Abstract
Transcription intermediary factor 1γ (Tif1γ), a ubiquitous nuclear protein, is a regulator of transforming growth factor-β (TGF-β)/Smad signaling. Tif1γ can function as an oncogene and as a tumor suppressor. In the present study, Tif1γ levels were measured in the plasma of patients with breast cancer in order to investigate the association of Tif1γ with overall survival (OS). The results indicated that Tif1γ is an independent prognostic and predictive factor in breast cancer, and thus, a promising target protein for use in diagnostics and patient follow-up. Plasma levels of Tif1γ were measured in samples obtained from 110 patients with operable breast cancer and in 110 healthy volunteers at the Breast Cancer Department of Yangpu Hospital between 2008 and 2016. The association between Tif1γ levels and clinicopathologic parameters, and the OS in a follow-up period of 98 months was evaluated. The prognostic significance was assessed using the Kaplan-Meier method. The levels of Tif1γ were significantly lower in patients with breast cancer compared with healthy controls. The average concentration of 18.40 ng/ml was used to discriminate between Tif1γ-positive (52) and Tif1γ-negative patients (58). Tif1γ-positive patients had a significantly improved OS compared with Tif1γ-negative patients. In the multivariate analysis, Tif1γ was an independent predictor of a favorable OS in a prospective follow-up setting; thus, Tif1γ plasma levels are an independent prognostic factor for patients with breast cancer. These findings support the potential of using measurements of Tif1γ plasma levels to guide breast cancer therapy and monitoring. Further studies are required to validate Tif1γ as an easily detectable, non-invasive prognostic biomarker for breast cancer.
Keywords: transcription intermediary factor 1γ, circulating tumor cells, prognosis, breast cancer, biomarker
Introduction
Transcription intermediary factor 1γ (Tif1γ), also known as tripartite motif containing 33 (TRIM33), is a ubiquitous nuclear protein (1) that regulates transforming growth factor-β (TGF-β)/Smad signaling. It is part of the transcriptional intermediary factor 1 family, which includes four identified Tif1 members in mammals (α to δ), with modulatory roles in the innate immune response and inflammation processes. Tif1γ was initially described a decade ago (2); however, this protein has recently gained increasing interest in oncology due to its dual function as an oncogene and a tumor suppressor gene (3,4).
Similar to its homolog transcriptional cofactors (Tif1α and Tif1β), Tif1γ contains multiple domains, including the RING finger, B boxes, coiled coil and a PHD-bromodomain that stabilizes Tif1γ chromatin occupancy and enhances its E3 ligase activity (5–7). Despite structural similarity with other family members, Tif1γ exhibits very specific functions. Tif1γ regulates TGF-β signaling pathways, potentially via SMAD family member 4 (Smad4) mono-ubiquitination, targeting Smad4 for degradation and inhibiting the TGF-β/Smad signaling pathway. TGF-β signaling is initiated by the dimerization of TGF-β I and II receptors, which results in Smad2 and Smad3 phosphorylation, and binding with Smad4. As a complex, Smad4 migrates to the nucleus, where it interacts with various transcription factors (8–11). Activation of mitogen-activated protein kinases (MAPKs) is another Smad-independent TGF-β signaling mechanism (12,13). Certain studies have suggested that Tif1γ competes with Smad2/Smad3 for binding to Smad4 (14–16). Tif1γ interacts with the Smad1/Smad4 complex in vivo, inhibiting transcriptional activity of the Smad1/Smad4 complex via its PHD domain. In blood stem cell lines, Tif1γ interacts with the transcription factor stem cell leukemia (SCL)/T-cell acute lymphocytic leukemia protein 1 to promote mRNA transcript elongation. In other cells, Tif1γ forms repressive complexes with SCL that inhibit transcriptional activation (17–19).
A previous study has demonstrated that Tif1γ expression is a biomarker of the response of chronic myelomonocytic leukemia to demethylating agents, and that Tif1γ is an epigenetically-regulated tumor suppressor gene (20,21). Tif1γ is involved in the process of vertebrate hematopoietic development by regulating the TGF-β1 receptor and mRNA elongation during transcription (22), and loss of Tif1γ expression or protein malfunction promotes hematopoietic stem cells to differentiate to the myelomonocytic lineage (17–23). In B cell neoplasms, Tif1γ acts as an oncogene, as it suppresses the apoptosis of B lymphoblastic leukemia cells. Notably, Tif1γ performs this function by associating with a single DNA cis element (24–26).
As for solid tumors, in vitro studies demonstrated that in benign and malignant pancreatic cell lines, overexpression of Tif1γ was associated with a reduced level of Smad4. However, both overexpression and knockdown of Tif1γ lead to an inhibition in tumor growth, and Tif1γ knockdown reduces tumor invasion. Tif1γ has also been reported to be a tumor suppressor in non-small cell lung cancer (NSCLC), as mRNA and protein expression in NSCLC cell lines is significantly decreased. By contrast, tissue microarray analysis revealed that Tif1γ was overexpressed in colorectal cancer and there was an absence of Smad4 expression in neoplastic samples. The levels of Tif1γ overexpression were stage dependent (higher in stage III compared with stage I and II) (27–29).
Breast cancer is the most common non-cutaneous cancer and the leading cause of cancer-associated mortality in females worldwide (30–33). Despite advances in diagnostics and therapeutics, breast cancer incidence rates are rising, due to demographical aging, hormone replacement use, manifestation of cancer risks in modern lifestyles and other factors (30,34). Breast cancer is one of the most heterogeneous diseases and, thus, the development of personalized cancer management is crucial. Personalized treatment plans may use established predictive factors, including receptor status, clinicopathological factors, urokinase-type plasminogen activator/plasminogen activator inhibitor 1 (PAI1), tumor size, lymph node stage, histological grade or lymphovascular invasion, and novel prognostic factors to determine the required therapy regimen (35–37). Prognostic markers are essential for decision-making as they attempt to foresee the outcome of patients, irrespective of the treatment received. Efforts are made to develop novel markers that are independently associated with the overall and disease-free survival (38–41). Screening techniques remain a crucial part of breast cancer prevention and reducing breast cancer-associated mortality. Currently, novel biomarkers are required for developing new treatment algorithms and prognosis evaluation.
In breast cancer, the prognostic significance of Tif1γ has not been established. TGF-β has been demonstrated to have tumor suppressive (early stages) and oncogenic [later stages; pro-metastatic and pro-epithelial-to-mesenchymal transition (EMT)] effects (42–44). The isoform TGF-β1 is an inhibitor of mammary gland epithelial cell proliferation and has a particularly important role in breast carcinogenesis (45–48). Certain studies have reported that lower levels of circulating TGF-β1 were associated with a poor disease prognosis (49,50); however, other studies have indicated the opposite (51–53).
It may be assumed that Tif1γ also has a paradoxical role in breast cancer development and outcome, as Tif1γ is involved in regulating TGF-β/Smad signaling. Tumor suppressor and tumorigenic roles of Tif1γ have been suggested in various cancer types (4,20,54). Recent data demonstrated that Tif1γ reduces Smad4 activity and, thus, inhibits TGF-β-induced EMT in mammary epithelial cells, terminal differentiation of mammary alveolar epithelial cells and lactation (55–58). Furthermore, Tif1γ binds to and represses the PAI1 promoter, directly regulating TGF-β-dependent gene expression. As a negative regulator of Smad4, Tif1γ is crucial for the regulation of TGF-β signaling (58–61). Kassem et al (62) suggested that Tif1γ and the TGF-β1/Smad4 signaling have a significant effect on the outcome of patients with operable breast cancer, and if measured in combination they can be used for prognostic evaluation.
The concept of circulating tumor cells (CTCs) was first proposed in 1896; however, the isolation methods and prognostic significance were established only recently (63–65). CTCs are tumor cells that are released from a solid tumor (the primary tumor or metastases) into the peripheral blood circulation, spontaneously or during treatment, where they form a tumor thrombus. CTCs have been suggested as biomarkers, as they are precursors of breast cancer metastases (66–68). In 2004, Cristofanilli et al (69) reported that the number of CTCs in therapy-naïve patients with metastatic breast cancer is an independent predictor of progression-free survival (PFS) and overall survival (OS). Further reports soon confirmed these findings and also demonstrated the prognostic significance in non-metastatic breast cancer (70–72). Despite advances in CTC capture technology, it remains an expensive and difficult method, which limits its use as a daily clinical diagnostic application.
The potential prognostic use of Tif1γ has not been well investigated in breast cancer to date. Therefore, the rationale of the current study was to elucidate the role of Tif1γ in breast cancer tumorigenesis, cancer progression and metastasis, and to determine whether its expression is an independent prognostic marker. CTCs were also analyzed to compare the prognostic use of CTCs with the prognostic value of Tif1γ, which can be easily and rapidly detected, thus allowing direct translation to the clinic without methodological constraints.
Patients and methods
Study population
Patients with breast cancer (n=110) were prospectively and randomly recruited between January 2008 and April 2016 at the Department of Breast Surgery, Yangpu Hospital, Tongji University (Shanghai, China). The patients were aged 33–74 years, with a median age of 51. An associated clinicopathological database was established and long-term clinical follow-up was performed. Written consent forms were obtained from all patients involved in the current study (approval no. LL-2016-WSJ-002). The ethics review board of Tongji University approved the study design a priori. All patients underwent surgical tumor excision in which tissue samples were collected. All participants provided their consent for the use of tumor samples in academic research. Preoperatively, neoadjuvant chemotherapy, endocrine therapy, radiation or other therapies were received, as needed. As a control group, blood samples were collected from 110 healthy subjects between January 2008 and April 2016 at the Medical Examination Center, Yangpu Hospital, Tongji University. Healthy volunteers (30–67 years old; median age, 49 years) were females without breast cancer, as indicated by pathological diagnostics. Individuals were followed until the end of the follow-up period of 98 months.
Main reagents and instruments
Rabbit anti-Tif1γ antibody (cat. no. ab47062), mouse anti-TGF-β1 (cat. no. ab64715), mouse anti-β-actin (cat. no. ab8226) and goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated antibody (cat. no. ab6721) were purchased from Abcam (Cambridge, MA, USA). Human Tif1γ ELISA kit (cat. no. GEN2413000) was purchased from Gentaur (Paris, France). The ChemiDoc MP imaging system was purchased from Bio-Rad Laboratories, Inc. (Hercules, CA, USA).
SDS-PAGE and western blot analysis
For analysis of protein expression, four samples of breast cancer tissue with paired adjacent normal breast tissue were obtained from Yangpu Hospital of Tongji University (Shanghai, China). Informed consent was provided by all patients and all samples were histologically confirmed prior to analysis. To confirm the expression of the protein of interest, western blot analysis was performed as previously described (73). In brief, the protein concentration of the crude tissue extracts was measured using the Bradford method. SDS-PAGE was used to separate samples, with an equal amount of total protein content loaded in each lane of 10% gels, then transferred to polyvinylidene difluoride membranes using a semi-dry apparatus. Nonspecific binding was blocked using PBS containing 3% bovine serum albumin. Membranes were probed with Tif1γ and TGF-β1 primary antibodies (1:1,000). β-actin (1:1,000) served as a loading control. The immune complexes were visualized using HRP-conjugated secondary antibody (1:1,500), according to the manufacturer's protocols. Blots were digitally imaged using the ChemiDoc MP imaging system. ImageJ version 1.51p software (National Institutes of Health, Bethesda, MD, USA) was used to quantify protein expression.
ELISA detection of Tif1γ in the plasma
Tif1γ levels were measured with the Human Tif1γ ELISA kit (cat. no. GEN2413000) according to the manufacturer's protocols. Serum was diluted in a 5- to 20-fold range to obtain values falling within the linear range of the standard curve. The qualitative absorbance analyses were performed using a Varioskan Flash multifunctional microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 450 nm minus 550 nm, according to the manufacturer's guidelines.
Detection of CTCs in peripheral blood
Peripheral venous blood of each patient was collected (1 ml) and placed in a CellSave tube (containing EDTA and cell preservative) at room temperature. The blood was diluted with PBS and a Percoll density gradient-based method was used to separate the mononuclear cell layer from the blood (74). Following removal of the mononuclear cells, CTC capture was performed using non-antibody-dependent specific magnetic beads (Fe3O4 inner cores) (75).
Statistical analysis for overall survival and prognostic calculations
Data were analyzed using SPSS (version 20.0; IBM Corp., Armonk, NY, USA). The Kaplan-Meier method was used to estimate OS and multivariate analysis was performed with the Cox proportional hazards model. OS was calculated from the date of diagnosis to the date of last follow-up. Analysis of the differences between groups was calculated using a two-tailed Student's t-test and χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
Of the 110 recruited patients, none was missed during follow-up or withdrew, and all successfully completed the study. All patients were Chinese females. The patient characteristics are summarized in Table I. Patients were followed for 98 months to assess the association between Tif1γ levels in serum and OS. The majority of the patients presented with human epidermal growth factor receptor 2 (Her2)-negative, grade 2 ductal type carcinoma. The distribution of clinicopathological features was representative and the median age at diagnosis was close to the population median age.
Table I.
Clinicopathologic variables of breast cancer patient cohort.
| Variables | Number of patients | % |
|---|---|---|
| Age | ||
| <50 years | 31 | 28.2 |
| ≥50 years | 79 | 71.8 |
| Tumor size | ||
| <2 cm | 46 | 41.8 |
| ≥2 cm | 64 | 58.2 |
| Tumor stage | ||
| T1 | 34 | 30.9 |
| T2 | 76 | 69.1 |
| Histologic grade | ||
| G1 | 5 | 4.5 |
| G2 | 86 | 78.2 |
| G3 | 19 | 17.3 |
| Node status | ||
| Negative | 68 | 61.8 |
| Positive | 42 | 38.2 |
| Histologic type | ||
| Ductal | 93 | 84.5 |
| Others | 17 | 15.5 |
| Molecular subtypes | ||
| Luminal | 30 | 27.3 |
| Others | 80 | 72.7 |
| Her2 status | ||
| Negative | 91 | 82.7 |
| Positive | 19 | 17.3 |
| CTC | ||
| Negative | 47 | 42.7 |
| Positive | 63 | 57.3 |
| Tif1γ status | ||
| Negative | 58 | 52.7 |
| Positive | 52 | 47.3 |
Her2, human epidermal growth factor receptor 2; CTCs, circulating tumor cells; Tif1γ, transcription intermediary factor 1γ.
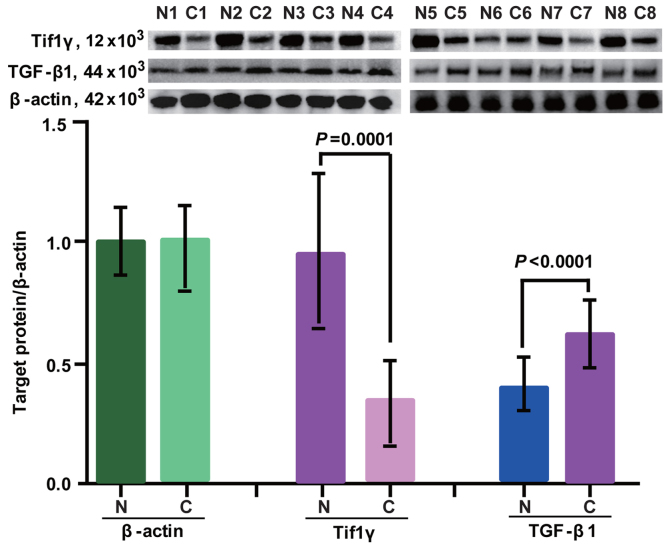
Expression of Tif1γ and TGF-β1 in breast cancer tissues
Protein expression of Tif1γ and TGF-β1 in cancer and healthy tissues was determined by western blot analysis. β-actin was used as a loading control. The expression profiles are presented in Fig. 1. Tif1γ and TGF-β1 were detected in adjacent normal control tissues and cancer tissues. In cancer tissues, the expression of Tif1γ was significantly lower compared with adjacent normal control tissues (Fig. 1). By contrast, TGF-β1 expression was higher in cancer tissues compared with the adjacent control samples (Fig. 1).
Figure 1.
Western blot analysis of Tif1γ and TGF-β1 expression levels in eight breast cancer tissues and eight adjacent normal control tissues. β-actin was used as the loading control. Tif1γ, transcription intermediary factor 1 γ; TGF-β1, transforming growth factor-β1; N, normal tissue; C, cancer tissue.
Tif1γ levels in plasma
Indirect ELISA was used to analyze the human Tif1γ levels in plasma samples from 110 patients with breast cancer and 110 healthy controls. Tif1γ levels were significantly higher in the healthy controls compared with the patients with breast cancer (Fig. 2). The average Tif1γ value in the breast cancer group was 13.89 ng/ml, while in the control group it was 22.90 ng/ml. The average concentration of 18.40 ng/ml was therefore used to divide the patients with breast cancer into Tif1γ-positive (n=52) and Tif1γ-negative (n=58; P=0.0008) groups, for subsequent analyses.
Figure 2.
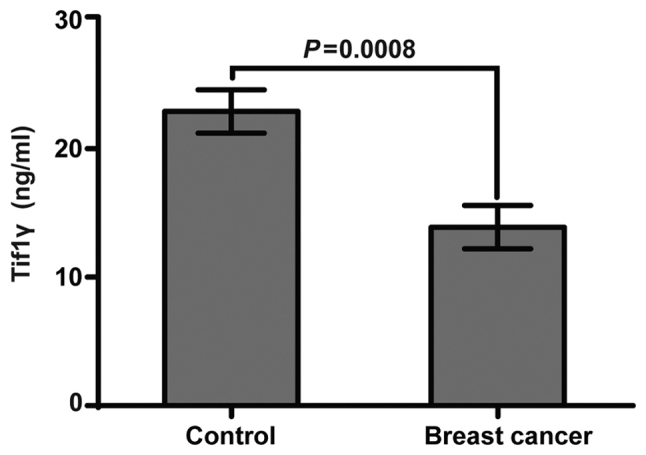
Detection of Tif1γ plasma levels by ELISA. Tif1γ was quantified in plasma samples from patients with breast cancer (n=110) and healthy controls (n=110). The average concentration was 22.90 ng/ml in control participants, and 13.89 ng/ml in patients with breast cancer. P=0.0008. Tif1γ, transcription intermediary factor 1 γ.
Comparing clinicopathological characteristics and CTC count with the Tif1γ plasma level
Tif1γ plasma levels were measured by ELISA. CTC detection was performed using non-antibody-dependent specific magnetic beads. The clinicopathological characteristics were collected upfront. χ2 correlations were performed and presented in Fig. 3 Statistically significant correlations were observed between the CTC count (Fig. 3A), tumor size (Fig. 3B) and tumor stage (Fig. 3C). The number of CTC count-positive patients was 67.2±6.2% in the Tif1γ-negative group and 46.2±7.0% in the Tif1γ-positive group (Fig. 3A). No correlation was observed with histological grade, tumor subtype, lymph node and Her2 status (Fig. 3D-I).
Figure 3.
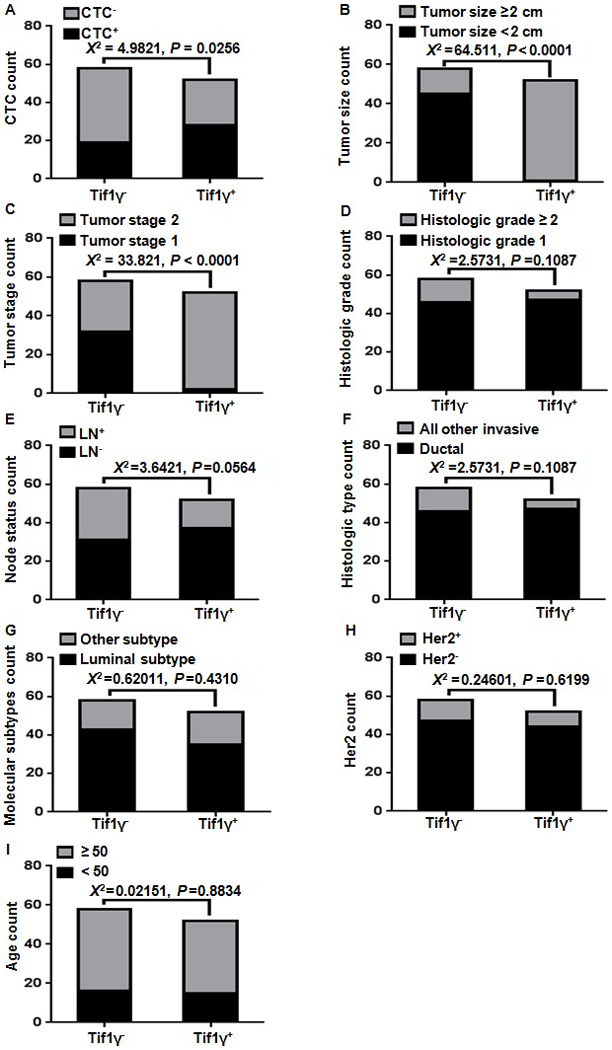
χ2 correlation of clinicopathological characteristics and CTC count with the Tif1γ plasma levels. (A) A significant correlation was observed between the CTC count and the Tif1γ plasma levels in the breast cancer patients. The number of CTC count-positive patients was 67.2±6.2% in the Tif1γ-negative group and 46.2±7.0% in the Tif1γ-positive group. (B) Statistically significant correlations were observed between the tumor size and (C) tumor stage and the Tif1γ plasma levels. (D-I) No significant correlation was observed with histological grade, lymph node status, ductal/invasive type, molecular subtype, Her2 status and age. CTC, circulating tumor cell; Tif1γ, transcription intermediary factor 1 γ; Her2, human epidermal growth factor receptor 2.
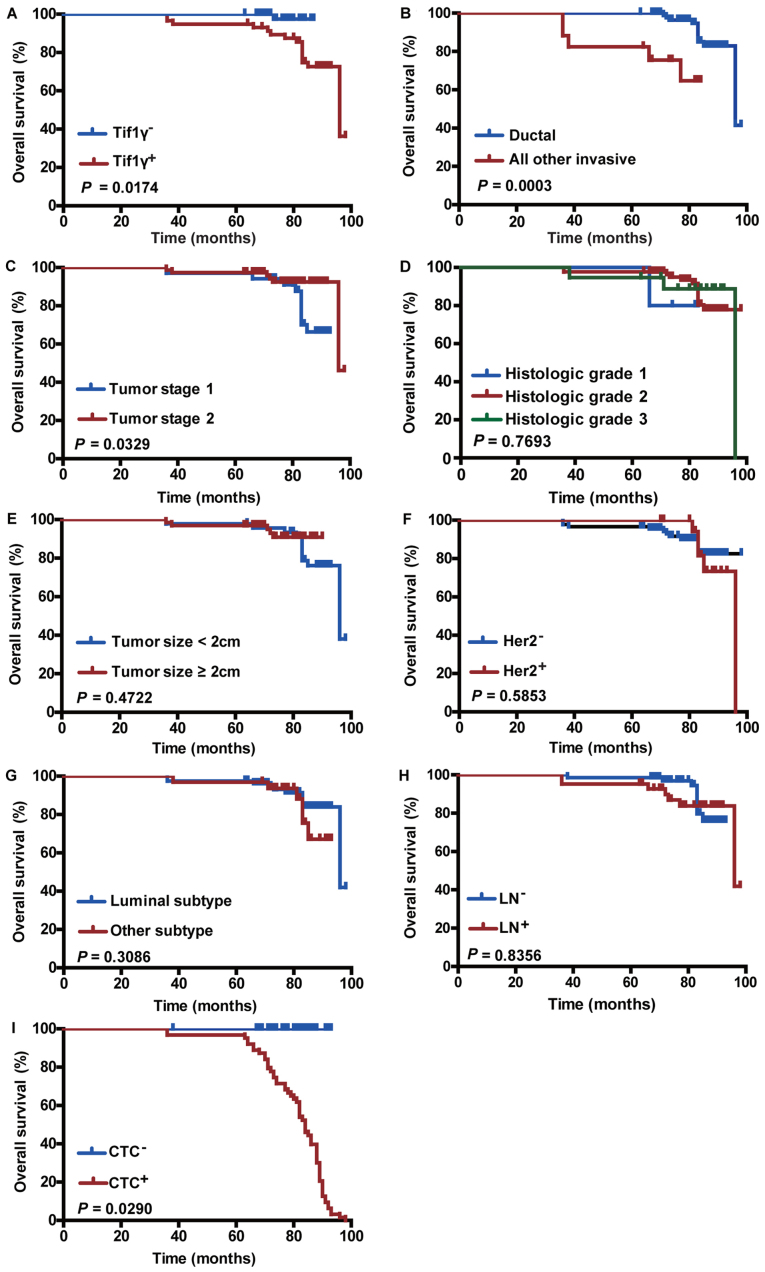
Association between Tif1γ plasma levels and OS
The clinical prognostic value of the Tif1γ serum levels in patients with breast cancer was investigated. Patients were categorized into high and low serum level groups. The high Tif1γ serum level group had improved OS compared with the low-level group (Kaplan-Meier method; 98 months; P=0.0174; Fig. 4A). This was further confirmed using univariate Cox analysis (Table II). The Tif1γ serum level was significantly associated with survival in patients with breast cancer [hazard ratio (HR)=0.125; P=0.046; Table II]. Furthermore, multivariate Cox analysis confirmed that high Tif1γ serum level was a significant independent prognostic factor in the patients with breast cancer (HR=0.046; P=0.011; Table II).
Figure 4.
Effect of Tif1γ plasma levels and other clinicopathological factors on overall survival. Kaplan-Meier survival analysis curves representing the association of overall survival and (A) Tif1γ serum levels, (B) ductal or other invasive types, (C) tumor stage, (D) histologic grade, (E) tumor size, (F) Her2 status, (G) tumor subtype, (H) LN status and (I) CTC count. Tif1γ, transcription intermediary factor 1 γ; Her2, human epidermal growth factor receptor 2; LN, lymph node; CTC, circulating tumor cell.
Table II.
Univariate and multivariate analyses of overall survival by the Cox proportional hazards model.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Clinicopathologic variables | HR | CI | P-value | HR | CI | P-value |
| Age | 1.062 | 0.991–1.138 | 0.087 | 1.060 | 0.978–1.150 | 0.157 |
| Tumor size | 0.661 | 0.221–1.974 | 0.458 | 2.733 | 0.544–13.726 | 0.222 |
| Tumor stage | 0.329 | 0.111–0.976 | 0.045 | 0.477 | 0.107–2.117 | 0.330 |
| Histologic grade | 0.837 | 0.262–2.672 | 0.763 | 1.122 | 0.402–3.131 | 0.826 |
| Node status | 0.773 | 0.365–1.639 | 0.502 | 0.556 | 0.171–1.800 | 0.327 |
| Histologic type | 2.878 | 1.282–6.464 | 0.010 | 2.328 | 0.543–9.972 | 0.255 |
| Molecular subtypes | 0.772 | 0.264–2.262 | 0.638 | 0.927 | 0.232–3.695 | 0.914 |
| Her2 status | 0.796 | 0.502–1.261 | 0.331 | 0.657 | 0.342–1.263 | 0.208 |
| CTC | 0.934 | 0.330–2.645 | 0.898 | 1.372 | 0.302–6.247 | 0.682 |
| Tif1γ status | 0.125 | 0.016–0.964 | 0.046 | 0.047 | 0.004–0.501 | 0.011 |
Significant P-values are denoted in bold font. HR, hazard ratio; CI, confidence interval; Her2, human epidermal growth factor receptor 2; CTC, circulating tumor cell; Tif1γ, transcription intermediary factor 1γ.
Association between clinicopathological features and CTCs on OS
As expected, OS was significantly improved in patients with ductal type cancer (P=0.0003; Fig. 4B) and early tumor stage (stage 1; P=0.0329; Fig. 4C), and in patients that were CTC count-negative (P=0.0290; Fig. 4I). In addition, these factors displayed prognostic significance in univariate Cox analysis (Table II). Her2 positivity, tumor size (≥2 cm), tumor subtype, histological grade and lymph node positivity were not significantly associated with the patient survival outcome (Fig. 4D-H), and had no prognostic significance (Table II).
Discussion
Breast cancer is one of the most well-studied diseases with constant novel insights into the molecular basis of the disease and identification of novel therapy targets (40,76,77). The subclassification of breast cancer is typically based on estrogen receptor, progesterone receptor and Her2 status, which is used for predicting prognosis and guiding treatment strategies (78–82). Given the heterogeneity and alterability of breast cancer, there is a constant need for novel biological markers with predictive power, such as Tif1γ. The findings of the current study demonstrated that Tif1γ is promising as a simple, efficient and effective outcome predictor in patients with breast cancer. Considering the increasing incidence of breast cancer, easily applicable methods used for prognosis assessment and early detection are crucial (83–86). Furthermore, in the era of individualized cancer care, diagnostic tumor markers allow oncologists to identify high-risk patients, select and monitor treatment, and screen for disease recurrence (40,76,87–90).
In the current study, the plasma levels of Tif1γ were significantly higher in healthy controls than in patients with breast cancer (Fig. 2). This supports the general expectations, as Tif1γ is part of the transcription intermediary factor 1 family and has been previously reported to inhibit the TGF-β/Smad signaling pathway (60,62,91). TGF-β itself is associated with tumor invasion and progression, as it acts as a potent inducer of EMT; thus, lower levels or depletion of Tif1γ increases the EMT process (pro-oncogenic) (55,58).
Previous microarray analysis has revealed that certain Tif1γ target genes are associated with EMT. A deficiency in Tif1γ expression has been reported in several cancer types, including NSCLC and hepatocellular, pancreatic and colorectal cancer. Thus, Tif1γ potentially has tumor suppressor gene activity that is lost during cancer development. The majority of research into Tif1γ in cancer has focused on the molecular functions and interactions, whereas in vivo and clinical outcome data are limited.
The aim of the present study was to confirm the association between Tif1γ and breast cancer by examining the expression of Tif1γ in the plasma of patients and healthy controls, and to investigate the potential prognostic use of Tif1γ. Western blot analysis was performed to determine the expression of Tif1γ, compared with TGF-β, in adjacent noncancerous and cancerous tissues. Tif1γ expression was reduced in breast cancer tissues compared with adjacent noncancerous tissues. As expected, reciprocal results were observed for TGF-β (Fig. 1). Serum Tif1γ levels were significantly lower in the patients with breast cancer compared with the healthy control samples (Fig. 2). This suggests that Tif1γ is active in normal cells, exerting a tumor suppression role, and is less active in cancer cells.
ELISA was used in order to quantify and to obtain objective numerical values of the Tif1γ expression levels (Tif1γ concentration), rather than biased, immunohistochemistry-based observations. In addition, using plasma as the clinical sample material has a multitude of benefits, the most important being the simplicity and minimal invasiveness of sampling. Using the plasma levels of Tif1γ, an average concentration was calculated, which was then selected as a cut-off value (18.40 ng/ml) to divide patients into Tif1γ-positive and Tif1γ-negative groups. The groups had a representative number (52 Tif1γ-positive, 58 Tif1γ-negative), so that a long-term follow-up (98 months) could be conducted. Patients with low plasma Tif1γ had significantly shorter OS, while Tif1γ-positive patients had improved OS compared with Tif1γ-negative patients.
In order to further validate the clinical significance of these results, the association of OS and well-established clinicopathological characteristics, including lymph node status or tumor grade, was assessed by univariate and multivariate Cox proportional hazards analysis. The results confirmed the prognostic significance of Tif1γ plasma levels.
In addition, the CTC count was measured, as CTC number has been established as a strong independent prognostic factor for OS and PFS in patients with breast cancer. Although not all CTCs produce metastases, their spread is an important prerequisite for clinical metastasis. CTCs with certain biological characteristics are more prone to develop micrometastases. The association between CTC number and poor OS was confirmed in the cohort of the current study. Additionally, the results were similar to those produced by measuring Tif1γ, which suggested that using Tif1γ is as accurate as detecting CTCs for prognostic evaluation, but simpler and more efficient.
Notably, the Tif1γ plasma levels maintained significance in univariate and multivariate analyses, with low Tif1γ being an independent negative prognostic factor for OS in patients with breast cancer. The results are consistent with reports in hepatocellular cancer, NSCLC and several other types of cancer. However, the findings of the present study do not coincide with those of Kassem et al (62), where high expression of TGF-β1 and Tif1γ was associated with poorer outcome.
Overall, low Tif1γ was identified as an independent and significant risk factor for survival following curative resection in treatment-naïve patients. These results suggest that the serum levels of Tif1γ can be used as an accessible and feasible outcome predictor. To the best of our knowledge, this is the first prospective, long-term study on the clinical impact of Tif1γ in patients with breast cancer. The measurement of Tif1γ serum levels is translatable into oncological practice as a simple, cost-effective and rapid method, which may be implemented for diagnosis, therapy decision-making, prognostic determination and disease monitoring.
There are several limitations to the present study. Although the patient cohort of 110 cases is the most comprehensive data composition assembled in the existing literature thus far, it is a relatively small sample. Further studies and longer follow-ups are required to validate the findings. Additionally, investigating the regulatory mechanisms of Tif1γ in tumor growth and metastasis via inhibition of TGF-β/Smad signaling is crucial.
The current prospective study with a long-term follow-up demonstrated that analyzing Tif1γ serum expression may be useful for determining the prognosis of patients with breast cancer. Further elaboration and validation are required to establish Tif1γ as an easily detectable, non-invasive, novel biomarker with predictive power that can be implemented in breast cancer management and disease monitoring.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 30972633), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (grant no. 2015-311), the Shanghai Health and Family Planning Commission Project (grant nos. 20134298 and 201640253), the Shanghai Health and Family Planning Commission Fund for Qing Nian Yi Shi Training Project (grant no. 2014118), the Shanghai Yangpu District Science and Technology Commission Project (grant nos. 2016-2017, YP17ZM02), the Shanghai Yangpu District Health and Family Planning Commission Project (grant nos. 2011-2013 and 2016–2017, YP17ZM02), the Shanghai Yangpu District Health and Family Planning Commission Fund for Bai Yi Deng Gao Training Project (grant no. 2014-2016), the Shanghai Yangpu District Health and Family Planning Commission Fund for Hao Yi Shi Training Project (grant no. 201742), the Academic Leader in Climbing Program from Yangpu Center Hospital (grant nos. YE2201608 and YE2201703), the Science Program from Yangpu Center Hospital (grant no. SE1201746), the Natural Science Foundation of Shanghai (grant no. 18ZR1436000), and the Fundamental Research Funds for the Central Universities (grant no. 22120180286).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
FC and XP made substantial contributions to the design of the experiments and revised the manuscript. LC, ZZ and XP performed the experiments, XP, MW, SC, MAFL, CC and EB analyzed and interpreted the data.
Ethics approval and consent to participate
Yangpu Hospital, Tongji University School of Medicine approved the project under ‘Prognostic Significance of Tif1γ Expression and Circulating Tumor Cells influence in Patients with Breast Cancer (approval no. LL-2016-WSJ-002).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Herquel B, Ouararhni K, Davidson I. The TIF1α-related TRIM cofactors couple chromatin modifications to transcriptional regulation, signaling and tumor suppression. Transcription. 2011;2:231–236. doi: 10.4161/trns.2.5.17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, de Thé H. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 3.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 4.Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, Béchade G, Van Dorsselaer A, Sanglier-Cianférani S, Hamiche A, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:8212–8217. doi: 10.1073/pnas.1101544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Dent SY. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massagué J, Xi Q. TGF-β control of stem cell differentiation genes. FEBS Lett. 2012;586:1953–1958. doi: 10.1016/j.febslet.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heldin CH, Moustakas A. A new twist in Smad signaling. Dev Cell. 2006;10:685–686. doi: 10.1016/j.devcel.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 10.Heldin CH, Moustakas A. Role of Smads in TGFβ signaling. Cell Tissue Res. 2012;347:21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 11.Attisano L, Lee-Hoeflich ST. The Smads. Genome Biol. 2001;2:REVIEWS3010. doi: 10.1186/gb-2001-2-8-reviews3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith AP, Verrecchia A, Fagà G, Doni M, Perna D, Martinato F, Guccione E, Amati B. A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 2009;28:422–430. doi: 10.1038/onc.2008.395. [DOI] [PubMed] [Google Scholar]
- 13.Padgett RW. TGFbeta signaling pathways and human diseases. Cancer Metastasis Rev. 1999;18:247–259. doi: 10.1023/A:1006325408261. [DOI] [PubMed] [Google Scholar]
- 14.Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Morsut L, Yan KP, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 16.Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1g to Chromatin via Its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Kusy S, Gault N, Ferri F, Lewandowski D, Barroca V, Jaracz-Ros A, Losson R, Romeo PH. Adult hematopoiesis is regulated by TIF1γ, a repressor of TAL1 and PU.1 transcriptional activity. Cell Stem Cell. 2011;8:412–425. doi: 10.1016/j.stem.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Bai RY, Koester C, Ouyang T, Hahn SA, Hammerschmidt M, Peschel C, Duyster J. SMIF, a Smad4-interacting protein that functions as a co-activator in TGFbeta signalling. Nat Cell Biol. 2002;4:181–190. doi: 10.1038/ncb753. [DOI] [PubMed] [Google Scholar]
- 19.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 Branches of the TGFbeta Pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 20.Aucagne R, Droin N, Paggetti J, Lagrange B, Largeot A, Hammann A, Bataille A, Martin L, Yan KP, Fenaux P, et al. Transcription intermediary factor 1γ is a tumor suppressor in mouse and human chronic myelomonocytic leukemia. J Clin Invest. 2011;121:2361–2370. doi: 10.1172/JCI45213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emanuel PD. Juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia. Leukemia. 2008;22:1335–1342. doi: 10.1038/leu.2008.162. [DOI] [PubMed] [Google Scholar]
- 22.Quéré R, Saint-Paul L, Carmignac V, Martin RZ, Chrétien ML, Largeot A, Hammann A, Pais de Barros JP, Bastie JN, Delva L. Tif1γ regulates the TGF-β1 receptor and promotes physiological aging of hematopoietic stem cells. Proc Natl Acad Sci USA. 2014;111:10592–10597. doi: 10.1073/pnas.1405546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindley LE, Briegel KJ. Generation of mice with a conditional Lbh null allele. Genesis. 2013;51:491–497. doi: 10.1002/dvg.22390. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, You MJ, Young KH, Lin P, Lu G, Medeiros LJ, Bueso-Ramos CE. Advances in the molecular pathobiology of B-lymphoblastic leukemia. Hum Pathol. 2012;43:1347–1362. doi: 10.1016/j.humpath.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Wang E, Kawaoka S, Roe JS, Shi J, Hohmann AF, Xu Y, Bhagwat AS, Suzuki Y, Kinney JB. The transcriptional cofactor TRIM33 prevents apoptosis in B lymphoblastic leukemia by deactivating a single enhancer. Elife. 2015;4:e06377. doi: 10.7554/eLife.06377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan N, Song L, Zhang S, Lin W, Cao Y, Xu F, Fang Y, Wang Z, Zhang H, Li X, et al. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica. 2015;100:345–356. doi: 10.3324/haematol.2014.113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain S, Singhal S, Francis F, Hajdu C, Wang JH, Suriawinata A, Wang YQ, Zhang M, Weinshel EH, Francois F, et al. Association of overexpression of TIF1γ with colorectal carcinogenesis and advanced colorectal adenocarcinoma. World J Gastroenterol. 2011;17:3994–4000. doi: 10.3748/wjg.v17.i35.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain S, Yu M, Singhal S, Francis F, Hajdu C, Suriawinata S, Zhang M, Aladhamy N, Chiriboga L, Pan R, et al. Overexpression of transcription intermediary factor 1 γ (TIF1γ) is an early event in colorectal carcinogenesis and inversely related to Smad4 inactivation in colorectal carcinoma. Lab Investig. 2010;90:149A. [Google Scholar]
- 29.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 30.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics: 2011. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 31.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 33.Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Kuzma CS, Pluard TJ, Somlo G, Port RT, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance) J Clin Oncol. 2015;33:13–21. doi: 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 35.Sharma P, Stecklein SR, Kimler BF, Sethi G, Petroff BK, Phillips TA, Tawfik OW, Godwin AK, Jensen RA. The prognostic value of BRCA1 promoter methylation in early stage triple negative breast cancer. J Cancer Ther Res. 2014;3:1–11. doi: 10.7243/2049-7962-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller HM, Fiegl H, Widschwendter A, Widschwendter M. Prognostic DNA methylation marker in serum of cancer patients. Annals of the New York Academy of Sciences. 2004:pp44–49. doi: 10.1196/annals.1318.008. [DOI] [PubMed] [Google Scholar]
- 37.Duffy MJ. Serum tumor markers in breast cancer: Are they of clinical value? Clin Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 38.Cai FF, Kohler C, Zhang B, Wang MH, Chen WJ, Zhong XY. Epigenetic therapy for breast cancer. Int J Mol Sci. 2011;12:4465–4487. doi: 10.3390/ijms12074465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2009;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar R. Breast cancer tumor markers. J Solid Tumors. 2012;2:43. doi: 10.5430/jst.v2n1p43. [DOI] [Google Scholar]
- 41.Payne SJ, Bowen RL, Jones JL, Wells CA. Predictive markers in breast cancer-the present. Histopathology. 2008;52:82–90. doi: 10.1111/j.1365-2559.2007.02897.x. [DOI] [PubMed] [Google Scholar]
- 42.Wakefield LM, Roberts AB. TGF-beta signaling: Positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/S0959-437X(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 43.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikushima H, Miyazono K. TGFbeta signalling: A complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 45.Faure E, Heisterkamp N, Groffen J, Kaartinen V. Differential expression of TGF-beta isoforms during postlactational mammary gland involution. Cell Tissue Res. 2000;300:89–95. doi: 10.1007/s004410000183. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen AV, Pollard JW. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Development. 2000;127:3107–3118. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 47.Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS, Hennighausen L, Furth PA. Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc Natl Acad Sci USA. 1997;94:3425–3430. doi: 10.1073/pnas.94.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bierie B, Gorska AE, Stover DG, Moses HL. TGF-beta promotes cell death and suppresses lactation during the second stage of mammary involution. J Cell Physiol. 2009;219:57–68. doi: 10.1002/jcp.21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panis C, Herrera AC, Victorino VJ, Aranome AM, Cecchini R. Screening of circulating TGF-beta levels and its clinicopathological significance in human breast cancer. Anticancer Res. 2013;33:737–742. [PubMed] [Google Scholar]
- 50.Paiva CE, Drigo SA, Rosa FE, Moraes Neto FA, Caldeira JR, Soares FA, Domingues MA, Rogatto SR. Absence of transforming growth factor-beta type II receptor is associated with poorer prognosis in HER2-negative breast tumours. Ann Oncol. 2010;21:734–740. doi: 10.1093/annonc/mdp518. [DOI] [PubMed] [Google Scholar]
- 51.Richardsen E, Uglehus RD, Johnsen SH, Busund LT. Immunohistochemical expression of epithelial and stromal immunomodulatory signalling molecules is a prognostic indicator in breast cancer. BMC Res Notes. 2012;5:110. doi: 10.1186/1756-0500-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desruisseau S, Palmari J, Giusti C, Romain S, Martin PM, Berthois Y. Determination of TGFbeta1 protein level in human primary breast cancers and its relationship with survival. Br J Cancer. 2006;94:239–246. doi: 10.1038/sj.bjc.6602920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koumoundourou D, Kassimatis T, Zolota V, Tzorakoeleftherakis E, Ravazoula P, Vassiliou V, Kardamakis D, Varakis J. Prognostic significance of TGFbeta-1 and pSmad2/3 in breast cancer patients with T1-2,N0 tumours. Anticancer Res. 2007;27:2613–2620. [PubMed] [Google Scholar]
- 54.Vincent DF, Yan KP, Treilleux I, Gay F, Arfi V, Kaniewsky B, Marie JC, Lepinasse F, Martel S, Goddard-Leon S, et al. Inactivation of TIF1gamma cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5:e1000575. doi: 10.1371/annotation/7941c465-532f-4b42-b541-72d0810943b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fattet L, Ay AS, Bonneau B, Jallades L, Mikaelian I, Treilleux I, Gillet G, Hesling C, Rimokh R. TIF1γ requires sumoylation to exert its repressive activity on TGFβ signaling. J Cell Sci. 2013;126:3713–3123. doi: 10.1242/jcs.126748. [DOI] [PubMed] [Google Scholar]
- 56.Mascle XH, Germain-Desprez D, Huynh P, Estephan P, Aubry M. Sumoylation of the transcriptional intermediary factor 1beta (TIF1beta), the Co-repressor of the KRAB multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J Biol Chem. 2007;282:10190–10202. doi: 10.1074/jbc.M611429200. [DOI] [PubMed] [Google Scholar]
- 57.Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Yang H, Lei Z, Zhao J, Chen Y, Chen P, Li C, Zeng Y, Liu Z, Liu X, Zhang HT. Repression of TIF1γ by SOX2 promotes TGF-β-induced epithelial-mesenchymal transition in non-small-cell lung cancer. Oncogene. 2016;35:867–877. doi: 10.1038/onc.2015.141. [DOI] [PubMed] [Google Scholar]
- 59.Mikaelian I, Malek M, Gadet R, Viallet J, Garcia A, Girard-Gagnepain A, Hesling C, Gillet G, Gonzalo P, Rimokh R, Billaud M. Genetic and pharmacologic inhibition of mTORC1 promotes EMT by a TGF-β-independent mechanism. Cancer Res. 2013;73:6621–6631. doi: 10.1158/0008-5472.CAN-13-0560. [DOI] [PubMed] [Google Scholar]
- 60.Hesling C, Fattet L, Teyre G, Jury D, Gonzalo P, Lopez J, Vanbelle C, Morel AP, Gillet G, Mikaelian I, Rimokh R. Antagonistic regulation of EMT by TIF1γ and Smad4 in mammary epithelial cells. EMBO Rep. 2011;12:665–672. doi: 10.1038/embor.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hesling C, Lopez J, Fattet L, Gonzalo P, Treilleux I, Blanchard D, Losson R, Goffin V, Pigat N, Puisieux A, et al. Tif1γ is essential for the terminal differentiation of mammary alveolar epithelial cells and for lactation through SMAD4 inhibition. Development. 2013;140:167–175. doi: 10.1242/dev.085068. [DOI] [PubMed] [Google Scholar]
- 62.Kassem L, Deygas M, Fattet L, Lopez J, Goulvent T, Lavergne E, Chabaud S, Carrabin N, Chopin N, Bachelot T, et al. TIF1γ interferes with TGFβ1/SMAD4 signaling to promote poor outcome in operable breast cancer patients. BMC Cancer. 2015;15:453. doi: 10.1186/s12885-015-1471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massagué J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang PH, Truica CI, Drabick JJ, El-Deiry WS, Dao M, et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci USA. 2015;112:4970–4975. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dive C, Brady G. SnapShot: Circulating tumor cells. Cell. 2017;168:P742–P742.e1. doi: 10.1016/j.cell.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 66.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 67.Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9:311–320. [PubMed] [Google Scholar]
- 68.Kuo AH, Clarke MF. Identifying the metastatic seeds of breast cancer. Nat Biotechnol. 2013;31:504–505. doi: 10.1038/nbt.2599. [DOI] [PubMed] [Google Scholar]
- 69.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 70.Giuliano M, Giordano A, Jackson S, Hess KR, de Giorgi U, Mego M, Handy BC, Ueno NT, Alvarez RH, De Laurentiis M, et al. Circulating tumor cells as prognostic and predictive markers in metastatic breast cancer patients receiving first-line systemic treatment. Breast Cancer Res. 2011;13:R67. doi: 10.1186/bcr2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pierga JY, Bidard FC, Mathiot C, Brain E, Delaloge S, Giachetti S, de Cremoux P, Salmon R, Vincent-Salomon A, Marty M. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin Cancer Res. 2008;14:7004–7010. doi: 10.1158/1078-0432.CCR-08-0030. [DOI] [PubMed] [Google Scholar]
- 72.Bidard FC, Proudhon C, Pierga JY. Circulating tumor cells in breast cancer. Mol Oncol. 2016;10:418–430. doi: 10.1016/j.molonc.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pan X, Zeng SL, Yu D, Liang XL, Ji C, Pan B, Cai J, Wang Y, Min Y, Fang W, Liao WQ. Variable domain of the heavy chain of heavy-chain antibody of the Rv0733 antigen of mycobacterium tuberculosis panned and identified from a nonimmune llama VHH phage display library. Int J Clin Exp Pathol. 2016;9:2869–2878. [Google Scholar]
- 74.Lin XY, Cai FF, Wang MH, Pan X, Wang F, Cai L, Cui RR, Chen S, Ewelina B. Mammalian sterile 20-like kinase 1 expression and its prognostic significance in patients with breast cancer. Oncology Lett. 2017;14:5457–5463. doi: 10.3892/ol.2017.6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen B, Le W, Wang Y, Li Z, Wang D, Ren L, Lin L, Cui S, Hu JJ, Hu Y, et al. Targeting negative surface charges of cancer cells by multifunctional nanoprobes. Theranostics. 2018;6:1887–1898. doi: 10.7150/thno.16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banin Hirata BK, Oda JM, Losi Guembarovski R, Ariza CB, de Oliveira CE, Watanabe MA. Molecular markers for breast cancer: Prediction on tumor behavior. Dis Markers. 2014;2014:513158. doi: 10.1155/2014/513158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiong R, Zhao J, Gutgesell LM, Wang Y, Lee S, Karumudi B, Zhao H, Lu Y, Tonetti DA, Thatcher GR. Novel selective estrogen receptor downregulators (SERDs) developed against treatment-resistant breast cancer. J Med Chem. 2017;60:1325–1342. doi: 10.1021/acs.jmedchem.6b01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: Classification, prognostication, and prediction. Lancet. 2011;378:1812–1823. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 81.Assi HA, Khoury KE, Dbouk H, Khalil LE, Mouhieddine TH, El Saghir NS. Epidemiology and prognosis of breast cancer in young women. J Thorac Dis. 2013;5(Suppl 1):S2–S8. doi: 10.3978/j.issn.2072-1439.2013.05.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hefti MM, Hu R, Knoblauch NW, Collins LC, Haibe-Kains B, Tamimi RM, Beck AH. Estrogen receptor negative/progesterone receptor positive breast cancer is not a reproducible subtype. Breast Cancer Res. 2013;15:R68. doi: 10.1186/bcr3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cancer Research UK. Breast cancer incidence statistics. Cancer Res UK. 2012 [Google Scholar]
- 84.Ban KA, Godellas CV. Epidemiology of breast cancer. Surg Oncol Clin N Am. 2014;23:409–422. doi: 10.1016/j.soc.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 85.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist. 2004;9:606–616. doi: 10.1634/theoncologist.9-6-606. [DOI] [PubMed] [Google Scholar]
- 86.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Fr Int Agency Res Cancer. Vol. 11. Lyon: 2013. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase. No. 11 [Internet] [Google Scholar]
- 87.Yoneda A, Lendorf ME, Couchman JR, Multhaupt HA. Breast and ovarian cancers: A survey and possible roles for the cell surface heparan sulfate proteoglycans. J Histochem Cytochem. 2012;60:9–21. doi: 10.1369/0022155411428469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Esteva FJ, Hortobagyi GN. Prognostic molecular markers in early breast cancer. Breast Cancer Res. 2004;6:109–118. doi: 10.1186/bcr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Payne SJL, Bowen RL, Jones JL, Wells CA. Predictive markers in breast cancer-the present. Histopathology. 2008;52:82–90. doi: 10.1111/j.1365-2559.2007.02897.x. [DOI] [PubMed] [Google Scholar]
- 91.Andrieux G, Fattet L, Le Borgne M, Rimokh R, Théret N. Dynamic regulation of Tgf-B signaling by Tif1γ: A computational approach. PLoS One. 2012;7:e33761. doi: 10.1371/journal.pone.0033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.