Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) gene is an attractive target for gene editing approaches, which may yield novel therapeutic approaches for genetic diseases such as cystic fibrosis (CF). However, for gene editing to be effective, aspects of the three-dimensional (3D) structure and cis-regulatory elements governing the dynamic expression of CFTR need to be considered. In this review, we focus on the higher order chromatin organization required for normal CFTR locus function, together with the complex mechanisms controlling expression of the gene in different cell types impaired by CF pathology. Across all cells, the CFTR locus is organized into an invariant topologically associated domain (TAD) established by the architectural proteins CCCTC-binding factor (CTCF) and cohesin complex. Additional insulator elements within the TAD also recruit these factors. Although the CFTR promoter is required for basal levels of expression, cis-regulatory elements (CREs) in intergenic and intronic regions are crucial for cell-specific and temporal coordination of CFTR transcription. These CREs are recruited to the promoter through chromatin looping mechanisms and enhance cell-type-specific expression. These features of the CFTR locus should be considered when designing gene-editing approaches, since failure to recognize their importance may disrupt gene expression and reduce the efficacy of therapies.
Keywords: CFTR, cis-regulatory elements, enhancers, chromatin architecture, transcription factors, gene editing, CRISPR/Cas9
1. Introduction
The cystic fibrosis transmembrane conductance regulator (CFTR) gene was one of the first genes that was shown to be regulated by elements outside its promoter [1,2,3,4]. At the time, the concept of critical cis-regulatory elements located in introns and intergenic regions was not widely accepted and still challenged dogma that introns contained unimportant DNA sequence. Of course, the Encyclopedia of DNA Elements (ENCODE) project [5,6,7] and many other advances, driven in large part by new technologies based on next generation sequencing protocols, have dramatically altered understanding of genome organization. CFTR is a large gene encompassing 189 kb at chromosome 7q31.2 [8]. Although necessary to drive basal levels of gene expression, the CFTR promoter is relatively weak and appears to lack tissue-specific control elements. The sequence is CpG-rich, contains no TATA box, has multiple transcription start sites (TSS) and has many binding sites for the transcription factor specificity protein 1 (Sp1) [9,10,11]. Despite this, CFTR expression is tightly regulated both during development and within different tissue types [12,13,14,15]. CFTR transcript levels are highly variable between different cell types, suggesting that the mechanisms controlling CFTR expression may diverge between them.
Cystic fibrosis transmembrane conductance regulator expression was initially thought to be restricted to epithelial cells, specifically epithelial cells within the organs affected by cystic fibrosis (CF) pathology such as the lung, intestine, pancreas, and reproductive tract [13,16,17,18,19]. However, many studies have shown that CFTR may also be expressed in non-epithelial cells [20,21]. Additionally, CFTR is transcribed in the central, peripheral, and enteric nervous systems [22,23,24,25,26,27,28,29,30,31,32]. Also, Schwann cells were reported to express CFTR and CFTR-deficient pigs were suggested to have peripheral nervous system (PNS) deficiencies [30]. Although CFTR is expressed in many different cell types, both epithelial and non-epithelial, this review will focus on the regulatory mechanisms controlling expression of the gene in epithelial cells as they are best studied. Here, we discuss both older seminal data and more recent advances that define the chromatin architecture of the CFTR locus, reveal multiple cell-type selective cis-regulatory elements within and adjacent to the locus, and identify key activating and repressive transcription factors (TFs). These data have renewed importance at a time when gene editing and replacement are being considered among novel therapeutic approaches for CF. Although CFTR is also regulated by post-transcriptional mechanisms including microRNAs, some of which directly target sequences in the 3′ untranslated region (UTR) of the gene, these will not be considered further here as they are reviewed elsewhere [14].
2. Common Features of the CFTR Locus in All Cell Types
2.1. The CFTR Locus Is Organized Within a Topologically Associating Domain
The three-dimensional (3D) chromatin structure has a dynamic and essential role in the regulation of gene expression. On a fine scale, gene regulation occurs at least in part through the physical looping of regulatory elements, such as enhancers to their gene promoters. These looping interactions are thought to be cell-type and locus-specific [33]. On a broader scale, chromatin is organized into topologically associating domains (TADs). TADs are self-associating genomic regions; cis-regulatory elements within one TAD have little to no interaction with genes in neighboring TADs. Therefore, TAD boundaries may represent physical insulators for the genes and regulatory elements contained between them [34,35,36]. These long-range chromatin interactions are measured by many techniques: chromosome conformation capture (3C) [37], circular chromosome conformation capture (4C) and deep sequencing [38], chromosome conformation capture carbon copy (5C) and deep sequencing [39], HiC [40], and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) [41]. The 3D interactions at the CFTR locus were first shown by 3C [42,43,44,45]. Building on these data, 4C-seq demonstrated that the CFTR locus is organized into a single TAD with boundaries at −80.1 kb 5′ to the translational start site and +48.9 kb from the translational stop site [46]. These data were confirmed independently by 5C-seq [47,48] and the TAD boundaries were shown to be invariant between cell types [46,47]. Consistent with other TAD boundaries, significant occupancy of the CCCTC-binding factor (CTCF) was observed at the −80.1 kb and +48.9 kb sites [49]. CTCF is an architectural protein involved in chromatin organization that binds to insulator elements and marks TAD boundaries [50,51,52].
2.2. The CFTR Locus Contains CTCF-Bound Insulator Elements
In addition to its role in the TAD structure, CTCF may occupy several insulator elements at the CFTR locus. These elements, which can block the interactions between an enhancer and a gene promoter, are located at −20.9 kb relative to the translational start site and at +6.8 kb and +15.6 kb to the translational stop site. The sites containing the CFTR insulators were initially identified using DNase I hypersensitivity mapping and DNase-seq [42,43,44,53]. CTCF was shown by chromatin immunoprecipitation (ChIP) to occupy the −20.9 kb and +6.8 kb insulators, but not to bind at +15.6 kb [42,53]. The insulator function of +15.6 kb may involve nuclear hormone receptors [53]. The cohesin complex, which occupies a subset of CTCF sites, was also seen to bind at several of the CFTR insulator elements, using an antibody specific for the Rad21 component of the complex [42,43,44]. Of note, both the +6.8 kb site and other more distal 3′ elements were cell-type selective [54], consistent with a subset of variant CTCF sites genome wide. Like CTCF, the cohesin complex is involved in chromatin looping and organization. CTCF works in concert with cohesin at ~60%–70% of its sites across the genome [55,56,57]. To determine the significance of CTCF and/or cohesin occupancy in the organization of the CFTR locus, small interfering RNA (siRNA)-mediated depletion of both architectural proteins was performed in an intestinal epithelial cell line (Caco2). Loss of CTCF greatly reduced both CTCF and cohesin complex occupancy across the CFTR locus, whereas depletion of Rad21 had little impact on CTCF binding. These results suggest the architectural proteins may not always function together, as is observed elsewhere in the genome. Furthermore, CTCF was shown to have a dominant effect on mediating higher order looping of the CFTR locus, while cohesin complex was crucial in maintaining stability of the 3D looping at the locus [49]. Interestingly, clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated deletion of −20.9 enhanced recruitment of CTCF at adjacent sites and thus had little effect on CFTR expression [46].
3. Cell-Type-Selective CFTR Regulatory Mechanisms
The TAD encompassing the CFTR locus is seen in all cells. However, CFTR expression is tightly regulated in a cell-type-specific manner due to the recruitment of different cis-regulatory elements (CREs) within and nearby the locus. The interaction between these CREs and the gene promoter generates cell-type-selective 3D conformations of the locus (Figure 1). Here, we discuss some of the known regulatory elements and their activating TFs in different cell types and also consider the significance of the CREs in the context of potential novel therapeutics. Of note, based on our extensive analysis of open chromatin and histone modifications in CF-relevant epithelial cells, there are also other CREs (Figure 2), which are not yet fully understood.
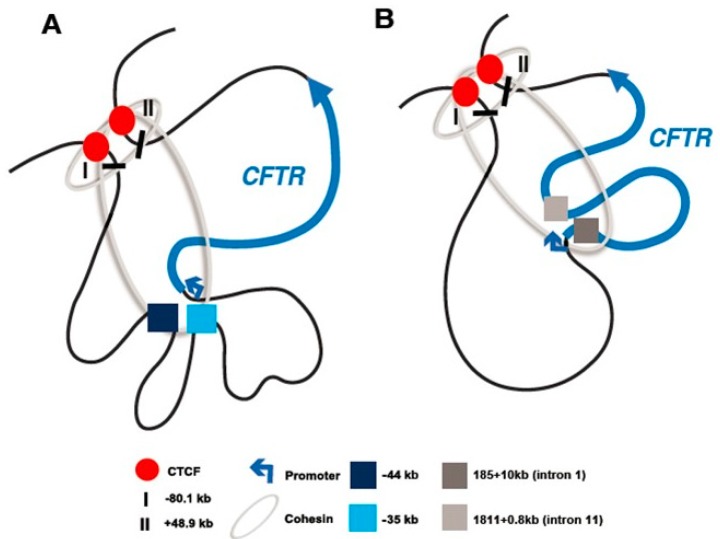
Figure 1.
Schematic of cystic fibrosis transmembrane conductance regulator (CFTR) topologically associated domain (TAD) in airway and intestinal cell types.CFTR promoter (blue arrow) and gene are shown. TAD boundaries are denoted as “I” (−80.1 kb) and “II” (+48.9 kb). Occupancy of CCCTC-binding factor (CTCF) at the TAD boundaries is shown as red circles. Cohesin complex is shown as gray rings. (A) Airway-selective enhancer elements at −44 kb and −35 kb are shown by dark and light blue boxes, respectively. (B) Intestine-selective enhancer elements at 185 + 10kb (intron 1) and 1811 + 0.8kb (intron 11) are shown by dark and light gray boxes, respectively. Figures are not drawn to scale.
Figure 2.
CFTR locus and important functional elements. University of California Santa Cruz (UCSC) genome browser displaying CFTR and nearby genes. Airway-selective enhancer elements at −35 kb and −44 kb are shown in dark purple. Intestinal-selective enhancer elements 185 + 10kb (intron 1) and 1811 + 0.8kb (intron 11) are shown in medium purple. Insulator elements at −20.9 kb and + 15.6 kb are shown in dark blue. TAD boundaries at −80.1 kb and + 48.9 kb are shown in medium blue. CFTR promoter is shown in light purple. Other key DNase I hypersensitive sites (DHS) are shown in black.
3.1. Airway
The major cause of reduced lifespan in CF patients is lung disease, hence the gene-editing protocols that are currently in development are likely to be targeted primarily at this tissue. In this context, understanding the regulation of CFTR expression in the airway epithelium, and how this might be disrupted by gene editing protocols, may be critical. Using DNase-chip and subsequently DNase-seq, many airway-selective DNase I hypersensitive sites (DHS) were discovered in lung cell lines that were also seen in primary human airway epithelial cells. These include: DHS at −44 kb, −35 kb, −3.4 kb, in intron 18, 19 and 23, and at +21.5 kb and +36.6 kb (3′ to last exon) [43,44,54]. The DHS at −44 kb and −35 kb were studied in detail to reveal the functions of the CREs they contain. Both have enhancer activity on the CFTR promoter in luciferase reporter gene assays and they appear to function cooperatively [43,44,54,58,59], but their enhancer activities are driven by different mechanisms. The core of the −35 kb DHS was mapped using DNase I footprinting and subsequently shown by ChIP to bind the immune mediators interferon regulatory factor 1 and 2 (IRF1/2) and the nuclear factor Y (NF-Y) TF [58]. Additionally, ChIP-seq revealed that this cis-element is enriched for the active histone mark histone 3 lysine 4 monomethylation (H3K4me1) [60]. NF-Y occupancy is required for maintenance of the H3K4me1 modification and so is likely necessary for the enhancer activity of the −35 kb CRE [58]. In contrast, the −44 kb enhancer element was uniquely activated by oxidative stress. It contains an antioxidant response element (ARE), which under normal conditions is occupied by the repressor BTB and CNC homology 1, basic leucine zipper TF (Bach1), and v-Maf avian musculoaponeurotic fibrosarcoma oncogene homolog K (MafK) heterodimers. However, upon exposure to oxidative stress, these repressive factors were displaced by the nuclear factor erythroid 2-like 2 (Nrf2) and CFTR expression was activated [59]. This is particularly relevant to CF, as oxidative stress is a hallmark of lung disease pathology [61].
Although there are at least two airway-selective enhancers of CFTR, expression in the majority of cells in lung epithelium is significantly lower (~1000–10,000 fold) than that in the intestinal and pancreatic duct epithelium [43,44,62]. This limited transcript abundance in the airway makes designing therapeutics to correct the defective protein quite challenging. A potential explanation for these low levels of CFTR transcript in the lung is that repressive TFs are being recruited to the locus in an airway-selective manner. In order to identify transcriptional repressors of CFTR in the lung, a siRNA screen was used to deplete ~1500 TFs in a lung adenocarcinoma cell line (Calu-3) [63]. About 50 TFs were, upon depletion, found to elevate CFTR transcript levels by at least 2-fold in replicate screens. Among these, knockdown of bromodomain-containing protein 8 (BRD8), ets homologous factor (EHF), krüppel-like factor 5 (KLF5), inhibitor of growth protein 2 (ING2), and nuclear receptor subfamily 2 group F member 2 (NR2F2) had the most robust impact on CFTR transcript and CFTR protein levels. Moreover, several of these TFs were also shown to repress CFTR in primary human bronchial epithelial (HBE) cells. Of note, both EHF and KLF5 were subsequently shown by ChIP to occupy the −35 kb enhancer element and so may directly repress CFTR expression through this site [63]. Understanding these repressive factors is essential, as potential therapeutics could also target their interactions at the CFTR CREs.
In contrast to the majority of cells in the airway surface epithelium discussed above, one rare cell type expresses very high levels of CFTR. Though they were observed many years ago by mRNA in situ hybridization and immunofluorescence [64,65,66], the likely function of these cells was recently revealed by single-cell RNA sequencing (scRNA-seq). ScRNA-seq of primary human bronchial epithelial (HBE) cells and mouse tracheal epithelial cells documented a high-CFTR expressing cell type named “pulmonary ionocytes”. Although these cells only comprise 0.5–1.5% of the airway epithelium, pulmonary ionocytes were shown to be responsible for the majority of CFTR activity (~54% in mouse, ~60% in human). Additionally, these cells were shown to co-express the transcription factor forkhead box I1 (FOXI1). FOXI1 expression was suggested to be necessary for CFTR expression and loss of FOXI1 was also associated with recognizable CF phenotypes such mucus viscosity and altered fluid composition [67,68]. However, the regulatory mechanisms driving high CFTR expression in ionocytes are currently unknown, though in silico predictions (unpublished) suggest they are divergent from other airway epithelial cells. Further experimental data are required to confirm this.
3.2. Intestine
Multiple organs in the digestive system are profoundly impaired by loss of CFTR function including the pancreas, bile duct, and intestinal epithelia, among others. As noted above, CFTR expression in all these cell types is significantly higher than in the majority of lung epithelial cells. This differential expression is achieved by cell-type selective cis-regulatory elements and their activating TFs. The two best-characterized CREs controlling CFTR expression in the intestinal epithelium are intronic: intron 1 (185 + 10 kb) and intron 11 (1811 + 0.8 kb), though other elements both within and outside the gene are also involved.
The 185 + 10 kb intronic CRE is located in a DHS ~10 kb 3′ to end of exon 1 (185 is the last coding base of exon 1) and was found by classical DHS mapping using Southern blots. Its enhancer activity was shown by luciferase assays in the colon carcinoma cell line Caco2 [3]. This CRE was then assayed in the genomic context using a yeast artificial chromosome (YAC) that contained the entire CFTR gene [2]. When introduced into Caco2 cells, CFTR expression from a YAC lacking the 185 +10 kb DHS (deleted by recombineering), was significantly reduced compared to a control YAC containing the intact CFTR gene [2]. To identify the transcription factors governing the enhancer activity of this CRE, DNase I footprinting and electromobility shift assays (EMSA) were performed [69]. These experiments and subsequent ChIP assays showed hepatocyte nuclear factor 1α (HNF1α) binding to the intron 1 185 + 10 kb CRE both in vitro and in vivo [43]. Furthermore, 3C analysis confirmed direct interaction of this enhancer with the CFTR promoter [43].
Another critical intestinal-specific enhancer of CFTR expression is located in intron 11 (legacy nomenclature) at 1811 + 0.8 kb (1811 is the last coding base in exon 11). This enhancer was first identified by DNase-chip within a 1.5 kb DHS in intron 11 [44]. It was also shown to recruit p300, cooperate with other intestinal enhancer elements within CFTR and interact with the gene promoter through direct chromosomal looping [44]. Among activating TFs for the intron 11 (1811 + 0.8 kb) CRE are forkhead box protein A1/A2 (FOXA1/A2), hepatocyte nuclear factor 1 homeobox A (HNF1α), and caudal type homeobox 2 (CDX2) [70,71]. These factors were shown to be essential for maintaining high levels of CFTR expression in Caco2 cells [70].
Although the enhancer elements governing intestinal-selective CFTR expression were extensively studied in colon carcinoma cell lines, these may be somewhat influenced by the properties of cancer cells. However, our recent studies (Yin et al., unpublished) suggest there is substantial overlap between the cell line CREs and sites of open chromatin in intestinal organoids, which provide a robust in vitro model of the normal intestinal epithelium [65,72].
3.3. Pancreas and Liver
CFTR expression is perhaps more abundant in the pancreatic duct epithelium than in any other cell type, consistent with loss of CFTR being associated with profound pancreatic dysfunction. However, very few pancreatic adenocarcinoma cell lines express the CFTR gene and so detailed analysis of CREs in this cell type has lagged behind the airway and intestinal epithelium. Earlier work in Capan 1 cells identified DHS in introns 16, 17a, 18, and 20 [4]. In vitro experiments (DNase I footprinting and EMSAs) suggested HNF1, CDX2, and PBX1 bound to the CREs in intron 16 and 17a, however, only PBX1 was shown by ChIP to occupy these sites in Capan 1 cells in vivo [73]. Pancreatic duct cell-specific regulatory mechanisms for CFTR are currently being re-examined using functional genomics protocols. Cystic fibrosis liver disease (CFLD) is the third leading cause of mortality in CF patients. In the liver, CFTR is expressed at the apical membrane of cholangiocytes within the bile ducts [74] and defective CFTR results in impaired biliary secretion and ductal cholestasis [75]. To date, cholangiocyte-selective CREs for CFTR expression have not been investigated.
3.4. Male Reproductive Tract
Nearly all (~97%) men with CF are infertile, though not sterile. Loss of CFTR is associated with absence of intact genital ducts, which may be due to early duct obstruction, or a developmental defect impairing duct formation. Congenital bilateral absence of the vas deferens (CVAD) is a common CF-associated diagnosis. The epithelial lining of the epididymis maintains an appropriate luminal environment that is crucial for sperm maturation [76] and CFTR is integral to its function. To determine CREs for CFTR in epididymis epithelial cells, we performed DNase-chip on immature human epididymis epithelial cells [44] and subsequently DNase-seq of adult primary human epididymis epithelial (HEE) cells [46] and immortalized, immature epididymis epithelial (REP) cells [77]. These data showed peaks of open chromatin at the CFTR locus at a subset of both intestinal and airway CREs, together with novel sites. CFTR transcripts are abundant in epididymis epithelial cells [44,78,79] and possibly multiple enhancers are being recruited to drive these high expression levels. Of note, the TFs driving these enhancers may also be different in epididymis cells from the same elements in the intestinal and airway cells. For example, the main form of hepatocyte nuclear factor 1 in intestinal epithelial cells is HNF1α, which enhances CFTR expression through multiple cis-elements. In HEE cells, HNF1β is the dominant form and HNF1β ChIP-seq data showed its occupancy at multiple CFTR CREs [80]. CFTR expression in HEE cells may also be under the control of the androgen receptor [81]. Hormonal control of the CFTR locus is not extensively studied to date [82,83].
4. Regulation of CFTR Expression and its Impact on Gene Editing
Recent advances in the field of gene editing have suggested these protocols as potential treatments for cystic fibrosis, among other diseases. The CRISPR/Cas9 system, which can make targeted double-stranded cuts in DNA, allows for the modification or deletion of any site in the genome [84,85,86]. More recently, the use of base editors is rapidly expanding the field [87,88,89,90,91,92,93]. However, it is not yet known which aspects of the critical 3D structure and interactions at the CFTR locus could be disrupted by direct therapeutic modification of the locus. CRISPR/Cas9 protocols have already provided important insights into the functions of CREs and CTCF sites at the locus [46]. The potential to use gene editing to target the CFTR gene, CF modifier genes [94,95,96,97,98], or their regulatory elements opens new therapeutic avenues [99]. These may be targeted to accessible sites and specific cell types primarily in the airway and perhaps in the future to other organs affected by CF. Once safety and ethical concerns are overcome, gene editing approaches may also need to account for higher order chromatin structure and the cell-type-selective regulatory networks of CFTR, so therapeutic efficacy is not impaired.
Author Contributions
H.S. wrote the first draft of this review, which was subsequently edited by H.S. and A.H.
Funding
Research on regulation of CFTR gene expression in the Harris laboratory is currently supported by grants from the National Institutes of Health R01 HL095485 and the Cystic Fibrosis Foundation (Harris16G0, 17XX0, 18P0).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Nuthall H.N., Moulin D.S., Huxley C., Harris A. Analysis of DNase-I-hypersensitive sites at the 3′ end of the cystic fibrosis transmembrane conductance regulator gene (CFTR) Pt 3Biochem. J. 1999;341:601–611. doi: 10.1042/bj3410601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowntree R.K., Vassaux G., McDowell T.L., Howe S., McGuigan A., Phylactides M., Huxley C., Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001;10:1455–1464. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
- 3.Smith A.N., Barth M.L., McDowell T.L., Moulin D.S., Nuthall H.N., Hollingsworth M.A., Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 4.Smith D.J., Nuthall H.N., Majetti M.E., Harris A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics. 2000;64:90–96. doi: 10.1006/geno.1999.6086. [DOI] [PubMed] [Google Scholar]
- 5.The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 6.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rommens J.M., Iannuzzi M.C., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 9.Chou J.L., Rozmahel R., Tsui L.C. Characterization of the promoter region of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1991;266:24471–24476. [PubMed] [Google Scholar]
- 10.Koh J., Sferra T.J., Collins F.S. Characterization of the cystic fibrosis transmembrane conductance regulator promoter region. Chromatin context and tissue-specificity. J. Biol. Chem. 1993;268:15912–15921. [PubMed] [Google Scholar]
- 11.Yoshimura K., Nakamura H., Trapnell B.C., Dalemans W., Pavirani A., Lecocq J.P., Crystal R.G. The cystic fibrosis gene has a “housekeeping”-type promoter and is expressed at low levels in cells of epithelial origin. J. Biol. Chem. 1991;266:9140–9144. [PubMed] [Google Scholar]
- 12.Broackes-Carter F.C., Mouchel N., Gill D., Hyde S., Bassett J., Harris A. Temporal regulation of CFTR expression during ovine lung development: Implications for CF gene therapy. Hum. Mol. Genet. 2002;11:125–131. doi: 10.1093/hmg/11.2.125. [DOI] [PubMed] [Google Scholar]
- 13.Crawford I., Maloney P.C., Zeitlin P.L., Guggino W.B., Hyde S.C., Turley H., Gatter K.C., Harris A., Higgins C.F. Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc. Natl. Acad. Sci. USA. 1991;88:9262–9266. doi: 10.1073/pnas.88.20.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosalia N., Harris A. Chromatin Dynamics in the Regulation of CFTR Expression. Genes. 2015;6:543–558. doi: 10.3390/genes6030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trezise A.E., Buchwald M. In vivo cell-specific expression of the cystic fibrosis transmembrane conductance regulator. Nature. 1991;353:434–437. doi: 10.1038/353434a0. [DOI] [PubMed] [Google Scholar]
- 16.Denning G.M., Ostedgaard L.S., Cheng S.H., Smith A.E., Welsh M.J. Localization of cystic fibrosis transmembrane conductance regulator in chloride secretory epithelia. J. Clin. Investig. 1992;89:339–349. doi: 10.1172/JCI115582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernald G.W., Roberts M.W., Boat T.F. Cystic fibrosis: A current review. Pediatr. Dent. 1990;12:72–78. [PubMed] [Google Scholar]
- 18.Harris A., Chalkley G., Goodman S., Coleman L. Expression of the cystic fibrosis gene in human development. Development. 1991;113:305–310. doi: 10.1242/dev.113.1.305. [DOI] [PubMed] [Google Scholar]
- 19.Quinton P.M. Physiological basis of cystic fibrosis: A historical perspective. Physiol. Rev. 1999;79:S3–S22. doi: 10.1152/physrev.1999.79.1.S3. [DOI] [PubMed] [Google Scholar]
- 20.Kulka M., Gilchrist M., Duszyk M., Befus A.D. Expression and functional characterization of CFTR in mast cells. J. Leukoc. Biol. 2002;71:54–64. [PubMed] [Google Scholar]
- 21.Yoshimura K., Nakamura H., Trapnell B.C., Chu C.S., Dalemans W., Pavirani A., Lecocq J.P., Crystal R.G. Expression of the cystic fibrosis transmembrane conductance regulator gene in cells of non-epithelial origin. Nucleic Acids Res. 1991;19:5417–5423. doi: 10.1093/nar/19.19.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y., Su M., McNutt M.A., Gu J. Expression and distribution of cystic fibrosis transmembrane conductance regulator in neurons of the human brain. J. Histochem. Cytochem. 2009;57:1113–1120. doi: 10.1369/jhc.2009.953455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y., Su M., Su M., McNutt M.A., Gu J. Expression and distribution of cystic fibrosis transmembrane conductance regulator in neurons of the spinal cord. J. Neurosci. Res. 2009;87:3611–3619. doi: 10.1002/jnr.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johannesson M., Bogdanovic N., Nordqvist A.C., Hjelte L., Schalling M. Cystic fibrosis mRNA expression in rat brain: Cerebral cortex and medial preoptic area. Neuroreport. 1997;8:535–539. doi: 10.1097/00001756-199701200-00031. [DOI] [PubMed] [Google Scholar]
- 25.Mulberg A.E., Resta L.P., Wiedner E.B., Altschuler S.M., Jefferson D.M., Broussard D.L. Expression and localization of the cystic fibrosis transmembrane conductance regulator mRNA and its protein in rat brain. J. Clin. Investig. 1995;96:646–652. doi: 10.1172/JCI118080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulberg A.E., Weyler R.T., Altschuler S.M., Hyde T.M. Cystic fibrosis transmembrane conductance regulator expression in human hypothalamus. Neuroreport. 1998;9:141–144. doi: 10.1097/00001756-199801050-00028. [DOI] [PubMed] [Google Scholar]
- 27.Mulberg A.E., Wiedner E.B., Bao X., Marshall J., Jefferson D.M., Altschuler S.M. Cystic fibrosis transmembrane conductance regulator protein expression in brain. Neuroreport. 1994;5:1684–1688. doi: 10.1097/00001756-199408150-00035. [DOI] [PubMed] [Google Scholar]
- 28.Niu N., Zhang J., Guo Y., Yang C., Gu J. Cystic fibrosis transmembrane conductance regulator expression in human spinal and sympathetic ganglia. Lab. Investig. 2009;89:636–644. doi: 10.1038/labinvest.2009.28. [DOI] [PubMed] [Google Scholar]
- 29.Pan J., Bear C., Farragher S., Cutz E., Yeger H. Cystic fibrosis transmembrane conductance regulator modulates neurosecretory function in pulmonary neuroendocrine cell-related tumor cell line models. Am. J. Respir. Cell Mol. Biol. 2002;27:553–560. doi: 10.1165/rcmb.4843. [DOI] [PubMed] [Google Scholar]
- 30.Reznikov L.R., Dong Q., Chen J.H., Moninger T.O., Park J.M., Zhang Y., Du J., Hildebrand M.S., Smith R.J., Randak C.O., et al. CFTR-deficient pigs display peripheral nervous system defects at birth. Proc. Natl. Acad. Sci. USA. 2013;110:3083–3088. doi: 10.1073/pnas.1222729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su M., Guo Y., Zhao Y., Korteweg C., Gu J. Expression of cystic fibrosis transmembrane conductance regulator in paracervical ganglia. Biochem. Cell Biol. 2010;88:747–755. doi: 10.1139/O10-016. [DOI] [PubMed] [Google Scholar]
- 32.Xue R., Gu H., Qiu Y., Guo Y., Korteweg C., Huang J., Gu J. Expression of Cystic Fibrosis Transmembrane Conductance Regulator in Ganglia of Human Gastrointestinal Tract. Sci. Rep. 2016;6:30926. doi: 10.1038/srep30926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 38.Splinter E., de Wit E., van de Werken H.J., Klous P., de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods. 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H., et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blackledge N.P., Ott C.J., Gillen A.E., Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott C.J., Suszko M., Blackledge N.P., Wright J.E., Crawford G.E., Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell Mol. Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. USA. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang R., Kerschner J.L., Gosalia N., Neems D., Gorsic L.K., Safi A., Crawford G.E., Kosak S.T., Leir S.H., Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–3094. doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith E.M., Lajoie B.R., Jain G., Dekker J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. Am. J. Hum. Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moisan S., Berlivet S., Ka C., Le Gac G., Dostie J., Ferec C. Analysis of long-range interactions in primary human cells identifies cooperative CFTR regulatory elements. Nucleic Acids Res. 2016;44:2564–2576. doi: 10.1093/nar/gkv1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gosalia N., Neems D., Kerschner J.L., Kosak S.T., Harris A. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014;42:9612–9622. doi: 10.1093/nar/gku648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonev B., Cavalli G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 51.Ghirlando R., Felsenfeld G. CTCF: Making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ong C.T., Corces V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blackledge N.P., Carter E.J., Evans J.R., Lawson V., Rowntree R.K., Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z., Ott C.J., Lewandowska M.A., Leir S.H., Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell Mol. Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z., Leir S.H., Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol. Cell Biol. 2013;33:2843–2853. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z., Leir S.H., Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am. J. Respir. Cell Mol. Biol. 2015;52:387–396. doi: 10.1165/rcmb.2014-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 61.Hector A., Griese M., Hartl D. Oxidative stress in cystic fibrosis lung disease: An early event, but worth targeting? Eur. Respir. J. 2014;44:17–19. doi: 10.1183/09031936.00038114. [DOI] [PubMed] [Google Scholar]
- 62.Riordan J.R., Rommens J.M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 63.Mutolo M.J., Leir S.H., Fossum S.L., Browne J.A., Harris A. A transcription factor network represses CFTR gene expression in airway epithelial cells. Biochem. J. 2018;475:1323–1334. doi: 10.1042/BCJ20180044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engelhardt J.F., Yankaskas J.R., Ernst S.A., Yang Y., Marino C.R., Boucher R.C., Cohn J.A., Wilson J.M. Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat. Genet. 1992;2:240–248. doi: 10.1038/ng1192-240. [DOI] [PubMed] [Google Scholar]
- 65.Ameen N.A., Ardito T., Kashgarian M., Marino C.R. A unique subset of rat and human intestinal villus cells express the cystic fibrosis transmembrane conductance regulator. Gastroenterology. 1995;108:1016–1023. doi: 10.1016/0016-5085(95)90198-1. [DOI] [PubMed] [Google Scholar]
- 66.Trezise A.E., Chambers J.A., Wardle C.J., Gould S., Harris A. Expression of the cystic fibrosis gene in human foetal tissues. Hum. Mol. Genet. 1993;2:213–218. doi: 10.1093/hmg/2.3.213. [DOI] [PubMed] [Google Scholar]
- 67.Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J., et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plasschaert L.W., Zilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., Jaffe A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mouchel N., Henstra S.A., McCarthy V.A., Williams S.H., Phylactides M., Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem. J. 2004;378:909–918. doi: 10.1042/bj20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerschner J.L., Harris A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem. J. 2012;446:203–212. doi: 10.1042/BJ20120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kerschner J.L., Gosalia N., Leir S.H., Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9:557–565. doi: 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 73.McCarthy V.A., Ott C.J., Phylactides M., Harris A. Interaction of intestinal and pancreatic transcription factors in the regulation of CFTR gene expression. Biochim. Biophys. Acta. 2009;1789:709–718. doi: 10.1016/j.bbagrm.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohn J.A., Strong T.V., Picciotto M.R., Nairn A.C., Collins F.S., Fitz J.G. Localization of the cystic fibrosis transmembrane conductance regulator in human bile duct epithelial cells. Gastroenterology. 1993;105:1857–1864. doi: 10.1016/0016-5085(93)91085-V. [DOI] [PubMed] [Google Scholar]
- 75.Colombo C., Battezzati P.M., Strazzabosco M., Podda M. Liver and biliary problems in cystic fibrosis. Semin. Liver Dis. 1998;18:227–235. doi: 10.1055/s-2007-1007159. [DOI] [PubMed] [Google Scholar]
- 76.Cornwall G.A. New insights into epididymal biology and function. Hum. Reprod. Updat. 2009;15:213–227. doi: 10.1093/humupd/dmn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bischof J.M., Gillen A.E., Song L., Gosalia N., London D., Furey T.S., Crawford G.E., Harris A. A genome-wide analysis of open chromatin in human epididymis epithelial cells reveals candidate regulatory elements for genes coordinating epididymal function. Biol. Reprod. 2013;89:104. doi: 10.1095/biolreprod.113.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leir S.H., Browne J.A., Eggener S.E., Harris A. Characterization of primary cultures of adult human epididymis epithelial cells. Fertil. Steril. 2015;103:647–654.e641. doi: 10.1016/j.fertnstert.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Browne J.A., Yang R., Leir S.H., Eggener S.E., Harris A. Expression profiles of human epididymis epithelial cells reveal the functional diversity of caput, corpus and cauda regions. Mol. Hum. Reprod. 2016;22:69–82. doi: 10.1093/molehr/gav066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Browne J.A., Yang R., Eggener S.E., Leir S.H., Harris A. HNF1 regulates critical processes in the human epididymis epithelium. Mol. Cell. Endocrinol. 2016;425:94–102. doi: 10.1016/j.mce.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang R., Browne J.A., Eggener S.E., Leir S.H., Harris A. A novel transcriptional network for the androgen receptor in human epididymis epithelial cells. Mol. Hum. Reprod. 2018;24:433–443. doi: 10.1093/molehr/gay029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rochwerger L., Buchwald M. Stimulation of the cystic fibrosis transmembrane regulator expression by estrogen in vivo. Endocrinology. 1993;133:921–930. doi: 10.1210/endo.133.2.7688293. [DOI] [PubMed] [Google Scholar]
- 83.Rochwerger L., Dho S., Parker L., Foskett J.K., Buchwald M. Estrogen-dependent expression of the cystic fibrosis transmembrane regulator gene in a novel uterine epithelial cell line. Pt 9J. Cell Sci. 1994;107:2439–2448. doi: 10.1242/jcs.107.9.2439. [DOI] [PubMed] [Google Scholar]
- 84.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hess G.T., Tycko J., Yao D., Bassik M.C. Methods and Applications of CRISPR-Mediated Base Editing in Eukaryotic Genomes. Mol. Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rees H.A., Komor A.C., Yeh W.H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eid A., Alshareef S., Mahfouz M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marx V. Base editing a CRISPR way. Nat. Methods. 2018;15:767–770. doi: 10.1038/s41592-018-0146-4. [DOI] [PubMed] [Google Scholar]
- 92.Rees H.A., Liu D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wright F.A., Strug L.J., Doshi V.K., Commander C.W., Blackman S.M., Sun L., Berthiaume Y., Cutler D., Cojocaru A., Collaco J.M., et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat. Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corvol H., Blackman S.M., Boelle P.Y., Gallins P.J., Pace R.G., Stonebraker J.R., Accurso F.J., Clement A., Collaco J.M., Dang H., et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fossum S.L., Mutolo M.J., Tugores A., Ghosh S., Randell S.H., Jones L.C., Leir S.H., Harris A. Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J. Biol. Chem. 2017;292:10938–10949. doi: 10.1074/jbc.M117.775304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fossum S.L., Mutolo M.J., Yang R., Dang H., O’Neal W.K., Knowles M.R., Leir S.H., Harris A. Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res. 2014;42:13588–13598. doi: 10.1093/nar/gku1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stolzenburg L.R., Yang R., Kerschner J.L., Fossum S., Xu M., Hoffmann A., Lamar K.M., Ghosh S., Wachtel S., Leir S.H., et al. Regulatory dynamics of 11p13 suggest a role for EHF in modifying CF lung disease severity. Nucleic Acids Res. 2017;45:8773–8784. doi: 10.1093/nar/gkx482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strug L.J., Stephenson A.L., Panjwani N., Harris A. Recent advances in developing therapeutics for cystic fibrosis. Hum. Mol. Genet. 2018;27:R173–R186. doi: 10.1093/hmg/ddy188. [DOI] [PMC free article] [PubMed] [Google Scholar]