labeling Cell Surface GPIs and GPI-Anchored Proteins through Metabolic Engineering with Artificial Inositol Derivatives. Angew Chem Int Ed Engl 54, 9679–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Pulsipher A, Dutta D, Lamb BM, and Yousaf MN (2014). Remote Control of Tissue Interactions via Engineered Photo-switchable Cell Surfaces 4, 6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Westcott N, Dutta D, Pulsipher A, Rogozhnikov D, Chen J, and Yousaf MN (2015). A Dual Receptor and Reporter for Multi-Modal Cell Surface Engineering. ACS Chem Biol 10, 2219–2226. [DOI] [PubMed] [Google Scholar]
- Merzaban JS, Imitola J, Starossom SC, Zhu B, Wang Y, Lee J, Ali AJ, Olah M, Abuelela AF, Khoury SJ, et al. (2015). Cell surface glycan engineering of neural stem cells augments neurotropism and improves recovery in a murine model of multiple sclerosis. Glycobiology 25, 1392–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A (2014). FDA approves first bispecific. Nature Reviews Drug Discovery 14, 7. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Luo W, Rogozhnikov D, Chen J, and Yousaf MN (2015). Spheroid and Tissue Assembly via Click Chemistry in Microfluidic Flow. Bioconjug Chem 26, 1939–1949. [DOI] [PubMed] [Google Scholar]
- Palte MJ, and Raines RT (2012). Interaction of nucleic acids with the glycocalyx. J Am Chem Soc 134, 6218–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, et al. (2014). The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511, 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher A, Dutta D, Luo W, and Yousaf MN (2014). Cell-surface engineering by a conjugation-and-release approach based on the formation and cleavage of oxime linkages upon mild electrochemical oxidation and reduction. Angew Chem Int Ed Engl 53, 9487–9492. [DOI] [PubMed] [Google Scholar]
- Rabuka D, Forstner MB, Groves JT, and Bertozzi CR (2008). Noncovalent cell surface engineering: incorporation of bioactive synthetic glycopolymers into cellular membranes. J Am Chem Soc 130, 5947–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozhnikov D, O’Brien PJ, Elahipanah S, and Yousaf MN (2016). Scaffold Free Bio-orthogonal Assembly of 3-Dimensional Cardiac Tissue via Cell Surface Engineering. Sci Rep 6, 39806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts WJ, and Ploemacher RE (2003). Primary murine MSC show highly efficient homing to the bone marrow but lose homing ability following culture. Leukemia 17, 160–170. [DOI] [PubMed] [Google Scholar]
- Sadelain M, Rivière I, and Riddell S (2017). Therapeutic T cell engineering. Nature 545, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Spencer JA, Phillips JA, Zhao W, Schafer S, Spelke DP, Mortensen LJ, Ruiz JP, Vemula PK, Sridharan R, et al. (2011). Engineered cell homing. Blood 118, e184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Vemula PK, Zhao W, Gupta A, Karnik R, and Karp JM (2010). Engineered mesenchymal stem cells with self-assembled vesicles for systemic cell targeting. Biomaterials 31, 5266–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selden NS, Todhunter ME, Jee NY, Liu JS, Broaders KE, and Gartner ZJ (2012). Chemically programmed cell adhesion with membrane-anchored oligonucleotides. J Am Chem Soc 134, 765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Ju E, Yan Z, Gao N, Wang J, Hou J, Zhang Y, Ren J, and Qu X (2016). Spatiotemporal control of cell-cell reversible interactions using molecular engineering. Nat Commun 7, 13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, and Sampaio JL (2011). Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol 3, a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S, Moriyama R, Mori T, and Iwasaki Y (2015). Surface engineering of macrophages with nucleic acid aptamers for the capture of circulating tumor cells. Chem Commun (Camb) 51, 17428–17430. [DOI] [PubMed] [Google Scholar]
- Swee LK, Lourido S, Bell GW, Ingram JR, and Ploegh HL (2015). One-Step Enzymatic Modification of the Cell Surface Redirects Cellular Cytotoxicity and Parasite Tropism. ACS Chemical Biology 10, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo M, Li C, Matsuda M, Nagai H, Hatanaka W, Yamamoto T, Kishimura A, Mori T, and Katayama Y (2015). Optimum design of amphiphilic polymers bearing hydrophobic groups for both cell surface ligand presentation and intercellular cross-linking. J Biomater Sci Polym Ed 26, 353–368. [DOI] [PubMed] [Google Scholar]
- Todhunter ME, Jee NY, Hughes AJ, Coyle MC, Cerchiari A, Farlow J, Garbe JC, LaBarge MA, Desai TA, and Gartner ZJ (2015). Programmed synthesis of three-dimensional tissues. Nat Methods 12, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Cheng H, Peng H, Zhou H, Li PY, and Langer R (2015). Non-genetic engineering of cells for drug delivery and cell-based therapy. Advanced Drug Delivery Reviews 91, 125–140. [DOI] [PubMed] [Google Scholar]
- Weber RJ, Liang SI, Selden NS, Desai TA, and Gartner ZJ (2014). Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules 15, 4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, and von Heijne G (2005). Transmembrane helices before, during, and after insertion. Curr Opin Struct Biol 15, 378–386. [DOI] [PubMed] [Google Scholar]
- Won YW, Patel AN, and Bull DA (2014). Cell surface engineering to enhance mesenchymal stem cell migration toward an SDF-1 gradient. Biomaterials 35, 5627–5635. [DOI] [PubMed] [Google Scholar]
- Woods EC, Kai F, Barnes JM, Pedram K, Pickup MW, Hollander MJ, Weaver VM, and Bertozzi CR (2017). A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods EC, Yee NA, Shen J, and Bertozzi CR (2015). Glycocalyx Engineering with a Recycling Glycopolymer that Increases Cell Survival In Vivo. Angew Chem Int Ed Engl 54, 15782–15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Liu H, Zhao Z, Altman MB, Lopez-Colon D, Yang CJ, Chang L-J, Liu C, and Tan W (2013). DNA Aptamer-Mediated Cell Targeting. Angewandte Chemie International Edition 52, 1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Loh W, Droujinine IA, Teo W, Kumar N, Schafer S, Cui CH, Zhang L, Sarkar D, Karnik R, et al. (2011). Mimicking the inflammatory cell adhesion cascade by nucleic acid aptamer programmed cell-cell interactions. Faseb j 25, 3045–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]