Abstract
We have synthesized quinoxaline analogs (1–25), characterized by 1H-NMR and HREI-MS and evaluated for thymidine phosphorylase inhibition. Among the series, nineteen analogs showed better inhibition when compared with the standard inhibitor 7-Deazaxanthine (IC50 = 38.68 ± 4.42 µM). The most potent compound among the series is analog 25 with IC50 value 3.20 ± 0.10 µM. Sixteen analogs 1, 2, 3, 4, 5, 6, 7, 12, 13, 14, 15, 16, 17, 18, 21 and 24 showed outstanding inhibition which is many folds better than the standard 7-Deazaxanthine. Two analogs 8 and 9 showed moderate inhibition. A structure-activity relationship has been established mainly based upon the substitution pattern on the phenyl ring. The binding interactions of the active compounds were confirmed through molecular docking studies.
Keywords: quinoxaline analogs, synthesis, thymidine phosphorylase inhibition, molecular docking
1. Introduction:
Thymidine phosphorylase (TP), an enzyme involved in catabolism, exists in both prokaryotic and eukaryotic organisms [1,2,3]. TP speeds up the initial step in catabolism and converts thymidine nucleoside into thymine and 2-deoxy-d-ribose-1-phosphate by cleaving glycoside bond [4,5]. The intermediate obtained through dephosphorylation is 2-deoxy-d-ribose, which plays a significant role in prompting the tumor angiogenesis and hence favors cancer metastasis [6,7,8]. With respect to tumor angiogenesis, TP plays a major role, in that it helps in the proliferation process of endothelial cells throughout the body in cancer metastasis [9,10]. TP performs the same function as platelet endothelial cell growth factor (PD-ECGF) [11,12]. TP belonging to mammalians shares 39% sequence similarity with TP of E. coli, while the enzyme of mammalians also shares 65% resemblance with the active sites of residues of E. coli enzyme [13]. The production of 2′-deoxy-d-ribose can be limited through TP inhibitors which in turn suffocate the growth of tumor cells [14,15]. Therefore, medicinal chemists have tried to synthesize novel inhibitors of thymidine phosphorylase which have the potential to overcome the formation of new blood vessels and arrest the growth of tumor cells. Various attempts have been made to developed TP inhibitors [16,17,18,19,20,21,22,23]. The most potent inhibitor belonging to human TP known up to now is 5-chloro-6-[1-(2-iminopyrrolidinyl)methyl] uracil hydrochloride (TPI), while 7-deazaxanthine (7DX) is the first purine analog labeled as a TP inhibitor [24,25,26].
Nitrogen-containing heterocycles have attracted considerable attention due to their wide range of pharmacological importance [27,28]. Quinoxaline has a six-membered cyclic ring with two nitrogen atoms inside the cyclic ring. Quinoxaline and their analogs have attracted medicinal chemists over the decades and are used as antimicrobial [29], antibacterial [30], antifungal [31,32], anti-protozoan [33], anti-inflammatory, antianalgesic [34], anti-cancer [35,36], antidiabetic, and anti-proliferative agents [37,38]. Our research group has been working on the design and synthesis of heterocyclic compounds in search of potential lead compounds for many years and has found promising results [39,40,41,42,43,44,45,46,47,48,49].
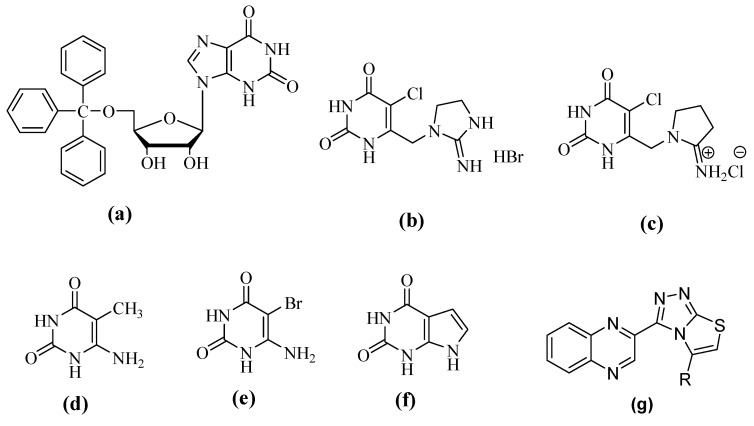
In the past, several derivatives having six-member ring with two nitrogen reported to showed excellent inhibition of TP such as (a) to (f) in Figure 1 [9]. They showed outstanding activity which induced us to synthesize compounds having similar type of structure with low cast synthesis and simple chemistry to make synthesis adaptable for large scale synthesis. We report in this study new derivatives of quinoxalines with fused triazole and thiadiazole ring VII. The structure of our compounds is very close to the standard drug Deazaxanthine but our compounds have fused triazole and thiadiazole ring as well, which show much better activity than the standard.
Figure 1.
Structures of some thymidine phosphorylase inhibitors (TPIs) (a–f) along with quinoxalines with fused triazol and thiazole ring (g).
2. Results and Discussion
2.1. Chemistry
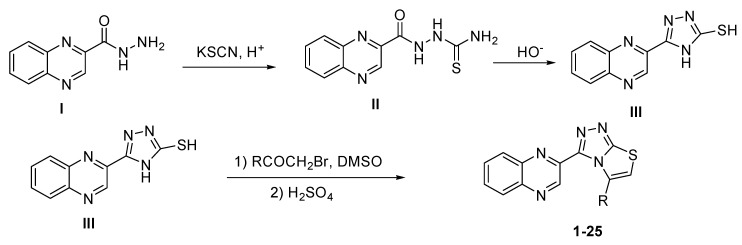
Synthesis of quinoxaline derivatives (1–25) started with treating quinoxaline-2-carbohydrazide (I) with potassium thiocyanate in the presence of acid to form quinoxaline thiosemicarbazone (II) which was treated with a basic solution to cyclize and form 5-(quinoxalin-3-yl)-4H-1,2,4-triazole-3-thiol (III) which was treated with different substituted phenacyl bromide to afford (1–25) target compounds. The crude product was washed with water and recrystallized in methanol to afford pure product in 80–75%. All synthesized compounds (Scheme 1) were characterized by different spectroscopic methods (see Supplementary Materials for full structures with activities).
Scheme 1.
Synthesis of 2-(5-arylthiazolo[2,3-c][1,2,4]triazol-3-yl)quinoxaline 1–25 derivatives.
2.2. In vitro Thymidine Phosphorylase Inhibitory Activity
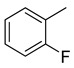
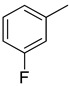
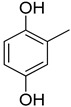
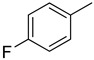
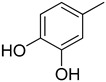
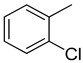
We have synthesized 25 analogs of 5-phenyl-3-quinoxalin (1–25) and screened for inhibitory potential against thymidine phosphorylase enzyme. With respect to inhibitory potential, many analogs of the series showed a variable degree of inhibition with IC50 values ranging between 3.50 ± 0.20 to 56.40 ± 1.20 μM when compared with standard 7-Deazaxanthine (IC50 = 38.68 ± 1.12 μM). The analogs 1, 2, 3, 4, 5, 6, 7, 12, 13, 14, 15, 16, 17, 18, 21, 24, and 25 showed excellent inhibitory potential with IC50 values 13.60 ± 0.4, 26.10 ± 0.70, 18.10 ± 0.50, 27.40 ± 0.60, 33.40 ± 0.80, 24.40 ± 0.60, 34.70 ± 0.80, 33.20 ± 0.75, 18.30 ± 0.55, 13.20 ± 0.40, 15.20 ± 0.50, 3.50 ± 0.20, 24.20 ± 0.70, 16.90 ± 0.60, 26.20 ± 0.50, 13.10 ± 0.30 and 3.20 ± 0.10 μM respectively by comparing with standard 7-Deazaxanthine. Two analogs 8 and 9 showed moderate inhibitory activity with IC50 values 47.50 ± 0.90 and 56.40 ± 1.20 μM respectively, while six analogs 10, 11, 19, 20, 22, and 23 were found inactive. Structure activity relationship has been established for all compounds, mainly based on substituents pattern of phenyl ring.
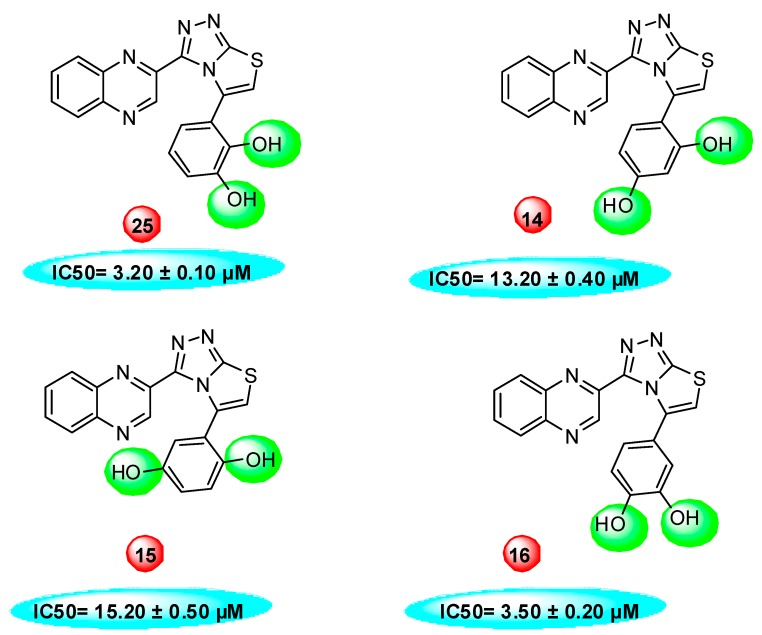
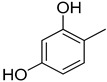
Compound 25, a 2,3-dihydroxy analog was found to be the most active analog among the series with IC50 value 3.20 ± 0.10 μM. When comparing analog 25 with other dihydroxy analogs like 14, a 2,4-dihydroxy analog (IC50 = 13.20 ± 0.40 μM) 15, a 2,5-dihydroxy analog (IC50 = 15.20 ± 0.50 μM) and 16, a 2,4-dihydroxy analog (IC50 = 3.50 ± 0.20 μM), analog 25 was found to be superior. Although all the four analogs have two hydroxyl groups at the phenyl ring, the position of attachment on phenyl ring are different. The difference in inhibitory activity of these four analogs seems due to the different position of the hydroxyl group on the phenyl ring, as seen in Figure 2.
Figure 2.
Dihydroxy substitutions at different positions affect their activity.
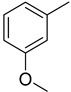
When comparing dihydroxy analogs with monohydroxy analog like 12, 13, 17, 18, 21, and 24 the dihydroxy analogs were found to be more potent. This greater potential seems to be due to the greater number of hydroxy groups on the phenyl ring.
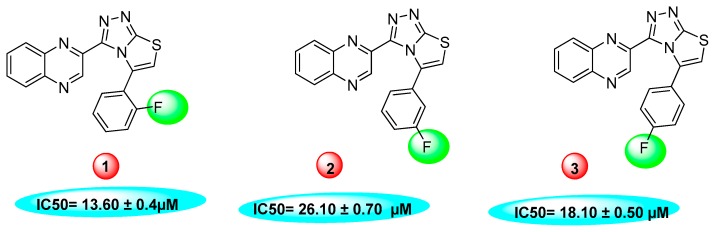
Similarly, a pattern was also observed in flourine substituted analogs like 1, 2, and 3 IC50 value 13.60 ± 0.4, 26.10 ± 0.70 and 18.10 ± 0.50 μM respectively. All three analogs possess flouro group at the phenyl ring, but the analog 1 shows greater potential than analogs 2 and 3. The difference in the inhibitory potential in analog 1, 2 and 3 seems due to attachment of flouro group at various positions on the phenyl part, as seen in Figure 3.
Figure 3.
Activity of fluorine substitutions at different positions.
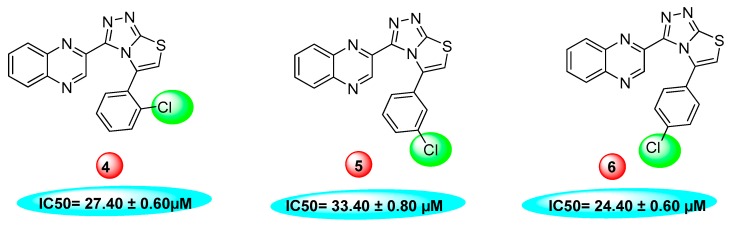
The same trend of difference in inhibitory activity was found in chloro substituted analogs 4, 5 and 6 with IC50 value 27.40 ± 0.60, 33.40 ± 0.80 and 24.40 ± 0.60 μM respectively. All three analogs have chloro group but their attachment on phenyl ring differs from each other, and the difference in inhibitory activity seems to be due to the attachment of chloro group at variable positions on the phenyl part Figure 4.
Figure 4.
Activity of chlorine substitutions at different positions.
So, it was concluded that in our designed molecules the position, nature, and number of substituent play critical role in thymidine phosphorylase inhibition.
2.3. Molecular Docking
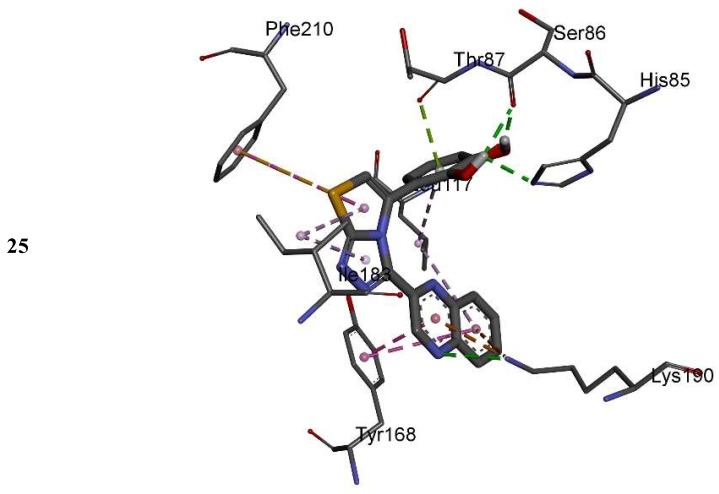
The IC50 values of quinoxaline derivatives as thymidine phosphorylase inhibitors are shown in Table 1. The thymidine phosphorylase inhibition by the synthesized derivatives is mainly due to the type, number, and positions of the functional group in the substitute group R of the basic skeleton (Table 1). For a better understanding of the enzyme inhibition by the synthesized compounds, molecular docking study has been carried out to shed light on the established binding modes of the four selected synthesized compounds 14, 15, 16, and 25. The selected compounds differ by the substitution position of the hydroxyl group in the aromatic ring (Table 1). Compounds 16 and 25 with OH groups in meta position to each other show higher activity than 14 and 15 where OH groups are in para position to each other (Table 1). Table 2 summarizes the calculated binding energies of the stable complex’s ligand thymidine phosphorylase, number of established intermolecular hydrogen bonding between the synthesized compounds (14, 15, 16 and 25) and active site residues of thymidine phosphorylase.
Table 1.
Different substituent of quinoxaline derivatives (1–25) and their thymidine phosphorylase inhibitory activity.
| S.No. | R | IC50 (µM ± SEM a) | S.No. | R | IC50 (mM ± SEM a) |
|---|---|---|---|---|---|
| 1 |
|
13.60 ± 0.4 | 14 |
|
13.20 ± 0.40 |
| 2 |
|
26.10 ± 0.70 | 15 |
|
15.20 ± 0.50 |
| 3 |
|
18.10 ± 0.50 | 16 |
|
3.50 ± 0.20 |
| 4 |
|
27.40 ± 0.60 | 17 |
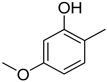
|
24.20 ± 0.70 |
| 5 |
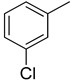
|
33.40 ± 0.80 | 18 |
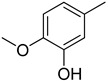
|
16.90 ± 0.60 |
| 6 |
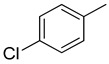
|
24.40 ± 0.60 | 19 |
|
N. A. |
| 7 |
|
34.70 ± 0.80 | 20 |
|
N. A. |
| 8 |
|
47.50 ± 0.90 | 21 |
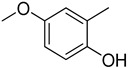
|
26.20 ± 0.50 |
| 9 |
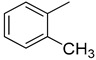
|
56.40 ± 1.20 | 22 |
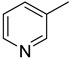
|
N. A. |
| 10 |
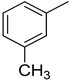
|
N. A. | 23 |
|
N. A. |
| 11 |
|
N. A. | 24 |
|
13.10 ± 0.30 |
| 12 |
|
33.20 ± 0.75 | 25 |
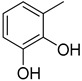
|
3.20 ± 0.10 |
| 13 |
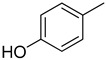
|
18.30 ± 0.55 | - | - | - |
| 7-Deazaxanthine (7DX) | 38.68 ± 4.42 µM | ||||
N.A. = Not active; SEM a = Standard error mean.
Table 2.
IC50, docking binding energies, hydrogen bonding, and the number of closest residues to the docked ligands in the active site of the selected quinoxaline derivatives 14, 15, 16, and 25 within the active binding site of thymidine phosphorylase.
| No. of Compound | Free Binding Energy (kcal/mol) | H-Bonds (HBs) | Number of Closest Residues to the Docked Ligand in The Active Site | IC50 ± SEM |
|---|---|---|---|---|
| 14 | −7.71 | 5 | 10 | 13.20 ± 0.40 |
| 15 | −7.61 | 3 | 8 | 15.20 ± 050 |
| 16 | −8.05 | 3 | 8 | 3.50 ± 0.20 |
| 25 | −8.25 | 4 | 8 | 3.20 ± 0.10 |
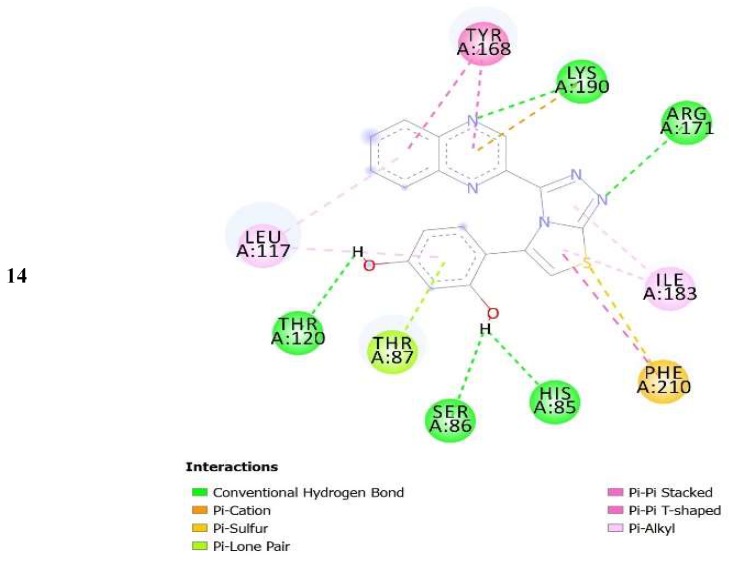
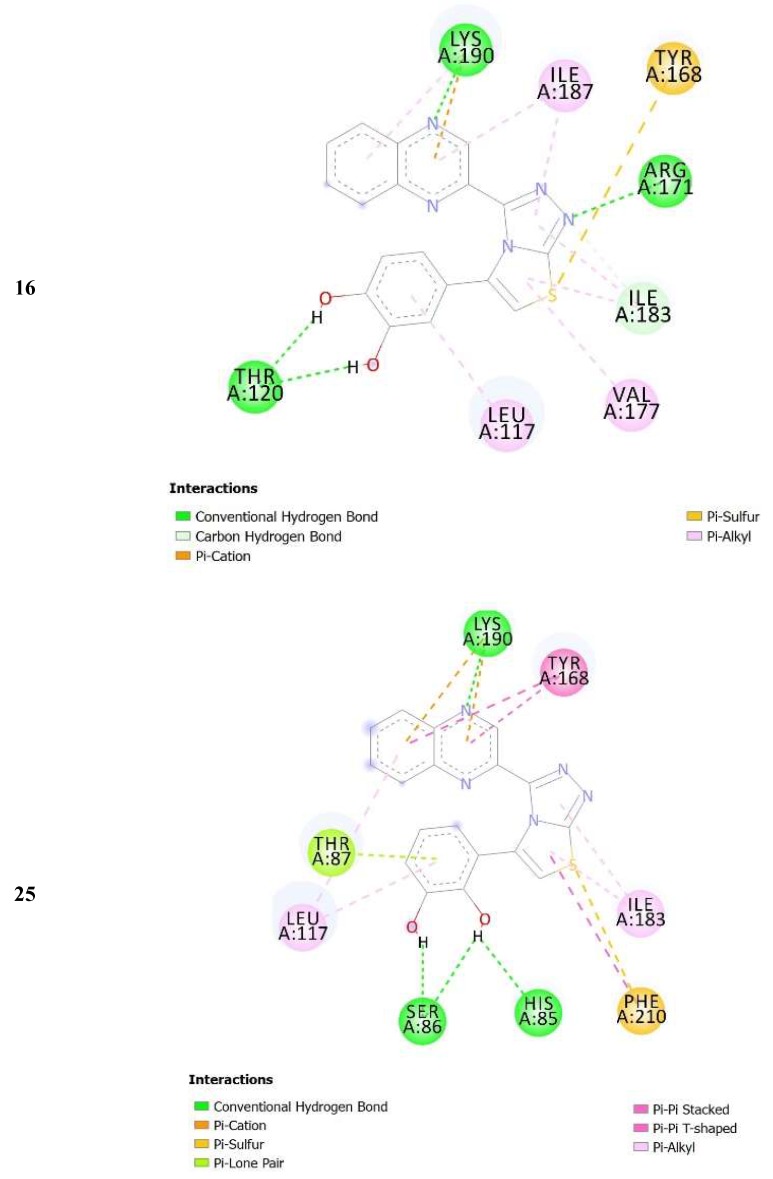
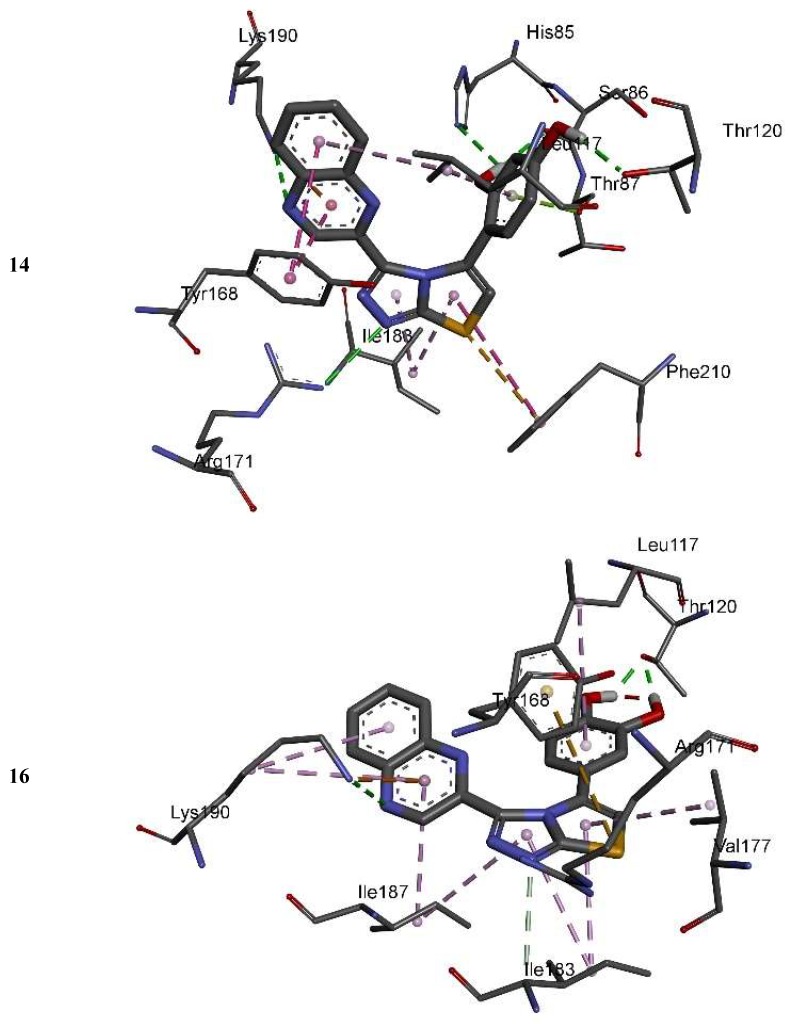
The complexes formed between the docked selected compounds and amino acids of the binding active site of thymidine phosphorylase exhibited negative binding energies, which is a signpost that the inhibition of thymidine phosphorylase by the selected compounds is thermodynamically favorable (Table 2). As can be seen from the docking results in Table 2 and Figure 5 and Figure 6, the highest activity of 16 and 25 compared with 14 and 15 mainly refers to the stability of the formed complexes between the docked compounds (16 and 25) and thymidine phosphorylase compared with the formed complexes between the docked compounds (14 and 15) and thymidine phosphorylase ones. The higher activity of 25 compared with 16 may refer to the number of intermolecular hydrogen bonding formed with substituted OH groups in the complex 25-receptor compared to 16-receptor one. Indeed, three hydrogen bonds are formed between OH groups of 25 and SER 86 and HIS 85 of the active site of thymidine phosphorylase of 1.71, 2.24, and 3.38 Å, respectively. However, two hydrogen bonds are formed between OH groups of 16 and THR 120 of the active site of thymidine phosphorylase of 1.26 and 1.78 Å, respectively.
Figure 5.
2D closest interactions between active site residues of thymidine phosphorylase and synthesized compounds 14, 16, and 25.
Figure 6.
3D closest interactions between active site residues of thymidine phosphorylase and synthesized compounds 14, 16, and 25.
3. Experimental Section
3.1. General Methods
All nuclear magnetic resonance experiments were carried out using on Avance Bruker 500 MHz (Wissembourg, Switzerland)). Electron impact mass spectra (EI–MS) were recorded on a Finnigan MAT-311A (Bremen, Germany). Thin layer chromatography (TLC) was performed on pre-coated silica gel aluminum plates (Kieselgel 60, 254, E. Merck, Darmstadt, Germany). Chromatograms were visualized by UV at 254 and 365 nm.
3.1.1. Thymidine Phosphorylase Assay
Since human TP is not easy to obtain, we used commercially available recombinant E. coli TP. The primary sequence of TP is frequently preserved throughout evolution as mammalian TP is reported to share 39% sequence resemblance with the TP of E. coli. The mammalian enzyme also shared up to 70% resemblance with the active site residues, and three-dimensional structure of E. coli TP enzyme [49]. The thymidine phosphorylase/PD-ECGF (E. coli) activity was determined by measuring the absorbance at 290 nm spectrophotometrically. The method was described in [50,51]. In brief, the total reaction mixture of 200 µL contained 145 µL of potassium phosphate buffer (pH 7.4), 30 µL of enzyme (human and E. coli) at concentration 0.05 and 0.002 U, respectively, were incubated with 5 µL of test materials for 10 min at 25 °C in a microplate reader. After incubation, a pre-read at 290 nm was taken to deduce the absorbance of substrate particles. The substrate (20 µL, 1.5 mM), dissolved in potassium phosphate buffer, was immediately added to the plate and continuously read after 10, 20, and 30 min in a microplate reader (SpectraMax, Molecular Devices, CA, USA). All assays were performed in triplicate.
3.1.2. Calculations
Reactions for above mentioned biological activities were carried out in triplicate. Results were then processed using SoftMax Pro 4.8 software (Molecular Devices, San Jose, CA, USA) and then by Microsoft Excel. The percent inhibition for above mentioned biological activities was calculated by following formula:
| Percent Inhibition = 100 − (ODtest compound/ODcontrol) × 100 | (1) |
3.1.3. Synthesis of Quinoxaline Thiosemicarbazone (II)
Quinoxaline-2-carbohydrazide (5 g, 26.60 mmol), potassium thiocyanate (2.61 g, 26.90 mmol), and 4 mL of conc. HCl in 40 mL of water were refluxed for 4 h. The reaction progress was monitored by TLC. After completion of the reaction, the reaction mixture was left for cooling and white solid ppts appeared then the solid was filtered and dried. Yield 5.62 g (85.6%); m.p. 289–290 °C.
3.1.4. Synthesis of 5-(quinoxalin-3-yl)-4H-1,2,4-triazole-3-thiol (III)
The 5-(quinoxalin-3-yl)-4H-1,2,4-triazole-3-thiol (III) was synthesized as reported in [52].
3.2. General Procedure for Synthesis of Quinoxaline Derivatives (1–25)
5-(quinoxalin-3-yl)-4H-1,2,4-triazole-3-thiol (III) (1 mmol) was refluxed with appropriate arylacyl bromide (1 mmol) in 15 mL ethanol for 12 h. The reaction was monitored by TLC. After completion of the reaction, the product was left for cooling. The solid was filtered and the crude products were recrystallized from methanol (all synthesized compounds with their SMILE structures and activities are provided in Supplementary Materials).
3.2.1. 5-(2-flourophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 81%. m.p.: 299–300 °C; 1H-NMR (500 MHz, DMSO-d6): δ 8.32 (s, 1H), 8.02 (d, J = 8.0 Hz, 1H), 7.88 (d, J = 7.0 Hz, 1H), 7.70–7.67 (m, 3H), 7.52–7.48 (m, 2H), 7.30–7.26 (m, 2H). 13C-NMR (150 MHz, DMSO -d6): δ 158.5, 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 130.5, 129.8, 129.7, 129.5, 129.4, 129.3, 124.10, 123.7, 115.7, 114.9. HR-ESI-MS: m/z calcd for C18H10FN5S, [M]+ 347.0641; Found 347.0623.
3.2.2. 5-(3-flourophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 80%. m.p.: 304–305 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.10 (s, 1H), 7.90 (d, J = 7.5 Hz, 1H), 7.67 (d, J = 7.5 Hz, 2H), 7.61 (d, J = 8.0 Hz, 1H), 7.56 (d, J = 7.5 Hz, 1H), 7.50–7.46 (m, 2H), 7.22 (t, J = 8.0 Hz, 1H), 7.18–7.16 (m, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 162.2, 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 134.8, 129.8, 129.7, 129.5, 129.4, 127.7, 115.7, 123.3, 115.10, 115.7. HR-ESI-MS: m/z calcd for C18H10FN5S, [M]+ 347.0641; Found 347.0625.
3.2.3. 5-(4-flourophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 77%. m.p.: 308–309 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.11(s, 1H), 7.90 (d, J = 7.5 Hz, 1H), 7.80 (d, J = 8.0 Hz, 2H), 7.72–7.68 (m, 2H), 7.50–7.45 (m, 2H), 7.24 (d, J = 7.5 Hz, 2H); 13C-NMR (150 MHz, DMSO -d6): δ 162.9, 155.5, 145.9, 145.5, 144.5, 142.4, 142.3, 141.11, 130.8, 130.7, 129.8, 129.7, 129.5, 129.4, 128.8, 116.3, 116.2, 115.7. HR-ESI-MS: m/z calcd for C18H10FN5S, [M]+ 347.0641; Found 347.0617.
3.2.4. 5-(2-chlorophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 82%. m.p.: 280–281 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.50 (s, 1H), 8.12 (d, J = 7.0 Hz, 1H), 7.90 (d, J = 7.0 Hz, 1H), 7.72–7.68 (m, 3H), 7.54–7.49 (m, 2H), 7.42–7.38 (m, 2H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 132.7, 132.4, 130.8, 130.3, 129.9, 129.8, 129.7, 129.5, 129.4, 128.9, 115.7. HR-ESI-MS: m/z calcd for C18H10ClN5S, [M]+ 363.0345; Found 363.0319.
3.2.5. 5-(3-chlorophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 80%. m.p.: 285–286 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.90 (s, 1H), 8.57 (d, J = 6.5 Hz, 1H), 8.16 (d, J = 7.5 Hz, 1H), 8.05 (s, 1H), 7.86 (d, J = 8.0 Hz, 1H), 7.70 (d, J = 7.5 Hz, 2H), 7.50–7.47 (m, 2H), 7.42 (d, J = 8.0 Hz, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 134.9, 134.6, 129.9, 129.8, 129.7, 129.6, 129.5, 129.4, 128.9, 125.8, 115.7. HR-ESI-MS: m/z calcd for C18H10ClN5S, [M]+ 363.0345; Found 363.0323.
3.2.6. 5-(4-chlorophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 89%. m.p.: 249–250 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.08 (s, 1H), 7.88 (d, J = 7.5 Hz, 1H), 7.70 (d, J = 8.5 Hz, 2H), 7.69–7.64 (m, 2H), 7.52–7.45 (m, 4H). 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 134.5, 131.3, 129.9, 129.8, 129.7, 129.6, 129.5, 129.4, 128.9, 127.9, 115.7. HR-ESI-MS: m/z calcd for C18H10ClN5S, [M]+ 363.0345; Found 363.0327.
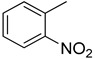
3.2.7. 5-(2-nitrophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 83%. m.p.: 310–311 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.26 (d, J = 7.0 Hz, 1H), 7.98 (d, J = 7.5 Hz, 1H), 7.90 (d, J = 7.5 Hz, 1H), 7.74 (d, J = 7.0 Hz, 1H), 7.70 (d, J = 7.0 Hz, 1H), 7.63 (d, J = 7.0 Hz, 1H), 7.61 (d, J = 7.0 Hz, 1H), 7.50–7.46 (m, 2H), 7.43 (s, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 148.9, 145.9, 145.5, 144.5, 142.4, 142.3, 141.11, 135.5, 132.8, 129.9, 129.8, 129.7, 129.5, 129.4, 125.4, 124.6,115.7. HR-ESI-MS: m/z calcd for C18H10N6O2S, [M]+ 374.0586; Found 374.0568.
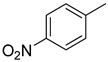
3.2.8. 5-(4-nitrophenyl)-3(Quinoxalin-2yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 81%. m.p.: 315–316 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.22 (d, J = 8.3 Hz, 2H), 8.18 (s, 1H), 7.98 (d, J = 8.0 Hz, 2H), 7.90 (d, J = 7.5 Hz, 1H), 7.70–7.65 (m, 2H), 7.54 (d, J = 7.0 Hz, 1H), 7.44 (s, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 147.9, 145.9, 145.4, 144.5, 142.4, 142.3, 142.1, 141.9, 141.8, 139.3, 129.8, 129.7, 129.5, 129.4, 126.4, 126.3, 124.6,124.5,115.7. HR-ESI-MS: m/z calcd for C18H10N6O2S, [M]+ 374.0586; Found 374.0559.
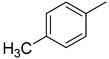
3.2.9. 3-(quinoxalin-2-yl)-5-o-tolylthiazolo[2,3-c][1,2,4]triazole
Yield: 88%. m.p.: 265-266 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.40 (s, 1H), 7.89 (d, J = 8.0 Hz, 2H), 7.68 (d, J = 7.0 Hz, 2H), 7.48–7.42 (m, 2H), 7.35–7.22 (m, 3H), 2.49 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 136.9, 130.1, 129.9, 129.8, 129.7, 129.5, 129.4, 128.8, 126.4, 122.9, 115.7, 18.9. HR-ESI-MS: m/z calcd for C19H13N5S, [M]+ 343.0892; Found 343.0864.
3.2.10. 3-(quinoxalin-2-yl)-5-m-tolylthiazolo[2,3-c][1,2,4]triazole
Yield: 84%. M.P.: 270–271 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.08 (s, 1H), 7.90 (d, J = 8.0 Hz, 1H), 7.72–7.68 (m, 2H), 7.57 (s, 1H), 7.52 (d, J = 7.0 Hz, 1H), 7.43 (s, 1H), 2.46 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 139.1, 133.1, 130.6, 129.9, 129.8, 129.7, 129.5, 129.4, 128.9, 124.6, 115.7, 21.8. HR-ESI-MS: m/z calcd for C19H13N5S, [M]+ 343.0892; Found 343.0871.
3.2.11. 3-(quinoxalin-2-yl)-5-p-tolylthiazolo[2,3-c][1,2,4]triazole
Yield: 81%. m.p.: 280–281 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.10 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.71–7.65 (m, 4H), 7.47–7.42 (m, 2H), 7.27 (d, J = 7.9 Hz, 2H), 2.49 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.9, 145.4, 145.3, 144.5, 142.4, 142.3, 141.11, 130.2, 131.9, 129.9, 129.8, 129.7, 129.5, 129.4, 129.3, 125.9, 125.7, 115.7, 21.5. HR-ESI-MS: m/z calcd for C19H13N5S, [M]+ 343.0892; Found 343.0873.
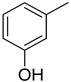
3.2.12. 3-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)phenol
Yield: 85%. m.p.: 289–290 °C. 1H-NMR (500 MHz, DMSO-d6): δ 9.05 (s, 1H, OH), 8.04 (s, 1H), 7.90 (d, J = 7.5 Hz, 1H), 7.70–7.65 (m, 2H), 7.49–7.43 (m, 2H), 7.25 (d, J = 7.5 Hz, 1H), 7.19–7.15 (m, 1H), 7.15 (d, J = 7.0 Hz, 1H), 6.81 (d, J = 7.0 Hz, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 157.7, 155.5, 145.9, 145.4, 144.5, 142.4, 142.3, 141.11, 134.6, 130.8, 129.8, 129.7, 129.5, 129.4, 120.3, 115.10, 115.9, 115.7. HR-ESI-MS: m/z calcd for C18H11N5OS, [M]+ 345.0684; Found 345.0666.
3.2.13. 4-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)phenol
Yield: 82%. m.p.: 297–298 °C. 1H-NMR (500 MHz, DMSO-d6): δ 9.82 (s, 1H, OH), 8.03 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.65–7.62 (m, 2H), 7.58 (d, J = 7.5 Hz, 2H), 7.46–7.40 (m, 2H), 6.80 (d, J = 8.0 Hz, 2H). 13C-NMR (150 MHz, DMSO -d6): δ 158.7, 155.5, 145.9, 145.5, 144.5, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 128.9, 128.7, 125.7, 116.6, 116.4,115.7. HR-ESI-MS: m/z calcd for C18H11N5OS, [M]+ 345.0684; Found 345.0661.
3.2.14. 4-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)benzene-1,3-diol
Yield: 79%. m.p.: 299–300 °C. 1H-NMR (500 MHz, DMSO-d6): δ 10.70 (s, 1H, OH), 9.80 (s, 1H, OH), 8.18 (s, 1H), 7.86 (d, J = 8.0 Hz, 1H), 7.71–7.65 (m, 3H), 7.50–7.43 (m, 2H), 6.32 (d, J = 7.0, 6.0 Hz, 2H). 13C-NMR (150 MHz, DMSO -d6): δ 160.1, 156.2, 155.5, 145.9, 145.5, 144.5, 142.4, 142.3, 141.11, 133.4, 129.8, 129.7, 129.5, 129.4, 113.3, 109.0, 105.8, 115.7. HR-ESI-MS: m/z calcd for C18H11N5O2S, [M]+ 361.0633; Found 361.0615.
3.2.15. 2-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)benzene-1,4-diol
Yield: 87%. m.p.: 301–302 °C. 1H-NMR (500 MHz, DMSO-d6): δ 9.72 (s, 2H, OH), 8.30 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.74–7.65 (m, 2H), 7.52–7.40 (m, 2H), 7.17 (d, J = 6.0 Hz, 1H), 6.75 (d, J = 8.0 Hz, 1H), 6.68 (d, J = 7.0 Hz, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 150.3, 147.9, 145.9, 145.5, 144.5, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 122.1, 117.9, 117.5, 115.7, 114.5. HR-ESI-MS: m/z calcd for C18H11N5O2S, [M]+ 361.0633; Found 361.0617.
3.2.16. 4-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)benzene-1,2-diol
Yield: 83%. m.p.: 293–294 °C. 1H-NMR (500 MHz, DMSO-d6): δ 9.30 (s, 1H, OH), 9.10 (s, 1H, OH), 7.96 (s, 1H), 7.82 (d, J = 8.0 Hz, 1H), 7.68(d, J = 6.0 Hz, 2H), 7.53–7.40 (m, 2H), 7.20 (s, 1H), 6.95 (d, J = 7.0 Hz, 1H), 6.70 (d, J = 8.0 Hz, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 147.5, 145.9, 145.7, 145.5, 144.5, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 127.2, 121.7, 116.4, 115.7, 114.5. HR-ESI-MS: m/z calcd for C18H11N5O2S, [M]+ 361.0633; Found 361.0612.
3.2.17. 5-methoxy-2-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)phenol
Yield: 86%. m.p.: 305–306 °C. 1H-NMR (500 MHz, DMSO-d6): δ 10.87 (s, 1H, OH), 8.30 (s, 1H), 7.82 (d, J = 7.5 Hz, 1H), 7.62 (s, 2H), 7.55 (d, J = 8.0 Hz, 1H), 7.46–7.40 (s, 2H), 6.51 (d, J = 8.0 Hz, 1H), 6.42 (d, J = 6.0 Hz, 1H), 3.75 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 162.2, 156.2, 155.5, 145.9, 145.5, 144.5, 142.4, 142.3, 141.11, 132.9, 129.8, 129.7, 129.5, 129.4, 115.7, 112.9, 107.6, 104.4, 55.9. HR-ESI-MS: m/z calcd for C19H13N5O2S, [M]+ 375.0790; Found 375.0772.
3.2.18. 2-methoxy-5-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)phenol
Yield: 83%. m.p.: 293–294 °C. 1H-NMR (500 MHz, DMSO-d6): δ 9.22 (s, 1H, OH), 8.04 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.76–7.69 (m, 2H), 7.46–7.41 (m, 2H), 7.28 (d, J = 6.0Hz, 1H), 7.04 (d, J = 8.0 Hz, 1H), 6.94 (d, J = 8.0 Hz, 1H), 3.77 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.7, 147.5, 147.4, 145.5, 144.5, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 126.9, 121.7, 114.1, 115.7, 111.6, 56.3. HR-ESI-MS: m/z calcd for C19H13N5O2S, [M]+ 375.0790; Found 375.0768.
3.2.19. 5-(3-methoxyphenyl)-3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 75%. m.p.: 179–180 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.10 (s, 1H), 7.90 (d, J = 8.0 Hz, 1H), 7.70–7.59 (m, 3H), 7.46–7.40 (m, 2H), 7.30 (d, J = 6.5 Hz, 2H), 6.96–6.89 (m, 1H), 3.80 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 161.2, 155.5, 145.7, 145.5, 144.5, 142.4, 142.3, 141.11, 134.2, 130.3, 129.8, 129.7, 129.5, 129.4, 119.9, 115.7, 114.1, 113.8, 55.9. HR-ESI-MS: m/z calcd for C19H13N5OS, [M]+ 359.0841; Found 359.0843.
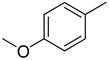
3.2.20. 5-(4-methoxyphenyl)-3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 87%. m.p.: 301–302 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.07 (s, 1H), 7.88 (d, J = 8.0 Hz, 2H), 7.70 (d, J = 8.0 Hz, 2H), 7.62 (d, J = 7.0 Hz, 1H), 7.47–7.42 (s, 2H), 7.02 (d, J = 8.0 Hz, 2H), 3.81 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 160.8, 155.5, 145.7, 145.5, 144.5, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 128.7, 128.5, 125.5, 115.7, 114.9, 114.7, 55.9. HR-ESI-MS: m/z calcd for C19H13N5OS, [M]+ 359.0841; Found 359.0819.
3.2.21. 4-methoxy-2-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)phenol
Yield: 83%. m.p.: 308–309 °C. 1H-NMR (500 MHz, DMSO-d6): δ 9.98 (s, 1H, OH), 8.40 (s, 1H), 7.88 (d, J = 8.2 Hz, 1H), 7.72–7.67 (m, 2H), 7.47–7.42 (m, 2H), 7.30 (s, 1H), 6.82 (d, J = 2.0 Hz, 2H), 3.79 (s, 3H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 153.9, 145.7, 147.5, 145.4, 144.5, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 121.7, 117.6, 115.7, 115.5, 112.9, 55.9. HR-ESI-MS: m/z calcd for C19H13N5O2S, [M]+ 375.0790; Found 375.0772.
3.2.22. 5-(pyridin-3-yl)-3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazole
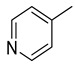
Yield: 82%. m.p.: 276–277 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.42 (s, 1H), 8.12 (d, J = 8.0 Hz, 1H), 7.90 (d, J = 7.0 Hz, 1H), 7.70–7.65 (m, 4H), 7.51 (d, J = 7.0 Hz, 1H), 7.47–7.42 (m, 2H). 13C-NMR (150 MHz, DMSO -d6): δ 147.9, 145.7, 147.5, 145.4, 144.5, 143.4, 142.4, 142.3, 141.11, 134.2, 133.2, 129.8, 129.7, 129.5, 129.4, 124.2,120.3. HR-ESI-MS: m/z calcd for C17H10N6S, [M]+ 330.0688; Found 330.0667.
3.2.23. 5-(pyridin-4-yl)-3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazole
Yield: 86%. m.p.: 276–277 °C. 1H-NMR (500 MHz, DMSO-d6): δ 8.60 (d, J = 7.0 Hz, 2H), 8.13 (s, 1H), 7.92 (d, J = 7.9 Hz, 1H), 7.74–7.66 (m, 4H), 7.50–7.42 (m, 2H). 13C-NMR (150 MHz, DMSO -d6): δ 149.9, 149.8, 145.7, 145.5, 144.5, 143.4, 142.4, 142.3, 141.11, 140.5, 129.8, 129.7, 129.5, 129.4, 121.5, 121.5, 120.3. HR-ESI-MS: m/z calcd for C17H10N6S, [M]+ 330.0688; Found 330.0670.
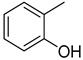
3.2.24. 2-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)phenol
Yield: 82%. m.p.: 286–287 °C. 1H-NMR (500 MHz, DMSO-d6): δ 10.54 (s, 1H, OH), 8.20 (s, 1H), 7.90 (d, J = 7.5 Hz, 1H), 7.72 (d, J = 7.0 Hz, 1H), 7.65 (d, J = 8.0 Hz, 2H), 7.52–7.44 (m, 2H), 7.25–7.20 (m, 2H), 6.90 (d, J = 6.5 Hz, 1H). 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 155.3, 145.7, 145.5, 144.5, 142.4, 142.3, 141.11, 131.7, 130.3, 129.8, 129.7, 129.5, 129.4, 120.7, 117.9, 121.7, 115.7. HR-ESI-MS: m/z calcd for C18H11N5OS, [M]+ 345.0684; Found 345.0668.
3.2.25. 3-(3-(quinoxalin-2-yl)thiazolo[2,3-c][1,2,4]triazol-5-yl)benzene-1,2-diol
Yield: 82%. m.p.: 309–310 °C. 1H-NMR (500 MHz, DMSO-d6): δ 10.08 (s, 1H, OH), 9.35 (s, 1H, OH), 8.40 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.64 (d, J = 6.0 Hz, 2H), 7.45–7.40 (m, 2H), 7.14 (d, J = 7.0 Hz, 1H), 6.80 (d, J = 7.0 Hz, 1H), 6.70 (t, J = 7.5 Hz, 1H); 13C-NMR (150 MHz, DMSO -d6): δ 155.5, 145.7, 145.5, 145.4, 144.5, 143.7, 142.4, 142.3, 141.11, 129.8, 129.7, 129.5, 129.4, 124.3, 123.4, 122.1, 117.5, 115.7. HR-ESI-MS: m/z calcd for C18H11N5O2S, [M]+ 361.0633; Found 361.0605.
3.3. Molecular Docking Details
The intermolecular binding modes between the docked selected synthesized quinoxaline derivatives and the active residues of thymidine phosphorylase have been explored using AutoDock package (The Scripps Research Institute, La Jolla, CA, USA) [53]. The geometries of thymidine phosphorylase and the original docked ligand 3′-azido-2′-fluoro-dideoxyuridine were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) data bank website (PDB code 4EAD) [54]. Water molecules were removed; polar hydrogen atoms and Kollman charge were added to the extracted receptor using the automated tool in AutoDock Tools 4.2. The active site is identified based on the co-crystallized receptor–ligand complex structure of thymidine phosphorylase. The re-docking of the original ligand 3′-azido-2′-fluoro-dideoxyuridine into the active site is well reproduced with an RMSD value less than 1.14 Å and a binding energy of −6.63 kcal/mol. The molecular structures geometries of quinoxaline derivatives were minimized at Merck molecular force field 94 (MMFF94) level44. The optimized structures were saved as PDB files. Nonpolar hydrogens were merged, and rotatable bonds were defined for each docked ligand. Docking studies were performed by the Lamarckian genetic algorithm, with 500 as a total number of the run for binding site for original ligand the synthesized derivatives. In each set, a population of 150 individuals with 27,000 generations and 250,000 energy evaluations were employed. Operator weights for crossover, mutation, and elitism were set to 0.8, 0.02, and 1, respectively. The binding site was defined using a grid of 35 × 35 × 35 points each with a grid spacing of 0.375 Å. The docking calculation has been carried out using an Intel (R) Core (TM) i5-3770 CPU @ 3.40 GHz workstation.
4. Conclusions
Twenty-five quinoxaline analogs (1–25) were synthesized. All synthesized compounds were reported first having novel structures. They were screened against thymidine phosphorylase. The result profile showed that dihydroxyl substituted compounds showed excellent activity along with some variation depending upon their position. The halogen groups, on other hand, showed a significant inhibitory potential against thymidine phosphorylase. The fluorine substituents showed better activity than the chlorines. The binding interactions of the most active analogs were determined by molecular docking study. That confirms the binding interactions of active compounds with enzyme.
Acknowledgments
The authors would like to Knowledge Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, for providing excellent lab facilities for research.
Supplementary Materials
The following are available online. Structures along with bioactivity.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “Conceptualization, M.T. and N.B.A.; Methodology, N.B.A.; Software, E.H.A and M.G.; Validation, A.A. S.A.A.S. and M.I.; Formal Analysis, R.K.F. and A.A.; Investigation, F.R.; Resources, M.N.; Data Curation, Q.U.A.; Writing-Original Draft Preparation, Z.A.Z.; Writing-Review & Editing, Z.A.Z and M.T.; Visualization, A.M.; Supervision, M.T. and Z.A.Z.; Project Administration, M.T.; Funding Acquisition, Z.A.Z”.
Funding
This research was funded by Deanship Scientific Research Imam Abdulrahman Bin Faisal university for this study; grant number [2018-066-IRMC].
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Esteban G.A., Balzarini J., Esnouf R., Clercq E.D., Camarasa M.J., Pérez-Pérez M.J. Design, Synthesis, and Enzymatic Evaluation of Multisubstrate Analogue Inhibitors of Escherichia coli Thymidine Phosphorylase. J. Med. Chem. 2000;43:971–983. doi: 10.1021/jm9911377. [DOI] [PubMed] [Google Scholar]
- 2.Friedkin M., Roberts D. Der Stoffwechsel der Purine und Pyrimidine. J. Biol. Chem. 1954;207:245–256. [PubMed] [Google Scholar]
- 3.Krenitsky T.A., Barclay M., Jacquez J.A. Specificity of mouse uridine phosphorylase. J. Biol. Chem. 1964;239:805–812. [PubMed] [Google Scholar]
- 4.Iltzsch M.H., el-Kouni M.H., Cha S. Kinetic studies of thymidine phosphorylase from mouse liver. Biochemistry. 1985;24:6799–6807. doi: 10.1021/bi00345a011. [DOI] [PubMed] [Google Scholar]
- 5.Brown N.S., Bicknell R. Thymidine phosphorylase, 2-deoxy-D-ribose and angiogenesis. Biochem. J. 1998;334:1–8. doi: 10.1042/bj3340001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa T., Yoshimura A., Sumizawa T., Haraguchi M., Akiyama S.I., Fukui K., Ishizawa M., Yamada Y. Angiogenic factor. Nature. 1992;356:668. doi: 10.1038/356668a0. [DOI] [PubMed] [Google Scholar]
- 7.Haraguchi M., Miyadera K., Uemura K., Sumizawa T., Furukawa T., Yamada K., Akiyama S.I., Yamada Y. Angiogenic activity of enzymes. Nature. 1994;368:198. doi: 10.1038/368198a0. [DOI] [PubMed] [Google Scholar]
- 8.Takao S., Akiyama S., Nakajo A., Yoh H., Kitazono M., Natsugoe S., Miyadera K., Fukushima M., Yamada Y., Aikou T. Suppression of metastasis by thymidine phosphorylase inhibitor. Cancer Res. 2000;60:5345–5348. [PubMed] [Google Scholar]
- 9.Bronckaers A., Gago F., Balzarini J., Liekens S. The dual role of thymidine phosphorylase in cancer development and chemotherapy. Med. Res. Rev. 2009;29:903–953. doi: 10.1002/med.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama S.I., Furukawa T., Sumizawa T., Takebayashi Y., Nakajima Y., Shimaoka S., Haraguchi M. The role of thymidine phosphorylase, an angiogenic enzyme, in tumor progression. Cancer Sci. 2004;95:851–857. doi: 10.1111/j.1349-7006.2004.tb02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sumizawa T., Furukawa T., Haraguchi M., Yoshimura A., Takeyasu A., Ishizawa M., Yamada Y., Akiyama S.I. Thymidine phosphorylase activity associated with platelet-derived endothelial cell growth factor. J. Biochem. 1993;114:9–14. doi: 10.1093/oxfordjournals.jbchem.a124146. [DOI] [PubMed] [Google Scholar]
- 12.Usuki K., Saras J., Waltenberger J., Miyazono K., Pierce G., Thomason A., Heldin C.H. Platelet-derived endothelial cell growth factor has thymidine phosphorylase activity. Biochem. Biophys. Res. Commun. 1992;184:1311–1316. doi: 10.1016/S0006-291X(05)80025-7. [DOI] [PubMed] [Google Scholar]
- 13.Abbas Y., Mansha M., Ullah N. The first total synthesis of potent antitumoral(±)- mafaicheenamine A, unnatural 6-fluoromafaicheenamine A and expedient synthesis of clausine E. RSC Adv. 2016;6:26104–26110. doi: 10.1039/C6RA03242G. [DOI] [Google Scholar]
- 14.Matsushita S., Nitanda T., Furukawa T., Sumizawa T., Tani A., Nishimoto K., Akiba S., Miyadera K., Fukushima M., Yamada Y., et al. The effect of a thymidine phosphorylase inhibitor on angiogenesis and apoptosis in tumors. Cancer Res. 1999;59:1911–1916. [PubMed] [Google Scholar]
- 15.Focher F., Spadari S. Thymidine phosphorylase: A two-face Janus in anticancer chemotherapy. Curr. Cancer Drug Targets. 2001;1:141–153. doi: 10.2174/1568009013334232. [DOI] [PubMed] [Google Scholar]
- 16.Miyadera K., Emura T., Suzuki N., Akiyama S., Fukushima M., Yamada Y. Novel functional antitumor nucleoside TAS-102, combined from of F3Rhd and its modulator (2): Inhibitory effect of TPI on tumor-derived angiogenesis and metasis. Proc. Am. Assoc. Cancer Res. 1998;39:609. [Google Scholar]
- 17.Pomeisl K., Votruba I., Holy A., Pohl R. Nucleosides, Syntheses of pyrimidine acyclic nucleoside phosphonates as potent inhibitors of thymidine phosphorylase (PD-ECGF) from SD-lymphoma. Nucleotides Nucleic Acids. 2007;26:1025–1028. doi: 10.1080/15257770701508679. [DOI] [PubMed] [Google Scholar]
- 18.Gbaj A., Edwards P.N., Reigan P., Freeman S., Jaffar M., Douglas K.T. Thymidine phosphorylase from Escherichia coli: Tight-binding inhibitors as enzyme active-site titrants. J. Enzym. Inhib. Med. Chem. 2006;21:69–73. doi: 10.1080/14756360500424010. [DOI] [PubMed] [Google Scholar]
- 19.Nencka R., Votruba I., Hrebabecky H., Jansa P., Tloustova E., Horska K., Masojidkova M., Holy A. Discovery of 5-substituted-6-chlorouracils as efficient inhibitors of human thymidine phosphorylase. J. Med. Chem. 2007;50:6016–6023. doi: 10.1021/jm070644i. [DOI] [PubMed] [Google Scholar]
- 20.McNally V.A., Rajabi M., Gbaj A., Stratford I.J., Edwards P.N., Douglas K.T., Bryce R.A., Jaffar M., Freeman S. Design, synthesis and enzymatic evaluation of 6-bridged imidazolyluracil derivatives as inhibitors of human thymidine phosphorylase. J. Pharm. Pharmacol. 2007;59:537–547. doi: 10.1211/jpp.59.4.0008. [DOI] [PubMed] [Google Scholar]
- 21.Liekens S., Balzarini J., De Hernández A.I., Clercq E., Priego E.M., Camarasa M.J., Pérez-Pérez M.J. Thymidine Phosphorylase is Noncompetitively Inhibited by 5′-O-Trityl-Inosine (KIN59) and Related Compounds. Nucleosides Nucleotides Nucleic Acids. 2006;25:975–980. doi: 10.1080/15257770600888925. [DOI] [PubMed] [Google Scholar]
- 22.Casanova E., Hernandez A.I., Priego E.M., Liekens S., Camarasa M.J., Balzarini J., Pérez-Pérez M.J. 5′-O-tritylinosine and analogues as allosteric inhibitors of human thymidine phosphorylase. J. Med. Chem. 2006;49:5562–5570. doi: 10.1021/jm0605379. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Pérez M.J., Priego E.M., Hernandez A.I., Camarasa M.J., Balzarini J., Liekens S. Thymidine phosphorylase inhibitors: Recent developments and potential therapeutic applications. Mini Rev. Med. Chem. 2005;5:1113–1123. doi: 10.2174/138955705774933301. [DOI] [PubMed] [Google Scholar]
- 24.Sun L., Bera H., Chui W.K. Synthesis of pyrazolo[1,5-a][1,3,5]triazine derivatives as inhibitors of thymidine phosphorylase. Eur. J. Med. Chem. 2013;65:1–11. doi: 10.1016/j.ejmech.2013.03.063. [DOI] [PubMed] [Google Scholar]
- 25.Yano S., Kazuno H., Sato T., Suzuki N., Emura T., Wierzba K., Yamashita J.I., Tada Y., Yamada Y., Fukushima M., et al. Synthesis and evaluation of 6-methylene-bridged uracil derivatives. Part 2: Optimization of inhibitors of human thymidine phosphorylase and their selectivity with uridine phosphorylase. Bioorg. Med. Chem. Lett. 2004;12:3443–3450. doi: 10.1016/j.bmc.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 26.Balzarini J., Gamboa A.E., Esnouf R., Liekens S., Neyts J., De Clercq E., Camarasa M.J., Pérez-Pérez M.J. 7-Deazaxanthine, a novel prototype inhibitor of thymidine phosphorylase, FEBS Lett. 1998, 438, 91–95. FEBS Lett. 1998;438:91–95. doi: 10.1016/S0014-5793(98)01271-X. [DOI] [PubMed] [Google Scholar]
- 27.Matwijczuk A., Karcz D., Pustuła K., Makowski M., Górecki A., Kluczyk D., Karpińska M.M., Niewiadomy A., Gagoś M. Spectroscopic and theoretical studies of fluorescence effects in bio-active: 4-(5-(methyl-1,3,4-thiadiazol-2-yl)) benzene-1,3-diol and 4-(5-(methylamino-1, 3, 4-thiadiazol-2-yl)) benzene-1, 3-diol compounds: Effect of molecular aggregation and amino group position. J. Fluoresc. 2018;201:44–56. [Google Scholar]
- 28.Tiwari S.V., Siddiqui S., Seijas J.A., Vazquez-Tato M.P., Sarkate A.P., Lokwani D.K., Nikalje A.P. Microwave-assisted facile synthesis, anticancer evaluation and docking study of N-((5-(substituted methylene amino)-1, 3, 4-thiadiazol-2-yl) methyl) benzamide derivatives. Molecules. 2017;15:995. doi: 10.3390/molecules22060995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh D.P., Deivedi S.K., Hashim S.R., Singhal R.G. Synthesis and antimicrobial activity of some new quinoxaline derivatives. Pharmaceuticals. 2010;3:2416–2425. doi: 10.3390/ph3082416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suresh M.L.P., Suchakar D., Vashu K., Rao C.V. Synthesis and biological activity of 8-chloro-[1,2,4]triazolo[4,3-a]quinoxalines. J. Chem. Pharm. Res. 2010;2:497–504. [Google Scholar]
- 31.Patidar A.J.M., Mobiya A., Selvam G. Internat. Exploring potential of quinoxaline moiety. J. Pharm. Tech. Res. 2011;3:386–392. [Google Scholar]
- 32.Vieira M., Pinheiro C., Fernandes R., Noronha J.P., Prudêncio C. Antimicrobial activity of quinoxaline 1,4-dioxide with 2-and 3-substituted derivatives. Microbiol. Res. 2014;169:287–293. doi: 10.1016/j.micres.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Abid M., Azam A. Synthesis, characterization and antiamoebic activity of 1-(thiazolo [4,5-b] quinoxaline-2-yl)-3-phenyl-2-pyrazoline derivatives. Bioorg. Med. Chem. Lett. 2006;16:2812–2816. doi: 10.1016/j.bmcl.2006.01.116. [DOI] [PubMed] [Google Scholar]
- 34.Abu-Hashem A.A., Gouda M.A., Badria F.A. Synthesis of some new pyrimido [2′, 1′: 2,3] thiazolo [4,5-b] quinoxaline derivatives as anti-inflammatory and analgesic agents. Euro. J. Med. Chem. 2010;45:1976–1981. doi: 10.1016/j.ejmech.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 35.Zarranz B., Jaso A., Aldana I., Monge A. Synthesis and anticancer activity evaluation of new 2-alkylcarbonyl and 2-benxoyl-3-trifluoromethyl-quinoxaline 1,4-do-N-oxide derivatives. Bioorg. Med. Chem. 2004;12:3711–3721. doi: 10.1016/j.bmc.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Husain A. Madhesia, Recent advances in pharmacological activities of quinoxaline derivates. J. Pharm. Res. 2011;4:924–929. [Google Scholar]
- 37.Deepika Y., Surendra P., Sachin N.K., Shewta S. Biological activity of quinoxaline derivatives. Int. J. Curr. Pharm. Rev. Res. 2011;1:33–46. [Google Scholar]
- 38.Chung H.J., Jung O.J., Chae M.J., Hong S.Y., Chung K.H., Lee S.K., Ryu C.K. Synthesis and biological evaluation of quinoxaline-5, 8-diones that inhibit vascular smooth muscle cell proliferation. Bioorg. Med. Chem. Lett. 2005;15:3380–3384. doi: 10.1016/j.bmcl.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Noreen T., Taha M., Imran S., Chigurpati S., Rahim F., Selvaraj M., Ismail N.H., Mohammad J.I., Ullah H., Javid M.T., et al. Synthesis of Alpha Amylase Inhibitors Based on Privileged Indole Scaffold. Bioorg. Chem. 2017;72:248–255. doi: 10.1016/j.bioorg.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Rahim F., Ali M., Ullah S., Rashid U., Ullah H., Taha M., Javed M.T., Rehman W., Abid O.U.R., Khan A.A., et al. Development of bis-Thiobarbiturates as Successful Urease Inhibitors and their Molecular Modeling Studies. Chin. Chem. Lett. 2016;27:693–697. doi: 10.1016/j.cclet.2015.12.035. [DOI] [Google Scholar]
- 41.Taha M., Sultan S., Nuzar H.A., Rahim F., Imran S., Ismail N.H., Naz H., Ullah H. Synthesis and biological evaluation of novel N-arylidenequinoline-3-carbohydrazides as potent β-glucuronidase inhibitors. Bioorg. Med. Chem. 2016;24:3696–3704. doi: 10.1016/j.bmc.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Rashid U., Rahim F., Taha M., Arshad M., Ullah H., Mahmood T., Ali M. Synthesis of 2-Acylated and Sulfonated 4-hydroxycoumarins: In vitro Urease Inhibition and Molecular Docking Studies. Bioorg. Chem. 2016;66:111–116. doi: 10.1016/j.bioorg.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Taha M., Imran S., Ismail N.H., Selvaraj M., Rahim F., Chigurupati S., Ullah H., Khan F., Salar U., Javid M.T., et al. Biology-oriented drug synthesis (BIODS) of 2-(2-methyl-5-nitro-1Himidazol-1-yl)ethyl aryl ether derivatives, in vitro α-amylase inhibitory activity and in silico studies. Bioorg. Chem. 2017;74:1–9. doi: 10.1016/j.bioorg.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Rahim F., Ullah K., Ullah H., Wadood A., Taha M., Rehman A., din I.U., Ashraf M., Shaukat A., Rehman W., et al. Triazinoindole analogs as potent inhibitors of α-glucosidase: Synthesis, biological evaluation and molecular docking studies. Bioorg. Chem. 2015;58:81–87. doi: 10.1016/j.bioorg.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Taha M., Ismail N.H., Imran S., Rahim F., Wadood A., Khan H., Ullah H., Salar U., Khan K.M. Synthesis, β-Glucuronidase Inhibition and Molecular Docking Studies of Hybrid Bisindole-Thiosemicarbazides Analogs. Bioorg. Chem. 2016;68:56–63. doi: 10.1016/j.bioorg.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Rahim F., Ullah H., Javid M.T., Wadood A., Taha M., Ashraf M., Shaukat A., Junaid M., Hussain S., Rehman W., et al. Synthesis, in vitro evaluation and molecular docking studies of thiazole derivatives as new inhibitors of α-glucosidase. Bioorg. Chem. 2015;62:15–21. doi: 10.1016/j.bioorg.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Rahim F., Javid M.T., Ullah H., Wadood A., Taha M., Ashraf M., Aine Q.U., Khan M.A., Khan F., Mirza S., et al. Synthesis, Molecular Docking, Acetylcholinesterase and Butyrylcholinesterase Inhibitory Potential of Thiazole Analogs as New Inhibitors for Alzheimer Disease. Bioorg. Chem. 2015;62:106–116. doi: 10.1016/j.bioorg.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Rahim F., Malik F., Ullah H., Wadood A., Khan F., Javid M.T., Taha M., Rehman W., Rehman A.U., Khan K.M. Isatin based Schiff bases as inhibitors of α-glucosidase: Synthesis, characterization, in vitro evaluation and molecular docking studies. Bioorg. Chem. 2015;60:42–48. doi: 10.1016/j.bioorg.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Rahim F., Ullah H., Taha M., Wadood A., Javid M.T., Rehman W., Nawaz M., Ashraf M., Ali M., Sajid M., et al. Synthesis and in vitro Acetylcholinesterase and Butyrylcholinesterase Inhibitory Potential of Hydrazide based Schiff Bases. Bioorg. Chem. 2016;68:30–40. doi: 10.1016/j.bioorg.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 50.Uddin I., Taha M., Rahim F., Wadood A. Synthesis and molecular docking study of piperazine derivatives as potent inhibitor of thymidine phosphorylase. Bioorg. Chem. 2018;78:324–331. doi: 10.1016/j.bioorg.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 51.Taha M., Rashid U., Imran S., Ali M. Rational design of bis-indolylmethane-oxadiazole hybrids as inhibitors of thymidine phosphorylase. Bioorg. Med. Chem. 2018;26:3654–3663. doi: 10.1016/j.bmc.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 52.Barbuceanu S.F., Almajan G.L., Saramet I., Draghici C., Tarcomnicu A.I., Bancescu G. Synthesis, characterization and evaluation of antibacterial activity of some thiazolo [3,2-b][1,2,4] triazole incorporating diphenylsulfone moieties. Eur. J. Med. Chem. 2009;44:4752–4757. doi: 10.1016/j.ejmech.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 53.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thymidine Phosphorylase from E. coli with 3′-Azido-2′-Fluoro-Dideoxyuridine. [(accessed on 27 March 2013)]; Available online: http://www.rcsb.org/structure/4EAD.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.