Abstract
The par genes, identified by their role in the establishment of anterior-posterior polarity in the Caenorhabditis elegans zygote, subsequently have been shown to regulate cellular polarity in diverse cell types by means of an evolutionarily conserved protein complex including PAR-3, PAR-6, and atypical protein kinase C (aPKC). The Drosophila homologs of par-1, par-3 (bazooka, baz), par-6 (DmPar-6), and pkc-3 (Drosophila aPKC, DaPKC) each are known to play conserved roles in the generation of cell polarity in the germ line as well as in epithelial and neural precursor cells within the embryo. In light of this functional conservation, we examined the potential role of baz and DaPKC in the regulation of oocyte polarity. Our analyses reveal germ-line autonomous roles for baz and DaPKC in the establishment of initial anterior-posterior polarity within germ-line cysts and maintenance of oocyte cell fate. Germ-line clonal analyses indicate both proteins are essential for two key aspects of oocyte determination: the posterior translocation of oocyte specification factors and the posterior establishment of the microtubule organizing center within the presumptive oocyte. We demonstrate BAZ and DaPKC colocalize to belt-like structures between germarial cyst cells. However, in contrast to their regulatory relationship in the Drosophila and C. elegans embryos, these proteins are not mutually dependent for their germ-line localization, nor is either protein specifically required for PAR-1 localization to the fusome. Therefore, whereas BAZ, DaPKC, and PAR-1 are functionally conserved in establishing oocyte polarity, the regulatory relationships among these genes are not well conserved, indicating these molecules function differently in different cellular contexts.
Oocyte differentiation of Drosophila depends on molecular mechanisms that govern the establishment and maintenance of cellular polarity. Oogenesis is initiated when a germ-line stem cell (GSC) divides asymmetrically to produce a daughter GSC and a differentiating daughter termed the cystoblast (reviewed in refs. 1 and 2). The cystoblast subsequently undergoes four rounds of division with incomplete cytokinesis, generating a 16-cell germ-line cyst in which each cyst cell is interconnected by means of stable cytoplasmic bridges termed ring canals. During the four cystocyte mitoses, one pole of each mitotic spindle is anchored by the fusome, which is asymmetrically partitioned and thought to polarize the cyst and thus ensure the stereotyped pattern of interconnections within the cyst (3, 4). The pattern of ring canal connections reflects the inherent asymmetry of the cyst as only one of the two cells with four ring canals will differentiate as the oocyte, whereas the remaining 15 cells differentiate as nurse cells (2). The process of oocyte determination depends, at least in part, on the differential accumulation of oocyte specification factors such as oo18 RNA binding protein (ORB) (5), Bicaudal-D (BIC-D) (6), and dynein heavy chain 64C (DHC64C) (7) within a single posterior cell of the cyst. The directional transport of such factors has been demonstrated to depend on the establishment of a polarized microtubule network that emanates from a posteriorly localized microtubule organizing center (MTOC) within the future oocyte (8), thus implicating the microtubule cytoskeleton in regulating oocyte differentiation. However, oocyte determination is more complex than a simple microtubule-based transport mechanism because the synaptonemal complex, which marks the meiotic pro-oocyte, becomes restricted to a single cell of the cyst in a microtubule-independent manner (9).
The par genes, initially characterized in the Caenorhabditis elegans zygote (10–13), are known to regulate anterior-posterior (A-P) axis determination and cellular polarity in diverse cell types including embryonic blastomeres (14,15), epithelial cells (16–20), and neural precursor cells (refs. 21–24; reviewed in refs. 25 and 26). Drosophila par-1 represents the first fusomal component identified with a specific role in oocyte cell fate maintenance (27, 28). In addition, par-1 is required in late oogenesis to regulate A-P axis formation (29, 30). Given the striking functional conservation of the par genes in mediating cellular polarity in diverse tissue types of evolutionarily distant organisms (reviewed in ref. 31), the demonstrated requirement of par-1 function in oogenesis suggests that other par gene homologs and their effectors also may function in germ-line cyst polarization and oocyte differentiation. The C. elegans PAR-3 homolog, Bazooka (BAZ), has been demonstrated to localize to the apical side of epithelial cells and neuroblasts of the embryo where it functions in mediating epithelial polarity and the asymmetric localization of cell fate determinants during neuroblast cell divisions (21–23). The atypical protein kinase C (aPKC) gene product has been shown to localize with PAR-3 and PAR-6 in a functional complex in C. elegans, Drosophila, and mammalian cells (17–19, 24, 32–34). Furthermore, like BAZ and DmPAR-6 (20, 22, 23), DaPKC is required to maintain epithelial polarity as well as regulate the differential localization of cell fate determinants during neuroblast divisions in the embryo (24). A recent study (35) implicated both BAZ and DmPAR-6 in the regulation of oocyte differentiation in Drosophila. Here we report our germ-line mosaic analyses of null alleles of DaPKC and baz, which reveal germ-line-autonomous roles for DaPKC and BAZ in oocyte determination. These analyses both confirm and extend our mechanistic understanding of how germ-line baz, as well as DaPKC, regulate oocyte differentiation.
Materials and Methods
Drosophila Strains and Genetic Clonal Analyses.
The bazXi106 (16) and bazEH171 null alleles were recombined onto the [FRT]9–2 chromosome, and the DaPKCk06403 null allele was recombined onto the [FRT]42D chromosome. The following strains were used to generate germ-line clones by the FLP/FRT technique (36): yw bazXi106 P[mini-w+, FRT]9–2/FM6, yw B; pr pwn P[ry+; hsFLP]38/CyO, yw bazEH171 P[mini-w+, FRT]9–2/FM6, yw B; pr pwn P[ry+; hsFLP]38/CyO, yw P[w+; ubi-nls-GFP] P[mini-w+, FRT]9–2, yw; P[ry+, FRT]42D DaPKCK06403, yw P[mini-w+, hsFLP]1; P[ry+, FRT]42D P[mini-w+; ubi-nls-GFP]2R/CyO, yw P[w+, hsFLP]1; P[mini-w+, FRT]G13 P[w+; ubi-nls-GFP], yw; P[mini-w+, FRT]G13 par-1Δ16/CyO (29). The P[w+; ubi-nls-GFP] FRT chromosomes bear a polyubiquitin promoter that drives ubiquitous nuclear green fluorescent protein (GFP) expression. Germ-line clones were induced by administering a 1-h heat shock at 37°C on 2 consecutive days during the third instar stage. Adult ovaries were subsequently processed for immunofluorescence as described (27) in which germ-line clones were identified by the absence of nuclear GFP expression.
Immunohistochemistry.
Wild-type and mosaic ovaries were dissected, fixed, and stained as described (27). The following antisera were used: rabbit anti-BAZ (1:1,500; A. Wodarz, Heinrich Heine Universitaet, Dusseldorf); rabbit anti-PKCζ C20 (1:1,000; Santa Cruz Biotechnology); rabbit anti-PAR-1 (1:5,000; ref. 29); mouse anti-1B1 [1:10; Developmental Studies Hybridoma Bank (DSHB)]; mouse anti-ARM (1:200; DSHB); mouse anti-ORB (1:30; DSHB); mouse anti-BIC-D (1:20; ref. 8); mouse anti-DHC64C (1:500; ref. 7); rat anti-α-tubulin (1:10; Accurate Chemicals); and rat anti-DE-cadherin (1:20; ref. 37). Rhodamine-conjugated phalloidin (Sigma) was used at 1:50 to visualize F actin. Cy2-, rhodamine red X-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch; Molecular Probes) were used at 1:200 to 1:500. Fluorescently labeled samples were counterstained with either propidium iodide or 4′,6′-diamidino-2-phenylindole to visualize DNA. Micrograph images were collected and processed as described (27).
Results
baz and DaPKC Function Germ Line Autonomously in Regulating Oocyte Differentiation.
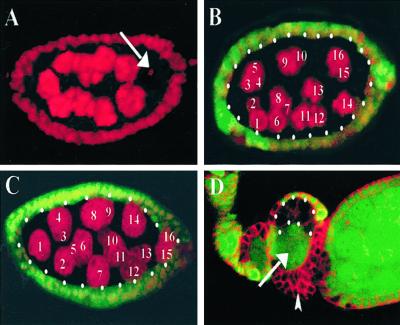
To examine the potential oogenic function of baz and DaPKC, we generated protein null germ-line mutant clones for both baz and DaPKC. Germ-line clones, identified by the absence of nuclear GFP expression, were counterstained with the chromatin marker propidium iodide to examine the number and ploidy of the germ-line nuclei (Fig. 1). In contrast to wild-type egg chambers, which invariably contain 15 nurse cells and a single oocyte (Fig. 1A; n = 50; 100% penetrance), baz mutant germ-line clones, while containing the normal complement of germ-line nuclei, fail to differentiate an oocyte, resulting in a 16-nurse cell phenotype as revealed by the polyploid state of all 16 germ-line nuclei (Fig. 1B; n = 150; 97.1% penetrance). Similarly, DaPKCk06403 germ-line clones also fail to differentiate an oocyte as indicated by the presence of 16 polyploid nurse cell nuclei in germ-line mutant egg chambers (Fig. 1C; n = 50; 92% penetrance). These results reveal a germ-line autonomous requirement for DaPKC and confirm the role of BAZ (35) in oocyte differentiation and/or maintenance.
Figure 1.
baz and DaPKC are cell-autonomously required in the germ line for oocyte differentiation. (A) Wild-type stage 6 egg chamber labeled with the chromatin marker propidium iodide reveals the presence of 15 nurse cells and a single posterior oocyte (arrow). (B) Stage 6 baz germ-line mutant egg chamber stained with propidium iodide (red) reveals all 16 nuclei develop as polyploid nurse cells, indicating a failure in oocyte differentiation. (C) Stage 6 DaPKC mutant egg chamber stained with propidium iodide (red) reveals the presence of 16 nurse cell nuclei and no oocyte. (D) baz mosaic egg chambers double-labeled with α-tubulin (red) and nuclear GFP (green). Mispartitioning of baz+ nurse cell nuclei (arrow) into an otherwise baz null germ-line clone caused by multilayering of baz null follicle cell clones (arrowhead). Mutant cysts are encircled by white dots. Anterior is left. (Magnifications: A–C, ×40; D, ×20.)
Our analyses further reveal that germ-line depletion of either DaPKC (data not shown) or baz (Fig. 1D, arrow) function from the follicle cells leads to their multilayering, which disrupts the normal partitioning of germ-line nuclei to successively mature egg chambers caused by mispositioning of mutant follicle cells (Fig. 1D). These mispartitioned baz+ nurse cell nuclei, into an otherwise baz null germ-line clone, may provide the threshold of germ-line BAZ required to rescue the oocyte differentiation defect and allow production of a mature egg. Alternatively, the BAZ protein may exhibit a long perdurance after mitotic clone induction, which depletes over a period of days, resulting in the cessation of egg production in these mosaic females. Our results, however, in no way invalidate the previous conclusions that maternally provided BAZ masks the severity of the embryonic polarity phenotype in both epithelial cells and neuroblasts (16, 22, 23). Rather, our results indicate that these embryos are not likely to represent a complete maternal depletion for BAZ.
BAZ and DaPKC Function in Establishing Initial A-P Polarity within the Oocyte.
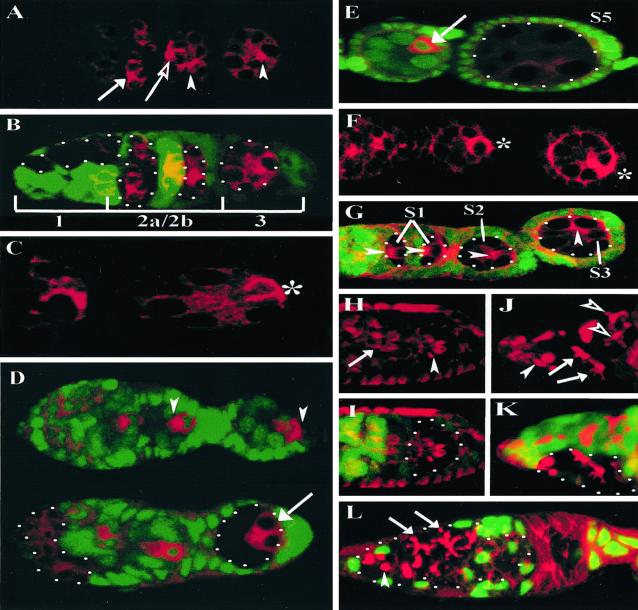
Oocyte differentiation requires the polarized accumulation of oocyte specification factors within a single cell of the germ-line cyst (5–7). To analyze the role of baz or DaPKC in the localization of these factors, we generated mutant germ-line clones for both genes and examined the expression of the oocyte specification factors ORB, BIC-D, and the microtubule motor protein DHC64C at early and late stages of oogenesis. In wild-type germarial cysts, both ORB (Fig. 2A, arrow) and BIC-D (data not shown) are initially uniformly distributed among the cyst cells in region 2a, and then both molecules are targeted first to the two pro-oocytes and ultimately to the fated oocyte by late region 2a (5) (Fig. 2A, open arrow and arrowhead, for wild-type and mutant cysts, respectively). Furthermore, whereas ORB protein initially concentrates at the anterior of the oocyte, it translocates to the posterior pole of the oocyte and condenses into a posterior crescent in region 3 (5) (Fig. 2C, *). In contrast, ORB fails to translocate from the anterior to a posterior crescent in both baz (Fig. 2A, arrowhead) and DaPKC (data not shown) null germ-line cysts in germarial region 3 and rather remains at the anterior margin of the presumptive oocyte. We observed an identical defect in A-P BIC-D translocation in baz and DaPKC null germ-line clones in germarial region 3 (data not shown). The defect in the translocation of ORB and BIC-D to the posterior of the oocyte at this early stage is subsequently manifest by a failure to accumulate these proteins in later-stage oocytes (Fig. 2E and data not shown).
Figure 2.
BAZ and DaPKC are required to establish A-P polarity in the oocyte. (A and B) Germarium containing wild-type and baz null germ-line cysts double-labeled for ORB (red) and nuclear GFP (green) to mark baz clones (B, merge). (A) In region 2, both wild-type (open arrow) and baz null (arrowhead) germ-line cysts accumulate ORB in the two pro-oocytes. In contrast to wild-type (C, *), the baz null germ-line cyst in region 3 fails to translocate ORB from the anterior to a posterior crescent in the presumptive oocyte (A, arrowhead). (D) Wild-type and DaPKC mosaic germaria double-labeled for DHC64C (red) and nuclear GFP (green). In contrast to wild-type cysts (Upper) in which DHC64C localizes to a single posterior cell (arrowheads), in DaPKC mutant cysts (Lower) DHC64C fails to accumulate in a single cell but rather is localized to the two posterior-most cyst cells (arrow). (E) Wild-type and DaPKC mutant egg chambers double-labeled for DHC64C (red) and nuclear GFP (green) reveal that in contrast to wild-type (arrow), DaPKC mutant egg chambers fail to accumulate DHC64C. (F and G) In contrast to a wild-type germarium (F, *), analyses of baz null germ-line cysts up to stage 3 (S3) reveals a defect in the A-P MTOC transition (G, arrowheads). (H and I) Germarium double-labeled with rhodamine phalloidin (red) and nuclear GFP (green) reveals no defect in ring canal formation or spatial distribution within baz null germ-line cysts (H, arrowhead) as compared with a wild-type cyst (H, arrow). (J and K) Germarium double-labeled with 1B1 (J, red) and nuclear GFP (K, green, merge) indicates that baz mutant cysts display relatively normal fusome morphology albeit a somewhat weaker elaboration of fusome branches (J, open arrowhead), when compared with wild-type cysts (J, arrows). Spectrosome formation (J, filled arrowhead) is likewise unaffected in baz mutant germ-line clones. (L) DaPKC mosaic germarium double-labeled with 1B1 (red) and nuclear GFP (green) reveals no defect in spectrosome (arrowhead) or fusome assembly (arrows) in DaPKC mutant cysts. (Magnifications: A, B, D, and F–L, ×40; C, ×63; E, ×25.)
Furthermore, in contrast to wild-type germ-line cysts in which DHC64C localizes to a single posterior cell, in DaPKC null germ-line clones DHC64C fails to localize to a single cell posteriorly, but rather accumulates in the two posterior-most presumptive pro-oocytes of the mutant germ-line cyst (Fig. 2D, arrow). Therefore, baz (35) and DaPKC display essentially identical phenotypes in germ-line mutant clones with regards to oocyte differentiation and the establishment of initial A-P polarity within the oocyte. The failure to maintain oocyte identity in either baz or DaPKC mutant cysts can therefore be directly correlated with defects in the A-P translocation of oocyte specification factors within a single posterior cell of a germ-line cyst, suggesting oocyte differentiation depends on this early polarization event.
The posterior assembly of a functional MTOC has been directly implicated in the differential segregation of oocyte specification factors within developing germ-line cysts (8), suggesting that the failure to translocate these factors to a posterior crescent in region 3 baz or DaPKC mutant cysts (Fig. 2 A–E and data not shown) may result from a defect in microtubule reorganization within these mutant cysts. In contrast to wild-type (Fig. 2F, *), baz (Fig. 2G, arrowheads) and DaPKC (data not shown) mutant cysts display a parallel defect in the A-P transition of the MTOC within the presumptive oocyte. These results support the conclusion that the defects observed in posterior translocation of oocyte specification factors in these mutants are likely caused, at least in part, by the observed disruption in the A-P transition of the oocyte MTOC.
In addition to the microtubule network, both ring canals and the fusome play critical roles in cyst polarization and oocyte differentiation. We therefore analyzed the formation and spatial distribution of ring canals as well as fusome morphogenesis in both baz and DaPKC mutant germ-line cysts. In baz mutant cysts (Fig. 2H, arrowheads), ring canal formation and spatial distribution are indistinguishable from wild-type germ-line cysts (Fig. 2H, arrow). Furthermore, the wild-type spatial arrangement of ring canals in baz null germ-line cysts suggests there is no apparent disruption in germ cell adhesion within the cyst. Similarly, we observed no defects in ring canal formation or spatial distribution in DaPKC mutant cysts (data not shown).
Consistent with previous studies (35), our analyses indicate baz mutant cysts display relatively normal fusome morphology, although mutant fusome branches appear slightly thinner (Fig. 2J, arrows) when compared with wild-type fusomes within the same germarium (Fig. 2J, open arrowheads). Similarly, we observed no apparent defect in fusome morphology in DaPKC null germ-line cysts, indicating DaPKC is dispensable in the germ line for proper fusome morphogenesis (Fig. 2L, arrows).
BAZ and DaPKC Colocalize to Belt-Like Structures Between Germ-Line Cyst Cells and to Apical Junctions in the Follicular Epithelium.
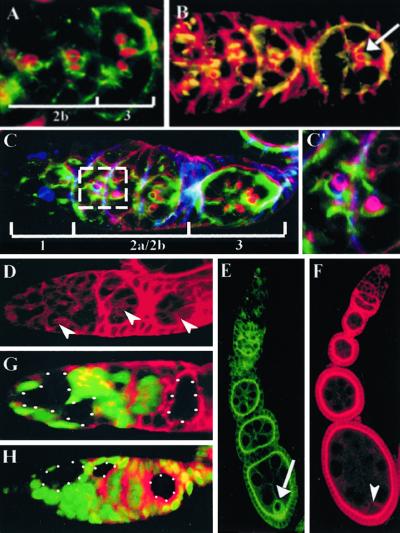
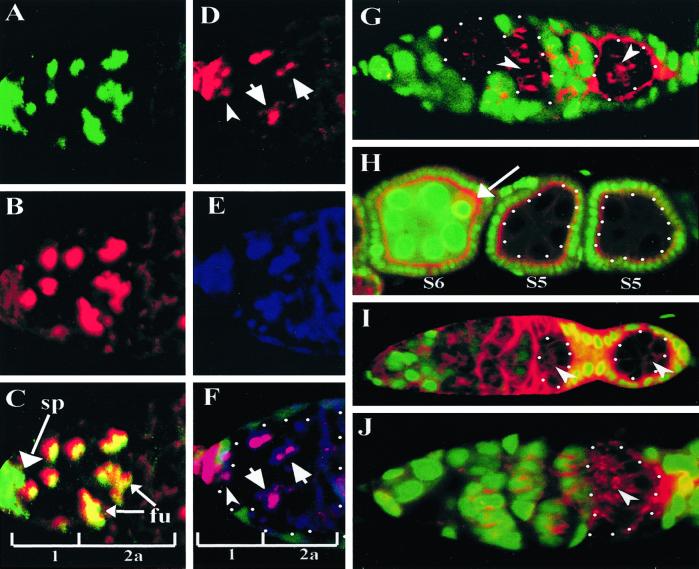
To investigate the mechanism by which BAZ and DaPKC exert their effects on oocyte differentiation, we analyzed the localization of these proteins in the germ line and soma during oogenesis (Fig. 3). The specificity of the BAZ and DaPKC antibodies were verified by using mosaic ovaries containing baz or DaPKC null mutant clones (Fig. 3 G and H). BAZ is first detected in the germarium as a belt-like specialization on germ cell membranes at sites of germ cell interconnection (Fig. 3A). These belt structures are reminiscent of ring canals with respect to their position between germ-line cyst cells; however, in contrast to ring canals these “BAZ belts” are approximately 2-fold greater in diameter. Germaria double-labeled for BAZ and rhodamine phalloidin reveal the BAZ belts localize adjacent to ring canals and further reveal an approximate 1:1 ratio between the two structures within germ-line cysts (Fig. 3A). We observed the microtubule cytoskeleton (Fig. 3B, arrow) as well as the fusome (Fig. 3 C and C′) projecting through individual cystocytes coincident with the site of BAZ belt expression on the germ cell membrane. Later in oogenesis, BAZ is transiently enriched in the oocyte cytoplasm at stages 5–6 before the onset of vitellogenesis (Fig. 3E, arrow) and is subsequently undetectable in the germ line of vitellogenic egg chambers. As with BAZ localization to the apical junctional zone in the embryonic epithelium (22, 23), we likewise observed tight apical localization of BAZ in follicular epithelia (Fig. 3E).
Figure 3.
BAZ and DaPKC localize in the germ line to belt-like structures in germaria and to the apical junctional zone in follicle cells. (A) Wild-type germarium double-labeled for BAZ (green) and rhodamine phalloidin (red) reveals an approximate 1:1 ratio of BAZ belts (green) to ring canals (red). (B) Wild-type germarium double-labeled for BAZ (green) and α-tubulin (red) reveals microtubules project through the region of BAZ belt expression between germ cells (arrow). (C) Wild-type germarium triple-labeled for BAZ (green), rhodamine phalloidin (red), and 1B1 (blue) reveals the fusome projects through both BAZ belts and ring canals (C′, high magnification view of boxed area in C). (D) Wild-type germarium stained for DaPKC reveals localization to BAZ belt structures and to apical membranes of somatic cells, which interdigitate between germ-line cyst cells. (E) Wild-type ovariole stained for BAZ reveals transient localization to the oocyte in stage 5–6 egg chambers (arrow) and apical localization in follicle cells. (F) Wild-type ovariole labeled for DaPKC indicates an enrichment on the oocyte cortex (arrowheads) and an apical concentration in follicular epithelia. (G and H) Antibody specificity for BAZ (G) and DaPKC (H) antisera was assayed by labeling baz and DaPKC germ-line mosaic germaria with nuclear GFP (green) and BAZ or DaPKC, respectively. Protein null germ-line clones are encircled by white dots. (Magnifications: A, ×60; B–D, G, and H, ×40; C, ×100; E and F, ×20.)
Consistent with previous studies demonstrating the colocalization of BAZ and DaPKC in embryonic epithelia and neural precursor cells, we find DaPKC localizes to the BAZ belts in the germarium (Fig. 3D, arrows), whereas in follicle cells DaPKC is apically constricted consistent with BAZ localization in these cells (Fig. 3F).
BAZ and DaPKC Colocalize with DE-Cadherin and Armadillo (ARM) in Germarial Cysts.
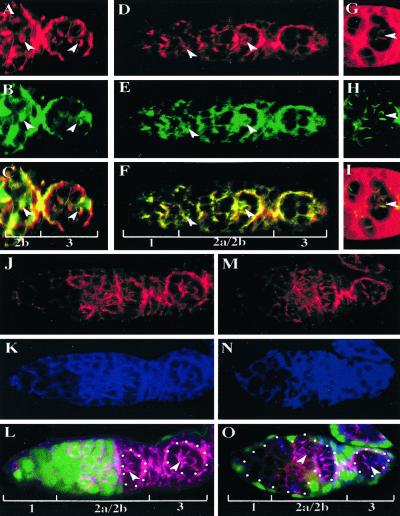
In embryonic epithelia, BAZ, DmPAR-6, and DaPKC apically colocalize and partially overlap with the apico-laterally enriched ARM and DE-cadherin proteins in the region of the apical zonula adherens. Furthermore, these proteins are required to maintain epithelial cell polarity and are mutually dependent for their proper localization (ref. 19, reviewed in ref. 38). Interestingly, previous studies (39, 40) revealed DE-cadherin is localized to belt-like structures adjacent to ring canals in region 2 germarial cysts reminiscent of BAZ belt localization, suggesting that these adherens junction components may similarly colocalize in the germ line. To further investigate the molecular and functional nature of BAZ belt expression in the germarium, we labeled germaria with anti-BAZ, anti-ARM, and anti-DE-cadherin antibodies and examined their potential colocalization within the germarium (Fig. 4). Our analyses reveal BAZ colocalizes with both DE-cadherin (Fig. 4 A–C, arrowheads) and ARM (Fig. 4 D–F, arrowheads) to the BAZ belts within the germarium. Furthermore, consistent with the colocalization of BAZ and DaPKC to BAZ belts within the germarium, we observed partial colocalization of DaPKC with DE-cadherin in wild-type germarial cysts (Fig. 4 G–I, arrowheads). To assay whether these proteins are mutually dependent for their germ-line localization, we examined the localization of DE-cadherin and ARM in baz null germ-line clones. In contrast to their mutual dependence in embryonic epithelia, our analyses indicate germ-line BAZ function is dispensable for the localization of either DE-cadherin or ARM to the BAZ belts in the germarium (Fig. 4 J–L). We also find that DaPKC is dispensable for the localization of either DE-cadherin or ARM to these structures (Fig. 4 M–O). These results indicate BAZ and DaPKC function are not required for the formation of these structures because both ARM and DE-cadherin localization to these belts in baz or DaPKC mutant cysts (Fig. 4 L and O, arrowheads) is indistinguishable from that observed in wild-type cysts (Fig. 4L, arrow). Previous studies revealed that germ-line clones of a strong allele of shotgun (shgIG29), the gene encoding DE-cadherin, disrupt the arrangement of germ cells in region 2b germarial cysts (40), suggesting DE-cadherin may mediate germ cell adhesion. In contrast, our analyses of ring canal spatial distribution in baz and DaPKC mosaic cysts (Fig. 2H, arrowheads and data not shown) strongly suggests BAZ and DaPKC function are dispensable for normal germ cell adhesion. Furthermore, germ-line clonal analyses of either shg or arm reveal neither gene is required for oocyte differentiation (refs. 39–41; reviewed in ref. 38), whereas both baz and DaPKC are essential in oocyte determination. These results suggest the components of the BAZ belts likely mediate diverse cellular functions essential for germ-line cyst development, which may include cell adhesion, cell signaling, and cyst polarization.
Figure 4.
BAZ and DaPKC colocalize with DE-cadherin and ARM to BAZ belts of germarial cysts. (A–C) Wild-type germarium double-labeled for DE-cadherin (A, red) and BAZ (B, green) reveals colocalization of DE-cadherin and BAZ to BAZ belts (arrowheads in A–C) between germ cells (C, merge). (D–F) Wild-type germarium double-labeled for ARM (D, red) and BAZ (E, green) reveals colocalization of ARM and BAZ to BAZ belts (arrowheads in D–F) (F, merge). (G–I) Wild-type germarium double-labeled for DE-cadherin (G, red) and DaPKC (H, green) reveals partial colocalization of DE-cadherin and DaPKC (I, merge, arrowheads). (J–L) baz mosaic germarium triple-labeled for DE-cadherin (J, red), ARM (K, blue), and nuclear GFP (L, green, merge) reveals baz is dispensable for localization of DE-cadherin as well as ARM to the BAZ belts (L, arrowheads) as compared with wild-type cysts (L, arrow). (M–O) DaPKC mosaic germarium triple-labeled for DE-cadherin (M, red), ARM (N, blue), and nuclear GFP (O, green, merge) reveals DaPKC is dispensable for localization of DE-cadherin and ARM to BAZ belts (O, arrowheads). (Magnification: ×40.)
Localization of BAZ, DaPKC, and PAR-1 in the Germ Line Is Mutually Independent.
In the C. elegans zygote, the PAR-3/PAR-6/PKC-3 complex is localized to the anterior where it is required for the posterior localization of PAR-1 (reviewed in ref. 15). To investigate the potential regulatory relationship between baz and par-1 in the Drosophila germ line, we generated baz null germ-line clones and analyzed PAR-1 expression and localization. In wild-type germ-line cysts, PAR-1 is localized to the spectrosome and fusome in germarial regions 1 and 2a and subsequently is down-regulated on fusomes in regions 2b and 3 (Fig. 5 A–C). In baz mutant germ-line cysts, PAR-1 localization to the spectrosome is unaffected (Fig. 5 D–F, arrowhead) and is likewise present on fusomes although somewhat weaker localization is observed on fusome branches of baz mutant germ-line cysts (Fig. 5 D–F, arrows) when compared with wild-type germ-line cysts. Taken together, these results suggest germ-line baz is dispensable in the germarium for normal PAR-1 expression and localization.
Figure 5.
BAZ, DaPKC, and PAR-1 are mutually independent for their localization in the germ line. (A–C) Wild-type germarium double-labeled for PAR-1 (A, green) and the fusome marker 1B1 (B, red). PAR-1 is localized to the spectrosome (sp) and fusome (fu) in region 1 and 2a of the germarium (C, merge, arrows). (D–F) baz mosaic germ-line cysts triple-labeled for PAR-1 (D and F, red), 1B1 (E and F, blue), and nuclear GFP (F, green, merge). In baz mutant cysts, PAR-1 localization to either the spectrosome in region 1 (D and F, arrowhead) or the fusomes in regions 1 and 2a, is relatively unaffected although there appears to be a slight decrease in PAR-1 localization to fusomal branches in baz mutant cysts (D and F, arrows). (G) par-1 mosaic germ-line cysts double-labeled with BAZ (red) and nuclear GFP (green) indicates PAR-1 is dispensable in the germ line for BAZ belt expression (arrowheads). (H) Wild-type and par-1 mutant egg chambers double-labeled with BAZ (red) and nuclear GFP (green) reveals in contrast to wild-type stage 5–6 (S5, S6) egg chambers (arrow), there is no posterior localization of BAZ in similarly staged par-1 null germ-line clones (S5). (I) baz mutant germ-line cysts double-labeled for DaPKC (red) and nuclear GFP (green) reveals baz is dispensable for DaPKC localization within the germarium (arrowheads). (J) DaPKC mutant germ-line cysts double-labeled for BAZ (red) and nuclear GFP (green) reveals DaPKC is dispensable for BAZ localization within the germarium (arrowhead). (Magnifications: A–F, ×80; G, I, and J, ×40; H, ×20.)
To determine whether par-1 may regulate BAZ expression, we generated par-1 null germ-line clones and analyzed BAZ localization. We observed no defect in BAZ belt expression in par-1 mutant germ-line cysts (Fig. 5G, arrowheads). Furthermore, in contrast to wild-type stage 5–6 egg chambers in which BAZ is transiently enriched in the oocyte cytoplasm (Fig. 2D, arrow), in par-1 mutant egg chambers BAZ localization is abolished from the posterior presumably due to the defect in oocyte differentiation observed in par-1 mutant egg chambers (27, 28) (Fig. 5H). These results indicate germ-line par-1 is dispensable for normal BAZ belt expression and localization.
To assay the regulatory relationship between baz and DaPKC we analyzed the localization of DaPKC in baz mutant germ-line cysts (Fig. 5I), as well as the localization of BAZ in DaPKC mutant germ-line cysts (Fig. 5J). In contrast to their mutual dependence for localization in the embryo, we find that both BAZ and DaPKC localization within the germ line is mutually independent (Fig. 5 I and J, arrowheads). These results indicate that despite the apparent functional conservation of BAZ, DaPKC, and PAR-1 in generating oocyte polarity, the regulatory relationships among these genes are not conserved in the germ line.
Discussion
We demonstrate that both DaPKC and BAZ, like PAR-1, are cell-autonomously required in the germ line for the initial establishment of A-P polarity within the germ-line cyst and for the subsequent maintenance of oocyte fate during Drosophila oogenesis. The colocalization of BAZ, DaPKC, ARM, and DE-cadherin to the BAZ belt structures in the germarium taken together with the results of independent germ-line clonal analyses for each of these genes (present study, refs. 38–41) strongly suggests the components of the BAZ belts are capable of mediating a multiplicity of functions in germ-line cysts. In embryonic epithelia, these proteins function in the formation of the apical zonula adherens junction and are mutually dependent for their apical localization (refs. 16 and 19, reviewed in ref. 38). The germ-line colocalization of these molecules to the BAZ belts suggests these structures may represent a potential polarity cue on the germ cell plasma membrane. The restricted localization of BAZ belt components to germ cell membranes at points of cell–cell contact represents an asymmetry on the plasma membrane of germ-line cyst cells with regards to the A-P axis of the cyst and stage 1 oocyte. The asymmetric localization of these molecules to one side of the germ cell plasma membrane may act as a polarity cue in defining anterior versus posterior within individual cystocytes of the 16-cell germ-line cyst and thus contribute to the establishment of an initial A-P axis and to subsequent germ-line cyst polarization. Our results further suggest that BAZ and DaPKC likely function in a signaling capacity, rather than a structural one, to mediate oocyte differentiation, whereas DE-cadherin and ARM are more likely to function in maintaining germ cell adhesion and cyst integrity.
Consistent with par gene function in other systems, our results indicate that BAZ, DaPKC, and PAR-1 are required for the establishment and maintenance of cellular polarity in the Drosophila germ line; however, the regulatory relationships observed between these genes in the germ line versus that observed in embryonic blastomeres, epithelial cells, and neural precursor cells indicates that, while these molecules are functionally conserved, the mechanisms by which these genes act appear to be less well conserved. In contrast to their mutual dependence for localization in the embryo, BAZ, DaPKC, and PAR-1 each are mutually independent for their localization in the germ line. Taken together, these results underscore the functional utility of the par genes and their effectors as a molecular module for generating cellular polarity in diverse cell types. Furthermore, the independence of BAZ, DaPKC, and PAR-1 in their germ-line localization provides a unique opportunity to probe new mechanisms by which these highly conserved proteins function in regulating diverse processes such as cellular polarity, asymmetric cell division, or growth control.
Acknowledgments
We thank A. Wodarz, B. Suter, T. Uemura, and S. Luschnig for providing reagents. We thank S. Younger-Shephard for generating the bazEH171 FRT9–2 recombinant chromosome. D.N.C. is a Fellow of The Jane Coffin Childs Fund for Medical Research. S.A.S. was supported by a fellowship of the Deutsche Forschungsgemeinschaft and the Howard Hughes Medical Institute. L.Y.J. and Y-N.J. are Investigators of the Howard Hughes Medical Institute.
Abbreviations
- BAZ
Bazooka
- PKC
protein kinase C
- aPKC
atypical PKC
- DaPKC
Drosophila aPKC
- MTOC
microtubule organizing center
- A-P
anterior-posterior
- BIC-D
Bicaudal-D
- DHC64C
dynein heavy chain 64C
- GFP
green fluorescent protein
- ARM
Armadillo
- ORB
oo18 RNA binding protein
References
- 1.Spradling A C. In: Drosophila Development. Bate M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1–70. [Google Scholar]
- 2.de Cuevas M, Lilly M A, Spradling A C. Annu Rev Genet. 1997;31:405–428. doi: 10.1146/annurev.genet.31.1.405. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Spradling A C. Dev Genet. 1995;16:6–12. doi: 10.1002/dvg.1020160104. [DOI] [PubMed] [Google Scholar]
- 4.Deng W, Lin H. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- 5.Lantz V, Chang J, Horabin J, Bopp D, Schedl P. Genes Dev. 1994;8:598–613. doi: 10.1101/gad.8.5.598. [DOI] [PubMed] [Google Scholar]
- 6.Suter B, Romberg L, Steward R. Genes Dev. 1989;3:1957–1968. doi: 10.1101/gad.3.12a.1957. [DOI] [PubMed] [Google Scholar]
- 7.McGrail M, Hays T S. Development (Cambridge, UK) 1997;124:2409–2419. doi: 10.1242/dev.124.12.2409. [DOI] [PubMed] [Google Scholar]
- 8.Theurkauf W E, Alberts B M, Jan Y N, Jongens T A. Development (Cambridge, UK) 1993;118:1169–1180. doi: 10.1242/dev.118.4.1169. [DOI] [PubMed] [Google Scholar]
- 9.Huynh J R, St. Johnston D. Development (Cambridge, UK) 2000;127:2785–2794. doi: 10.1242/dev.127.13.2785. [DOI] [PubMed] [Google Scholar]
- 10.Guo S, Kemphues K J. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 11.Etemad-Moghadam B, Guo S, Kemphues K J. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- 12.Watts J L, Etemad-Moghadam B, Guo S, Boyd L, Draper B W, Mello C C, Priess J R, Kemphues K J. Development (Cambridge, UK) 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- 13.Hung T J, Kemphues K J. Development (Cambridge, UK) 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- 14.Guo S, Kemphues K J. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- 15.Bowerman B, Shelton C A. Curr Opin Genet Dev. 1999;9:390–395. doi: 10.1016/S0959-437X(99)80059-8. [DOI] [PubMed] [Google Scholar]
- 16.Muller H A, Wieschaus E. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joberty G, Petersen C, Gao L, Macara I G. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 18.Lin D, Edwards A S, Fawcett J P, Mbamalu G, Scott J D, Pawson T. Nat Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- 19.Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y-N, Stainier D Y R, Abdelilah-Seyfried S. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 20.Petronczki M, Knoblich J A. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 21.Kuchinke U, Grawe F, Knust E. Curr Biol. 1998;8:1357–1365. doi: 10.1016/s0960-9822(98)00016-5. [DOI] [PubMed] [Google Scholar]
- 22.Wodarz A, Ramrath A, Kuchinke U, Knust E. Nature (London) 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 23.Schober M, Schaefer M, Knoblich J A. Nature (London) 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- 24.Wodarz A, Ramrath A, Grimm A, Knust E. J Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jan Y N, Jan L Y. Cell. 2000;100:599–602. doi: 10.1016/s0092-8674(00)80695-9. [DOI] [PubMed] [Google Scholar]
- 26.Lu B, Jan L, Jan Y N. Annu Rev Neurosci. 2000;23:531–556. doi: 10.1146/annurev.neuro.23.1.531. [DOI] [PubMed] [Google Scholar]
- 27.Cox D N, Lu B, Sun T-Q, Williams L T, Jan Y N. Curr Biol. 2001;11:75–87. doi: 10.1016/s0960-9822(01)00027-6. [DOI] [PubMed] [Google Scholar]
- 28.Huynh J-R, Shulman J M, Benton R, St. Johnston D. Development (Cambridge, UK) 2001;128:1201–1209. doi: 10.1242/dev.128.7.1201. [DOI] [PubMed] [Google Scholar]
- 29.Shulman J M, Benton R, St. Johnston D. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- 30.Tomancak P, Piano F, Riechmann V, Gunsalus K C, Kemphues K J, Ephrussi A. Nat Cell Biol. 2000;7:458–460. doi: 10.1038/35017101. [DOI] [PubMed] [Google Scholar]
- 31.Doe C Q. Nat Cell Biol. 2001;3:E7–E9. doi: 10.1038/35050684. [DOI] [PubMed] [Google Scholar]
- 32.Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues K J, Ohno S. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabuse Y, Izumi Y, Piano F, Kemphues K J, Miwa J, Ohno S. Development (Cambridge, UK) 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- 34.Wu S L, Staudinger T, Olson E N, Rubin C S. J Biol Chem. 1998;273:1130–1143. doi: 10.1074/jbc.273.2.1130. [DOI] [PubMed] [Google Scholar]
- 35.Huynh J-R, Petronczki M, Knoblich J A, St. Johnston D. Curr Biol. 2001;11:901–906. doi: 10.1016/s0960-9822(01)00244-5. [DOI] [PubMed] [Google Scholar]
- 36.Chou T-B, Perrimon N. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, Takeichi M. Genes Dev. 1996;10:659–671. doi: 10.1101/gad.10.6.659. [DOI] [PubMed] [Google Scholar]
- 38.Muller H-A J. Dev Dyn. 2000;218:52–67. doi: 10.1002/(SICI)1097-0177(200005)218:1<52::AID-DVDY5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 39.Godt D, Tepass U. Nature (London) 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Reyes A, St. Johnston D. Development (Cambridge, UK) 1998;125:3635–3644. doi: 10.1242/dev.125.18.3635. [DOI] [PubMed] [Google Scholar]
- 41.Cox R T, Kirkpatrick C, Peifer M. J Cell Biol. 1996;134:133–148. doi: 10.1083/jcb.134.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]