Abstract
Sequencing of the Drosophila genome has revealed that there are “silent” homologues of many important genes—family members that were not detected by classic genetic approaches. Why have so many homologues been conserved during evolution? Perhaps each one has a different but important function in every system. Perhaps each one works independently in a different part of the body. Or, perhaps some are redundant. Here, we take one well known gene family and analyze how the individual members contribute to the making of one system, the tracheae. There are seven DWnt genes in the Drosophila genome, including wingless (wg). The wg gene helps to pattern the developing trachea but is not responsible for all Wnt functions there. We test each one of the seven DWnts in several ways and find evidence that wg and DWnt2 can function in the developing trachea: when both genes are removed together, the phenotype is identical or very similar to that observed when the Wnt pathway is shut down. DWnt2 is expressed near the tracheal cells in the embryo in a different pattern to wg but is also transduced through the canonical Wnt pathway. We find that the seven DWnt genes vary in their effectiveness in specific tissues, such as the tracheae, and, moreover, the epidermis and the tracheae respond to DWnt2 and Wg differently. We suggest that the main advantage of retaining a number of similar genes is that it allows more subtle forms of control and more flexibility during evolution.
Comparison of sequences within gene families shows that it is usually the genetically identified members of the family that are most conserved between different groups of organisms (1). An example is the Wnt gene family (2). Wnt genes act in many different developmental processes; in vertebrates, some Wnts are oncogenes (reviewed in refs. 3 and 4). The Wnt family is ancient and “underwent much of its expansion before the divergence of the arthropod and chordate lineages” (5), so that each lineage still has related groups of paralogues. Of the seven DWnt Drosophila genes, only one member, wg, is well known. In vertebrates (at least 15 Wnt genes in the human genome), the orthologue of wg, Wnt1 was identified as an oncogene by ectopic expression (6, 7). In Drosophila, none of the other homologues was discovered in screens that detect mutant phenotypes; instead, they were identified by means of their molecular homology (“reverse genetics”). Indeed, even now mutants for only DWnt2 are available; this gene is required for the development of the male reproductive tract (8). Understanding of the remaining DWnt genes has depended on patterns of expression or phenotypes caused by overexpression (9–16), and, therefore, it is not clear what functions they have in the wild type. For more information and for a sequence comparison, visit the Wnt page (http://www.stanford.edu/∼rnusse/wntwindow.html). Current experiments on Drosophila suggest that the wg gene is responsible for many of the Wnt functions (reviewed in refs. 2 and 17). If so, one can ask, are the silent homologues idle or redundant? And if they are ineffective, why have they survived unscathed during evolution?
Here, we use the tracheal system of Drosophila and assess its reaction to all members of the DWnt family. We confirm that wg is indispensable (18, 19) but find that one homologue, DWnt2, may assist Wg to specify the main tracheal trunk. We present evidence that tracheal cells are primed to respond differently to the seven DWnt proteins, of which Wg and DWnt2 both are made near the tracheal primordia at the appropriate time (18, 19). We find that DWnt2 affects the tracheal development but, apparently, has no effect on the cuticle, whereas Wg can influence both.
Materials and Methods
Drosophila Strains and Genetics.
The following amorphic or loss-of-function alleles were used: wgCX4 (ref. 20; referred to elsewhere as wg−); Df(2L)RF (21); armXM19 (22); fzH51 (23); fz2C1 (24); DWnt2EMSO, DWnt2EMSO-II (a cleaned DWnt2EMSO allele), DWnt2EMS80P(w+, 47A), DWnt2EMSK, DWnt2EMSI, and Df(2R)11 (8). The different DWnt2 alleles were crossed inter se (referred to elsewhere as Dwnt2− combinations). In embryos carrying these combinations, there is a mild tracheal phenotype: typically embryos (4–40%) show a gap in the most anterior part of the dorsal trunk (DT). However, and confusingly, gaps also are found in embryos only heterozygous for the same mutant alleles usually, but not always, in a lower percentage. Therefore, we were unable to decide whether this phenotype is caused by dysfunction in the DWnt2 gene. The phenotype of Dwnt2− was not significantly altered in wg−/+ background. The double mutants for wg and Dwnt2, referred to as wg−DWnt2−, were recombinants of wgCX4 and different DWnt2 alleles. Recombinants of wgCX4 and DWnt2EMSO-II, DWnt2EMS80P(w+, 47A) and Df(2R)11 in homozygous or transheterozygous conditions produced the phenotypes described in the text. However, recombinants of wgCX4 and DWnt2EMSO, DWnt2EMSI, and DWnt2EMSK produced embryos with no DT in 100% of hemisegments. Genetic analysis points to the presence of a dominant modifier in the original chromosome in which the alleles of the second EMS mutagenesis in (8) were induced.
The P(lacZ) trachealess enhancer trap line 1-eve-1 was used to follow the tracheal cells (25).
To remove the maternal contribution, germ-line clones were induced with the FRT/FLP/ovoD method (26). Females carrying a doubly mutant chromosome fzH51fz2C1FRT2A/ovoD1FRT2A were heat-shocked at 37°C for 1 h in second- or third-instar larvae.
The following UAS lines were used: UASwg (27), UASarm* (28), UASDWnt4 (12), andUASDWnt2, UASDWnt4, UASDWnt5, UASDWnt6, UASDWnt8, and UASDWnt10 (Gary Struhl, unpublished work).
The Gal4 lines used were: btlGal4 (which drives the expression of UAS constructs in the tracheal cells from stage 11; ref. 29); wgGal4 (30); ptcGal4 (31); 1407Gal4 (32); armGal4, and armFRTGal4VP16 (33). To maximize the efficacy of the Gal4/UAS system, the embryo collections were done at 29°C.
To identify mutant embryos, we used “blue balancers” of the first, second, and third chromosomes: FM7 ftz-lacZ; CyO hb-lacZ or CyO ftz-lacZ; and TM3 ftz-lacZ.
Embryo Fixation and Staining.
For horseradish peroxidase (HRP) histochemistry, embryos were fixed in 100 mM Pipes/2 mM EGTA/1 mM MgSO4-formaldehyde for 20–30 min and stained with the Vectastain-ABC kit (Vector Laboratories) according to standard protocols. The mouse monoclonal mAb2A12 (developed by N. Patel and C. Goodman and obtained from the Developmental Studies Hybridoma Bank) was used at 1:5 to stain the lumen of the tracheal system from stage 13/14 onwards. The rabbit anti-Spalt (from R. Schuh, Max Planck Institute, Goettingen, Germany) was used at 1:30. The rabbit anti-β galactosidase (Cappel) was used at 1:1,000 to 1:1,500 to detect tracheal markers and blue balancers. The purified mouse monoclonal anti-Wingless (Development Studies Hybridoma Bank, Iowa City) was used at 1:200. The guinea pig anti-Knirps (developed by J. Reivitz and provided by M. Ruiz-Gomez, Centro Biologia Molecular Severo Ochoa, Madrid) was used at 1:1,000. The rabbit anti-DWnt3 (kindly provided by F. Mourkioti and H. Jäkcle, Max Planck Institute) was used 1:50. Biotinylated or Cy3-, FITC-, and Cy5-secondary antibodies (Jackson ImmunoResearch) were used at 1:300. To optimize double stainings, embryos were first stained in black with NiCl2 and then in brown.
Antisense RNA probes were synthesized from cDNA clones of wg (from J. Bolivar, Univ. of Cadiz, Cadiz, Spain); DWnt2 (from R. Nusse, Stanford University, Stanford, CA); DWnt4, DWnt6, DWnt10 (from E. Wilder, Univ. of Pennsylvania, Philadelphia); DWnt3 (from F. Mourkioti, Max Planck Institute); and dpp (from I. Alvarez, Harvard University, Cambridge, MA). A DNA probe was synthesized from a cDNA clone of DWnt8 (from G. Struhl, Columbia University, College of Physicians and Surgeons, New York). Whole-mount in situ hybridization and antibody staining were performed as described (34) with minor modifications. Fluorescent in situ hybridization, with tyramide signal amplification (NEN Life Science), were performed according to (35) and followed by antibody staining.
Embryos were observed and photographed with a Zeiss Axiophot or with an MRC Bio-Rad 1024 confocal microscope.
Embryos were staged according to (36). Images were processed in Adobe PHOTOSHOP.
Results and Discussion
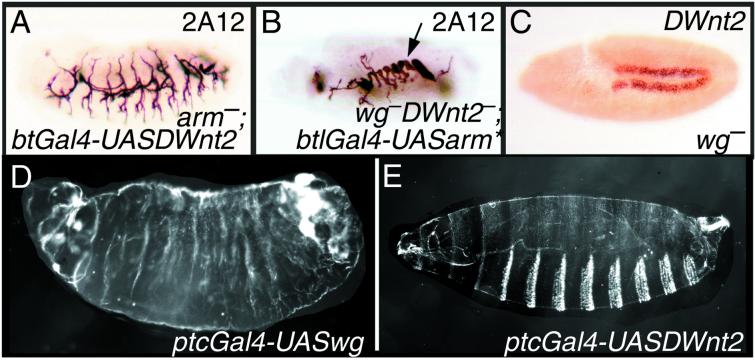
DWnt proteins bind as ligands to a family of receptor proteins {four Frizzled (Fz) homologues in Drosophila, of which Fz and Fz2 are the most important (24, 37) and act through a cascade of genes [e.g., disheveled, armadillo (arm), pangolin] on the nucleus (reviewed in refs. 38–43)}. If, therefore, Wg is the only ligand acting from the outside of the cell on the receptors, the wg− phenotype should be identical to the phenotype when fz and fz2 are removed—in some organs, this is so (24, 37). However, in the trachea, although removal of the two receptor proteins (Fig. 1B) or one of the intracellular proteins in the cascade eliminates all DT; removing only Wg leaves some DT intact (refs. 18 and 19; Fig. 1C). Therefore, it seems that another molecule, presumably a DWnt, acts through the canonical Wnt pathway to build DT. We now ask, which DWnt is responsible?
Figure 1.
The Wnt pathway is required for tracheal development. Lateral views of embryos at late stages of embryogenesis stained with mAb2A12 to highlight the lumen of the tracheae. In all of the figures, anterior is to the left and dorsal is above. (A) Wild-type embryo. DT, dorsal trunk (arrow); VB, visceral branch (arrowhead). (B) fz−fz2− germ-line clones. The DT is completely missing apart from minute vestiges of DT material found in the posterior part. (C) wg−, a substantial amount of DT is formed (arrow). (D) Df(2L)RF, a similar phenotype to wg− is observed (arrow points to DT). (E) Ectopic expression of wg in all tracheal cells—note hypertrophy of DT (arrow). (F) Ectopic expression of DWnt2 in all tracheal cells—note hypertrophy of DT (arrow).
Overexpression of Seven DWnts and Removal of Four of Them.
Overexpression of wg or other downstream elements of the Wnt pathway in the tracheal cells results in increased DT at the expense of the VB (refs. 18 and 19; Fig. 1E). To investigate further, we overexpressed each one of the seven DWnts locally in the embryonic trachea in a normal background. Overexpression of five DWnt genes (DWnt5, -4, -6, -8, and -10) had no detectable effects; indeed, the flies were viable, fertile, and seemed normal. This experiment suggests that the tracheae are not particularly sensitive to these five proteins. To check whether these proteins are made properly and can function, they were tested in other assays. DWnt6 and DWnt8 were able to affect tracheal development in a sensitized background (see below). DWnt5 produced a phenotype in the ventral nerve cord when expressed with the neural specific driver 1407Gal4, in agreement with the phenotype produced by an HS-DWnt5 line (11). Moreover, we detected protein expression in the tracheae when DWnt5 was expressed in tracheal cells (data not shown). DWnt4 produced ectopic denticles in the ventral epidermis when overexpressed with armGal4, and the flies died as pharate adults, showing several defects in the wings when crossed to ptcGal4 (data not shown). These phenotypes have been described by using a different UASDWnt4 line (12, 13). We have not been able to find any noticeable phenotype when overexpressing DWnt10 in several structures, and, thus, the activity of this line awaits confirmation. However, we removed DWnt10 together with three other DWnts (DWnt4, -6, and wg) in Df(2L)RF embryos (ref. 15; see Flybase at http://flybase.bio.indiana.edu) and found a similar phenotype to wg−, there still being some DT (ref. 18; Fig. 1D). This experiment argues that at least zygotic DWnt 4, -6 and -10 do not have a significant function in the trachea under normal conditions. However, overexpression of DWnt2 locally in the tracheal cells did affect its development in a similar way to that of wg, producing an excess of DT cells and DT material at the expense of the VB (Fig. 1F). These tracheae were defective; they failed to fill with air and the flies died as embryos and young larvae. This result suggests that both wg and DWnt2 act or can act in the developing trachea.
DWnt2 Is Produced Near the Tracheal Cells.
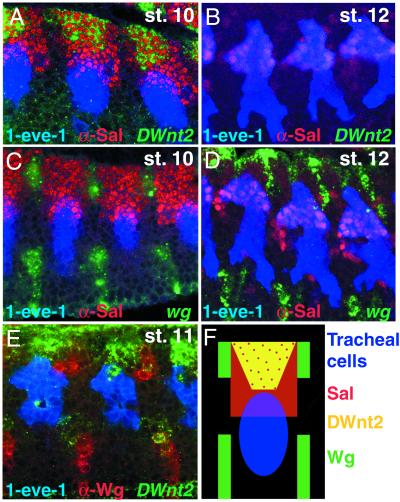
The expression pattern for DWnt2 has been described (16). However, we looked closely at this pattern with respect the tracheal cells. The tracheal placodes are specified by stage 10 in a specific part of the dorsal ectoderm and express several markers such as trachealess (44, 45). The results with DWnt2 are suggestive: it is expressed close to and dorsal to the tracheal placode by stage 10 and early stage 11 but later disappears (Fig. 2 A and B).
Figure 2.
Pattern of wg and DWnt2 expression with respect to Sal protein and the tracheal cells. (A and B) Three tracheal metameres of a 1-eve-1 embryo at stage 10 (A) or 12 (B) stained with a riboprobe for DWnt2 (green) and for Sal (in red) and β-Gal (blue) for the tracheal cells. Note that DWnt2 is expressed near the tracheal cells during stage 10 (A) in cells that also express sal. (C and D) Three tracheal metameres of a 1-eve-1 embryo at stage 10 (C) or 12 (D) stained with a riboprobe for wg (green) and for Sal (in red) and β-Gal (blue) for the tracheal cells. (E) Two tracheal metameres of a 1-eve-1 embryo at stage 11 stained with a riboprobe for DWnt2 (green) and for Wg (red) and β-Gal (blue) for the tracheal cells. Note that wg and DWnt2 differ in their pattern of expression. (F) Diagram summarizing the stainings in A–E. DWnt2 is expressed in the dorsal ectoderm near the tracheal cells that is in the most dorsal Sal-positive cells. wg expression alternates in stripes with Sal protein. Both genes are produced near the tracheal cells.
The spalt (sal) gene (coding for a transcription factor) is expressed in the dorsal ectoderm, including some tracheal cells, during stage 10 and persists later in those tracheal cells that form the DT (ref. 46; Fig. 2 A–D, F). sal is absolutely required for DT formation (46) and is thus a good marker for DT cell identity. The most dorsal cells that express sal also coexpress DWnt2 (Fig. 2A). The pattern of wg expression differs strikingly from that of DWnt2 (refs. 18, 19, and 47; Fig. 2 C–F), although both gene products are made near the tracheal cells. In arm mutants, Sal is not expressed in tracheal cells (18, 19) and no DT is formed, suggesting that sal expression in tracheal cells depends on activation of the Wnt pathway. Thus, sal could be induced in the tracheal cells wherever either Wg or DWnt2 proteins are received.
The Function of DWnt2.
The above results suggest that wg and DWnt2, made near the tracheal cells, together sponsor DT formation. We have more evidence supporting this hypothesis: in wg− embryos, some DT is still formed (Fig. 1C). However, the tracheal phenotype of wg−DWnt2− embryos is significantly different from that of wg− embryos: in 40–45% of hemisegments, the DT is completely missing (Fig. 3A), and in the remaining 55–60%, only some reduced and thin DT forms (Fig. 3B). Interestingly, in practically all hemisegments of Df(2L)RF DWnt2− embryos, the DT is completely missing (data not shown), indicating that other DWnts account for these traces of DT (see below). Nevertheless, wg−DWnt2− double-mutant embryos are very similar (Fig. 3B) or indistinguishable (Fig. 3A) from fz−fz2− embryos (Fig. 1B), suggesting that wg and DWnt2 sponsor virtually all DT formation.
Figure 3.
Tracheal requirements of wg and/or DWnt2. (A–C) Lateral views of embryos at late stages of embryogenesis stained with mAb2A12 to highlight the lumen of the tracheae. (A and B) wg−DWnt2− double mutants, the DT is missing (A) or very much reduced (arrow in B). The arrowhead in A points to an incomplete VB. (C) DWnt2 combination, no tracheal defects are observed. Arrow points to DT. (D–I) Sal distribution at stages 12 (D–F) and 14/15 (G–I) in lateral views of embryos of the indicated genotypes. Note the absence of Sal in wg−DWnt2− mutants (E, H, and Insets) as compared with WT (D, G, and Insets). Low levels of Sal still are observed in wg− mutants (F, I, and Insets). (J and K) dpp expression at stage 11 in lateral views of embryos of the indicated genotypes. (L and M) Kni distribution at stage 14/15 in lateral views of embryos of the indicated genotypes.
Removal of Wg and DWnt2 proteins (in wg−DWnt2− embryos) eliminates detectable expression of sal in the presumptive tracheal cells of the DT (Fig. 3 E and H), whereas in wg− embryos, very low levels of sal still can be detected in some embryos (Fig. 3 F and I). The early expression of sal in the dorsal ectoderm still is observed in both wg− and wg−DWnt2− embryos (data not shown). In wg−DWnt2− embryos, late kni expression in tracheal cells is normal (Fig. 3 L and M), as is the case in arm mutants (18, 19). In addition, dpp expression also is normal (Fig. 3 J and K)—Dpp has been shown to inhibit sal expression by activating kni in tracheal cells (48). Thus, the lack of sal must be caused by the absence of direct or indirect stimulation by the DWnt pathway and not to repression by the Dpp pathway.
Does DWnt2 act through the canonical Wnt pathway? It seems so, because the ectopic effects of DWnt2 protein are blocked in embryos that lack the arm gene (Fig. 4A). Moreover, in wg−DWnt2− embryos, the DT can be substantially rescued by expressing a constitutively active form of Arm in the tracheal cells (Fig. 4B). Also, the tracheal phenotype of porcupine (por) mutants (18) is very similar to that of wg−DWnt2− embryos, indicating that por also might be required for DWnt2 secretion.
Figure 4.
(A and B) Lateral views of embryos of the indicated genotypes at late stages of embryogenesis stained with mAb2A12. Arrow in B points to the rescued DT. (C) Pattern of DWnt2 expression in wg− embryo at stage 10. (D and F) Dark field images of larval cuticle preparations of the indicated genotypes. Overexpression of wg in the epidermis produces a naked cuticle phenotype, whereas overexpression of DWnt2 does not.
If DWnt2 sponsors at least part of DT formation, one might expect that loss of DWnt2 alone would affect trachea in some noticeable way. Surprisingly, DWnt2− embryos and larvae have normal trachea (Fig. 3C) and normal expression of sal (data not shown). However, the flies have reduced viability (our results) and the males are sterile (8).
Interdependence of DWnt2 and wg.
In normal embryos, the wg gene is expressed in a row of cells at the rear of the A compartment, whereas DWnt2 is expressed at the front. Wg protein spreads to make a gradient that patterns the anterior compartment (49, 50). DWnt2 protein is expressed where the concentration of Wg is low or absent; that is where the tracheal placodes form and where the cuticle secretes denticles (50). Thus, in wg− embryos, where there is no Wg protein and the denticles are continuous (20), one might expect the tracheal placodes (51) and DWnt2 (Fig. 4C) expression to form one continuous stripe and, indeed, they do.
This adventitious expression of DWnt2 in a broad domain in wg− embryos could compensate at least in part for the lack of wg itself. Indeed, in these embryos, it must be mainly DWnt2 that activates some sal and determines most or all of the DT found. We could not detect any change in the pattern of wg RNA or protein distribution in DWnt2 mutants.
DWnt Genes Vary in Effectiveness in Different Tissues.
We assayed the potency of DWnt2 and Wg in the tracheae: we took DWnt2−wg− double mutants and added back each of the two missing proteins in the normal pattern of expression for the wg gene. We found that DWnt2 and Wg both rescued some DT in the trachea (Fig. 5 B and D); however, only Wg can partially rescue the various embryonic defects in morphology found in wg− embryos. When either DWnt2 or Wg is expressed locally in the tracheal cells, each gives strong rescue, and more DT is made (Fig. 5 A and C).
Figure 5.
Rescue of DT in double mutants by DWnt proteins. Lateral views of embryos at late stages of embryogenesis stained with mAb2A12 to show the trachea. Each image shows a wg−DWnt2− double mutant in which the DWnt indicated has been added in the pattern of the Gal4 line used. Note the rescue of DT (arrows) when wg, DWnt2, or DWnt8 are added to the embryo (A–F). Conversely, DWnt5 does not rescue the DT (G and H); the arrowhead in H points to a piece of the VB.
We also expressed the DWnt2 gene in wild-type embryos either universally and strongly (arm VP16 Gal4) or in stripes (ptcGal4), and in both cases, the tracheae are altered to the same extent as when DWnt2 is expressed in the tracheal cells alone. However, DWnt2 fails to alter the cuticle pattern (Fig. 4E), whereas wg produces a naked cuticle phenotype (ref. 52; Fig. 4D). This lack of effect of DWnt2 on the epidermis is remarkable as both the drivers used are strong and, when wg is driven, are more than adequate to make a naked cuticle. Interestingly, when DWnt2 is missexpressed in the eye, it also does not emulate the phenotype produced by missexpression of wg (53). Moreover, the effects of overexpressing DWnt2 in the ovary are stronger than when overexpressing wg (54). All these results argue that the tracheal cells and other tissues, including the epidermis, the eye, and the ovary are differentially sensitive to the two DWnt molecules, the trachea and the ovary being particularly responsive to DWnt2.
There are several ways this difference could be achieved. Perhaps DWnt2 does not act through the canonical Wnt pathway in some tissues, such as the ectoderm or the eye. Perhaps DWnt2, on its way to the tracheal cells, could be secreted or processed differently. Perhaps the tracheal cells have something that allows efficient presentation of the ligand to the Fz receptors, or they lack a component that, in other tissues, impedes DWnt2 binding or transduction. One possibility is that glucosaminoglycans help breathless (btl, an FGF receptor expressed in tracheal cells; refs. 55–57) and are needed for Wnt signaling (58, 59). Maybe Btl helps to gather or modify the heparan sulfate glucosaminoglycans, thereby altering the presentation of DWnt2 to the two receptors, Fz and Dfz2. Whatever the explanation may be, the tracheal cells are more responsive than other tissues to the DWnt2 signal.
We next looked at the other DWnts. We drove DWnts5, -6, -8, and -10 in the epidermis of wild-type embryos with one copy of ptcGal4; none of these affected the cuticle pattern in a noticeable way. The effects of expressing DWnt4 have been described above. Are these DWnts able to affect tracheal development in the wg−DWnt2− double mutants? We added back each of these five DWnts to either the tracheal cells themselves or in the pattern of normal wg expression. We found that DWnts6 and -8 (Fig. 5 E and F) were each able to rescue DT partially, whereas -4, -5 (Fig. 5 G and H) and -10 did not. Note that DWnt6 and DWnt8 are not able to produce a tracheal phenotype when expressed in tracheal cells of normal embryos, but they can do so in a sensitized background.
Which DWnt Genes Help Make the DT?
The results indicate that wg and DWnt2 make the main contribution to DT formation, as the absence of both genes completely eliminates DT in many cases. However, traces of DT still are formed in about half the hemisegments of wg−DWnt2− embryos, indicating that contributions of other genes might help. Also, rescue experiments show that some other DWnts are able to activate the pathway. In agreement with this result, we find that in most Df(2L)RF DWnt2− embryos, all DT is missing, indicating that DWnt6 and/or -4 and/or -10 can compensate weakly for the absence of wg and DWnt2. However, expression of DWnt6 and DWnt10 (15) does not suggest that they act in tracheal development in the wild type. DWnt4 is expressed in a similar pattern to that of wg (14) but does not seem to assist wg during embryogenesis (9, 12). In addition, none of DWnt4, -6, or -10 affected tracheal development when expressed in tracheal cells of wild-type embryos. Most likely, they produce traces of DT in the wg−DWnt2− embryos, because those embryos offer a very sensitive test of stimulation of the Wnt pathway. It remains unclear whether these DWnts make any residual contribution to DT in the wild type.
However, several observations suggest that DWnt2 contributes to tracheal development in the wild-type fly. Notably, DWnt2 is expressed near the tracheal cells at the appropriate stage, and when overexpressed in tracheal cells, it mimics the effects of overexpressing wg or a constitutively activated Arm. But, most importantly, the phenotype of wg−DWnt2− embryos indicate that wg and DWnt2 together are responsible for virtually all DT formation. Thus, DWnt2 probably cooperates with Wg or reinforces its main action.
Nevertheless, DWnt2− embryos do not show a visible tracheal phenotype, indicating, at first sight, that the gene does not normally contribute to DT formation. This lack of abnormality suggests that wg alone (or with some help from different DWnts) is sufficient to sponsor normal development in these mutant flies. Nevertheless, it remains possible that DWnt2 could act in the wild-type. There are at least two alternative hypotheses that could explain the lack of tracheal phenotype in DWnt2− embryos.
First, the loss of DWnt2 could induce compensatory changes in the amount, distribution, or activity of the other DWnts. As in the case of DWnt6, -4, and -10 (see above), the expression of DWnt5 (11, 16) and DWnt8 (its pattern of expression has not been reported by others and we have detected expression only in the CNS at early stages; our unpublished results) does not suggest that they act in tracheal development, although we cannot discard contributions under the level of detection. Moreover, although we have detected some small changes in the expression of some DWnts in wg− and wg−DWnt2− embryos (e.g., the loss of DWnt5 expression in the labial segment at stage 10 as well as loss of expression in lateral clusters of the thoracic segments at stage 11), we have not detected any changes in the pattern of expression that might account for any strong tracheal rescue of DWnt2− embryos (our unpublished results). Therefore, it is not clear how other DWnts could contribute to the complete DT formation in DWnt2− embryos.
Second, let us suppose that all DWnts bind the receptor with different affinities, with Wg binding most strongly. In the wild type, the DWnts could compete, but Wg would be most effective—the contribution of DWnt2 to DT formation would be minor. However, in embryos lacking Wg, mainly DWnt2 (which is expressed in a broader domain in wg− embryos and is not now competing with Wg) could bind and partially substitute for Wg. In the absence of DWnt2, Wg (and maybe other DWnts) would have no competition from DWnt2 and would become even more efficient, compensating for the contribution to DT formation that DWnt2 has in the wild type. Finally, in the absence of both Wg and DWnt2, other DWnts, even if they did not act in the wild type, could now bind to the unoccupied receptors and have some tiny effect on DT formation.
Complications of this kind may bedevil attempts to analyze the precise wild type contributions of individual members of other gene families.
Conclusions
We have presented evidence that DWnt2 can act in tracheal development, whereas Wg acts in both developing epidermis and trachea. The other five DWnts do little for the trachea. As with the achaete/scute homologues (which are alike in structure and function but have different patterns of expression and, therefore, act in different places; ref. 60), it may be that the DWnts are preserved fundamentally because seven genes, even if they do similar things, can be regulated in a more sophisticated way than one. Perhaps, like DWnt2, they perform specialized tasks, acting locally to help Wg in ways that could not be provided by any additional regulatory control of wg itself. We also have shown that, in at least this one case, tissues can have differential sensitivity to specific homologues, a property that would allow even more intricate forms of control.
Acknowledgments
We thank Gary Struhl for his advice and generosity in giving us stocks carrying UASDWnts before publication. We also thank J. Casal, J. Casanova, M. Furriols, K. Kozopas, and J. P. Vincent for advice and M. Northcote for skilled assistance. K. Kozopas, C. Logan, and R. Nusse kindly gave us DWnt2 alleles and DWnt2 cDNA. We thank I. Alvarez, M. Bienz, J. Bolivar, H. Jäckle, F. Mourkioti, D. Page, R. Schuh, G. Struhl, M. Ruiz-Gomez, J. P. Vincent, E. Wilder, and the Developmental Studies Hybridoma Bank for fly stocks and reagents. M.L. has been supported by a Long-Term EMBO Fellowship.
Abbreviations
- DT
dorsal trunk
- VB
visceral branch
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ashburner M, Misra S, Roote J, Lewis S E, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 3.Moon R, Brown J, Torres M. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 4.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 5.Rubin G M, Yandell M D, Wortman J R, Gabor Miklos G L, Nelson C R, Hariharan I K, Fortini M E, Li P W, Apweiler R, Fleischmann W, et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusse R, Varmus H E. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 7.Nusse R, Varmus H E. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 8.Kozopas K M, Samos C H, Nusse R. Genes Dev. 1998;12:1155–1165. doi: 10.1101/gad.12.8.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratovich M A, Anderson S, Gieseler K, Pradel J, Wilder E L. Dev Genes Evol. 2000;210:111–119. doi: 10.1007/s004270050017. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg L M, Ingham P W, Brown A M C. Dev Biol. 1992;154:73–83. doi: 10.1016/0012-1606(92)90049-m. [DOI] [PubMed] [Google Scholar]
- 11.Fradkin L G, Noordermeer J N, Nusse R. Dev Biol. 1995;168:202–213. doi: 10.1006/dbio.1995.1072. [DOI] [PubMed] [Google Scholar]
- 12.Gieseler K, Graba Y, Mariol M C, Wilder E L, Martinez-Arias A, Lemaire P, Pradel J. Mech Dev. 1999;85:123–131. doi: 10.1016/s0925-4773(99)00097-0. [DOI] [PubMed] [Google Scholar]
- 13.Gieseler K, Wilder E, Mariol M-C, Buratrovich M, Bérenger H, Graba Y, Pradel J. Dev Biol. 2001;232:339–350. doi: 10.1006/dbio.2001.0184. [DOI] [PubMed] [Google Scholar]
- 14.Graba Y, Gieseler K, Aragnol D, Laurenti P, Mariol M C, Berenger H, Sagnier T, Pradel J. Development (Cambridge, UK) 1995;121:209–218. doi: 10.1242/dev.121.1.209. [DOI] [PubMed] [Google Scholar]
- 15.Janson K, Cohen E D, Wilder E L. Mech Dev. 2001;103:117–120. doi: 10.1016/s0925-4773(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 16.Russell J, Gennissen A, Nusse R. Development (Cambridge, UK) 1992;115:475–485. doi: 10.1242/dev.115.2.475. [DOI] [PubMed] [Google Scholar]
- 17.Klingensmith J, Nusse R. Dev Biol. 1994;166:396–414. doi: 10.1006/dbio.1994.1325. [DOI] [PubMed] [Google Scholar]
- 18.Llimargas M. Development (Cambridge, UK) 2000;127:4407–4417. doi: 10.1242/dev.127.20.4407. [DOI] [PubMed] [Google Scholar]
- 19.Chihara T, Hayashi S. Development (Cambridge, UK) 2000;127:4433–4442. doi: 10.1242/dev.127.20.4433. [DOI] [PubMed] [Google Scholar]
- 20.Baker N E. EMBO J. 1987;6:1765–1774. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiong S, Nash D. Genetics. 1990;124:889–897. doi: 10.1093/genetics/124.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox R T, Pai L M, Kirkpatrick C, Stein J, Peifer M. Genetics. 1999;153:319–332. doi: 10.1093/genetics/153.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones K H, Liu J, Adler P N. Genetics. 1996;142:205–215. doi: 10.1093/genetics/142.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Struhl G. Development (Cambridge, UK) 1999;126:5441–5452. doi: 10.1242/dev.126.23.5441. [DOI] [PubMed] [Google Scholar]
- 25.Perrimon N, Noll E, McCall K, Brand A. Dev Genet (Amsterdam) 1991;12:238–252. doi: 10.1002/dvg.1020120309. [DOI] [PubMed] [Google Scholar]
- 26.Chou T, Noll E, Perrimon N. Development (Cambridge, UK) 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence P, Bodmer R, Vincent J. Development (Cambridge, UK) 1995;121:4303–4308. doi: 10.1242/dev.121.12.4303. [DOI] [PubMed] [Google Scholar]
- 28.Pai L M, Orsulic S, Bejsovec A, Peifer M. Development (Cambridge, UK) 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- 29.Shiga Y, Tanaka-Matakatsu M, Hayashi S. Dev Growth Diff. 1996;38:99–106. [Google Scholar]
- 30.Pfeiffer S, Alexandre C, Calleja M, Vincent J. Curr Biol. 2000;10:321–324. doi: 10.1016/s0960-9822(00)00381-x. [DOI] [PubMed] [Google Scholar]
- 31.Hinz U, Giebel B, Campos-Ortega J. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 32.Luo L, Liao Y J, Jan L Y, Jan Y N. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 33.Sanson B, White P, Vincent J P. Nature (London) 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann R, Tautz D. In: Drosophila Melanogaster: Practical Uses in Cell and Molecular Biology. Goldstein L S B, Fyberg E A, editors. Vol. 44. New York: Academic; 1994. pp. 576–597. [Google Scholar]
- 35.Wilkie G, Davis I. Technical Tips Online. Vol. 1. August; 1998. http://research.bmn.com/tto . Available at http://research.bmn.com/tto. Access no. T01458. . Access no. T01458. [Google Scholar]
- 36.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila Melanogaster. Berlin: Springer; 1997. [Google Scholar]
- 37.Bhanot P, Fish M, Jemison J A, Nusse R, Nathans J, Cadigan K M. Development (Cambridge, UK) 1999;126:4175–4186. doi: 10.1242/dev.126.18.4175. [DOI] [PubMed] [Google Scholar]
- 38.Bienz M. Curr Opin Genet Dev. 1999;9:595–603. doi: 10.1016/s0959-437x(99)00016-7. [DOI] [PubMed] [Google Scholar]
- 39.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 40.Cavallo R, Rubenstein D, Peifer M. Curr Opin Genet Dev. 1997;7:459–466. doi: 10.1016/s0959-437x(97)80071-8. [DOI] [PubMed] [Google Scholar]
- 41.Cox R T, Peifer M. Curr Biol. 1998;8:R140–144. doi: 10.1016/s0960-9822(98)70081-8. [DOI] [PubMed] [Google Scholar]
- 42.Dierick H, Bejsovec A. Curr Top Dev Biol. 1999;43:153–190. doi: 10.1016/s0070-2153(08)60381-6. [DOI] [PubMed] [Google Scholar]
- 43.Willert K, Nusse R. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 44.Isaac D D, Andrew D J. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- 45.Wilk R, Weizman I, Shilo B Z. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- 46.Kuhnlein R P, Schuh R. Development (Cambridge, UK) 1996;122:2215–2223. doi: 10.1242/dev.122.7.2215. [DOI] [PubMed] [Google Scholar]
- 47.van den Heuvel M, Nusse R, Johnston P, Lawrence P A. Cell. 1989;59:739–749. doi: 10.1016/0092-8674(89)90020-2. [DOI] [PubMed] [Google Scholar]
- 48.Chen C K, Kuhnlein R P, Eulenberg K G, Vincent S, Affolter M, Schuh R. Development (Cambridge, UK) 1998;125:4959–4968. doi: 10.1242/dev.125.24.4959. [DOI] [PubMed] [Google Scholar]
- 49.Lawrence P A, Sanson B, Vincent J P. Development (Cambridge, UK) 1996;122:4095–4103. doi: 10.1242/dev.122.12.4095. [DOI] [PubMed] [Google Scholar]
- 50.Bejsovec A, Martinez Arias A. Development (Cambridge, UK) 1991;113:471–485. doi: 10.1242/dev.113.2.471. [DOI] [PubMed] [Google Scholar]
- 51.de Celis J F, Llimargas M, Casanova J. Development (Cambridge, UK) 1995;121:3405–3416. doi: 10.1242/dev.121.10.3405. [DOI] [PubMed] [Google Scholar]
- 52.Noordermeer J, Johnston P, Rijsewijk J, Nusse R, Lawrence P A. Development (Cambridge, UK) 1992;116:711–719. doi: 10.1242/dev.116.3.711. [DOI] [PubMed] [Google Scholar]
- 53.Wehrli M, Tomlinson A. Development (Cambridge, UK) 1998;125:1421–1432. doi: 10.1242/dev.125.8.1421. [DOI] [PubMed] [Google Scholar]
- 54.Willert K, Logan C, Arora A, Fish M, Nusse R. Development (Cambridge, UK) 1999;126:4165–4173. doi: 10.1242/dev.126.18.4165. [DOI] [PubMed] [Google Scholar]
- 55.Lin X, Buff E M, Perrimon N, Michelson A M. Development (Cambridge, UK) 1999;126:3715–3723. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- 56.Klambt C, Glazer L, Shilo B Z. Genes Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- 57.Glazer L, Shilo B Z. Genes Dev. 1991;5:697–705. doi: 10.1101/gad.5.4.697. [DOI] [PubMed] [Google Scholar]
- 58.Lin X, Perrimon N. Nature (London) 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- 59.Häcker U, Lin X, Perrimon N. Development (Cambridge, UK) 1997;124:3565–3573. doi: 10.1242/dev.124.18.3565. [DOI] [PubMed] [Google Scholar]
- 60.Campuzano S, Modolell J. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]