Abstract
Salt stress (SS) has become an important factor limiting afforestation programs. Because of their salt tolerance and fully sequenced genomes, poplars (Populus spp.) are used as model species to study SS mechanisms in trees. Here, we review recent insights into the physiological and molecular responses of Populus to SS, including ion homeostasis and signaling pathways, such as the salt overly sensitive (SOS) and reactive oxygen species (ROS) pathways. We summarize the genes that can be targeted for the genetic improvement of salt tolerance and propose future research areas.
Keywords: poplars (Populus), salt tolerance, molecular mechanisms, SOS, ROS
1. Introduction
Poplars (Populus spp.), which include about 100 species [1], are widely distributed across a variety of climatic regions [2] and have become important species for global afforestation and shelterbelt projects because of their rapid growth and high biomass yields [3]. These traits, in combination with other characteristics, such as extensive and deep root systems, considerable genetic variation, small genome size, convenient asexual propagation, genetic transformability, and economic significance, have led to the use of poplars as model tree species [4,5,6].
The increasing salinization of soils has greatly limited the planting of salt-sensitive Populus species [7]. Salt stress (SS) induces water deficiency, osmotic stress, ion toxicity, and oxidative damage [8] and thereby reduces photosynthesis, respiration, transpiration, metabolism, and growth in poplars. Like most plants, poplars can adapt to SS by maintaining their cellular ion homeostasis, accumulating osmotic-adjustment substances, and activating scavengers of reactive oxygen species (ROS) via the initiation of an efficient signal transduction network [9]. Desert poplar (Populus euphratica) is one of the most salt-tolerant Populus species [10] and is often used to study the salt-response mechanisms of trees. P. euphratica was reported to be tolerant to up to 450 mM NaCl (about 2.63%) under hydroponic conditions and showed high recovery efficiency when NaCl was removed from the culture medium [11]. A previous study has shown that P. euphratica could grow in soils with up to 2.0% salinity and can survive in soils with up to 5.0% salinity [12]. As a non-halophyte, P. euphratica could activate salt secretion mechanisms when soil salinity concentrations are greater than 20%, which may be one of the reasons for its high salt-tolerance [13]. Most other Populus species are relatively salt-sensitive, including the grey poplar (Populus × canescens), Populus × euramericana, and Populus popularis, which are usually used as a salt-sensitive control in salt-response studies. Populus alba has a wide variation of salinity tolerance within the species: for example ‘Guadalquivir F-21–38′, ‘Guadalquivir F-21–39′, and ‘Guadalquivir F-21–40′ clones show salt tolerance, while most other clones have a common salt sensitivity. Considering the wide variation within the species, P. alba could be used as a model species to understand the mechanisms of SS [2].
Previous research primarily focused on the anatomical, physiological, and biochemical changes in poplars during SS; many recent studies have focused on the molecular mechanisms using new techniques, such as genome-scale transcript analysis [14], high-throughput sequencing [15], metabolite profiling [16], bioinformatic analyses [17,18,19], and a non-invasive micro-test technique (NMT) [20].
Here, we review the recent progress in understanding the physiological and molecular responses of Populus to SS, including SS injuries, the main mechanisms of salt tolerance, and the genes targeted for the genetic improvement of salt tolerance in Populus, with a major focus on ion homeostasis, osmotic adjustment, ROS scavenging, salt overly sensitive (SOS) signaling pathways, potential candidate genes, and transcription factor-mediated regulation of the SS response.
2. SS Injury
2.1. Inhibition of Poplar Growth by Salinity Stress
Salinity affects all stages of Populus growth, including germination [21], vegetation growth [15,22], and sexual reproduction [23]. The percentage of seeds that germinate and the extent of leaf expansion were both reported to decline as salt concentrations increase [21]. In addition, shoot growth is more sensitive to salt than root growth [8]. When salt-sensitive white poplar (P. alba) clones were exposed to SS (0.6% NaCl), striking reductions were observed in their leaf elongation rate and internode lengths, while significant increases were observed in the numbers of short branches and bud numbers, as well as in the levels of leaf epinasty, necrosis, and abscission [7].
Long-term SS also induces early leaf maturity, early flowering, and early tree maturation in P. alba clones, which display a greater architectural modification when exposed to high SS (0.6%) than lower SS (0.3%) [22]. When exposed to 0.6% NaCl, the heights, ground diameter, and leaf numbers of Poplar 107 were significantly reduced, while plants exposed to 0.3% NaCl stress had relatively minor phenotypic changes [15]. When P. euphratica was exposed to 300 mM (1.76%) NaCl stress, the three growth indexes (plant height, ground diameter, and leaf number) were reduced to 31%, 45.5%, and 20% of the control plants, respectively. The mean leaf area of these stressed trees was reduced by up to 60%, and the leaves began to wither and yellow after 10 days. By contrast, a treatment of 50 mM (about 0.29%) NaCl did not cause a significant reduction in these traits in P. euphratica [24].
2.2. Salt-Induced Physiological and Cellular Changes
The adverse effects of SS also result in physiological and microscopic anatomical changes. P. euphratica and P. alba trees exposed to SS have significantly reduced stomatal area, aperture, and conductance, but increased stomatal density and hydraulic conductance [25,26,27]. The salt-induced reduction of leaf area in Populus may be one of the reasons for the increased stomatal density and decreased stomatal area [24,27]. The percentage loss of hydraulic conductivity (PLC%) in P. euphratica increased from 31.81% at 0 mM NaCl to 83.83% at 150 mM NaCl (0.88%), causing a 40–80% decrease in hydraulic conductivity and ensuring high hydraulic efficiency [27]. Populus trees thus reduce their transpiration by decreasing their stomatal apertures and conductance and increasing their PLC% values to cope with the salt-induced water deficit.
Populus may adjust their xylems to adapt to salinity stress [28]; for example, when exposed to SS, salt-sensitive P. × canescens produces narrower xylem vessels in which it stores sodium ions (Na+), reducing the effect of ion toxicity. In contrast, the salt-tolerant species P. euphratica produces narrow xylem vessels to reduce Na+ uptake even under normal conditions, the abundance of which remains largely unaltered under moderate SS [29]. Overall, this may indicate an evolutionary adaptation of the xylem structure in P. euphratica.
2.3. Salt-Induced Damage to the Photosynthetic System
Chlorophyll (Chl) is the main pigment used for photosynthesis in plants and is often studied in the evaluation of salt damage. The contents of Chla and Chlb, as well as the relative electron transport rate, decreased significantly in poplar 107 (a superior variety selected from Populus × euramericana cv. ’74/76′ hybrids) under 0.6% NaCl but increased under 0.3% NaCl [15].
Almost all poplar trees show a decreased net photosynthetic rate (Pn) under high SS; for example, the Pn decreased by 48.3% in P. euphratica under a 300 mM NaCl treatment, in comparison with the unstressed plants [24]. SS has a negative effect on growth, possibly due to the Pn affecting the accumulation of biomass. The stomatal conductance (Gs), transpiration rate, and internal CO2 (Ci) concentration of P. euphratica leaves also decreased by 48.5%, 42.1%, and 15.7% under 300 mM NaCl, indicating that the photosynthetic system was injured [24]. The fluorescence transient curve OJIP is highly sensitive to salinity stress; the OJ phase is the photochemical phase, leading to the reduction of QA to QA-. The I level is related to the heterogeneity in the filling up of the plastoquinone (PQ) pool. The P level is reached when all the PQ molecules are reduced to PQH2 [30]. While in poplars treated with 0.3% NaCl the OJIP curve follows the same trend as in the control plants, it significantly decreases at the J, I, and P phases in plants treated with 0.6% NaCl [15]. Large decreases in the J and I phases are also observed in P. euphratica treated with 300 mM NaCl [24]. As the electron transfer between Pheo and QA occurs during the J phase, these results indicate the higher salt concentration had a significant influence on electron transfer, which led to a great degree of salt damage [15]. Besides, F0 (minimal constant fluorescence of dark-adapted plants) and Fv/Fm (a surrogate of the maximum quantum efficiency of PSII) are also used as fluorescence parameters to identify salt tolerance, for example, to detect genotypic differences in the sensitivity of white poplar clones to SS [31].
3. Primary Mechanism of Salt Tolerance in Populus
3.1. Maintaining an Optimal K+/Na+ Ratio
Salt tolerance can be determined by the net Na+ efflux capacity in Populus. The net uptake of Na+ depends on its influx, exclusion, and sequestration, as well as on other Na+ regulation processes, such as xylem Na+ loading and unloading, and phloem Na+ recirculation [32]. Na+ moves into cells through non-selective cation channels (NSCCs) and high-affinity K+ transporter (HKT) [33], while excessive Na+ can be extruded into the apoplast across the plasma membrane (PM) by the Na+/H+ antiporter salt overly sensitive1 (SOS1; see Section 3.4) [34], which may also involve Na+ loading in the xylem [35]. The net Na+ efflux increased significantly in P. euphratica after 0.5–12 h under 100 mM NaCl but was reduced at 0.5 h in P. popularis, which showed an overall Na+ influx after 6–12 h of SS [36]. This is consistent with results of X-ray microanalysis showing that P. euphratica had lower concentration of Na+ in all subcellular compartments than salt-sensitive poplar species [37]. Moreover, salt tolerance can be improved by increasing Na+ efflux and the uptake of mineral nutrients via interactions between mycorrhizal fungi and the roots of salt-sensitive Populus [10,38].
It is crucial for Populus to maintain an optimal K+/Na+ ratio in the cytoplasm when exposed to high salinity. An excessive uptake of Na+ ions not only increases their abundance in plant cells, but also induces the loss of potassium ions (K+) by depolarizing the cellular membranes. Moreover, Na+ can compete with K+ for the binding sites of the uptake system, resulting in an imbalance of the K+/Na+ ratio that eventually causes ion toxicity [32]. Under high SS, P. euphratica maintained an optimal Na+/K+ ratio by restricting the net Na+ uptake and transport from roots to shoots and by maintaining higher K+ uptake and transport [39,40]. X-ray microanalysis showed that high salinity reduced the K+/Na+ ratio by 93% in P. popularis but only by 69% in P. euphratica [40].
Populus improves salt tolerance by accumulating Na+/K+ ions in the vacuoles. Na+ is sequestered into plant cell vacuoles by the tonoplast Na+/H+ antiporter NHX1 [41]. Besides detoxifying the cytoplasm, the accumulation of Na+ ions in the vacuoles is used as an osmoticum to draw water into the cells [42]. Vacuolar H+-ATPase and vacuolar H+-PPase generate an electrochemical gradient across the vacuolar membrane for the tonoplast Na+/H+ antiporters in P. euphratica cells [42]. In addition, expressing the Arabidopsis thaliana Na+/H+ antiporter gene AtNHX1 in transgenic poplars improves their salt resistance by improving their Na+/H+ exchange activity [43,44,45]. AtNHX3 was previously shown to act as the tonoplast K+/H+ antiporter for the transportation of K+ and the maintenance of ion homeostasis [46]; however, AtNHX1 was also recently found to enhance the accumulation of K+ in the vacuoles of transgenic poplars [47]. Yang reported that the constitutive expression of either AtNHX1 or AtNHX3 in transgenic Populus increased the vacuolar accumulation of Na+ and K+, leading to improved salt tolerance and drought tolerance [47].
K+ and Na+ uptake and transport mediated by HKT1 facilitate the rapid response of Populus to SS. The Na+/K+ transporter HKT1 is located on the plasma membrane and mediates the uptake and transport of K+ and Na+ in poplar. Furthermore, HKT is also involved in xylem Na+ unloading in tomato (Solanum lycopersicum) [48], as well as in the phloem-mediated recirculation of Na+ from the shoots to the roots of rice (Oryza sativa) [49], avoiding the excessive accumulation of Na+ in the leaves. The expression of HKT1 in P. euphratica is three times higher after 1 h of a 1% NaCl treatment, which facilitates its rapid response to SS by taking a certain amount of Na+ ions into the cells to maintain their osmotic balance [50].
Restricting the K+ efflux is important for the salt resistance of Populus. Plasma membrane H+-ATPase restricts K+ efflux, thus improving salt resistance of Populus. K+ can be transported into cells via an inward-rectifying K+ channel and the high-affinity K+ transporter HKT1, while K+ efflux from the roots is mediated by the activation (by depolarization) of outward-rectifying K+ channels (DA-KORCs) and NSCCs (DA-NSCCs), which can be induced by salt and inhibited by the plasma membrane H+-ATPase [51,52]. The concentration of K+ is markedly reduced in P. euphratica callus cells exposed to SS when pretreated with vanadate, an inhibitor of H+-ATPase, because of the enhanced efflux of K+ [53,54].
H+-ATPase activity plays a large role in salt tolerance of Populus. In addition to restricting K+ efflux, H+-ATPases can also maintain a proton gradient across the membrane, used by the Na+/H+ antiporters [8,55,56], which is closely associated with the salt sensitivity of Populus. The activity of plasma membrane H+-ATPase was higher in salt-tolerant genotypes than in salt-sensitive genotypes of P. alba [57]. Similarly, H+-ATPase genes were more highly expressed in the salt-tolerant species P. euphratica than in the salt-sensitive Populus trichocarpa [55]. This is further supported by the work of Ma et al., who showed that the P. euphratica genome contains more copies of the P-type H+-ATPase genes than P. trichocarpa [58].
Overall, except the above mechanisms, P. euphratica owns more effective mechanisms to respond to SS, for example, develop smaller vessel lumina than other salt sensitive poplars to limit ion loading into the xylem and develop leaf succulence after a long time of SS to dilute salt as a plastic morphological adaptation. Up to one-fifth of Na+ and one-third of Cl− are stored in foliage in the harvest season and are eliminated as leaves are ultimately shed. Excessive Na+( Cl−) can also be extruded via phloem retranslocating into the roots [59]. Sequestering Cl− in cortical vacuoles at high salinity is also important for restricting its transport into above-ground organs [60].
3.2. Accumulation of Osmotic-Adjustment Substances
The water potential of the soil and the availability of water to plant roots are lower in saline soils; therefore, salt-stressed plants first experience water deficiencies caused by osmotic stress [16]. Stress caused by 150 mM NaCl caused a drop of −0.68 MPa in the osmotic potential and a rapid decrease of 0.77 MPa in the shoot water potential in young P. euphratica trees [16]. Plant cells tend to accumulate soluble osmolytes to adjust their osmotic potential, such as proline, glycine betaine, soluble sugars, and proteins, which enable the plants to alleviate the osmotic stress and maintain cell turgor, water uptake, and metabolic activity [8,61]. Both salt-sensitive and salt-tolerant poplar species showed an accumulation of free amino acids under long-term SS [62]. Proline is an important osmotic-adjustment substance that exists in a free state in plant cells, has a low molecular weight, is highly soluble in water, is relatively non-toxic, and has no net charge in the physiological pH range [63]. Proline accumulation preserves the osmotic balance under salinity stress, and proline content can be used as a physiological index of plant resistance to SS [8,64]. Salt-tolerant P. euphratica increased proline accumulation by 50–90% when exposed to 150–300 mM NaCl [24], while salt-sensitive hybrid poplars, such as P. alba cv. Pyramidalis × P. tomentosa, showed no significant accumulation when exposed to 50 and 150 mM NaCl [65]. Sucrose and total soluble sugars increased with the elevation of foliar Na+ and Cl− concentrations in P. euphratica [59]. Except for Valine (Val) and Isoleucine (Ile), soluble carbohydrates, sugar alcohols, organic acids, and amino acids in P. euphratica leaves did not show significant changes after 24 h of SS. However, the changes of these amino acids were too low to significantly affect the total osmotic potential of leaves [16]. As a “cheap” osmolyte, the accumulation of sodium mainly contributes to osmotic recovery in P. euphratica [16,66].
3.3. ROS and Reactive Nitrogen Species (RNS)
ROS, including hydrogen peroxide (H2O2), superoxide anions (O2·−), hydroxyl radicals (·OH), and singlet oxygen (1O2), accumulate when plants are exposed to high SS [67,68]. At moderate levels, functioning as signaling molecules, ROS trigger signal transduction events and elicit specific cellular responses thus regulating plant growth and stress responses [69]. Some ROS can react with almost all the components of living cells leading to severe damage to lipids, proteins, and nucleic acids [70]. Excessive ROS can induce oxidative damage and might be detoxified through enzymatic and non-enzymatic antioxidant systems. Poplars expressing TaMnSOD show greatly improved tolerance to NaCl, with higher superoxide dismutase (SOD) activities, lower malondialdehyde (MDA) contents, and lower relative electrical conductivity (REC) than the wild-type lines [71]. The peroxidase (POD) activity of P. euphratica was 61.8% higher under 200 mM NaCl stress relative to the control [26], while 3,3′-diaminobenzidine (DAB) staining and H2O2 measurement demonstrated a sharply increased level of H2O2 in Chinese white poplar (Populus tomentosa) exposed to 200 mM NaCl for 24 h [72]. Most of the genes encoding glutathione peroxidases (GSH-Px), glutathione S-transferases (GST), and glutaredoxins were more highly expressed in salt-stressed P. tomentosa than in the control [72], while the transcription levels of genes encoding antioxidant enzymes were upregulated [69] in plants exposed to 150 mM NaCl for 24 h [55]. Genes encoding enzymes involved in the glutathione metabolism pathway, including GSH-Px, glucose-6-phosphate Dehydrogenase (G6PD), glucose phosphate dehydrogenase (GPD), and isocitrate dehydrogenase (IDH), were significantly upregulated in plants treated with NaCl, which facilitated the detoxification of the salinity-induced ROS [15].
Non-enzymatic antioxidants include ascorbate (AsA), glutathione (GSH), and carotenoids (Car). Carotenoids not only protect against active oxygen species by quenching the excited states of photosensitizing molecules and singlet oxygen and by scavenging free radicals, but also protect biomembranes against oxidative damage by modifying the structural and dynamic properties of lipid membranes [73]. Recently, a carotenoid-deficient mutant of bacteria Pantoea sp. YR343 was found, showing reduced colonization on Populus deltoids roots [74].
In addition to the antioxidant enzymes, heat-shock transcription factors (HSFs) play a role in scavenging ROS in plants under SS. The transgenic expression of PeHSF in tobacco enhanced the activities of ascorbate peroxidase, GSH-Px, and glutathione reductase [75], and PtHSP17.8 expression in Arabidopsis increased the activation levels of antioxidative enzymes under SS [76].
RNS includes nitric oxide (NO·), nitric dioxide (NO2·), nitrous acid (HNO2), and dinitrogen tetroxide (N2O4), which can be produced when plants are subjected to SS. Like ROS, RNS also function as signaling molecules in the response to abiotic stress. NO is involved in plant growth, development, senescence, as well as stress response [69]. NO was also reported to enhance salt tolerance in plants [77]. NO reacts with GSH, forming S-nitrosoglutathione (GSNO); NO, GSNO, and peroxynitrite (ONOO–) can produce covalent post-translational modifications (PTMs), such as S-nitrosylation and the protein nitration [78]. However, ONOO−, generated from nitric oxide NO and superoxide anion(O2·−), can produce tyrosine nitration of plant proteins and originate nitrosative damage in plant cells [69].
3.4. Poplar Salt Stress (SS) Signaling Pathways
Calcium ions (Ca2+) are an important secondary messenger in plants and mediate poplar salt tolerance by enhancing Na+ exclusion, restricting K+ efflux, and sustaining the selectivity of the cell membrane [40]. In higher plants, the Ca2+- dependent SOS signaling pathway helps maintain ion homoeostasis and thus confers salt tolerance under saline conditions [79,80]. Upon NaCl exposure, the resulting elevated cytosolic Ca2+ levels are sensed by SOS3, which activates SOS2 and stimulates the membrane-localized Na+/H+ antiporter SOS1, resulting in Na+ efflux into the apoplast of the root [79,81]. Similarly, in Populus, although SOS gene expression is generally ubiquitous, some studies have indicated that SOS2 functions upstream of SOS1 and downstream of SOS3 [82].
In addition to extruding Na+ from the roots, SOS1 controls the long-distance transport of Na+ and affects its partitioning in plant organs [34,35]. PeSOS1 (Salt overly sensitive 1 from P. euphratica) expression was upregulated 5- to 10-fold in P. euphratica leaves treated with 200 mM NaCl for 24 h relative to the untreated controls, and PeSOS1 partially suppressed salt sensitivity when transgenically expressed in the Escherichia coli mutant strain EP432 [83]. Similarly, PabSOS1 expression was about five times higher after 12 h of NaCl treatment [82]. SOS2 not only acts as a central regulator of Na+ extrusion but also is involved in the signaling node between the SOS pathway and other signaling pathways [80].
Tang identified two CBL10 homologs, PtCBL10A and PtCBL10B, in the western balsam poplar (P. trichocarpa) genome, which may interact with the salt tolerance component PtSOS2 and may help accumulate Na+ in vacuoles [84]. Like PtSOS3, PtCBL10s also interacts with PtSOS2 to stimulate the activity of PtSOS1. Whereas PtCBL10s primarily functions in green tissues such as the shoots and targets the downstream component PtSOS2 to the tonoplast, PtSOS3 functions in the roots and targets PtSOS2 to the plasma membrane [84].
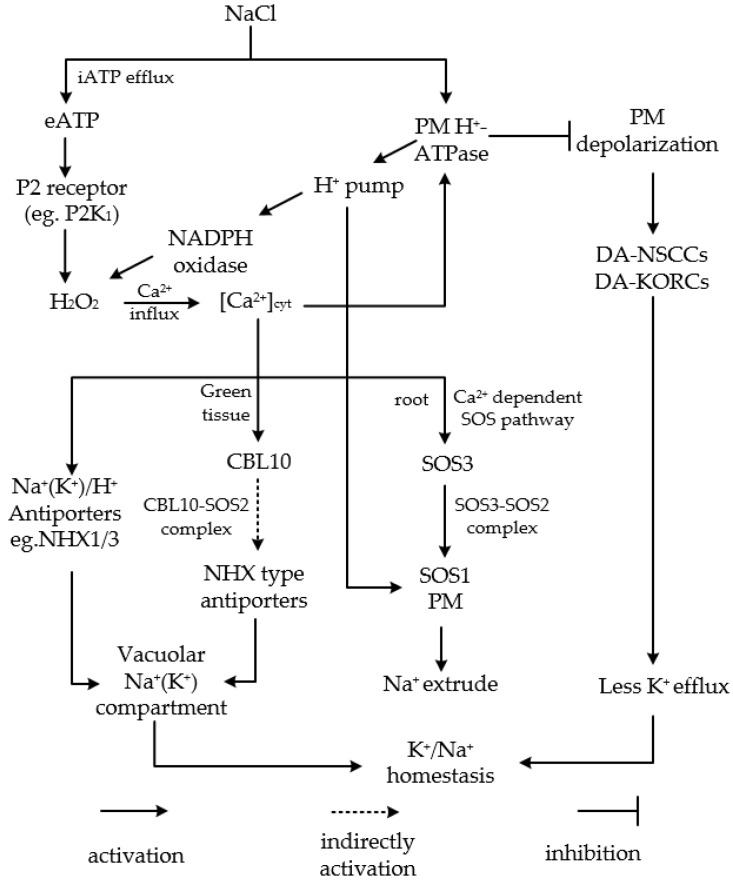
H+-ATPases not only provide the proton-motive force used to enhance Na+/H+ antiporter activity, but also can restrict the NaCl-induced efflux of K+ through DA-KORCs and DA-NSCCs (Figure 1). Genes encoding plasma membrane H+-ATPases are upregulated in P. euphratica [37], likely enhancing the exchange of Na+ and H+ across the plasma membrane.
Figure 1.
Schematic model showing multiple signaling networks active in Populus in response to NaCl stress. NaCl induces the efflux of intracellular ATP (iATP) and an increase in extracellular ATP (eATP), which is sensed by P2K1 in the plasma membrane (PM) and leads to the induction of H2O2 production. This stimulates the movement of Ca2+ into the cells via Ca2+-permeable channels. The elevated cytosolic Ca2+ concentration initiates the SOS pathway by stimulating Na+/H+ antiporters, such as SOS1, localized in the PM to extrude Na+, or activates CBL10, forming the CBL10–SOS2 complex, which may indirectly target the NHX type antiporters to the tonoplast to compartmentalize Na+ into vacuoles in green tissues. The elevated cytosolic Ca2+ also stimulates tonoplast-localized NHX1/3 to accumulate Na+(K+) into vacuoles. Besides, the elevated cytosolic Ca2+ increases H+-ATPase activity in the PM, which activates a H+ pump to supply a proton gradient for the Na+/H+ antiporters, stimulating the extrusion of Na+. A proton gradient supplied by the H+ pump contributes to the activation of NADPH oxidases, which leads to H2O2 production. H+-ATPases can also inhibit the efflux of K+ by further polarizing the PM. All these signaling components help to maintain K+/Na+ homeostasis in Populus cells.
As a hinge signal molecule for sensing and responding to SS, H2O2 is vital for K+/Na+ homeostasis. In response to SS, the salt-resistant species P. euphratica rapidly produces H2O2 in a process triggered by proton-coupled ion transporters such as the H+-pumps and the Na+/H+ antiporters in the plasma membrane [53]. This H2O2 accumulation causes a net Ca2+ influx by activating non-selective cation channels, which enhances Ca2+ concentration in the cytosol [54] and stimulates the SOS signaling pathway [81,85,86] (Figure 1). In addition, H2O2 signaling results in the upregulation of plasma membrane H+-ATPases, whose activity limits the NaCl-induced efflux of K+ [40,87]. Overall, H2O2 is involved in salt resistance in P. euphratica by controlling Na+ extrusion via the H2O2–cytosolic [Ca2+]–SOS pathway and by reducing K+ efflux to maintain ion homeostasis via the H2O2–Ca2+–PM H+-ATPases pathway (Figure 1).
NADPH oxidases are the main source of H2O2. During SS, plasma membrane H+-ATPases enhance H+ efflux, decreasing the pH and contributing to the activation of NADPH oxidases, which leads to H2O2 production and triggers the Ca2+-dependent SOS signaling pathway [53,54,87] (Figure 1). NaCl induces a transient increase in extracellular ATP (eATP), which is sensed by purinoceptors in the plasma membrane (e.g., P2K1) and causes a rapid H2O2 burst that in turn increases the concentration of Ca2+ in the cytosol of Populus cells [36,88,89]. Consequently, the salt-elicited eATP-H2O2-cytosolic [Ca2+] cascade contributes to enhancing Na+ extrusion through the Ca2+-dependent SOS pathways and to reducing K+ efflux by activating H+-ATPase, thus controlling cellular K+/Na+ homeostasis (Figure 1).
NaCl-induced expression of HSF (Heat shock transcription factor) in P. euphratica is markedly restricted by inhibitors of NADPH oxidase and Ca2+-permeable channels, suggesting that salt-induced H2O2 and cytosolic Ca2+ enhance the transcription of HSFs, which in turn upregulate genes encoding antioxidant enzymes for scavenging ROS under saline conditions [75].
In conclusion, eATP, Ca2+, H2O2, NADPH, H+-ATPase, and Na+ (K+)/H+ transporters play important roles in mediating salt tolerance in Populus trees.
4. Candidate Genes Used for the Genetic Improvement of Salt Tolerance
Currently, many studies are focused on introducing known salt-response signaling genes into Populus and testing the performance of the transgenic plants in high-salinity conditions. Many of these genes confer a significantly improved salt tolerance, as described below. Transferring transcription factors (TFs) genes into poplars is generally a more efficient approach than transferring structural genes, because transcription factors usually regulate the expression of many target genes in related pathways. A total of 59 ERF (Ethylene response factor) genes are associated with SS in Populus [90]. ERF76 from dihaploid P. simonii × P. nigra plants was transferred into the same Populus clone and significantly upregulated 16 genes encoding other transcription factors, as well as 45 stress-related genes [91]. When exposed to SS, ERF76-expressing transgenic plants were significantly taller and had increased root lengths, fresh weights, abscisic acid (ABA) and gibberellic acid (GA) contents compared to the control. Transgenic ERF76 expression enhanced salt tolerance by upregulating the expression of stress-related genes and increasing ABA and GA biosynthesis [91]. The PsnERF75 gene from P. simonii × P. nigra is induced by salt, drought, and ABA treatments [92] and confers salt tolerance when transgenically expressed in Arabidopsis [91].
The DREB (for dehydration-responsive element-binding protein) transcription factors, members of the ERF family, are vital regulatory nodes in the signaling pathways involved in the salt-stress response [93]. PeDREB2a, encoding a DREB transcription factor in P. euphratica, improved salt tolerance in Arabidopsis or birdsfoot trefoil (Lotus corniculatus) when transgenically expressed under the stress-inducible rd29A promoter [94]. The transgenic expression of LbDREB (a DREB gene from the halophyte Limonium bicolor) in Populus ussuriensis enhanced its resistance to salt, increasing its SOD and POD activities and the expression of the genes encoding these enzymes, reducing its MDA content, and enhancing its proline accumulation in the leaves [93]. The transgenic P. ussuriensis plants also had higher root/shoot ratios, higher relative water contents (RWC), and lower relative electrolytic leakage. Consistent with these changes, the genes encoding NAM (no apical meristem), GT-1(trihelix transcription factor), and WRKY70 (WRKY transcription factor 70) displayed inducible temporal expression patterns and are important components in the SS response signaling networks [93]. The LbDREB protein may inhibit the expression of NAM, GT-1, and WRKY70 and induce the expression of SOD and POD in response to high salinity stress, but this requires further verification.
The GTPase RabE is located in the Golgi apparatus and the plasma membrane, where it plays an important role in vesicle transport [95]. The overexpression of constitutively active PtRabE1b conferred salt tolerance in poplar [96]. This gene is directly co-expressed with many genes involved in salt tolerance, such as HSFA4a, SOS2, MPK19, and the Ca2+ signaling-related genes CAM7, CKL6, and calcium exchanger. HSFA4a expression is regulated by oxidative stress and MPK3/MPK6 and positively influenced salt tolerance in Arabidopsis [97]. CmHSFA4, a Chrysanthemum homologue of this gene, positively regulates salt tolerance by regulating the activities of SOS1, HKT2, and the ROS scavengers [98].
Recently, Yoon et al. identified a novel gene, PagSAP1(stress-associated proteins), from the hybrid poplar P. alba × Populus glandulosa. PagSAP1 negatively mediates salt-stress responses, and SS can in turn suppress the expression of this gene in poplar roots [99]. PagSAP1 overexpression resulted in enhanced sensitivity to SS, while PagSAP1 silencing via RNA interference (RNAi) significantly increased cytosolic Ca2+ in the roots. This increased cytosolic Ca2+ activated SOS signal transduction, resulting in high SOS3 transcript levels in the RNAi-derived plants. HKT1 expression is significantly reduced in all poplar genotypes under salt treatment; however, the lowest level is observed in the PagSAP1-overexpressing lines. HKT1 is responsible for Na+ influx and xylem-mediated Na+ recirculation from the shoot to the root [100]; therefore, the low HKT1 activity levels in the PagSAP1-overexpressing lines may explain the higher Na+ accumulation in the leaves and the lower Na+ levels in the roots compared with the control and RNAi-derived lines. By contrast, the excess Na+ in the roots of the PagSAP1-RNAi lines was eliminated by increased SOS1 activity, which resulted in lower Na+ levels in both the roots and the leaves of these lines. As a result, the salt tolerance of the PagSAP1-RNAi lines was improved through the upregulation of SOS3, SOS1, HKT1, H+-ATPase, AAA-type ATPase, and Arabidopsis K+ channel 2 (AKT2), all of which are essential for maintaining Na+/K+ homeostasis [99].
The poplars SOS proteins share high functional conservation with their Arabidopsis homologues [82]. SOS2 interacts with or regulates the activity of several tonoplast-localized transporters, such as the Ca2+/H+ antiporter [101], the vacuolar H+- ATPase [102], and the Na+/H+ exchanger [103,104]. PtSOS2.1, PtSOS2.2, and PtSOS2.3 (the PtSOS2 genes in P. trichocarpa) overexpression improves the salt tolerance of poplars and increases the concentrations of proline and photosynthetic pigments, relative water content, and the activity of their antioxidant enzymes, while significantly decreasing the levels of MDA [105].
The mutant SOS2 protein PtSOS2TD, generated by mutating the 169th amino acid in the activation loop of PtSOS2 from threonine (T) to aspartic acid (D), is more active than PtSOS2 and can sufficiently activate PtSOS1 in a PtSOS3-independent manner [80]. PtSOS2TD overexpression in poplars significantly increased salt tolerance, causing higher plasma membrane Na+/H+ exchange activity, greater Na+ efflux, decreased Na+ accumulation in the leaves, and improved ROS scavenging capacity [80].
The transgenic expression of the PtCBL10s (Calcineurin B-like from P. trichocarpa) conferred greater salt tolerance to poplars by maintaining shoot ion homeostasis under SS. The doubling of the CBL10 genes in poplar may represent an evolutionary adaptation to the adverse environment [84].
The genes that have been shown to increase the salt tolerance of transgenic Populus are presented in Table 1.
Table 1.
Candidate genes for improving salt tolerance in Populus.
| Candidate Genes and Source | Transgenic Species | Effect of SS in Transgenic Species Compared with WT | Reference |
|---|---|---|---|
| AtNHX1/3 (Vacuolar Na+/H+ antiporter from Arabidopsis thaliana) | Populus davidiana × Populus bolleana | 1 Normal growth and morphology; 2 Promoted vacuolar Na+ (K+)/H+ exchange activity; 3 Increased Na+ and K+ accumulation in the vacuoles; 4 Elevated the eaccumulation of proline. |
[47] |
| AtNHX1 (See above) |
Populus × euramericana ‘Neva’ |
1 Enhanced plant growth and photosynthetic capacity; 2 Lowerd MDA and REC; 3 Increased Na+ (K+) accumulation in roots and leaves. |
[44] |
|
Populus × euramericana ‘Neva’ |
1 Reduced decrease in Chl, Car, PSII, Fv/Fm, and qP; 2 Smaller reduction of Pn, Gs, Ci, CE; 3 Greater increase of stem and leaf, smaller increase in root. |
[43] | |
|
Populus deltoides × P. euramericana
CL ‘NL895′ |
1 Higher content of sodium ions; 2 Decreased MDA content. |
[45] | |
| PtSOS2TD (Salt overly sensitive 2 from Populus trichocarpa) |
P. davidiana × P. bolleana hybrid poplar clone Shanxin |
1 More vigorous growth; 2 Greater biomass produced; 3 Less Na+ in the leaves; 4 Higher Na+/H+ exchange activity and Na+ efflux; 5 More scavenging of ROS. |
[80] |
|
PtSOS2 (See above) |
Populus tremula × Populus tremuloides Michx clone T89 |
1 Improved PM Na+/H+ exchange activity, Na+ efflux; 2 Higher proline activity; 3 Higher RWC and sustained decrease of water loss; 4 Increased SOD, POD, CAT activity; 5 Decreased MDA concentration. |
[105] |
|
PtCBL10A and PtCBL10B (Calcineurin B-like from P. trichocarpa) |
P. davidiana × P. bolleana hybrid poplar clone Shanxin |
1 Less impairment by SS with higher stature and greater shoot biomass; 2 Lower Na+ in the leaves, more Na+ in the stem. |
[84] |
|
PeCBL6, PeCBL10 (Calcineurin B-like from P. euphratica) |
triploid white poplar | 1 Higher height growth rate; 2 Less wilted leaves; 3 Lower MDA content; 4 Higher chl content. |
[106] |
|
PtSOS3 (Salt overly sensitive 3 from P. trichocarpa) |
P. davidiana × P. bolleana hybrid poplar clone Shanxin |
1 Lower Na+ in the root; 2 Higher K+ content in the root; 3 More Na+ in the stem. |
[84] |
|
TaMnSOD (Mn-superoxide dismutases from Tamarix Androssowii) |
P. davidiana × P. bolleana hybrid poplar clone Shanxin |
1 Higher SOD activity; 2 Lower MDA contents; 3 Lower REC; 4 More weight gains. |
[71] |
|
TaLEA (Late embryogenesis abundant from T. androssowii) |
Populus simonii × Populus nigra Xiaohei poplar |
1 Decrease in MDA content; 2 Decrease in relative electrolyte leakage; 3 Improved salt and drought resistance. |
[107] |
| P. davidiana × P. bolleana | 1 Higher Survival percentages; 2 Higher Seedling height and photosynthetic capabilities; 3 Lower Na+ in young leaves but higher in yellow and withered leaves. |
[108] | |
|
ERF76 (Ethylene response factor from di-haploid P. simonii × P. nigra) |
P. simonii × P. nigra di-haploid | 1 Higher plant height, root length, fresh weight; 2 Higher in ABA and GA concentration. |
[91] |
|
JERFs (Jasmonic ethylene responsive factor from the tomato) |
Populus alba × Populus berolinensis | 1 Lower reductions of height, basal diameter, and biomass; 2 Lower reduction in leaf water content and increase in root/crown ratio; 3 Greater increase of foliar proline concentration; 4 Higher foliar Na+ concentration. |
[109] |
|
LbDREB (dehydration responsive element binding TF from Limonium bicolor) |
Populus ussuriensis Kom. Chinese Daqing poplar |
1 Higher SOD, POD activity; 2 Less MDA accumulation in the leaves; 3 More proline accumulation; 4 Increased root/shoot ratio; 5 Reduced decrease of RWC; 6 Lower increase of relative electrolytic leakage. |
[93] |
|
AhDREB1 (dehydration responsive element binding-like TF from the halophyte Atriplex hortensis) |
Populus tomentosa | 1 Higher survival rate; 2 High proline content. |
[110] |
|
AtSTO1 (Salt tolerant1 from Arabidopsis thaliana) |
P. tremula × P. alba Poplar 717-1B4 |
1 Higher aboveground biomass; 2 Higher root biomass; 3 Higher shoot height; 4 Higher chl content. |
[111] |
|
AtPLDα (Phospholipase Dα from A. thaliana) |
P. tomentosa | 1 Higher root rate and root length; 2 Reduced decrease of total chl content; 3 Lower REC and MDA content 4 Higher SOD, POD, and CAT activities. |
[112] |
|
AtSRK2C, AtGolS2 (Stress responses, SNF1-related protein kinase 2C, galactinol synthase 2 from A. thaliana) |
P. tremula × tremuloides | 1 Reduced decrease of dry weight; 2 Reduced decrease of total adventitious root length. |
[113] |
|
PtRabE1b(Q74L) (Rab GTPase from P. trichocarpa) |
P. alba × P. glandulosa clone 84 K |
1 More adventitious roots; 2 Greater root growth status in seedlings. |
[96] |
|
PagSAP1 (stress-associated proteins from P. alba × P. glandulosa) |
P. alba × P. glandulosa | RNAi plants accumulate more Ca2+, and K+ and less Na+. | [99] |
SS, salt stress; PM, plasma membrane; SOD, superoxide dismutase; POD, peroxidase; CAT, catalase; MDA, malondialdehyde; Chl, chlorophyll; Car, carotenoid; PSII, actual quantum yield of PSII; Fv/Fm, maximum photochemical efficiency; qP, photochemical quenching coefficient; Pn, net photosynthetic rate; Gs, stomatal conductance; Ci, internal CO2; CE, carboxylation efficiency concentration; ROS, reactive oxygen species; REC, relative electrical conductivity; RWC, relative water content; ABA, abscisic acid; GA, gibberellic acid.
5. Conclusions and Outlook
Soil salinization is increasingly problematic and is now a dominant factor limiting Populus growth [7]. Therefore, it is important to improve the salt tolerance of poplar trees. Plant salt tolerance is a typical quantitative trait affected by many physiological and biochemical factors [15]. Different Populus species, such as the salt-resistant poplar species P. euphratica and the salt-sensitive Populus species P. × canescens, have different SS responses [114,115]. Furthermore, trees such as the hybrid poplar 107 display different responses when exposed to different salinity levels [15]. Overall, Populus adapt to SS by maintaining suitable Na+/K+ ratios, accumulating osmotic-adjustment substances, activating antioxidative enzymes and antioxidants, and activating stress response signaling networks to reduce the negative effects of high salinity. With the innovation of transcriptomics technologies, a substantial number of stress-responsive and/or stress-regulated genes have been identified—in addition to the signal regulatory networks in which they function—and transferred between Populus and Arabidopsis or other species with great success. This has greatly elucidated the molecular mechanisms of the poplar stress responses [113].
ROS/RNS and hormones were identified as signaling molecules involved in the response to SS avoiding high salinity damage. The crosstalk between ROS, RNS, ABA, ethylene, and/or other hormones in poplar salt stress will be further studied. Molecular chaperones, especially dehydrins and osmotin, which contribute to protect proteins, are supposed key factors for coping with SS [16]. Nutrient fertilization with N and P was reported to reduce the accumulation of ROS (e.g., O3) and enhance membrane stability, thus protecting from oxidative stress by activating a cross-talk between antioxidant and osmotic mechanisms [116]. The role of mycorrhization and polymer amendment in enhancing mineral nutrition and improving salt tolerance is also a topic of future study. All of the above topics need to be more deeply studied in the future to improve salt tolerance in poplar species as well as in other tree species.
Determining the key transcription factors and molecular mechanisms underlying salt tolerance is an important goal for future research and will facilitate the enhancement of salt tolerance in Populus.
Acknowledgments
We are grateful to Li Liu in KIB for the kind discussion and guidance to the project.
Abbreviations
| SOS | salt overly sensitive |
| ROS | reactive oxygen species |
| SOD | superoxide dismutase |
| POD | peroxidase |
| MDA | malondialdehyde |
| GSH-Px | glutathione peroxidases |
| GST | glutathione S-transferases |
| GR | glutathione reductase |
| GPX | glutathione peroxidases |
| GPD | glucose phosphate dehydrogenase |
| G6PD | glucose-6-phosphate dehydrogenase |
| APX | ascorbate peroxidase |
| IDH | Isocitrate dehydrogenase |
| GA | gibberellic acid |
| ABA | abscisic acid |
| REC | relative electrical conductivity |
| RWC | relative water content |
| TFs | transcription factors |
| Chl | chlorophyll |
| Pn | net photosynthetic rate |
| Gs | stomatal conductance |
| Ci | internal CO2 |
| CE | carboxylation efficiency concentration; |
| Car | carotenoid |
| PSII | actual quantum yield of PSII |
| qP | photochemical quenching coefficient |
| NPQ | non photochemical quenching |
| Fv/Fm | maximum photochemical efficiency |
| eATP | extracellular ATP |
| iATP | intracellular ATP |
| DAB | 3,3′-diaminobenzidine |
| PM | plasma membrane |
| D A | depolarization activated |
| KORCs | K+ outward rectifying channels |
| NSCCs | non-selective cation channels |
| SS | salt stress |
Author Contributions
All authors worked on manuscript preparation.
Funding
This work was supported by the GuangXi Natural Science Foundation (2016GXNSFBA380224); the Department of Human Resources and Social Security of Guangxi Zhuang Autonomous Region, China (GuiCaiSheHan [2018]112), and the Fundamental Research Funds for Guangxi Forestry Research Institute No. LK201812).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Wu Z.Y., Raven P.H., editors. Flora of China. Volume 4. Beijing: Science Press; Beijing, China: Missouri Botanical Garden Press; St. Louis, MO, USA: 1999. pp. 139–162. [Google Scholar]
- 2.Sixto H. Response to sodium chloride in different species and clones of genus Populus L. Forestry. 2005;78:93–104. doi: 10.1093/forestry/cpi009. [DOI] [Google Scholar]
- 3.Jansson S., Douglas C.J. Populus: A model system for plant biology. Annu. Rev. Plant Biol. 2007;58:435–458. doi: 10.1146/annurev.arplant.58.032806.103956. [DOI] [PubMed] [Google Scholar]
- 4.Bradshaw H.D., Ceulemans R., Davis J., Stettler R. Emerging Model Systems in Plant Biology: Poplar (Populus) as A Model Forest Tree. J. Plant Growth Regul. 2000;19:306–313. doi: 10.1007/s003440000030. [DOI] [Google Scholar]
- 5.Polle A., Douglas C. The molecular physiology of poplars: Paving the way for knowledge-based biomass production. Plant Biol. 2010;12:239–241. doi: 10.1111/j.1438-8677.2009.00318.x. [DOI] [PubMed] [Google Scholar]
- 6.Taylor G. Populus: Arabidopsis for Forestry. Do We Need a Model Tree? Ann. Bot. 2002;90:681–689. doi: 10.1093/aob/mcf255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R.G., Chen S.L., Deng L., Fritz E., Hüttermann A., Polle A. Leaf photosynthesis, fluorescence response to salinity and the relevance to chloroplast salt compartmentation and anti-oxidative stress in two poplars. Trees. 2007;21:581–591. doi: 10.1007/s00468-007-0154-y. [DOI] [Google Scholar]
- 8.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 9.Hirayama T., Shinozaki K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen S.L., Hawighorst P., Sun J., Polle A. Salt tolerance in Populus: Significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole-plant nutrition. Environ. Exp. Bot. 2014;107:113–124. doi: 10.1016/j.envexpbot.2014.06.001. [DOI] [Google Scholar]
- 11.Gu R.S., Fonseca S., PuskÁs L.G., Hackler L.J., Zvara Á., Dudits D., Pais M.S. Transcript identification and profiling during salt stress and recovery of Populus euphratica. Tree Physiol. 2004;24:275–276. doi: 10.1093/treephys/24.3.265. [DOI] [PubMed] [Google Scholar]
- 12.Wang S., Chen B., Li H. Euphrates Poplar Forest. China Environmental Science Press; Beijing, China: 1996. pp. 43–52. [Google Scholar]
- 13.Fu A.H., Li W.H., Chen Y.N. The threshold of soil moisture and salinity influencing the growth of Populus euphratica and Tamarix ramosissima in the extremely arid region. Environ. Earth Sci. 2012;66:2519–2529. doi: 10.1007/s12665-011-1474-1. [DOI] [Google Scholar]
- 14.Qiu Q., Ma T., Hu Q.J., Liu B.B., Wu Y.X., Zhou H.H., Wang Q., Wang J., Liu J.Q. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 2011;31:452–461. doi: 10.1093/treephys/tpr015. [DOI] [PubMed] [Google Scholar]
- 15.Chen P.F., Zuo L.H., Yu X.Y., Dong Y., Zhang S., Yang M.S. Response mechanism in Populus × euramericana cv. ‘74/76’ revealed by RNA-seq under salt stress. Acta Physiol. Plant. 2018;40 doi: 10.1007/s11738-018-2676-x. [DOI] [Google Scholar]
- 16.Brinker M., Brosche M., Vinocur B., Abo-Ogiala A., Fayyaz P., Janz D., Ottow E.A., Cullmann A.D., Saborowski J., Kangasjarvi J., et al. Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 2010;154:1697–1709. doi: 10.1104/pp.110.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yer E.N., Baloglu M.C., Ayan S. Identification and expression profiling of all Hsp family member genes under salinity stress in different poplar clones. Gene. 2018;678:324–336. doi: 10.1016/j.gene.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 18.Zhao K., Li S.X., Yao W.J., Zhou B.R., Li R.H., Jiang T.B. Characterization of the basic helix-loop-helix gene family and its tissue-differential expression in response to salt stress in poplar. PeerJ. 2018;6:e4502. doi: 10.7717/peerj.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K., Zhang X.M., Cheng Z.H., Yao W.J., Li R.H., Jiang T.B., Zhou B.R. Comprehensive analysis of the three-amino-acid-loop-extension gene family and its tissue-differential expression in response to salt stress in poplar. Plant Physiol. Biochem. 2019;136:1–12. doi: 10.1016/j.plaphy.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y.N., Wang Y., Sa G., Zhang Y.H., Deng J.Y., Deng S.R., Wang M.J., Zhang H.L., Yao J., Ma X.Y., et al. Populus euphratica J3 mediates root K+/Na+ homeostasis by activating plasma membrane H+-ATPase in transgenic Arabidopsis under NaCl salinity. Plant Cell Tiss Organ Cult. 2017;131:75–88. doi: 10.1007/s11240-017-1263-y. [DOI] [Google Scholar]
- 21.Liu J.P., Li Z.J., He L.R., Zhou Z.L., Xu Y.L. Salt-tolerance of Populus euphratica and P. pruinosa seed during germination. Sci. Silvae Sin. 2004;40:165–169. doi: 10.11707/j.1001-7488.20040229. [DOI] [Google Scholar]
- 22.Abassi M., Mguis K., Béjaoui Z., Albouchi A. Morphogenetic responses of Populus alba L. under salt stress. J. For. Res.-JPN. 2014;25:155–161. doi: 10.1007/s11676-014-0441-6. [DOI] [Google Scholar]
- 23.Meilan R., Sabatti M., Ma C.P., Elena K. An Early-Flowering Genotype of Populus. J. Plant Biol. 2004;47:52–56. doi: 10.1007/BF03030228. [DOI] [Google Scholar]
- 24.Zhao C.Y., Si J.H., Feng Q., Deo R.C., Yu T.F., Li P.D. Physiological response to salinity stress and tolerance mechanics of Populus euphratica. Environ. Monit. Assess. 2017;189:533. doi: 10.1007/s10661-017-6257-z. [DOI] [PubMed] [Google Scholar]
- 25.Abbruzzese G., Beritognolo L., Muleo R., Piazzai M., Sabatti M., Mugnozza G.S., Kuzminsky E. Leaf morphological plasticity and stomatal conductance in three Populus alba L. genotypes subjected to salt stress. Environ. Exp. Bot. 2009;66:381–388. doi: 10.1016/j.envexpbot.2009.04.008. [DOI] [Google Scholar]
- 26.Rajput V.D., Chen Y., Ayup M. Effects of high salinity on physiological and anatomical indices in the early stages of Populus euphratica growth. Russ. J. Plant Physiol. 2015;62:229–236. doi: 10.1134/S1021443715020168. [DOI] [Google Scholar]
- 27.Rajput V.D., Chen Y.N., Ayup M., Minkina T., Sushkova S., Mandzhieva S. Physiological and hydrological changes in Populus euphratica seedlings under salinity stress. Acta Ecol. Sin. 2017;37:229–235. doi: 10.1016/j.chnaes.2017.02.005. [DOI] [Google Scholar]
- 28.Awad H., Barigah T., Badel E., Cochard H., Herbette S. Poplar vulnerability to xylem cavitation acclimates to drier soil conditions. Physiol. Plant. 2010;139:280–288. doi: 10.1111/j.1399-3054.2010.01367.x. [DOI] [PubMed] [Google Scholar]
- 29.Junghans U., Polle A., Duchting P., Weiler E., Kuhlman B., Gruber F., Teichmann T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006;29:1519–1531. doi: 10.1111/j.1365-3040.2006.01529.x. [DOI] [PubMed] [Google Scholar]
- 30.Strassert R.J., Srivastava A. Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem. Photobiol. 1995;61:32–42. doi: 10.1111/j.1751-1097.1995.tb09240.x. [DOI] [Google Scholar]
- 31.Sixto H., Aranda I., Grau J.M. Assessment of salt tolerance in Populus alba clones using chlorophyll fluorescence. Photosynthetica. 2006;44:169–173. doi: 10.1007/s11099-006-0002-0. [DOI] [Google Scholar]
- 32.Wu H.H. Plant salt tolerance and Na+ sensing and transport. Crop J. 2018;6:215–225. doi: 10.1016/j.cj.2018.01.003. [DOI] [Google Scholar]
- 33.Ward J.M., Hirschi K.D., Sze H. Plants pass the salt. Trends Plant Sci. 2003;8:200–201. doi: 10.1016/S1360-1385(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 34.Olias R., Eljakaoui Z., Li J., de Morales P.A., Marin-Manzano M.C., Pardo J.M., Belver A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009;32:904–916. doi: 10.1111/j.1365-3040.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- 35.Shi H.Z., Quintero F.J., Pardo J.M., Zhu J.K. The Putative Plasma Membrane Na+/H+ Antiporter SOS1 Controls Long-Distance Na+ Transport in Plants. Plant Cell Online. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao N., Wang S.J., Ma X.J., Zhu H.P., Sa G., Sun J., Li N.F., Zhao C.J., Zhao R., Chen S.L. Extracellular ATP mediates cellular K+/Na+ homeostasis in two contrasting poplar species under NaCl stress. Trees. 2015;30:825–837. doi: 10.1007/s00468-015-1324-y. [DOI] [Google Scholar]
- 37.Ma X.Y., Deng L., Li J.K., Zhou X.Y., Li N.Y., Zhang D.C., Lu Y.J., Wang R.G., Sun J., Lu C.F., et al. Effect of NaCl on leaf H+-ATPase and the relevance to salt tolerance in two contrasting poplar species. Trees. 2010;24:597–607. doi: 10.1007/s00468-010-0430-0. [DOI] [Google Scholar]
- 38.Li J., Bao S.Q., Zhang Y.H., Ma X.J., Mishra-Knyrim M., Sun J., Sa G., Shen X., Polle A., Chen S.L. Paxillus involutus strains MAJ and NAU mediate K+/Na+ homeostasis in ectomycorrhizal Populus × canescens under sodium chloride stress. Plant Physiol. 2012;159:1771–1786. doi: 10.1104/pp.112.195370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S.L., Li J.K., Wang S.S., Fritz E., Hüttermann A., Altman A. Effects of NaCl on shoot growth, transpiration, ion compartmentation, and transport in regenerated plants of Populus euphratica and Populus tomentosa. Can. J. For. Res. 2003;33:967–975. doi: 10.1139/x03-066. [DOI] [Google Scholar]
- 40.Sun J., Dai S.X., Wang R.G., Chen S.L., Li N.Y., Zhou X.Y., Lu C.F., Shen X., Zheng X.J., Hu Z.M., et al. Calcium mediates root K+/Na+ homeostasis in poplar species differing in salt tolerance. Tree Physiol. 2009;29:1175–1186. doi: 10.1093/treephys/tpp048. [DOI] [PubMed] [Google Scholar]
- 41.Mansour M.M.F., Salama K.H.A., Al Mutawa M.M. Transport proteins and salt tolerance in plants. Plant Sci. 2003;164:891–900. doi: 10.1016/S0168-9452(03)00109-2. [DOI] [Google Scholar]
- 42.Silva P., Façanha A.R., Tavares R.M., Gerós H. Role of Tonoplast Proton Pumps and Na+/H+ Antiport System in Salt Tolerance of Populus euphratica Oliv. J. Plant Growth Regul. 2010;29:23–34. doi: 10.1007/s00344-009-9110-y. [DOI] [Google Scholar]
- 43.Jiang C.Q., Zheng Q.S., Liu Z.P., Liu L., Zhao G.M., Long X.H., Li H.Y. Seawater-irrigation effects on growth, ion concentration, and photosynthesis of transgenic poplar overexpressing the Na+/H+ antiporter AtNHX1. J. Plant Nutr. Soil Sci. 2011;174:301–310. doi: 10.1002/jpln.201000033. [DOI] [Google Scholar]
- 44.Jiang C.Q., Zheng Q.S., Liu Z.P., Xu W.J., Liu L., Zhao G.M., Long X.H. Overexpression of Arabidopsis thaliana Na+/H+ antiporter gene enhanced salt resistance in transgenic poplar (Populus × euramericana ‘Neva’) Trees. 2012;26:685–694. doi: 10.1007/s00468-011-0635-x. [DOI] [Google Scholar]
- 45.Qiao G.R., Zhuo R.Y., Liu M.Y., Jiang J., Li H.Y., Qiu W.M., Pan L.Y., lin S., Zhang X.G., Sun Z.X. Over-expression of the Arabidopsis Na+/H+ antiporter gene in Populus deltoides CL × P. euramericana CL “NL895” enhances its salt tolerance. Acta Physiol. Plant. 2011;33:691–696. doi: 10.1007/s11738-010-0591-x. [DOI] [Google Scholar]
- 46.Liu H., Tang R.J., Zhang Y., Wang C.T., Lv Q.D., Gao X.S., Li W.B., Zhang H.X. AtNHX3 is a vacuolar K+/H+ antiporter required for low-potassium tolerance in Arabidopsis thaliana. Plant Cell Environ. 2010;33:1989–1999. doi: 10.1111/j.1365-3040.2010.02200.x. [DOI] [PubMed] [Google Scholar]
- 47.Yang L., Liu H., Fu S.M., Ge H.M., Tang R.J., Yang Y., Wang H.H., Zhang H.X. Na+/H+ and K+/H+ antiporters AtNHX1 and AtNHX3 from Arabidopsis improve salt and drought tolerance in transgenic poplar. Biol. Plant. 2017 doi: 10.1007/s10535-017-0724-9. [DOI] [Google Scholar]
- 48.Jaime-Perez N., Pineda B., Garcia-Sogo B., Atares A., Athman A., Byrt C.S., Olias R., Asins M.J., Gilliham M., Moreno V., et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017;40:658–671. doi: 10.1111/pce.12883. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi N.I., Yamaji N., Yamamoto H., Okubo K., Ueno H., Costa A., Tanoi K., Matsumura H., Fujii-Kashino M., Horiuchi T., et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017;91:657–670. doi: 10.1111/tpj.13595. [DOI] [PubMed] [Google Scholar]
- 50.Xu M., Sun Z.M., Liu S.A., Chen C.H., Xu L.A., Huang M.R. Cloning and Expression Analysis of Salinity Stress Related Peu HKT1 Gene from Populus euphratica. MPB. 2016 in press. [Google Scholar]
- 51.Britto D.T., Kronzucker H.J. Cellular mechanisms of potassium transport in plants. Physiol. Plant. 2008;133:637–650. doi: 10.1111/j.1399-3054.2008.01067.x. [DOI] [PubMed] [Google Scholar]
- 52.Shabala L., Zhang J., Pottosin I., Bose J., Zhu M., Fuglsang A.T., Velarde-Buendia A., Massart A., Hill C.B., Roessner U., et al. Cell-Type-Specific H+-ATPase Activity in Root Tissues Enables K+ Retention and Mediates Acclimation of Barley (Hordeum vulgare) to Salinity Stress. Plant Physiol. 2016;172:2445–2458. doi: 10.1104/pp.16.01347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun J., Li L.S., Liu M.Q., Wang M.J., Ding M.Q., Deng S.R., Lu C.F., Zhou X.Y., Shen X., Zheng X.J., et al. Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl-stressed callus cells of two contrasting poplars. Plant Cell Tiss Organ Cult. 2010;103:205–215. doi: 10.1007/s11240-010-9768-7. [DOI] [Google Scholar]
- 54.Sun J., Wang M.J., Ding M.Q., Deng S.R., Liu M.Q., Lu C.F., Zhou X.Y., Shen X., Zheng X.J., Zhang Z.K., et al. H2O2 and cytosolic Ca2+ signals triggered by the PM H+-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant Cell Environ. 2010;33:943–958. doi: 10.1111/j.1365-3040.2010.02118.x. [DOI] [PubMed] [Google Scholar]
- 55.Ding M.Q., Hou P.C., Shen X., Wang M.J., Deng S.R., Sun J., Xiao F., Wang R.G., Zhou X.Y., Lu C.F., et al. Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol. Biol. 2010;73:251–269. doi: 10.1007/s11103-010-9612-9. [DOI] [PubMed] [Google Scholar]
- 56.Polle A., Chen S. On the salty side of life: Molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 2015;38:1794–1816. doi: 10.1111/pce.12440. [DOI] [PubMed] [Google Scholar]
- 57.Beritognolo I., Piazzai M., Benucci S., Kuzminsky E., Sabatti M., Mugnozza G.S., Muleo R. Functional characterisation of three Italian Populus alba L. genotypes under salinity stress. Trees. 2007;21:465–477. doi: 10.1007/s00468-007-0139-x. [DOI] [Google Scholar]
- 58.Ma T., Wang J.Y., Zhou G.K., Yue Z., Hu Q.J., Chen Y., Liu B.B., Qiu Q., Wang Z., Zhang J., et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013;4:2797. doi: 10.1038/ncomms3797. [DOI] [PubMed] [Google Scholar]
- 59.Zeng F.J., Yan H.L., Arndt S.K. Leaf and whole tree adaptations to mild salinity in field grown Populus euphratica. Tree Physiol. 2009;29:1237–1246. doi: 10.1093/treephys/tpp055. [DOI] [PubMed] [Google Scholar]
- 60.Chen S.L., Li J.K., Fritzb E., Wang S.S., Huttermann A. Sodium and chloride distribution in roots and transport in three poplar genotypes under increasing NaCl stress. For. Ecol. Manag. 2002;168:217–230. doi: 10.1016/S0378-1127(01)00743-5. [DOI] [Google Scholar]
- 61.Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 62.Ottow E.A., Brinker M., Teichmann T., Fritz E., Kaiser W., Brosche M., Kangasjarvi J., Jiang X.N., Polle A. Populus euphratica displays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress. Plant Physiol. 2005;139:1762–1772. doi: 10.1104/pp.105.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang W.J., Ma X.L., Wan P., Liu L.Y. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018;495:286–291. doi: 10.1016/j.bbrc.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 64.Hasanuzzaman M., Alam M.M., Rahman A., Hasanuzzaman M., Nahar K., Fujita M. Exogenous proline and glycine betaine mediated upregulation of antioxidant defense and glyoxalase systems provides better protection against salt-induced oxidative stress in two rice (Oryza sativa L.) varieties. Biomed. Res. Int. 2014;2014:757219. doi: 10.1155/2014/757219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe S., Kojima K., Ide Y., Sasaki S. Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tiss Organ Cult. 2000;63:199–206. doi: 10.1023/A:1010619503680. [DOI] [Google Scholar]
- 66.Janz D., Polle A. Harnessing salt for woody biomass production. Tree Physiol. 2012;32:1–3. doi: 10.1093/treephys/tpr127. [DOI] [PubMed] [Google Scholar]
- 67.Apel K., Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 68.Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 69.Del Rio L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015;66:2827–2837. doi: 10.1093/jxb/erv099. [DOI] [PubMed] [Google Scholar]
- 70.Sies H. Role of metabolic H2O2 generation: Redox signaling and oxidative stress. J. Biol. Chem. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y.C., Qu G.Z., Li H.Y., Wu Y.J., Wang C., Liu G.F., Yang C.P. Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol. Biol. Rep. 2010;37:1119–1124. doi: 10.1007/s11033-009-9884-9. [DOI] [PubMed] [Google Scholar]
- 72.Zheng L.Y., Meng Y., Ma J., Zhao X.L., Cheng T.L., Ji J., Chang E.M., Meng C., Deng N., Chen L.Z., et al. Transcriptomic analysis reveals importance of ROS and phytohormones in response to short-term salinity stress in Populus tomentosa. Front Plant Sci. 2015;6:678. doi: 10.3389/fpls.2015.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gruszecki W.I., Strzalka K. Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta. 2005;1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 74.Bible A.N., Fletcher S.J., Pelletier D.A., Schadt C.W., Jawdy S.S., Weston D.J., Engle N.L., Tschaplinski T., Masyuko R., Polisetti S., et al. A Carotenoid-Deficient Mutant in Pantoea sp. YR343, a Bacteria Isolated from the Rhizosphere of Populus deltoides, Is Defective in Root Colonization. Front. Microbiol. 2016;7:491. doi: 10.3389/fmicb.2016.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen Z.D., Ding M.Q., Sun J., Deng S.R., Zhao R., Wang M.J., Ma X.J., Wang F.F., Zhang H.L., Qian Z.Y., et al. Overexpression of PeHSF mediates leaf ROS homeostasis in transgenic tobacco lines grown under salt stress conditions. Plant Cell Tiss Organ Cult. 2013;115:299–308. doi: 10.1007/s11240-013-0362-7. [DOI] [Google Scholar]
- 76.Li J.B., Zhang J., Jia H.X., Li Y., Xu X.D., Wang L.J., Lu M.Z. The Populus trichocarpa PtHSP17.8 involved in heat and salt stress tolerances. Plant Cell Rep. 2016;35:1587–1599. doi: 10.1007/s00299-016-1973-3. [DOI] [PubMed] [Google Scholar]
- 77.Poor P., Kovacs J., Borbely P., Takacs Z., Szepesi A., Tari I. Salt stress-induced production of reactive oxygen-and nitrogen species and cell death in the ethylene receptor mutant Never ripe and wild type tomato roots. Plant Physiol. Biochem. 2015;97:313–322. doi: 10.1016/j.plaphy.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 78.Corpas F.J., Palma J.M., Del Rio L.A., Barroso J.B. Protein tyrosine nitration in higher plants grown under natural and stress conditions. Front Plant Sci. 2013;4:29. doi: 10.3389/fpls.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji H.T., Pardo J.M., Batelli G., Van Oosten M.J., Bressan R.A., Li X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant. 2013;6:275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- 80.Yang Y., Tang R.J., Jiang C.M., Li B., Kang T., Liu H., Zhao N., Ma X.J., Yang L., Chen S.L., et al. Overexpression of the PtSOS2 gene improves tolerance to salt stress in transgenic poplar plants. Plant Biotechnol. J. 2015;13:962–973. doi: 10.1111/pbi.12335. [DOI] [PubMed] [Google Scholar]
- 81.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang R.J., Liu H., Bao Y., Lv Q.D., Yang L., Zhang H.X. The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 2010;74:367–380. doi: 10.1007/s11103-010-9680-x. [DOI] [PubMed] [Google Scholar]
- 83.Wu Y.X., Ding N., Zhao X., Zhao M.G., Chang Z.Q., Liu J.Q., Zhang L.X. Molecular characterization of PeSOS1: The putative Na+/H+ antiporter of Populus euphratica. Plant Mol. Biol. 2007;65:1–11. doi: 10.1007/s11103-007-9170-y. [DOI] [PubMed] [Google Scholar]
- 84.Tang R.J., Yang Y., Yang L., Liu H., Wang C.T., Yu M.M., Gao X.S., Zhang H.X. Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ. 2014;37:573–588. doi: 10.1111/pce.12178. [DOI] [PubMed] [Google Scholar]
- 85.Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 86.Zhu J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003;6:441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 87.Zhang F., Wang Y., Yang Y., Wu H., Wang D., Liu J. Involvement of hydrogen peroxide and nitric oxide in salt resistance in the calluses from Populus euphratica. Plant Cell Environ. 2007;30:775–785. doi: 10.1111/j.1365-3040.2007.01667.x. [DOI] [PubMed] [Google Scholar]
- 88.Choi J., Tanaka K., Cao Y.R., Qi Y., Qiu J., Liang Y., Lee S.Y., Stacey G. Identification of a Plant Receptor for Extracellular ATP. Science. 2014;343 doi: 10.1126/science.343.6168.290. [DOI] [PubMed] [Google Scholar]
- 89.Sun J., Zhang X., Deng S.R., Zhang C.L., Wang M.J., Ding M.Q., Zhao R., Shen X., Zhou X.Y., Lu C.F., et al. Extracellular ATP signaling is mediated by H2O2and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS ONE. 2012;7:e53136. doi: 10.1371/journal.pone.0053136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang S.J., Zhou B.R., Yao W.J., Jiang T.B. PsnERF75 Transcription Factor from Populus simonii × P. nigra Confers Salt Tolerance in Transgenic Arabidopsis. J. Plant Biol. 2018;61:61–71. doi: 10.1007/s12374-017-0450-z. [DOI] [Google Scholar]
- 91.Yao W.J., Wang S.J., Zhou B.R., Jiang T.B. Transgenic poplar overexpressing the endogenous transcription factor ERF76 gene improves salinity tolerance. Tree Physiol. 2016;36:896–908. doi: 10.1093/treephys/tpw004. [DOI] [PubMed] [Google Scholar]
- 92.Wang Z.L., Liu J., Guo H.Y., He X., Wu W.B., Du J.C., Zhang Z.Y., An X.M. Characterization of two highly similar CBF/DREB1-like genes, PhCBF4a and PhCBF4b, in Populus hopeiensis. Plant Physiol. Biochem. 2014;83:107–116. doi: 10.1016/j.plaphy.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 93.Zhao H., Zhao X.Y., Li M.Y., Jiang Y., Xu J.Q., Jin J.J., Li K.L. Ectopic expression of Limonium bicolor (Bag.) Kuntze DREB (LbDREB) results in enhanced salt stress tolerance of transgenic Populus ussuriensis Kom. Plant Cell Tiss Organ Cult. 2017;132:123–136. doi: 10.1007/s11240-017-1317-1. [DOI] [Google Scholar]
- 94.Zhou M.L., Ma J.T., Zhao Y.M., Wei Y.H., Tang Y.X., Wu Y.M. Improvement of drought and salt tolerance in Arabidopsis and Lotus corniculatus by overexpression of a novel DREB transcription factor from Populus euphratica. Gene. 2012;506:10–17. doi: 10.1016/j.gene.2012.06.089. [DOI] [PubMed] [Google Scholar]
- 95.Speth E.B., Imboden L., Hauck P., He S.Y. Subcellular Localization and Functional Analysis of the Arabidopsis GTPase RabE. Plant Physiol. 2009;149:1824–1837. doi: 10.1104/pp.108.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang J., Li Y., Liu B.L., Wang L.J., Zhang L., Hu J.J., Chen J., Zheng H.Q., Lu M.Z. Characterization of the Populus Rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 2018;18:124. doi: 10.1186/s12870-018-1342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perez-Salamo I., Papdi C., Rigo G., Zsigmond L., Vilela B., Lumbreras V., Nagy I., Horvath B., Domoki M., Darula Z., et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014;165:319–334. doi: 10.1104/pp.114.237891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li F., Zhang H., Zhao H., Gao T., Song A., Jiang J., Chen F., Chen S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018;16:1311–1321. doi: 10.1111/pbi.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoon S.K., Bae E.K., Lee H., Choi Y.L., Han M., Choi H., Kang K.S., Park E.J. Downregulation of stress-associated protein 1 (PagSAP1) increases salt stress tolerance in poplar (Populus alba × P. glandulosa) Trees. 2018;32:823–833. doi: 10.1007/s00468-018-1675-2. [DOI] [Google Scholar]
- 100.Xue S.W., Yao X., Luo W., Jha D., Tester M., Horie T., Schroeder J.I. AtHKT1;1 mediates nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS ONE. 2011;6:e24725. doi: 10.1371/journal.pone.0024725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng N.H., Pittman J.K., Zhu J.K., Hirschi K.D. The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 2004;279:2922–2926. doi: 10.1074/jbc.M309084200. [DOI] [PubMed] [Google Scholar]
- 102.Baxter A., Mittler R., Suzuki N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014;65:1229–1240. doi: 10.1093/jxb/ert375. [DOI] [PubMed] [Google Scholar]
- 103.Huertas R., Olias R., Eljakaoui Z., Galvez F.J., Li J., De Morales P.A., Belver A., Rodriguez-Rosales M.P. Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 2012;35:1467–1482. doi: 10.1111/j.1365-3040.2012.02504.x. [DOI] [PubMed] [Google Scholar]
- 104.Qiu Q.S., Guo Y., Quintero F.J., Pardo J.M., Schumaker K.S., Zhu J.K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 2004;279:207–215. doi: 10.1074/jbc.M307982200. [DOI] [PubMed] [Google Scholar]
- 105.Zhou J., Wang J.J., Bi Y.F., Wang L.K., Tang L.Z., Yu X., Ohtani M., Demura T., Zhu Ge Q. Overexpression of PtSOS2 Enhances Salt Tolerance in Transgenic Poplars. Plant Mol. Biol. Rep. 2014;32:185–197. doi: 10.1007/s11105-013-0640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li D.D., Song S.Y., Xia X.L., Yin W.L. Two CBL genes from Populus euphratica confer multiple stress tolerance in transgenic triploid white poplar. Plant Cell Tiss Organ Cult. 2012;109:477–489. doi: 10.1007/s11240-011-0112-7. [DOI] [Google Scholar]
- 107.Gao W., Bai S., Li Q., Gao C., Liu G., Li G., Tan F. Overexpression of TaLEA Gene from Tamarix androssowii improves salt and drought tolerance in transgenic Poplar (Populus simonii × P. nigra) PLoS ONE. 2013;8:e67462. doi: 10.1371/journal.pone.0067462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun Y.S., Chen S., Huang H.J., Jiang J., Bai S., Liu G.F. Improved salt tolerance of Populus davidiana × P. bolleana overexpressed LEA from Tamarix androssowii. J. For. Res.-JPN. 2014;25:813–818. doi: 10.1007/s11676-014-0529-z. [DOI] [Google Scholar]
- 109.Li Y.L., Su X.H., Zhang B.Y., Huang Q.J., Zhang X.H., Huang R.F. Expression of jasmonic ethylene responsive factor gene in transgenic poplar tree leads to increased salt tolerance. Tree Physiol. 2009;29:273–279. doi: 10.1093/treephys/tpn025. [DOI] [PubMed] [Google Scholar]
- 110.Du N.X., Liu X., Li Y., Chen S.Y., Zhang J.S., Ha D., Deng W.G., Sun C.K., Zhang Y.Z., Pijut P.M. Genetic transformation of Populus tomentosa to improve salt tolerance. Plant Cell Tiss Organ Cult. 2011;108:181–189. doi: 10.1007/s11240-011-0026-4. [DOI] [Google Scholar]
- 111.Lawson S.S., Michler C.H. Overexpression of AtSTO1 leads to improved salt tolerance in Populus tremula × P. alba. Transgenic Res. 2014;23:817–826. doi: 10.1007/s11248-014-9808-x. [DOI] [PubMed] [Google Scholar]
- 112.Zhang T.T., Song Y.Z., Liu Y.D., Guo X.Q., Zhu C.X., Wen F.J. Overexpression of phospholipase Dα gene enhances drought and salt tolerance of Populus tomentosa. Chin. Sci. Bull. 2008;53:3656–3665. doi: 10.1007/s11434-008-0476-1. [DOI] [Google Scholar]
- 113.Yu X., Ohtani M., Kusano M., Nishikubo N., Uenoyama M., Umezawa T., Saito K., Shinozaki K., Demura T. Enhancement of abiotic stress tolerance in poplar by overexpression of key Arabidopsis stress response genes, AtSRK2C and AtGolS2. Mol. Breed. 2017;37 doi: 10.1007/s11032-016-0618-0. [DOI] [Google Scholar]
- 114.Chen S., Polle A. Salinity tolerance of Populus. Plant Biol. 2010;12:317–333. doi: 10.1111/j.1438-8677.2009.00301.x. [DOI] [PubMed] [Google Scholar]
- 115.Janz D., Behnke K., Schnitzler J.P., Kanawati B., Schmitt Kopplin P., Polle A. Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biol. 2010;10:150. doi: 10.1186/1471-2229-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Podda A., Pisuttu C., Hoshika Y., Pellegrini E., Carrari E., Lorenzini G., Nali C., Cotrozzi L., Zhang L., Baraldi R., et al. Can nutrient fertilization mitigate the effects of ozone exposure on an ozone-sensitive poplar clone? Sci. Total Environ. 2019;657:340–350. doi: 10.1016/j.scitotenv.2018.11.459. [DOI] [PubMed] [Google Scholar]