Abstract
A novel dextranase was purified from Penicillium cyclopium CICC-4022 by ammonium sulfate fractional precipitation and gel filtration chromatography. The effects of temperature, pH and some metal ions and chemicals on dextranase activity were investigated. Subsequently, the dextranase was used to produce dextran with specific molecular mass. Weight-average molecular mass (Mw) and the ratio of weight-average molecular mass/number-average molecular mass, or polydispersity index (Mw/Mn), of dextran were measured by multiple-angle laser light scattering (MALS) combined with gel permeation chromatography (GPC). The dextranase was purified to 16.09-fold concentration; the recovery rate was 29.17%; and the specific activity reached 350.29 U/mg. Mw of the dextranase was 66 kDa, which is similar to dextranase obtained from other Penicillium species reported previously. The highest activity was observed at 55 °C and a pH of 5.0. This dextranase was identified as an endodextranase, which specifically degraded the α-1,6 glucosidic bonds of dextran. According to metal ion dependency tests, Li+, Na+ and Fe2+ were observed to effectively improve the enzymatic activity. In particular, Li+ could improve the activity to 116.28%. Furthermore, the dextranase was efficient at degrading dextran and the degradation rate can be well controlled by the dextranase activity, substrate concentration and reaction time. Thus, our results demonstrate the high potential of this dextranase from Penicillium cyclopium CICC-4022 as an efficient enzyme to produce specific clinical dextrans.
Keywords: Penicillium cyclopium, dextranase, purification, molecular mass, degradation, dextran
1. Introduction
Dextranase (1,6-α-d-glucan-6-glucanohydrolase; E.C. 3.2.1.11) is an inducible enzyme and can specially catalyse the endohydrolysis of α-(1,6)-d-glycoside linkages at random sites of dextran. The main products are isomaltose, isomaltotriose, small amounts of d-glucose and a series of low molecular-mass polysaccharides [1]. Dextranase has been widely used in the production of specific clinical dextrans [2,3]. Dextran molecules produced by dextranase with relatively low weight-average molecular mass (Mw) of 20–70 kDa can be used as blood extenders and those with Mw of 6–8 kDa can form complexes with iron to treat severe anaemia [4]. Since dextranase can effectively degrade dextran in dental plaques, the use of this enzyme has become an effective way to treat dental caries [5,6,7,8].
Currently, bacteria and fungi are two of the main sources to produce dextranases [9]. Dextranases from bacteria usually have favorable thermal stability but relatively low dextranase activity. Hide et al. [10] obtained a dextranase from Paenibacillus sp. mutant with the optimal temperature of 60 °C. Rashida et al. [11] purified a dextranase from Bacillus licheniformis KIBGE-IB25, of which the specific activity was 1405 U/mg. Compared with dextranases from bacteria, those from fungi may achieve higher activities. Wu et al. [4] obtained a dextranase from Hypocrea lixii F1002, of which the specific enzyme activity was 2782 U/mg. Siwames et al. [12] acquired a dextranase from Aspergillus allahabadii X26, of which the specific activity reached 3009 U/mg. Dextranases from Penicillium have been found to have high dextranase activity as well as excellent temperature and pH stability. Zhang et al. [13] screened Talaromyces pinophilus H6 from the soil, which was found to be stable at 35–60 °C and pH of 3.0–10.0. This special dextranase displayed activity as high as 14,894 U/mg.
Previous research about dextranases has mostly focused on the mutation [9], screening [14], construction of genetically engineered bacteria [15,16], the optimization of fermentation medium [17] and fermentation conditions [18] and the purification and enzymatic properties [19,20,21]. To the best of our knowledge, there are few studies on how to regulate molecular mass of dextran during production. Since the molecular mass of dextrans largely determines their applications, it is essential to find an efficient way to control this parameter. Besides, we found that the determination of the molecular mass of dextrans was inaccurate in most reports [4,13]. In these reports, weight-average molecular weight (Mw) was detected and determined by a refractive index detector and its accuracy relies on the flow rate and the standard curve. However, the standard curves were plotted according to the relationship between retention time and peak molecular weight (Mp). This means that the determination of Mw depended on the calculation of measured Mp. Since the molecular mass distribution of dextran was uneven during the degradation, Mp can hardly match Mw. This might cause great errors in the measurement. Moreover, it is also worth mentioning that in most of the previous studies, the molecular mass distribution of dextrans was not measured, despite that this parameter is important in that it directly reflects the quality of dextran. The polydispersity index, which is defined as the ratio of weight-average molecular weight and number-average molecular weight (Mw/Mn), is an index to describe the molecular mass distribution.
In this work, a novel dextranase was purified from Penicillium cyclopium CICC-4022 and its properties were characterized. The effects of temperature, pH and metal ions on dextranase activity were evaluated. The dextranase was then applied to degrade dextran. We found that Mw of the degraded dextrans could be effectively controlled by adjusting the reaction time, concentrations of the dextranase and the substrate concentration during the degradation. Mw and Mw/Mn of the degraded dextrans was analysed using gel permeation chromatography (GPC) coupled with multiple-angle laser light scattering (GPC-MALS).
2. Results and Discussions
2.1. Purification of Dextranase
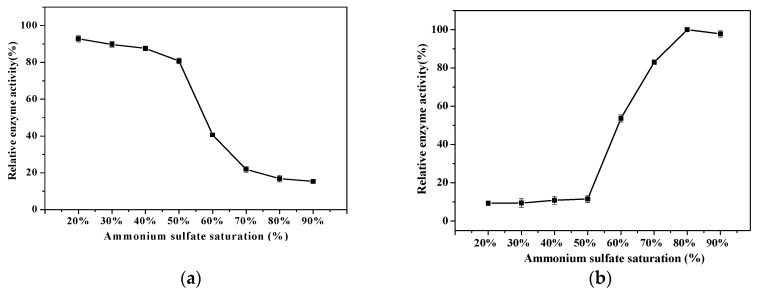
It can be seen from Figure 1, when ammonium sulfate saturation was less than 50%, the relative dextranase activity of the supernatant was above 80.73% (taking the dextranase activity of the crude enzyme as 100%) and that of the precipitate was no more than 11.50% (taking the highest dextranase activity as 100%), only a small amount of the dextranase and miscellaneous proteins were salted out and most of the enzyme was in the supernatant. When ammonium sulfate saturation was 80%, the relative dextranase activity of the supernatant was 16.89%. In this latter case, most of the dextranase has been precipitated and the relative activity of the precipitate was high. To increase the recovery rate of the dextranase and not bring extra miscellaneous proteins during the process, the precipitate of 50–80% saturation should be collected. Dextranase was purified to 1.71-fold concentration and its yield was 87.35%. The specific activity of the purified enzyme was 111.06 U/mg (Table 1).
Figure 1.
Relative dextranase activity of the supernatant (a) and the precipitate (b) with different ammonium sulphate saturation.
Table 1.
Parameters related to purification of the dextranase from Penicillium cyclopium.
| Purification Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Purification (Fold) | Yield (%) |
|---|---|---|---|---|---|
| Culture dextranase | 7.08 | 533.15 | 75.32 | 1.00 | 100.00 |
| (NH4)2SO4 precipitation | 4.19 | 465.71 | 111.06 | 1.71 | 87.35 |
| Sepharose 6B | 0.44 | 155.54 | 350.29 | 16.09 | 29.17 |
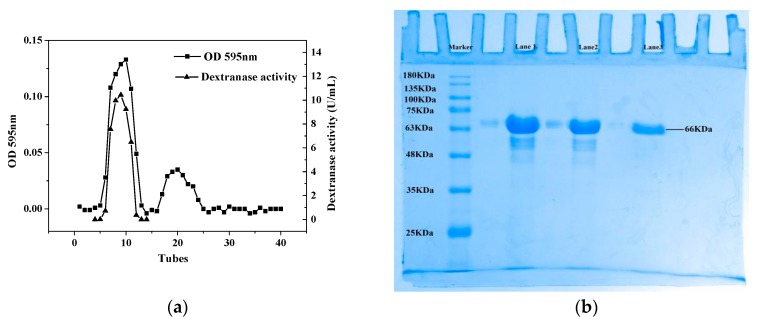
It can be seen from Figure 2a that the dextranase has been effectively eluted and separated using Sepharose 6B chromatography. Fractions with high specific activity were collected from Tube 4 to 14. Dextranase was purified to 16.09-fold concentration and its yield was 29.17%. The specific activity of the purified enzyme reached 350.29 U/mg (Table 1).
Figure 2.
(a) Gel filtration chromatography results of the dextranase on Sepharose 6B; (b) SDS-PAGE results of the dextranase. For (b), lane markers represent marker proteins; Lane 1: the crude enzyme; Lane 2: the ammonium sulphate precipitate; Lane 3: the purified dextranase, collected from gel filtration chromatography.
The samples from Tube 7, 8, 9 and 10 were merged and analysed by SDS-PAGE. As shown in Figure 2b, only one band appeared in Lane 3, suggesting the dextranase from Penicillium cyclopium was a monomer and the purification process was effective. Mw of purified dextranase was about 66 kDa, which was similar to dextranases from other fungi such as Aspergillus allahabadii X26 (66 kDa) [12] and Talaromyces pinophilus H6 (58kDa) [13] as reported previously.
2.2. Dextranase Enzymatic Properties
2.2.1. Effect of Temperature on Dextranase Activity and Thermal Stability
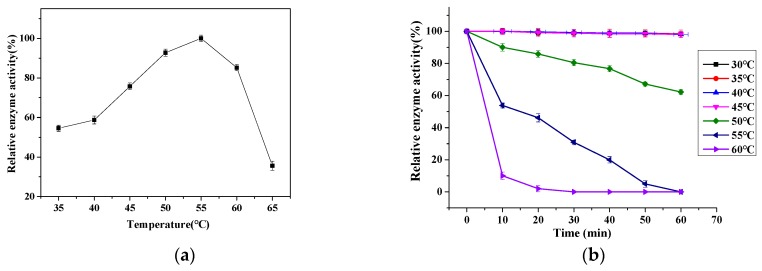
Figure 3a shows the relative enzyme activity of the dextranase as affected by temperature. With increasing temperature, the relative activity firstly increased and then decreased. When temperature varying from 35 to 60 °C, the relative activity was always higher than 54.55%; and the highest relative activity was reached at 55 °C. It is worth mentioning that this optimal temperature of 55 °C was higher than those dextranase from Hypocrea lixii F1002 [4] (25 °C) and Bacillus sp [22] (50 °C) reported previously. This demonstrates the higher thermal stability of our dextranase from Penicillium cyclopium than that of dextranase from other sources.
Figure 3.
Relative enzymatic activity of the dextranase as a function of temperature (a) and time (b).
Figure 3b shows the changes in relative enzymatic activity of the dextranase stored at pH of 5 from 45 °C to 60 °C for 10–60 min. After stored at 45 °C and 50 °C for 60 min, the dextranase activity remained relatively stable with the values being 98.09% and 62.21%, respectively. This suggests that the dextranase from Penicillium cyclopium had higher thermal stability than those from Catenovulum sp DP03 [15] and Arthrobacter oxydans [23].
2.2.2. Effect of pH on Dextranase Activity and pH Stability
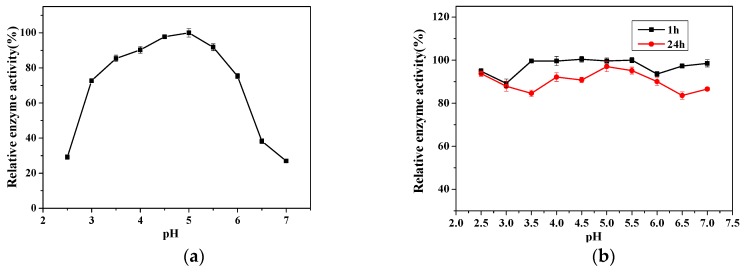
Figure 4a shows the relative enzyme activity at different pH values. With increasing pH, the relative activity firstly increased and then decreased and the highest relative activity was achieved at a pH of 5. With pH ranged from 3.0 to 6.0, the relative enzyme activity was higher than 72.67%. It is worth noting that with pH of 3.0, the relative enzyme activity of our dextranase from Penicillium cyclopium was more than 72.67%, while the dextranase from Talaromyces pinophilus H6 [13] was only 45%. This indicates that our dextranase has better acid-resistance compared to other dextranases obtained from Penicillium.
Figure 4.
Relative enzymatic activity of the dextranase as a function of pH (a) and as a function of pH for 1 h and 24 h (b).
Figure 4b shows the changes in the relative enzymatic activity of dextranase stored at a pH of 2.5–7 for 1 h and 24 h, respectively. It can be seen that the relative activity for both storage times was stable. The relative activity was more than 89.22% after 1 h and more than 84.58% after 24 h. This performance was better than the dextranase from Chaetomium globosum [24]. Therefore, our dextranase from Penicillium cyclopium has greater acid-resistance and suitable to be applied in a wide pH range.
2.2.3. Substrate Specificity and Final Products Analysis of Enzyme
Dextranase activity during the catalysed hydrolysis of substrates with diverse glucosidic bonds was measured to evaluate the substrate specificity of the dextranase (Table 2). The substrates containing α-1,6 glucosidic bonds had higher relative dextranase activity. This means that our dextranase specifically degraded the α-1,6 glucosidic bonds of dextran and the specificity is similar to dextranases from other sources [4,25,26]. With an increase in dextran Mw, the relative enzyme activity firstly increased and then decreased and dextran T70 was the optimum substrate for the dextranase. The dextranase was less effective for soluble starch, which contains most α-1,4 glucosidic linkages and limited amounts of a-1,6 linkages [2,27]. Moreover, the dextranase showed no effects on cellulose, sucrose, chitin, chitosan and β-cyclodextrin. Thus, our dextranase showed high specificity toward dextrans that contain mostly α-1,6 glucosidic bond and it was inactive with the α-1,4, β-1,2, β-1,4 glucosidic bonds of the substrate.
Table 2.
Effects of dextranase on diverse carbohydrates.
| Substrate | Main Linkage | Relative Activity (%) |
|---|---|---|
| DextranT3 | α-1,6 | 65.17 ± 0.7 |
| DextranT5 | α-1,6 | 65.54 ± 0.6 |
| DextranT20 | α-1,6 | 90.64 ± 0.4 |
| DextranT40 | α-1,6 | 98.50 ± 0.2 |
| DextranT70 | α-1,6 | 100.00 ± 0.5 |
| DextranT500 | α-1,6 | 99.62 ± 0.1 |
| DextranT2000 | α-1,6 | 97.00 ± 0.4 |
| Soluble starch | α-1,4; α-1,6 | 20.83 ± 0.8 |
| Cellulose | β-1,4 | 0 |
| Sucrose | β-1,2 | 0 |
| Chitin | β-1,4 | 0 |
| Chitosan | β-1,4 | 0 |
| β-Cyclodextrin | β-1,4 | 0 |
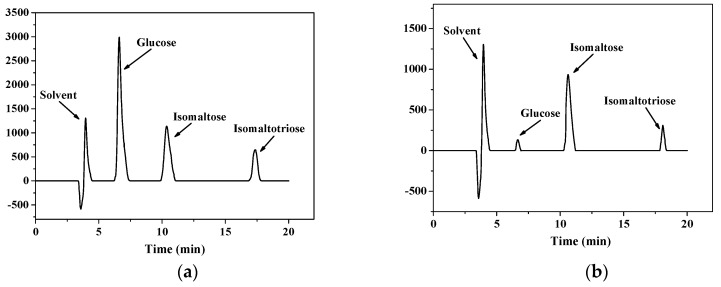
HPLC results show that the peaks of glucose, isomaltose and isomaltotriose appeared at 6.6, 10.4 and 17.4 min, respectively (Figure 5a). The peaks of our final products of dextranase were consistent with the peaks of glucose, isomaltose and isomaltotriose (Figure 5a). It is known that the final products of endodextranase were mainly isomaltose, isomaltotriose and a small amount of glucose [2]. Therefore, it can be concluded that the dextranase from Penicillium cyclopium was an endodextranase.
Figure 5.
HPLC results for glucose, isomaltose and isomaltotriose (a); and for the final products of dextranase (b).
2.2.4. Effects of Metal Ions and Compounds on Enzyme Activity
The effects of several metal ions and compounds on dextranase activity are listed in Table 3. The results show that Li+, Na+, Fe2+ and Tris could improve the dextranase activity at all concentrations, especially in 0.001 M Li+, where the activity could reach 116.28%. In addition, NH4+, Tris and urea slightly enhanced the enzymolysis. Fe3+, Ca2+ and EDTA slightly reduced the dextranase activity [13]. Regarding SDS, low concentrations (0.001 M) had positive effects on dextranase activity, while high concentrations had negative effects. Cu2+, Zn2+, Cr2+, Pb2+ and Ni2+ all inhibited the enzymolysis. Furthermore, with increasing concentration, this inhibition effect became stronger. The dextranase activity was only 16.41% when Cu2+ was 0.01 M.
Table 3.
Effects of metal ions and compounds on the activity of dextranase.
| Compound | Relative Activity (%) | Relative Activity (%) | Relative Activity (%) |
|---|---|---|---|
| (0.001 M) | (0.005 M) | (0.01 M) | |
| Control | 100 | 100 | 100 |
| Cu2+ | 24.02 ± 0.15 | 22.53 ± 0.05 | 16.41 ± 0.2 |
| Fe3+ | 99.55 ± 0.10 | 64.61 ± 0.07 | 67.27 ± 0.07 |
| Zn2+ | 90.95 ± 0.02 | 59.35 ± 0.13 | 16.85 ± 0.05 |
| Ca2+ | 101.81 ± 0.03 | 93.30 ± 0.04 | 40.73 ± 0.06 |
| Cr2+ | 90.95 ± 0.08 | 72.74 ± 0.12 | 48.25 ± 0.06 |
| K+ | 97.74 ± 0.06 | 114.83 ± 0.10 | 105.75 ± 0.05 |
| Li+ | 116.28 ± 0.05 | 115.78 ± 0.09 | 103.98 ± 0.12 |
| Na+ | 104.98 ± 0.07 | 111.48 ± 0.02 | 105.75 ± 0.05 |
| Pb2+ | 59.29 ± 0.05 | 22.53 ± 0.03 | 20.39 ± 0.15 |
| Ni2+ | 95.48 ± 0.15 | 97.61 ± 0.05 | 67.71 ± 0.16 |
| NH4+ | 107.58 ± 0.04 | 105.21 ± 0.07 | 99.53 ± 0.15 |
| Fe2+ | 104.74 ± 0.06 | 100.95 ± 0.02 | 109.95 ± 0.12 |
| Tris | 103.79 ± 0.08 | 100.00 ± 0.07 | 100.47 ± 0.10 |
| SDS | 106.16 ± 0.03 | 46.94 ± 0.05 | 34.15 ± 0.13 |
| Urea | 112.79 ± 0.04 | 109.00 ± 0.11 | 99.05 ± 0.04 |
| EDTA | 102.37 ± 0.07 | 80.58 ± 0.14 | 81.05 ± 0.05 |
2.2.5. Kinetic Parameters of Dextranase
The Km, Vm, Kcat, Kcat/Km values of dextran T5, T70 and T2000 are listed in Table 4. Km is a parameter to evaluate the affinity with the substrate; higher Km indicates lower affinity. It can be seen that with an increase in Mw of the substrate, Km decreased, indicating stronger affinity of the dextranase to the dextran with high Mw; The Km value of our dextranase for dextran T2000 was 2.45 mM, which was smaller than those of dextranases from Arthrobacter oxydans (4.73 mM) [28], Aspergillus allahabadii X26 (14.29 mM) [12] and Streptomyces sp. NK458 (94.30 mM) [29]. This shows the stronger affinity of the dextranase from Penicillium cyclopium than that of the dextranases reported previously.
Table 4.
Kinetic constants of purified dextranase from Penicillium cyclopium.
| Dextran T5 b | Dextran T70 b | Dextran T2000 b | |
|---|---|---|---|
| Km (mM) | 2.72 ± 0.04 | 2.61 ± 0.02 | 2.45 ± 0.01 |
| Vmax (µmol Glucose min−1 mg−1 Protein) | 189.50 ± 0.03 | 192.70 ± 0.04 | 203.21 ± 0.01 |
| Kacat (s−1) | 208.45 ± 0.01 | 211.97 ± 0.02 | 223.53 ± 0.03 |
| Kcat/Km (M−1 s−1) | 76636.0 ± 0.06 | 81214.6 ± 0.03 | 91236.7 ± 0.05 |
aKcat value was calculated assuming a molecular mass of 66 kDa for native dextranase. b The results reported were the means of three replications ± SD.
The Kcat and Kcat/Km value of the dextranase for dextran increased with increasing Mw of the dextran, which showed that the dextranase from Penicillium cyclopium has higher catalytic efficiency for high-Mw dextran.
2.3. Determination of Mw and Mw/Mn of Dextran
2.3.1. Standard Curve of Dextran of GPC
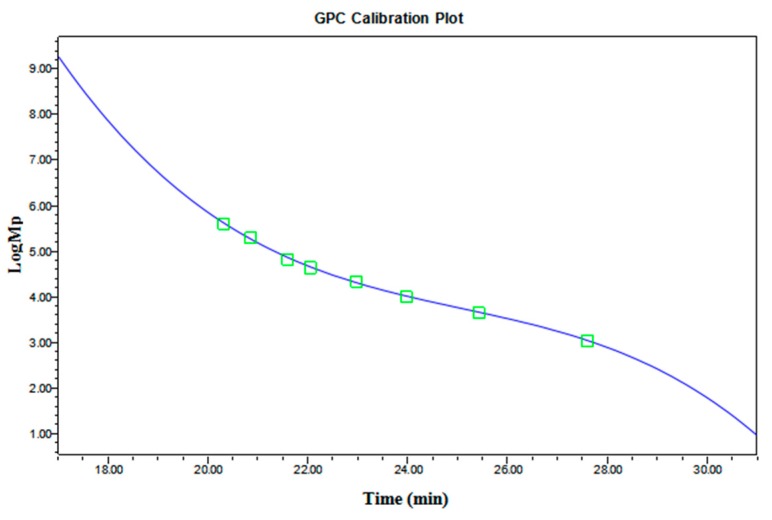
GPC is the most commonly used technique for determining the molecular mass of polymers including dextran. Using this technique, the relative Mw value of dextran can be obtained based on the standard curve of dextran (Figure 6). According to Section 3.7.1, the retention time was taken as the abscissa, the logarithmic values of Mp were taken as the ordinate and the standard curve is obtained by fitting using Waters Breeze 2. The standard curve of dextran was calculated to be: log(Mp) = s − 1.31t + 0.512t2 − 0.0068t3, R2 = 0.9993, σ = 2.88%. The third-order function fits well with minimal error.
Figure 6.
The standard curve of dextran.
2.3.2. Comparison of GPC-MALS with GPC and Error Analysis
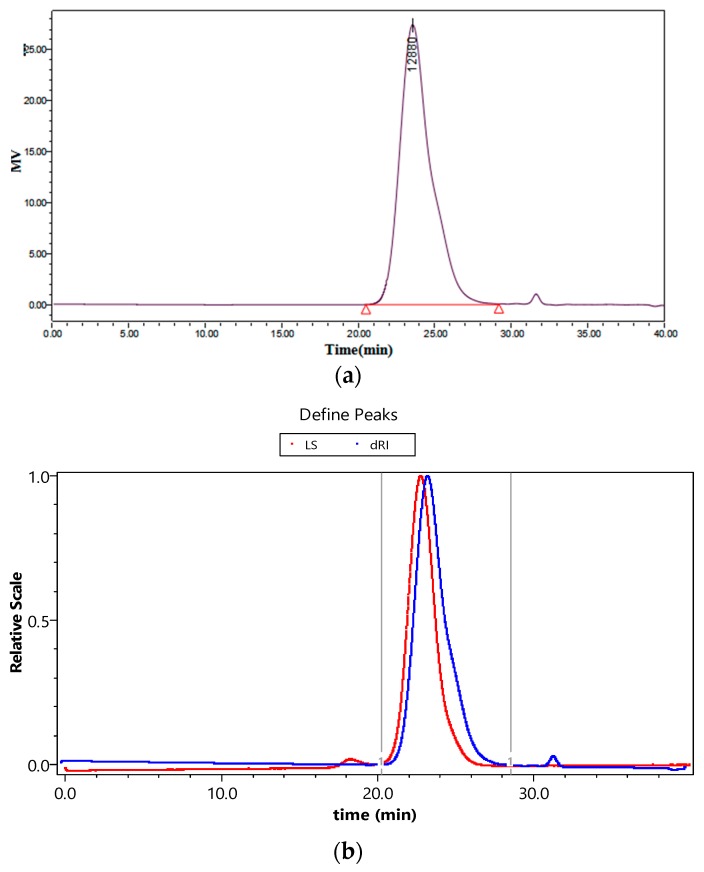
Figure 7 presents the chromatograms of GPC (a) and GPC-MALS (b), both of which show a well-defined peak. For both techniques, the sample peaks display a symmetrical normal distribution. Figure 7a has a signal from the refractive index detector and Mp, Mw and Mw/Mn were calculated using Breeze 2 software (Waters) based on the standard curves. Figure 7b has two signal peaks from the refractive index detector and MALS respectively and the curves were generated by ASTRA 7.1.3 software of MALS (Wyatt). The differential signal should be used for the occurrence of the peak while the laser signal should be taken for the conclusion of the peak. In this way, we measured Mp, Mn, Mz, Mw and Mw/Mn of dextrans.
Figure 7.
Chromatograms of GPC (a) and GPC-MALS (b).
Table 5 shows the values of Mw and Mw/Mn and relative errors of Mw and Mw/Mn of dextrans measured by GPC and GPC-MALS. The relative errors of Mw of dextran measured by GPC-MALS were less than 7%, while those measured by GPC were more than 10% and even up to 40%. The relative errors of Mw/Mn of dextran measured by GPC-MALS were less than 8%, while the maximum relative error measured by GPC was 160.7%. Thus, compared to GPC, GPC-MALS produced much smaller relative errors of Mw and Mw/Mn of dextrans. The reason could be that the relative Mw values measured by GPC mainly depends on the standard curve and pump speed. Moreover, Mw must be within the linear range of the standard curve; for generating the standard curve, the standard samples with larger Mw (>1000 kDa) was difficult to obtain and store. On the contrary, these factors are not related to GPC-MALS, which could be considered as a more accurate method to measure Mw and Mw/Mn of dextrans.
Table 5.
Error analysis of GPC and GPC-MALS.
| Standard | Mw (kDa) | Mw δ (%) | Mw/Mn | Mw/Mn δ (%) | ||||
|---|---|---|---|---|---|---|---|---|
| GPC | GPC-MALS | GPC | GPC-MALS | GPC | GPC-MALS | GPC | GPC-MALS | |
| Dextran (Mw ≈ 1.27 kDa) | 0.84 | 1.32 | −33.86 | 3.94 | 1.72 | 1.20 | 36.51 | −4.76 |
| Dextran (Mw ≈ 5.52 kDa) | 3.09 | 5.4 | −44.02 | −2.17 | 3.03 | 1.57 | 89.38 | −1.88 |
| Dextran (Mw ≈ 23.8 kDa) | 21.4 | 23.99 | −10.08 | 0.80 | 1.28 | 1.229 | 24.62 | 7.69 |
| Dextran (Mw ≈ 48.6 kDa) | 31.57 | 49.98 | −35.04 | 2.84 | 1.28 | 1.24 | −5.88 | −5.15 |
| Dextran (Mw ≈ 80.9 kDa) | 59.26 | 82.26 | −26.74 | 1.68 | 1.64 | 1.28 | 12.33 | −5.48 |
| Dextran (Mw ≈ 667.8 kDa) | 827.80 | 621.5 | 23.96 | −6.93 | 5.24 | 2.049 | 160.70 | 1.94 |
2.3.3. Degradation of Dextran Polymers by Dextranase
The changes in Mw and Mn/Mw of dextran with time under different enzyme activity and substrate concentrations are shown in Table 6 and Table 7, respectively. It can be seen that MW decreased rapidly during the first 4 min and did slowly, suggesting that the enzyme had higher affinity with higher-MW dextrans [24]. When MW was 100–1000 kDa, Mn/Mw was higher, which might be contributed by dextran molecules of diverse Mw in the system. With the reaction proceeded, Mw decreased and gradually reached to a plateau, and, thus, Mn/Mw became smaller. For example, when the enzyme activity was 0.2 U/mL, Mw and Mn/Mw of the dextran after 8 min of hydrolysis were 720.6 kDa and 4.524, respectively, which decreased to 51.16 kDa and 1.966 respectively after 30 min.
Table 6.
Mw and Mn/Mw of dextran under different dextranase activity and time.
| Reaction Time (min) | 0.2 U/mL | 0.4 U/mL | 0.6 U/mL | |||
|---|---|---|---|---|---|---|
| Mw (kDa) | Mn/Mw | Mw (kDa) | Mn/Mw | Mw (kDa) | Mn/Mw | |
| 0 | 1869 | 3.236 | 1859 | 1.523 | 1859 | 3.358 |
| 4 | 1553 | 2.322 | 865.9 | 3.048 | 251 | 5.891 |
| 8 | 720.6 | 4.524 | 242.3 | 4.272 | 83.69 | 3.991 |
| 12 | 426.5 | 5.329 | 69.25 | 3.12 | 48.34 | 2.198 |
| 16 | 216 | 4.837 | 40.74 | 2.504 | 24.13 | 2.133 |
| 20 | 119.5 | 4.373 | 29.54 | 2.071 | 20.78 | 1.917 |
| 30 | 51.16 | 1.966 | 16.77 | 1.82 | 11.6 | 1.731 |
| 40 | 32.41 | 1.618 | 11.35 | 1.672 | 9.247 | 1.507 |
| 50 | 22.26 | 2.187 | 9.685 | 1.542 | 7.156 | 1.502 |
| 60 | 13.79 | 1.627 | 8.052 | 1.433 | 6.262 | 1.36 |
| 90 | 11.39 | 1.494 | 6.362 | 1.306 | ||
| 120 | 8.239 | 1.375 | 5.954 | 1.238 | ||
| 150 | 7.358 | 1.396 | ||||
Table 7.
Mw and Mn/Mw of dextran under different substrate concentrations and time.
| Reaction Time (min) | Dextran (1%) | Dextran (3%) | Dextran (5%) | |||
|---|---|---|---|---|---|---|
| Mw (kDa) | Mn/Mw | Mw (kDa) | Mn/Mw | Mw (kDa) | Mn/Mw | |
| 0 | 1726 | 2.995 | 1759 | 3.358 | 1750 | 3.829 |
| 4 | 49.94 | 2.534 | 251 | 5.891 | 487.8 | 11.383 |
| 8 | 16.06 | 1.702 | 83.69 | 3.991 | 194.4 | 6.03 |
| 12 | 10.65 | 1.39 | 48.34 | 2.198 | 107.1 | 5.032 |
| 16 | 7.196 | 1.388 | 24.13 | 2.133 | 63.9 | 3.519 |
| 20 | 5.893 | 1335 | 20.78 | 1.917 | 42.4 | 2.964 |
| 30 | 4.993 | 1.234 | 11.6 | 1.731 | 23.88 | 2.237 |
| 40 | 4.434 | 1.53 | 9.247 | 1.507 | 16.54 | 1.979 |
| 50 | 7.156 | 1.502 | 12.8 | 1.693 | ||
| 60 | 6.262 | 1.36 | 10.05 | 1.651 | ||
Dextranase activity and dextran concentration are two important factors that can affect the degradation rate. Higher dextranase activity could lead to an increased degradation rate of dextrans. As listed in Table 6, when the dextranase activity was increased from 0.2 U/mL to 0.6 U/mL, MW of dextran after 8 min of hydrolysis decreased from 720.6 kDa to 83.69 kDa. On the other hand, increasing dextran concentration could decrease the degradation rate. As listed in Table 7, when the dextran concentration was increased from 1% to 5%, MW of dextran after 16 min of hydrolysis increased from 7.196 kDa to 63.9 kDa. Based on these results, we can conclude that MW of dextran can be effectively controlled by adjusting the dextranase activity, dextran concentration and reaction time [24,30].
3. Materials and Methods
3.1. Materials
Dextrans T3, T5, T20, T40, T70, T500 and T2000 (Mw ≈ 3, 5, 20, 40, 70, 500 and 2000 kDa) and a series of standards (dextran 1.27, 5.52, 11.6, 23.8, 48.6, 80.9, 273.0, 667.8 kDa) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sepharose 6B was a product from Henghui (Beijing, China). The protein marker was obtained from ShineGene Molecular Biotech (Shanghai, China). Penicillium cyclopium (CICC-4022) was purchased from the China Centre of Industrial Culture Collection (Beijing, China). All other chemicals were of analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China).
3.2. Preparation of Crude Dextranase
According to our preliminary experiments with dextran, sucrose, glucose, maltose and fructose for optimized carbon sources, we have assumed that that α-1,6 glycosidic bonds in dextran played an inducing role in the production of dextranase. A Czapek Dox medium was used as the solid medium and seed culture medium for Penicillium cyclopium. The fermentation medium was prepared by dextran T20 30 g/L, yeast extract 4 g/L, KCl:FeSO4·7H2O = 10:1 (concentration ratio) and K2HPO4 1.0 g/L. The mixture had an initial pH of 6.1. 60 mL of the fermentation medium was transferred into a 250mL flask, which was then sterilized at 121 °C for 30 min. Inoculum concentration was set as 3% and the fermentation medium was incubated in a rotary shaker at 160 r/min and 30 °C for 72 h. The thalli of Penicillium cyclopium were separated by centrifugation for 20 min at 4 °C and 10,000× g. The supernatant was stored at 4 °C for further dextranase purification.
3.3. Dextranase Activity Assay
Dextranase activity was assayed by the determination of the amount of substance of reducing sugar from Dextran T70. The diluted dextranase of 1.0 mL was incubated with 1.0 mL 3% dextran T70 (preparation with 0.02 M acetate buffer of pH 5) at 50 °C for 10 min, the amount of reducing sugar was obtained using the 3,5-dinitrosalicylic acid method as reported previously [31]. One unit (1 U) of dextranase activity was defined as the amount of enzyme that degrades dextran T70 to produce 1 µmol glucose equivalent per min at 50 °C and pH of 5 [32] and calculated according to Equation (1).
| Dextranase activity (U/mL) = [Amount of reducing sugar (µmol) × Dilution multiple of enzyme solution]/[Volume of enzyme solution (mL) × Time (min)] |
(1) |
3.4. Determination of Protein Concentration
Protein concentration (mg/mL) was measured by the Bradford method [33]. Crystalline bovine serum albumin was used as the protein standard.
3.5. Purification of Dextranase
The cell-free filtrate (CFF) was subjected to ammonium sulfate fractionation with a saturation of 20–90%. The precipitate was collected by centrifugation at 10,000× g for 20 min at 4 °C. Then the precipitate was dissolved and dialyzed in 0.02 M acetate buffer (pH 5.0) overnight at 4 °C. The concentrated dextranase was loaded on a Sepharose 6B column (1.5 cm i.d. × 50 cm) and the dextranase was eluted with 0.02 M acetate buffer (pH 5.0) at a flow rate of 0.2 mL/min. The absorbance of each fraction (3 mL) was determined at 595 nm to monitor the proteins during chromatographic separation. The fractions with high dextranase activity were gathered to measure their dextranase activity and protein concentration as described in Section 3.3 and Section 3.4. Specific activity was calculated according to the above-measured results. The purity of the dextranase and its Mw were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The unstained protein marker was used as control samples, Mw of the dextranase was analysed by a 5% concentrated gel and a 12% separated gel. Following electrophoresis, the gel was stained with Coomassie blue R-250 and decolorized several times with a destainer.
3.6. Dextranase Enzymatic Properties
3.6.1. Effect of Temperature and pH on Dextranase Activity and Stability
For investigating the effect of temperature, dextran T70 (30 mg/mL) and the dextranase were prepared with 0.02M acetate buffer (pH = 5.0). The effect of temperature on dextranase activity was measured at 35–65 °C. Relative activity was expressed as percentage of the highest activity while the highest dextranase activity is 100%. The thermal stability of dextranase was measured by exposing the dextranase in 0.02M acetate buffer (pH = 5.0) for 10–60 min at 45–65 °C without any substrate.
For studying the effect of pH, dextranase activity was determined at pH of 2.5–7.0 at 55 °C. Relative activity was expressed as percentage of the highest activity while the highest dextranase activity was 100%. The pH stability of the dextranase was evaluated by incubating the dextranase in reaction buffers at pH 2.5–7.5 at 4 °C for 1 h and 24 h respectively without any substrate.
For both thermal and pH stability, dextran T70 was added to the dextranase to measure the relative dextranase activity. The activity of non-heated and non-stored dextranase was taken as 100% dextranase activity, respectively.
3.6.2. Substrate Specificity and Analysis of Hydrolysis Products
Dextran T3, T5, T20, T40, T70, T500 and T2000, soluble starch, cellulose, sucrose, chitin, chitosan and β-cyclodextrin (30 mg/mL) were prepared with 0.02M acetate buffer (pH = 5.0) and the dextranase activity was measured with these different substrates. Relative activity was expressed as percentage of the highest activity so that the highest dextranase activity is 100%.
The final hydrolysis products of dextran T70 were analysed by high-performance liquid chromatography (HPLC) with a mobile phase of 80% acetonitrile. Glucose, isomaltose and isomaltotriose were used as the standards. The hydrolysates were diluted to 10-fold with the mobile phase and then centrifuged at 11,000× g for 20 min. The supernatant was filtered with a 0.22 μm filter and analysed by HPLC, in which a Polaris NH2 (4.6 mm × 250 nm, Agilent Technologies, Santa Clara, CA, USA) column was used. The chromatograph was operated under a flow rate of 1.0 mL/min at 35 °C and then connected to an RID-10A refractive index detector (Shimadzu Corp. Kyoto, Japan).
3.6.3. Effects of Metal ions and other Compounds on Dextranase Activity
Metal ions and reagents (namely, Cu2+ (CuSO4), Fe3+ (FeCl3), Zn2+ (ZnSO4), Ca2+ (CaCl2), Co2+ (CoCl2), K+ (KCl), Li+ (LiCl), Na+ (NaCl), Pb2+ (Pb(NO3)2), Ni2+ (NiCl2), NH4+ (NH4Cl), Fe2+ (FeSO4), Tris, SDS, urea and EDTA) were dissolved with 0.02 M acetate buffer (pH = 5.0). The concentrations of these solutions were 0.001, 0.005 and 0.01 M, respectively. The effects of different ions and reagents on dextranase activity were measured. Dextranase activity without any compounds was considered as 100% to calculate the relative activity of the above compounds.
3.6.4. Enzyme Kinetics
In order to determine the kinetic constants, the initial velocity (v) was measured with various concentrations (0.1–0.6%) of Dextran T5, T70 and T2000 in 0.02M acetate buffer (pH = 5.0) at 55 °C for 6 min. The kinetic constants were calculated from Lineweaver–Burk plots [34].
3.7. Determination of Mw and Mw/Mn
3.7.1. Gel permeation chromatography (GPC)
The chromatographic conditions used were as follows: the mobile phases: 0.03% NaN3 and 0.1 M NaNO3; the columns: UltrahydrogelTM 2000 (7.8 mm × 300 mm, Shimadzu, Japan), UltrahydrogelTM 250 (7.8 mm × 300 mm, Shimadzu, Japan) and UltrahydrogelTM DP120A (7.8 mm × 300 mm, Shimadzu, Japan); flow rate: 1 mL/min; the detector: Waters 2414 refractive index detector (Waters Corporation, Milford, MA, USA); column temperature: 35 °C; and detector temperature: 35°C.
The dextran standards with Mw of 1.27, 5.52, 11.6, 23.8, 48.6, 80.9, 273.0 and 667.8 kDa (Mp = 1.08, 4.44, 9.89, 21.4, 43.5, 66.7, 196.3 and 401.3 kDa, respectively) were prepared with a concentration of 1 mg/mL using the mobile phase. The samples were injected after filtration with a 0.22 μm water filter, GPC was performed to detect the retention time for different Mw. Waters Breeze 2 software was used to generate the standard curve of dextran.
Both GPC and GPC-MALS techniques were used to determine Mw and Mw/Mn of dextran. The relative errors of Mw and Mw/Mn determined by the two techniques were analysed and compared. Except that GPC-MALS had a refractive index detector and a laser detector, the two techniques used the same chromatographic conditions.
3.7.2. Degradation of dextrans by dextranase
GPC-MALS was used to measure Mw and Mw/Mn of dextrans. Mw, Mn and Mw/Mn can be obtained without depending on the pump speed and the standard curve. Pawcenis et al. [35] have used this method to measure Mw of cellulose.
Dextran T2000 (1%, 3%, 5%) and Penicillium cyclopium dextranase (dextranase activity: 0.2, 0.4 and 0.6 U/mL) were prepared with 0.02 M acetate buffer (pH = 5.0). The dextranase was incubated with the substrate at 50 °C. Samples were obtained at intervals, boiled for at least 3 min to stop the reaction and diluted to 10 folds with the mobile phase. The samples were filtered with a 0.22 μm water filter. Mw and Mw/Mn of dextran were analysed by GPC-MALS (Waters Corporation, Milford, MA, USA; Wyatt Technology Corporation, Santa Barbara, CA, USA) according to the conditions stated in Section 3.7.1.
4. Conclusions
In this work, we obtained a novel dextranase from Penicillium cyclopium CICC-4022. After purification to 16.09-fold concentration, the dextranase showed specific enzymatic activity of 3780.13 U/mg and molecular weight of 66 kDa. For this dextranase, the optimum temperature and pH were 55 °C and 5.0, respectively. The dextranase was stable below 45 °C under pH from 3.5 to 7. We found that this dextranase could specifically degrade α-1,6 glucosidic bonds of dextran and could be identified as an endodextranase. Besides, some metal ions such as Li+, Na+ and Fe2+ could effectively promote the dextranase activity. The obtained dextranases were then applied to degrade dextrans with excellent performance. Moreover, we found Mw of dextran could be effectively controlled by adjusting the dextranase activity, substrate concentration and reaction time during degradation. Therefore, our findings show the high potential of this dextranase from Penicillium cyclopium CICC-4022, which can be potentially applied to the production of specific dextrans for clinical applications.
Acknowledgments
L.Z. would like to thank the Xiangsihu Young Scholars Innovative Research Team of Guangxi University for Nationalities for the support.
Author Contributions
Conceptualization, R.H. and A.L.; Measurement, R.H., L.W., L.G. and X.W.; Data analysis, R.H.; Funding acquisition, A.L.; Methodology, R.H.; Project administration, A.L.; Writing–original draft, L.Z.; Writing–review & editing, L.Z. and F.X.
Funding
This research was funded by the Guangxi Biological Polysaccharide Separation, Purification and Modification Research Platform (grant No. GKZY18076005) and the Guangxi Key Laboratory of Polysaccharide Materials and Modification, Guangxi University for Nationalities (grant No. GXPSMM18ZD-02). The APC was also funded by the Guangxi Biological Polysaccharide Separation, Purification and Modification Research Platform (grant No. GKZY18076005).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bashari M., Eibaid A., Wang J.P., Tian Y.Q., Xu X.M., Jin Z.Y. Influence of low ultrasound intensity on the degradation of dextran catalyzed by dextranase. Ultrason. Sonochem. 2013;20:155–161. doi: 10.1016/j.ultsonch.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Ren W., Cai R.H., Yan W.L., Lyu M.S., Fang Y.W., Wang S.J. Purification and characterization of a biofilm-degradable dextranase from a marine bacterium. Mar. Drugs. 2018;16:51. doi: 10.3390/md16020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahid F., Aman A., Nawaz M.A., Karim A., Qader S.A. Chitosan hydrogel microspheres: An effective covalent matrix for crosslinking of soluble dextranase to increase stability and recycling efficiency. Bioprocess Biosyst. Eng. 2017;40:451–461. doi: 10.1007/s00449-016-1713-7. [DOI] [PubMed] [Google Scholar]
- 4.Wu D.T., Zhang H.B., Huang L.J., Hu X.Q. Purification and characterization of extracellular dextranase from a novel producer, Hypocrea lixii F1002, and its use in oligodextran production. Process Biochem. 2011;46:1942–1950. doi: 10.1016/j.procbio.2011.06.025. [DOI] [Google Scholar]
- 5.Walker G.V., Heng N.C.K., Carne A., Tagg J.R., Wescombe P.A. Salivaricin E and abundant dextranase activity may contribute to the anti-cariogenic potential of the probiotic candidate Streptococcus salivarius JH. Microbiology. 2016;162:476–486. doi: 10.1099/mic.0.000237. [DOI] [PubMed] [Google Scholar]
- 6.Qiu Y.X., Mao M.Y., Jang D., Hong X., Yang Y.M., Hu T. Co-operative effect of exogenous dextranase and sodium fluoride on multispecies biofilms. J. Dent. Sci. 2016;11:41–47. doi: 10.1016/j.jds.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marotta M., Martino A., Rosa A.D., Farina E., Carteni M., Rosa M.D. Degradation of dental plaque glucans and prevention of glucan formation using commercial enzymes. Process Biochem. 2002;38:101–108. doi: 10.1016/S0032-9592(02)00058-4. [DOI] [Google Scholar]
- 8.Ren W., Wang S.J., Lü M.S., Wang X.B., Fang Y.W., Jiao Y.L., Hu J.E. Optimization of four types of antimicrobial agents to increase the inhibitory ability of marine Arthrobacter oxydans KQ11 dextranase mouthwash. Chin. J. Oceanol. Limn. 2016;34:354–366. doi: 10.1007/s00343-015-4376-3. [DOI] [Google Scholar]
- 9.Wang X.B., Lu M.S., Wang S.J., Fang Y.W., Wang D.L., Ren W., Zhao G.M. The atmospheric and room-temperature plasma (ARTP) method on the dextranase activity and structure. Int. J. Biol. Macromol. 2014;70:284–291. doi: 10.1016/j.ijbiomac.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Hild E., Brumbley S.M., O’Shea M.G., Nevalainen H., Bergquist P.L. A Paenibacillus sp. dextranase mutant pool with improved thermostability and activity. Appl. Microbiol. Biotechnol. 2007;75:1071–1078. doi: 10.1007/s00253-007-0936-6. [DOI] [PubMed] [Google Scholar]
- 11.Zohra R.R., Aman A., Ansari A., Haider M.S., Qader S.A.U. Purification, characterization and end product analysis of dextran degrading endodextranase from Bacillus licheniformis, KIBGE-IB25. Int. J. Biol. Macromol. 2015;78:243–248. doi: 10.1016/j.ijbiomac.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Netsopa S., Niamsanit S., Araki T., Kongkeitkajorn M.B., Milintawisamai N. Purification and Characterization Including Dextran Hydrolysis of Dextranase from Aspergillus allahabadii X26. Sugar Tech. 2018;8:1–12. doi: 10.1007/s12355-018-0652-9. [DOI] [Google Scholar]
- 13.Zhang Y.Q., Li R.H., Zhang H.B., Wu M., Hu X.Q. Purification, characterization, and application of a thermostable dextranase from Talaromyces pinophilus. J. Ind. Microbiol. Biotechnol. 2017;44:317–327. doi: 10.1007/s10295-016-1886-8. [DOI] [PubMed] [Google Scholar]
- 14.Cai R.H., Lu M.S., Fang Y.W., Jiao Y.L., Zhu Q., Liu Z.P., Wang S.J. Screening, production, and characterization of dextranase from Catenovulum sp. Ann. Microbiol. 2014;64:147–155. doi: 10.1007/s13213-013-0644-7. [DOI] [Google Scholar]
- 15.Klahan P., Okuyama M., Jinnai K., Ma M., Kikuchi A., Kumagai Y., Tagami T., Kimura A. Engineered dextranase from Streptococcus mutans enhances the production of longer isomaltooligosaccharides. Biosci. Biotechnol. Biochem. 2018;82:1–8. doi: 10.1080/09168451.2018.1473026. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y.M., Kim D. Characterization of novel thermostable dextranase from Thermotoga lettingae TMO. Appl. Microbiol. Biotechnol. 2010;85:581–587. doi: 10.1007/s00253-009-2121-6. [DOI] [PubMed] [Google Scholar]
- 17.Li K., Lu H.Q., Hang F.X., Li S.B., Liu J.D. Improved dextranase production by Chaetomium gracile through optimization of carbon source and fermentation parameters. Sugar Tech. 2017;19:432–437. doi: 10.1007/s12355-016-0476-4. [DOI] [Google Scholar]
- 18.Zohra R.R., Aman A., Zohra R.R., Ansari A., Ghani M., Qader S.A.U. Dextranase: Hyper production of dextran degrading enzyme from newly isolated strain of Bacillus licheniformis. Carbohydr. Polym. 2013;92:2149–2153. doi: 10.1016/j.carbpol.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 19.Gozu Y., Ishizaki Y., Hosoyama Y., Miyazaki T., Nishikawa A., Tonozuka T. A glycoside hydrolase family 31 dextranase with high transglucosylation activity from Flavobacterium johnsoniae. Biosci. Biotechnol. Biochem. 2016;80:1562–1567. doi: 10.1080/09168451.2016.1182852. [DOI] [PubMed] [Google Scholar]
- 20.Sufiate B.L., Soares F.E.F., Moreira S.S., Gouveia A.S., Cardoso E.F., Braga F.R., Araújo J.V., Queiroz J.H. In vitro and in silico characterization of a novel dextranase from Pochonia chlamydosporia. Biotech. 2018;8:1–9. doi: 10.1007/s13205-018-1192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatia S., Bhakri G., Arora M., Batta S.K., Uppal S.K. Kinetic and thermodynamic properties of partially purified dextranase from Paecilomyces lilacinus and its application in dextran removal from cane juice. Sugar Tech. 2016;18:1–7. doi: 10.1007/s12355-015-0378-x. [DOI] [Google Scholar]
- 22.Khalikova E., Susi P., Usanov N., Korpela T. Purification and properties of extracellular dextranase from a Bacillus sp. J. Chromatogr. B. 2003;796:315–326. doi: 10.1016/j.jchromb.2003.08.037. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y.M., Ko E.A., Kang H.K., Kim D. Construction, expression and characterization of fusion enzyme from Arthrobacter oxydans dextranase and Klebsiella pneumoniaeamylase. Biotechnol Lett. 2009;31:1019–1024. doi: 10.1007/s10529-009-9967-7. [DOI] [PubMed] [Google Scholar]
- 24.Yang L., Zhou N.D., Tian Y.P. Purification, characterization, and biocatalytic potential of a novel dextranase from Chaetomium globosum. Biotechnol Lett. 2018;40:1407–1418. doi: 10.1007/s10529-018-2599-z. [DOI] [PubMed] [Google Scholar]
- 25.Abdelnaby M.A., Ismail A.M.S., Abdelfattah A.M. Preparation and some properties of immobilized Penicillium funiculosum 258 dextranase. Process Biochem. 1999;34:391–398. doi: 10.1016/S0032-9592(98)00127-7. [DOI] [Google Scholar]
- 26.Zhang L.X., Duan R., Chen L.J., Ling X.P., Lu Y.H. Expression of α-1,6-dextranase from Penicillium minioluteum in P.pastoris. Sci. Technol. Food Ind. 2011;11:194–197. [Google Scholar]
- 27.Wynter C.V.A., Chang M., Jersey J.D., Patel B., Inkerman P.A., Hamiton S. Isolation and characterization of a thermostable dextranase. Enzym. Microb. Technol. 1997;20:242–247. doi: 10.1016/S0141-0229(96)00118-4. [DOI] [Google Scholar]
- 28.Lee J.H., Nam S.H., Park H.J., Kim Y.M., Kim N., Kim G., Seo E.S., Kang S.S., Kim D. Biochemical characterization of dextranase from Arthrobacter oxydans and its cloning and expression in Escherichia coli. Food Sci. Biotechnol. 2010;19:757–762. doi: 10.1007/s10068-010-0106-y. [DOI] [Google Scholar]
- 29.Purushe S., Prakash D., Nawani N.N., Dhakephalkar P., Kapadnis B. Biocatalytic potential of an alkalophilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manufacture. Bioresour. Technol. 2012;115:2–7. doi: 10.1016/j.biortech.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Galvez M.A., Lopez M.A. Production and characterization of a dextranase from an isolated Paecilomyces lilacinus strain. Appl. Microbiol. Biotechnol. 1991;36:327–331. [Google Scholar]
- 31.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 32.Abdelwahed N.A.M., Ahmed E.F., El-Gammal E.W., Hawas U.W. Application of statistical design for the optimization of dextranase production by a novel fungus isolated from Red Sea sponge. Biotech. 2014;4:533–544. doi: 10.1007/s13205-013-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsson N., Borde A., Wölfel S., Kerman B., Larsson A. Quantification of protein concentration by the Bradford method in the presence of pharmaceutical polymers. Anal. Biochem. 2011;411:116–121. doi: 10.1016/j.ab.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 34.Lineweaver H., Burk D. The determination of enzyme dissociation constant. J. Am. Chem. Soc. 1934;56:658–666. doi: 10.1021/ja01318a036. [DOI] [Google Scholar]
- 35.Pawcenis D., Thomas J.L., Łojewski T., Milczarek J.M., Łojewska J. Towards determination of absolute molar mass of cellulose polymer by size exclusion chromatography with multiple angle laser light scattering detection. J. Chromatogr A. 2015;1409:53–59. doi: 10.1016/j.chroma.2015.06.042. [DOI] [PubMed] [Google Scholar]