Abstract
Transposable elements inhabiting eukaryotic genomes are generally regarded either as selfish DNA, which is selectively neutral to the host organism, or as parasitic DNA, deleterious to the host. Thus far, the only agreed-upon example of beneficial eukaryotic transposons is provided by Drosophila telomere-associated retrotransposons, which transpose directly to the chromosome ends and thereby protect them from degradation. This article reports the transposon content of the genome of the protozoan Giardia lamblia, one of the earliest-branching eukaryotes. A total of three non-long terminal repeat retrotransposon families have been identified, two of which are located at the ends of chromosomes, and the third one contains exclusively dead copies with multiple internal deletions, nucleotide substitutions, and frame shifts. No other reverse transcriptase- or transposase-related sequences were found. Thus, the entire genome of this protozoan, which is not known to reproduce sexually, contains only retrotransposons that are either confined to telomeric regions and possibly beneficial, or inactivated and completely nonfunctional.
According to the model of transposon proliferation presented by Hickey (1), deleterious transposons are not expected to persist in long-term asexuals. Results consistent with this expectation were obtained in recent experiments testing 24 eukaryotic phyla for the presence of known transposons by nested PCR (2). There are two classes of autonomous eukaryotic transposons encoding conserved enzymes necessary for transposition: (i) retrotransposons, which transpose by means of an RNA intermediate copied into DNA by an element-encoded reverse transcriptase (RTase); and (ii) DNA transposons, which transpose as DNA by a cut and paste mechanism, using an element-encoded transposase (3, 4). In PCR assays, the only group that tested negative for the presence of RTase-related sequences, although positive for mariner-like DNA transposases, were rotifers of the class Bdelloidea, a monophyletic group which apparently lost sexual reproduction many millions of years ago (5). Of those tested in ref. 2, the only other species in which no sexual process is known is Giardia lamblia (or G. intestinalis), which tested positive for RTases. Cloning and sequencing of the corresponding PCR products suggested the existence of two non-long terminal repeat [also called long interspersed nuclear element (LINE)-like] retrotransposon families in the G. lamblia genome, a finding seemingly at odds with the indication from the Bdelloidea that such elements would not persist in long-term asexuals. Only in sexuals, but not long-term asexuals, can deleterious transposons be expected to go to fixation (6).
G. lamblia is a protozoan, parasitizing the intestines of mammals and birds, which has two morphologically identical nuclei in each cell and a polyploid genome (reviewed in refs. 7–9). Its genome can be divided into five major groups displaying physical linkage of markers, and five chromosome-like bodies can be visualized in each nucleus (7, 8). Each linkage group, however, can be represented by several size variants detectable by pulse-field gel electrophoresis, with an invariant central core and a significant degree of variability toward the ends of the chromosomes (7–11). The variable ends undergo frequent rearrangements, but the central regions do not.
Because all of the eukaryotic genomes previously sequenced to completion are those of sexually reproducing organisms, it was of particular interest to evaluate the G. lamblia genome for abundance and activity status of these retrotransposons, in light of their apparent absence from bdelloids. An ongoing G. lamblia genome sequencing project (12) is approaching completion and is currently at the gap-closure stage, making it possible to assess the transposon content of this genome and to analyze internal and flanking sequences of all identified transposon copies with respect to their degrees of divergence, intactness of ORFs, and insertional specificities.
Methods
Primers, DNA sources, and amplification conditions for PCR reactions were as described (2). Single-pass sequencing reads from the G. intestinalis (strain WB) genome-sequencing project were obtained from the high-throughput genomic sequence (HTGS) subdivision of GenBank (see www.mbl.edu/Giardia). Consensus sequences were assembled from 216, 66, and 588 individual sequencing reads, averaging 800 bp in length, for GilM, GilT, and GilD, respectively. For gap closure in GilT, additional sequencing of G. lamblia genomic clones was performed with the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) and analyzed on an Applied Biosystems Prism 310 genetic analyzer. Sequence assembly and analysis was done with WISCONSIN PACKAGE VERSION 10.0 (GCG). RTase sequences chosen for phylogenetic analysis represent a subset of the seed alignment PF00078 rvt (PFAM RELEASE 6.4), which includes only the seven conserved domains that are common to all RTases. After removal of the most prominent gaps, a total of 304 amino acids was included in the analysis. Inference of phylogenetic relationships was performed by using MRBAYES2.01 (13), using the JTT substitution matrix and a mixed (invariable plus gamma) model of rate heterogeneity, with rates inferred from the data set. Four Markov chains were initiated at random, and the program was allowed to run for 100,000 generations with sample frequency of 10. On average, 30,000 generations were required for likelihood convergence, with the first 3,000 less likely trees discarded as burn-in, and the remaining 7,000 trees used to build a consensus tree.
Results
PCR Experiments.
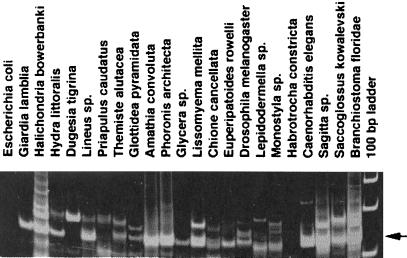
Nested PCR amplification of G. lamblia genomic DNA with highly degenerate primers specific for the superfamily of LINE-like RTases typically yielded a single band of high intensity (Fig. 1). This result indicates that representatives of multiple LINE-like clades, which usually have different characteristic distances between B and C motifs of the RTase gene, are not likely to be present in the G. lamblia genome, in contrast to most other eukaryotes (ref. 2; Fig. 1), as was confirmed by analysis of genome sequence (see below). Cloning and sequencing of 120-bp PCR products from G. lamblia revealed that their sequences fell into two different groups, which were assigned to two transposon families hereafter named GilM and GilD (G. intestinalis LINEs). It was not possible, however, to assign these fragments with confidence to the superfamily of LINE elements, because their homology with known RTases in GenBank was insufficient. Therefore, full-length RTase sequences for each family had to be determined to reliably establish their relationship to known RTases.
Figure 1.
PCR amplification of total genomic DNA isolated from representatives of 20 animal phyla, plus G. lamblia and Escherichia coli, with nested primers specific for LINE-like RTases. An arrow corresponds to the position of prominent sequence-specific amplification products about 120 bp in length, typically representing members of the most abundant CR1 clade; larger products correspond to members of other clades such as L1, jockey, etc. (2). Amplification products are not visible in E. coli and in Habrotrocha constricta, a bdelloid rotifer (2).
Sequence Assembly and Database Analysis.
Genomic shotgun reads were used to assemble full-length consensus sequences for GilM and GilD by extending sequence homology in both directions from the RTase fragments obtained in PCR experiments. Conceptual translations of ORFs from the consensus sequences were used in blastx searches to detect any other related elements in the G. lamblia genome. This search revealed a third family of LINE-like elements, designated GilT. This family has a much lower copy number than the other two, and the sequences present in the database could not be assembled into a single consensus without gaps, which were closed by targeted sequencing. Analysis of the resulting full-length ORFs from the three families demonstrates that they can be unambiguously assigned to the LINE-like superfamily of RTases, with E values of blastx matches to the conserved pfam00078 RTase domain ranging from 10−19 to 10−22.
No other known transposon types, such as retrovirus-like or DNA transposons, could be detected in database searches by using profiles corresponding to RTases and transposases from all currently known superfamilies. Moreover, analysis of total repetitive DNA sequences in the assembled contigs demonstrated that none of the repeats correspond to any DNA or RNA transposons other than those described here. Because the G. lamblia project has already achieved the target 4-fold coverage (12), and other multicopy elements are not likely to be present exclusively in the unsequenced portion of the genome (estimated at about 5%), it may be concluded that the identified families represent the only autonomous transposons that are repeated in the genome of this protozoan, barring the presence of unknown transposon classes.
Telomere-Associated LINE Families.
GilT and GilM are potentially active elements, because they are mostly represented by intact sequences. The nucleotide sequence identity within each family exceeds 99%, which is indicative of recent retrotransposition activity. Remarkably, both families are confined to immediate subtelomeric regions, because any GilT or GilM sequence is flanked at its 5′ end either by reverse complement of G. lamblia telomeric repeats (TACCC)n (14) or by another copy of the same element in the same orientation. Members of such a tandem array are separated from each other by the (A)n stretch (n = 10–16) and arranged in a strictly head to tail orientation, with no target site duplications. The most distal member in the array is truncated at its 5′ end and capped by telomeric repeats. Interestingly, (A)n stretches are always followed by an intact 5′ end of another copy and never by a 5′-truncated copy (Fig. 2B).
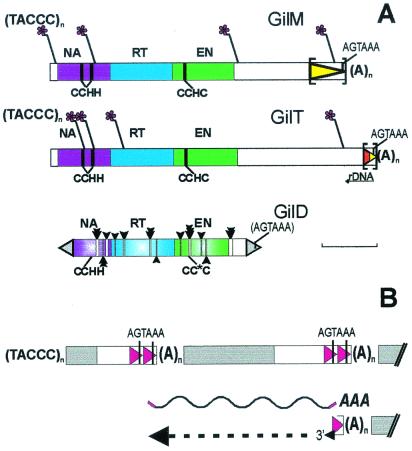
Figure 2.
(A) Structure of three LINE-like retrotransposon families from G. lamblia. Asterisks designate sites of telomeric repeat addition to 5′-truncated copies of GilM and GilT. Oligo(A) tracts are designated by (A)n and are immediately followed by the intact 5′ end of another copy of the same element. Protein domains are designated as NA, nucleic acid binding (purple); RT, RTase (blue); EN, REL-endonuclease (green). Also shown are the Zn-finger motifs (CCHH and CCHC, vertical lines) and the polyadenylation signal (AGTAAA), which is included in parentheses in GilD because it can be identified only in a subset of copies. Noncoding regions are in white. The region in the 3′ UTR, which may also exist as a tandem duplication, is shown by a triangle in square brackets; the segment in GilT originally incorporated from rDNA is shown in red. The sites of deletion tracts in GilD are indicated by empty bars with vertical arrows, the number and size of arrows corresponding to the number and size of deletions at a particular site, and the adjacent inverted repeat is shown as a gray triangle. [Bar = 1 kb.] (B) Model of telomere formation by LINE elements based on features shared between telomere-associated retrotransposons of Giardia and Drosophila (ref. 25; this study). Transcription from a nontruncated member of a tandem array can result in a full-length polyadenylated transcript (wavy line), which gets attached to the 3′ end of a chromosome serving as a primer for reverse transcription. Note that there is a potential to use annealing of the 3′ RNA end to a homologous segment at the DNA termini. Coding sequences are in blue; the 3′ segment may or may not be tandemly duplicated; telomeric repeats are not added in Drosophila. Not to scale. The telomere is on the left.
The coding region of GilT and GilM consists of a long ORF about 1,000 amino acids in length, which is preceded by a short 55-bp 5′ untranslated region (UTR), has 54% overall identity and 67% similarity between the two families, and may be subdivided into three distinct domains, with RTase in the middle (Fig. 2A; alignment in Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). A total of nine conserved RTase motifs, characteristic for other members of the LINE superfamily (15), can be identified. The N terminus contains two Zn finger motifs of the C2H2 type. The C terminus consists of a CCHC finger followed by the so-called REL-ENDO domain previously identified in a small group of LINEs from trypanosomatids (CRE, SLACS, and CZAR), arthropods (R2 ribosomal insertions), and Caenorhabditis elegans (NeSL-1) (refs. 16 and 17). In arthropods, this domain was shown to encode an rDNA-specific endonuclease (16), and in trypanosomatids and the nematode, these LINEs insert sequence-specifically into spliced leader exons (17–20).
About half of the copies from each family carry a frameshift between the C2H2 fingers and the RTase domain. Such frameshifts are not uncommon in retroelements and are usually thought to reduce the expression level of RTase relative to the upstream gag-like proteins (21). Its appearance in both GilM and GilT is intriguing, because the ORF of the HeT-A telomere-associated retrotransposon family in Drosophila can also be either with or without such a frameshift (22).
The 3′ UTRs of GilT and GilM are unusually long, being similar in length to the 3-kb ORF. The two UTRs have no sequence similarity other than the 110-bp segment preceding the polyadenylation signal AGTAAA (7, 8) and the (A)n stretch. The 3′ UTR of GilM ends in a 750-bp sequence that can also be present as a tandem repeat (including the polyadenylation signal but not the (A)n stretch; Fig. 2A). About half of these 750-bp segments are preceded by the coding region at the 5′ end, and another half appear in tandem. The GilT 3′ UTR also contains a 3′ terminal 270-bp segment that may be present in individual clones either as a single copy or as a tandem repeat. This segment is of composite origin; it consists of 160 bp originating from the 3′ end of the large subunit of ribosomal DNA in an antisense orientation, followed by 110 bp from the very 3′ end of GilM, including the polyadenylation signal. Capture of downstream sequences by read-through transcription and subsequent retrotransposition (termed 3′ transduction) is a known property of human LINE elements (23) and may have played a role in 3′ UTR formation in G. lamblia LINEs.
Most Telomeric Repeats Are Joined to LINE Elements.
Telomeric repeat-containing clones were extracted from the database in a blastn search, and the junctions between telomeric repeats and the rest of chromosomal DNA were inspected by blast comparisons or obtained by targeted sequencing. All of the (TAGGG)n stretches (n > 3) were joined to chromosomal DNA only on one side, indicating that there were no interstitial TAGGG sequences repeated more than three times. This finding agrees with results of Upcroft et al. (24), who reported no hybridization of the telomeric probe to internal chromosome fragments.
Remarkably, the analysis of junction DNA revealed that 8 of 11 (TAGGG)n sequences were adjacent to 5′-truncated copies of LINE elements (Table 1). Thus, these elements are the predominant components of telomeric junctions, in contrast to previous studies identifying sequences adjoining G. lamblia telomeric repeats mostly as rDNA (10, 11, 14). In the remaining three clones, telomeric repeats are joined to a variant-specific surface protein or to rDNA (Table 1).
Table 1.
Junctions between telomeric repeats and chromosomal DNA of G. lamblia
| Clone* | Junction‡ | Sequence¶ |
|---|---|---|
| EJ1336/ej2414 | GilM 826 | (taccc)5tcctcactgacgtattcacagagatggcgg |
| KJ4819/ki1170 | GilM 3374 | (taccc)9tactagcgcgaacggacccctgggcctgcg |
| LJ0347 | GilM 94 | (taccc)54tacaggcccctactaggggcactcccgatc |
| NF0311/ng2053 | GilM 5262 | (taccc)18tactctgtgccgtgacccgcgcgcccccgc |
| KJ1196 | GilT 5316 | (taccc)27tactctgccagcatagtcttctctcctccc |
| NJ1197 | GilT 1356 | (taccc)25tactctgtctgccccatagcgatacaagag |
| HG1254 | GilT 783 | (taccc)4tacatgatcgggatacgagcggcaacccca |
| nj3761 | GilT 547 | (taccc)47tactccatccgccgactcctctggctgccc |
| KJ6036/ei1613 | rDNA 5393 | (taccc)81tactytcycytcstktggaattaccccggc |
| NJ2364† | rDNA ← | (taccc)53tcactttcgtttcggtttggtccggcctcg |
| MJ3348/aj1354 | VSP ← | (taccc)49tactctggcgtatcagtatctgtagtagtg |
Clones sequenced by primer walking are in lowercase.
Junction is formed with a unique sequence, and the rDNA is at the opposite end, so that the direction of rDNA transcription is also toward the telomere (←). VSP, variant-specific surface protein.
Numbers specify nucleotides at which GilM or GilT is truncated and telomeric repeats are added. Numbering in the rDNA repeat unit corresponds to that in X52949, and the junction is in the middle of the 16S subunit. One more clone, EJ2167, contains (TACCC)n at one end and rDNA at the other but was not sequenced to completion, therefore the junction may or may not be identical to the ones shown here.
Nucleotides between (TACCC)n and the beginning of GilM/GilT or rDNA homology are underlined.
Most of the GilM arrays are followed at their 3′ ends by single-copy genes (e.g., ABC transporter, DMC1, WD-repeat protein, Zn-finger); some are adjacent to rDNA. GilT arrays are mostly followed by rDNA, which agrees with the presence of a short region of homology to rDNA in the 3′ UTR. The transcriptional orientation of adjacent chromosomal genes (toward the telomere) is always opposite to that of GilT and GilM (away from the telomere). The tandem arrangement of telomeric transposons protects proximal copies from terminal degradation and preserves their transcriptional capability (25). The 75% identity of the 3′-most sequence of GilM and GilT, together with the lack of conservation in the 5′ UTRs, suggests the presence of transcriptionally important elements such as promoters and terminators in the terminal segment of the 3′ UTR.
A Retrotransposon Inactivated by Multiple Deletions.
Assembly of a full-length consensus sequence of the high-copy-number family, GilD, was not a trivial task: it is represented exclusively by dead copies, none of which have preserved an intact ORF, and the process is complicated by the inability of common sequence-assembly programs to combine sequences with long deletions and a high degree of divergence into a multiple-sequence alignment. Therefore, in contrast to the easily defined ORFs of GilT and GilM, the ORF of GilD is a result of a multistep reconstruction, which included restoration of many deleted regions disrupting the ORF and substitution of frameshifts and stop codons with sequences present in other copies wherever necessary for reconstitution of the reading frame. The restored consensus ORF occupies most of the 3-kb element and can be aligned with the ORFs of the other two elements with an overall 25% identity and 39% similarity, except for the truncated N terminus lacking one of the C2H2 fingers and a short 3′ UTR.
There are 12 deletion tracts throughout the entire length of the element (Fig. 2A), and very few (only the shortest) clones have no internal deletions. Any of these deletions introduced into an intact element would abolish its function by disrupting conserved protein motifs. Deletions are apparently mediated by 3–5-bp repeats at the boundaries and range in size from 8 to 60 bp. Many deletions are shared between several copies but are not present in the others containing deletions in other places, suggesting that some of the deleted copies continued to proliferate in trans, using the enzymes provided by still-intact copies. The presence of deletions in some copies but not in the others implies that deletions are not induced by sequence per se, and also indicates that proliferation of deleted copies took place before accumulation of single-nucleotide polymorphisms, most of which are not shared between copies. No tandem arrangement, as for GilT and GilM, is observed nor are there any junctions with telomeric repeats.
GilD sequences with shared single-nucleotide polymorphisms (SNPs) are typically repeated 3–6 times in the database, each version representing a diverged unique copy and reflecting the current degree of genome coverage. The copy-number estimate for this family (about 30 per genome, as determined by the number of sequence clusters with shared SNPs) is about 2-fold higher than that of the two telomere-associated families combined. The divergence of GilD sequences from the consensus ranges from 6% to 13%, indicating its inactivation in the distant past. Single-nucleotide substitutions and indels are distributed uniformly and without any bias toward synonymous sites.
Adjacent to the coding region at the 5′ and 3′ ends are ≈200-bp imperfect inverted terminal repeats with a low degree of homology, alignments of which also exhibit a mosaic appearance indicative of frequent recombination or gene conversion. Such repeats are not typical of LINE-like elements and may not constitute an integral part of the element but might be present at both ends as a result of sequence-specific GilD insertion in either direction. A polyadenylation signal [but not the (A)n stretch] can be identified in at least some copies of the repeat, favoring the explanation of GilD insertion into its own UTR. GilD is often located near variant-specific surface protein genes/pseudogenes or other repetitive genes such as ankyrins. Recombination in these regions might contribute to generation of antigenic diversity, as described in other parasitic protozoans (26, 27).
Phylogenetic Placement of Giardia LINEs.
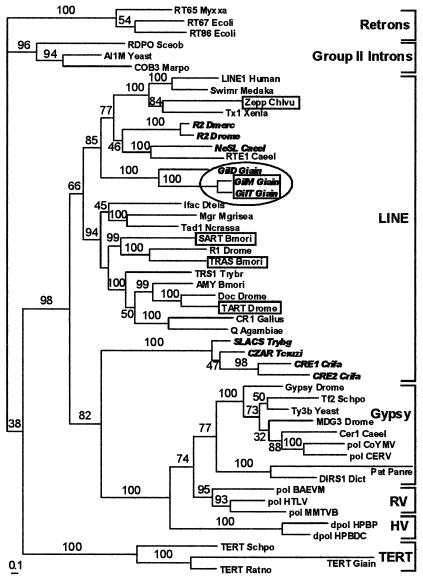
In the world of RTases (Fig. 3), LINE families from G. lamblia seem to be phylogenetically closest to those containing the REL-ENDO domain, such as the NeSL and R2 clades (15, 17). The dead GilD family occupies a more basal position than the two functional ones, and all of them form a distinct clade, indicating that they established residence in the G. lamblia genome a long time ago, with the active ones maintaining a high degree of sequence homogeneity.
Figure 3.
Phylogenetic analysis of RTase domains, including telomerase RTases (TERT), LINE-like retrotransposons, gypsy-like retrotransposons (including pararetroviruses), group II introns, bacterial retrons, retroviruses (RV), and hepadnaviruses (HV). The Giardia non-long terminal repeat retrotransposons identified in this study are enclosed in an oval. The tree was arbitrarily rooted with bacterial retrons, which have the simplest RTase domain structure. Telomere-associated LINE-like retrotransposons are boxed, and those containing the REL-ENDO domain appear in bold italic. Numbers above the branches are the clade credibility values or percentage of trees containing each bipartition. [Bar = 0.1 amino acid substitutions per site.]
It may also be seen that the property of telomeric localization was acquired during evolution by several LINE families (boxed) independently, because they definitely belong to different LINE clades. Although all LINE elements do not form a single clade, because of the CRE clade which groups together with virus-like elements, it should be noted that the most highly diverged RTases are not well resolved when analyzed by using other phylogenetic analysis programs. Telomerase RTases (TERTs) were probably separated from RTases of LINE-like transposons together with the appearance of the extended N-terminal domain responsible for recognition of an unlinked RNA template (see Discussion), which would not allow the RTase gene to multiply itself.
Discussion
Transposons and Telomeres.
Telomeric and subtelomeric regions are believed to represent a particularly suitable environment for harboring transposon insertions, because the latter would not cause much damage to the host by interfering with the function of nuclear genes, and might even confer benefits by expanding the buffer zone between the end of the chromosome and the nearby single-copy genes. Indeed, transposons with insertional specificities for telomeric or subtelomeric regions have been identified in such diverse organisms as Saccharomyces cerevisiae (Ty5), Bombyx mori (SART, TRAS), Chlorella vulgaris (Zepp), and Allium cepa (MP7) (28–31).
Only in Drosophila melanogaster (HeT-A and TART) (32, 33) have retrotransposons completely taken over the function of telomere maintenance. There are no telomeric repeats in this species, and the full genome sequence (34) does not contain coding regions homologous to telomerases. Associated with this function is the ability for terminal transposition, in which the RNA transcript of a retrotransposon attaches directly to the end of the chromosome by means of a poly(A) tail and undergoes reverse transcription in situ, initiated at the 3′ hydroxyl end of the chromosome (Fig. 2B). The process repeats itself, resulting in a chain of HeT-A and/or TART retrotransposons (often interspersed), with the same polarity for all members of the chain. Healing of chromosome breaks has been shown to occur by telomere formation associated with terminal transposition of HeT-A and TART (32, 35, 36). The mechanisms underlying direct attachment to chromosome termini and array formation are unknown.
Another distinctive feature of HeT-A and TART, found also in GilM and GilT, is an unusually long 3′ UTR. Long 3′ noncoding regions are highly atypical for retroelements, and their presence in the two Drosophila telomere-associated LINEs prompted speculations that they are required for telomeric chromatin structure and/or terminal transposition (33, 35, 37). HeT-A and TART can also carry tandemly duplicated segments at their 3′ ends (36).
Interestingly, the ORFs of telomere-associated retrotransposons from Giardia and Drosophila retain a coding capacity for an endonuclease-like protein, which is expected to provide recognition and cleavage of the target sequence. TART has an endonuclease domain belonging to the apurinic-apyrimidinic (AP)-like category (38), whereas the Giardia elements possess the REL-ENDO domain. It is unclear why these domains would be retained in otherwise unrelated families of telomeric transposons. Sequence-specific insertion into telomeric repeats may be excluded, because such repeats are never found at their 3′ flanks. Perhaps insertions into internal sites, if they do occur, are rapidly eliminated.
Also required for transposition in cis is the nucleic acid-binding capacity, which serves the nucleic acid chaperone function (39–41). This capacity is conserved not only in the autonomous TART element but also in HeT-A, which is nonautonomous because it does not code for its own RTase (22, 36). GilM and GilT are highly homologous in the putative NA-binding region, whereas only one of the C2H2 fingers can be identified in the reconstructed GilD.
Overall, structural comparisons show that GilT and GilM bear profound functional resemblance to the telomere-associated retrotransposons of Drosophila, including the ability to form tandem polar arrays at the chromosome ends, the coding capacity for RTase and nucleic acid-binding proteins, and an exceptionally long UTR with the 3′-most region prone to tandem duplications (Fig. 2B). It is quite remarkable that telomere-associated retrotransposons are found in Giardia, considered to be one of the earliest-branching eukaryotes on the basis of numerous molecular phylogenetic studies (e.g., refs. 42–45). It is also notable that in a tandem head to tail arrangement, which is characteristic of terminally transposing HeT-A as well as GilM/GilT, the transposon may no longer be regarded as selfish DNA when the promoter is located in the 3′ UTR, because it provides transcription of its downstream neighbor but not itself (25). Studies of GilM/GilT promoter activity will therefore be of significant interest.
A single-copy coding sequence for the telomerase catalytic subunit, which is a specialized RTase, has been identified in the G. lamblia genome (46). Its structure differs from other eukaryotic telomerase RTase genes because it lacks the conserved T motif. Its absence might interfere with proper telomerase function by affecting interaction with telomerase RNA (47, 48). It is possible that terminal transposition of LINE elements can to some extent compensate for such deficiencies in telomerase function, so that 5′ truncation occurs when LINEs are exposed to terminal degradation until telomerase starts adding TAGGG repeats to their ends. An alternative but less likely possibility is that GilM and GilT may insert sequence-specifically into their own 3′ UTRs in the same orientation, somehow losing oligo(A) after such insertion. The integration process does not proceed to completion in its usual sense, however; thus, target-site duplication is not observed and incomplete reverse transcription is followed not by template joining to the other end of the target, but by telomeric repeat addition to the truncated 5′ end. This explanation would imply direct coupling of telomerase action, and therefore telomere formation, with the transposition process.
It is only after complete assembly of the G. lamblia genome that we will know the degree of variability of its chromosome ends. The minimum number of telomeric repeat junction fragments would be 10; additional junctions may be present in minor chromosome variants, seen in many G. lamblia isolates. A total of 10 restriction fragments was reported to hybridize to the telomeric repeat probe in at least one G. lamblia isolate (24). Telomeric probe also hybridizes to two NotI fragments of chromosome 4 (11). Eleven telomeric junctions, eight of them being LINE elements, were identified in this study; the exact relationship between size variants of different chromosomes and terminal sequences remains to be determined as the assembly progresses.
Complete Inactivation of a High-Copy-Number LINE-Like Element.
The main structural differences between GilT/GilM and the reconstructed GilD are the absence of the N-terminal C2H2 finger and of the extended 3′ UTR region with the (A)n stretch in GilD. If these features are required for terminal transposition and they were initially present in the ancestral GilD transposon, their loss could have resulted in the inability to attach to the chromosome termini in a chain-like fashion. An alternative possibility is that GilM and GilT both acquired this ability, perhaps by means of addition of the long 3′ UTR, and therefore persisted in an active state, whereas GilD did not.
Short deletions involving regions of microhomology are known to occur during the error-prone Ku-independent nonhomologous end-joining double-strand break (DSB) repair backup pathway (49). In light of the apparent absence of Ku homologs from the G. lamblia genome, it seems plausible that such deletions could be generated as a result of such error-prone DSB repair. Interestingly, two or three deletion tracts of different sizes can occur in the same limited region, indicating possible DSB hotspots repaired independently by using different microhomologies in the same region.
Is Giardia Asexual?
Several unicellular organisms, once considered to be entirely asexual, have been found to undergo meiosis or at least form synaptonemal complexes (SC) after more careful examination (50, 51). A recent example is the pathogenic yeast Candida albicans, for which whole-genome sequencing data in combination with experimental stimulation revealed the potential to undergo at least part of a sexual cycle (52–54). Is it possible that Giardia, although thought to be asexual (7–9), might also have some form of sexual process? Its genome sequence does contain coding regions with homology to several genes known in other organisms to be involved in meiotic recombination (RAD51, RAD52, rec14, SPO11, and DMC1) or meiosis initiation and regulation (SME1/IME2, MEK1, and ran1 +) (www.mbl.edu/Giardia/Giardia-Total-BlastX/blastreport.html). ORFs similar to SC proteins (HOP1, ZIP1/SCP1) can also be identified; these, however, are most similar to other coiled-coil proteins (e.g., myosins and kinesins) and their function cannot be established at this time on the basis of sequence similarity alone. In addition, SC formation is not always required for a sexual cycle (55). It is worth noting that Candida, even if it has a potential to undergo sexual reproduction, is still mostly asexual, because its populations are primarily clonal in structure and genetic exchange is infrequent (56). This finding seems to be correlated with low active transposon content. In contrast to S. cerevisiae, which has mostly intact retrotransposons belonging to a few families with multiple members, the C. albicans genome contains 35 retrotransposon families, each having only a few highly rearranged and defective members. Only two or three copies appear intact (57). Thus, even when sexual reproduction is not completely excluded from the lifestyle of the organism, its prolonged absence may influence the transposon content of the genome. Because Giardia populations seem to be mostly clonal (58), it is likely that sexual reproduction, if any, did not play a major role in shaping its genome structure.
In conclusion, our previous studies of transposon content in sexual and anciently asexual rotifers and numerous other eukaryotes strongly suggested that the genomes of ancient asexuals do not retain RTase-related sequences detectable in PCR assays. The present analysis of transposon content of G. lamblia at the level of the entire genome sequence demonstrates that its two intact retroelements are confined to telomeric regions and therefore neither cause deleterious insertional mutations nor serve as sites for ectopic rearrangements in internal chromosomal locations. Because they are found on the majority of the chromosome ends, they could be beneficial to the host by providing additional protection from terminal degradation, even though they have not entirely replaced telomeric repeats, as has happened in Drosophila. A complete inactivation of the third family has occurred, as evidenced by multiple deletions and point mutations. Together with the absence of other autonomous transposon-related sequences, it may be concluded that the genome of this protozoan is free of active deleterious transposons. In combination with our earlier PCR experiments in bdelloid rotifers, this study establishes a connection between the mode of reproduction and the abundance, activity, and role of transposable elements in eukaryotic genomes.
Supplementary Material
Acknowledgments
We thank M. Meselson for encouragement and support; D. Mark Welch, J. Mark Welch, and M. Meselson for valuable comments; A. McArthur for critical reading and for performing hidden Markov searches of G. lamblia contigs to verify the absence of transposases and Ku homologs; M. Sogin for permission to use unpublished contig data and clones from the library; the National Science Foundation for supporting studies of transposable elements (MCB-9905998); and the National Institutes of Health for supporting the Giardia genome project (AI43273).
Abbreviations
- LINE
long interspersed nuclear element
- RTase
reverse transcriptase
- UTR
untranslated region
Footnotes
References
- 1.Hickey D. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkhipova I, Meselson M. Proc Natl Acad Sci USA. 2000;97:14473–14477. doi: 10.1073/pnas.97.26.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkhipova I, Lyubomirskaya N, Ilyin Y. Drosophila Retrotransposons. Austin, TX: Landes; 1995. [Google Scholar]
- 4.Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Austin, TX: Landes; 1998. [Google Scholar]
- 5.Mark Welch D, Meselson M. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T. Genetica. 1999;107:289–295. [PubMed] [Google Scholar]
- 7.Adam R. Int J Parasitol. 2000;30:475–484. doi: 10.1016/s0020-7519(99)00191-5. [DOI] [PubMed] [Google Scholar]
- 8.Adam R. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Upcroft J, Upcroft P. Protist. 2000;150:17–23. doi: 10.1016/S1434-4610(99)70005-2. [DOI] [PubMed] [Google Scholar]
- 10.Hou G, Le Blancq S, E Y, Zhu H, Lee M. Nucleic Acids Res. 1995;23:3310–3317. doi: 10.1093/nar/23.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Blancq S, Adam R. Mol Biochem Parasitol. 1998;97:199–208. doi: 10.1016/s0166-6851(98)00150-9. [DOI] [PubMed] [Google Scholar]
- 12.McArthur A, Morrison H, Nixon J, Passamaneck N, Kim U, Hinkle G, Crocker M, Holder M, Farr R, Reich C, et al. FEMS Microbiol Lett. 2000;189:271–273. doi: 10.1111/j.1574-6968.2000.tb09242.x. [DOI] [PubMed] [Google Scholar]
- 13.Huelsenbeck J, Ronquist F. Bioinformatics (Oxford) 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 14.Adam R, Nash T, Wellems T. Mol Cell Biol. 1991;11:3326–3330. doi: 10.1128/mcb.11.6.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik H, Burke W, Eickbush T. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Malik H, Eickbush T. Proc Natl Acad Sci USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik H, Eickbush T. Genetics. 2000;154:193–203. doi: 10.1093/genetics/154.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aksoy S, Williams S, Chang S, Richards F. Nucleic Acids Res. 1990;18:785–792. doi: 10.1093/nar/18.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabriel A, Yen T, Schwartz D, Smith C, Boeke J, Sollner-Webb B, Cleveland D. Mol Cell Biol. 1990;10:615–624. doi: 10.1128/mcb.10.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva M, Williams S, Beard C, Richards F, Aksoy S. Mol Cell Biol. 1991;11:6139–6148. doi: 10.1128/mcb.11.12.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatfield D, Oroszlan S. Trends Biochem Sci. 1990;15:186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardue M, Danilevskaya O, Lowenhaupt K, Wong J, Erby K. J Mol Evol. 1996;43:572–583. doi: 10.1007/BF02202105. [DOI] [PubMed] [Google Scholar]
- 23.Moran J, DeBerardinis R, Kazazian H. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 24.Upcroft P, Chen N, Upcroft J. Genome Res. 1997;7:37–46. doi: 10.1101/gr.7.1.37. [DOI] [PubMed] [Google Scholar]
- 25.Danilevskaya O, Arkhipova I, Traverse K, Pardue M. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 26.Borst P, Rudenko G. Science. 1994;264:1872–1873. doi: 10.1126/science.7516579. [DOI] [PubMed] [Google Scholar]
- 27.Freitas-Junior L, Bottius E, Pirrit L, Deitsch K, Scheidig C, Guinet F, Nehrbass U, Wellems T, Scherf A. Nature (London) 2000;407:1018–1021. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 28.Noutoshi Y, Arai R, Fujie M, Yamada T. Mol Gen Genet. 1998;259:256–263. doi: 10.1007/s004380050811. [DOI] [PubMed] [Google Scholar]
- 29.Pich U, Schubert I. Chromosome Res. 1998;6:315–321. doi: 10.1023/a:1009227009121. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Zou S, Wright D, Voytas D. Genes Dev. 1999;13:2738–2749. doi: 10.1101/gad.13.20.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anzai T, Takahashi H, Fujiwara H. Mol Cell Biol. 2001;21:100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biessmann H, Mason J, Ferry K, d'Hulst M, Valgeirsdottir K, Traverse K, Pardue M. Cell. 1990;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 33.Levis R, Ganesan R, Houtchens K, Tolar L, Sheen F. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 34.Adams M, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 35.Biessmann H, Valgeirsdottir K, Lofsky A, Chin C, Ginther B, Levis R, Pardue M. Mol Cell Biol. 1992;12:3910–3918. doi: 10.1128/mcb.12.9.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheen F, Levis R. Proc Natl Acad Sci USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biessmann H, Mason J. Chromosoma. 1997;106:63–69. doi: 10.1007/s004120050225. [DOI] [PubMed] [Google Scholar]
- 38.Martin F, Olivares M, Lopez M, Alonso C. Trends Biochem Sci. 1996;21:283–285. [PubMed] [Google Scholar]
- 39.Feng Q, Moran J, Kazazian H, Boeke J. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 40.Dawson A, Hartswood E, Paterson T, Finnegan D. EMBO J. 1997;16:4448–4455. doi: 10.1093/emboj/16.14.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin S, Bushman F. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sogin M, Gunderson J, Elwood H, Alonso R, Peattie D. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- 43.Henze K, Morrison H, Sogin M, Muller M. Gene. 1998;222:163–168. doi: 10.1016/s0378-1119(98)00499-5. [DOI] [PubMed] [Google Scholar]
- 44.Roger A, Morrison H, Sogin M. J Mol Evol. 1999;48:750–755. doi: 10.1007/pl00006519. [DOI] [PubMed] [Google Scholar]
- 45.Bouzat J, McNeil L, Robertson H, Solter L, Nixon J, Beever J, Gaskins H, Olsen G, Subramaniam S, Sogin M, Lewin H. J Mol Evol. 2000;51:532–543. doi: 10.1007/s002390010117. [DOI] [PubMed] [Google Scholar]
- 46.Malik H, Burke W, Eickbush T. Gene. 2000;251:101–108. doi: 10.1016/s0378-1119(00)00207-9. [DOI] [PubMed] [Google Scholar]
- 47.Friedman K, Cech T. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai C, Mitchell J, Collins K. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raikov I. Eur J Protistol. 1995;31:1–7. [Google Scholar]
- 51.Dacks J, Roger A. J Mol Evol. 1999;48:779–783. doi: 10.1007/pl00013156. [DOI] [PubMed] [Google Scholar]
- 52.Hull C, Raisner R, Johnson A. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 53.Magee B, Magee P. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 54.Tzung K, Williams R, Scherer S, Federspiel N, Jones T, Hansen N, Bivolarevic V, Huizar L, Komp C, Surzycki R, et al. Proc Natl Acad Sci USA. 2001;98:3249–3253. doi: 10.1073/pnas.061628798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohli J, Bahler J. Experientia. 1994;50:295–306. doi: 10.1007/BF01924013. [DOI] [PubMed] [Google Scholar]
- 56.Graser Y, Volovsek M, Arrington J, Schonian G, Presber W, Mitchell T, Vilgalys R. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodwin T, Poulter R. Genome Res. 2000;10:174–191. doi: 10.1101/gr.10.2.174. [DOI] [PubMed] [Google Scholar]
- 58.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere S, Darde M L, Ayala F. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.