Abstract
Buckwheat (Fagopyrum esculentum) is a valuable crop which can produce multiple human beneficial secondary metabolites, for example, the anthocyanins in sprouts and flowers. However, as the predominant group of visible polyphenols in pigmentation, little is known about the molecular mechanisms underlying the anthocyanin biosynthesis within buckwheat. In this study, a comparative transcriptome analysis of green and red common buckwheat cultivars was carried out through RNA sequencing. Overall, 3727 and 5323 differently expressed genes (DEGs) were identified in flowers and cotyledons, respectively. Through GO and KEGG analysis, we revealed that DEGs in flowers and cotyledons are predominately involved in biosynthesis of anthocyanin. A total of 42 unigenes encoding 11 structural enzymes of the anthocyanin biosynthesis were identified as DEGs. We also identified some transcription factor families involved in the regulation of anthocyanin biosynthesis. Real-time qPCR validation of candidate genes was performed in flowers and cotyledons, and the results suggested that the high expression level of structural genes involved in anthocyanin biosynthetic pathway promotes anthocyanin accumulation. Our results provide the insight understanding for coloration of red common buckwheat.
Keywords: buckwheat (Fagopyrum esculentum Moench), cotyledon, flower, transcriptome analysis, anthocyanin
1. Introduction
As one of the most important grain and honey crops, buckwheat (Fagopyrum esculentum Moench) is widely planted all over the world, especially in China, Japan, Canada, and Russia [1,2]. The harvested seeds of buckwheat are conventionally used to make whole grain foods, groats, noodles, and flour, as well as other cereal crops [3,4]. It has been reported that buckwheat can accumulate abundant nutrients in grain and hull, such as flavonoids, anthocyanin, essential amino acids, and tannin [5,6,7]. Generally, common buckwheat has two different flower colors, either white or pink. However, some cultivars have developed red flowers, which is believed to be related to the accumulation of anthocyanin, and the petals of the red flower buckwheat are traditionally used as materials in the manufacturing of several beverages, such as alcohol and tea after the detoxification of fagopyrin [8,9].
Anthocyanin is normally considered to be one of the water-soluble flavonoids which are widely distributed in plant vegetative tissues, and are synthesized in cytoplasm then accumulated in vacuoles. The ranging colors, from red to purple, reflected on higher plant tissues are mostly attributed to the level of anthocyanin accumulation [10,11]. Moreover, anthocyanins play important roles in plant development, such as providing protection against UV radiation, low temperature, drought stress, and pathogens [12,13,14], and their accumulation in flowers attracts pollinators [15,16]. More importantly, these pigments could also help humans to prevent several chronic diseases [17,18].
The anthocyanin biosynthetic pathway has been extensively studied in several plant species, including Petunia hybrid, Antirrhinum majus, and Zea mays [19,20,21], and the biosynthesis of anthocyanin is viewed as a branch of flavonoid production The structural genes encoding multiple enzymes of this pathway are classified as two groups: the early-stage biosynthetic genes (EBGs), including phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H); and the late-stage biosynthetic genes (LBGs) induced afterwards, dihydroflavonol reductase (DFR), anthocyanidin synthase/leucoanthocyanidin oxygenase (ANS/LDOX), and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) are involved in the LBGs [22,23]. Transcriptional factors regulating the expression of these genes are presently known as R2R3 MYB, basic helix-loop-helix (bHLH), and WD repeat (WDR), which can form a MYB-bHLH-WDR (MBW) complex binding to the anthocyanin structural gene promoter to control its transcription and affect the accumulation of anthocyanins [24,25,26]. Currently, with the development of the 3rd-generation sequencing technology, an increasing number of genes involved in the plant anthocyanin biosynthetic have been identified and annotated without reference genomes, such as Litchi chinensis [27], bougainvillea [28], and radish [29]. Nevertheless, the molecular mechanism of pigment accumulation in red buckwheat has yet to be further explored.
A previous study has reported that a Chinese buckwheat variety, Gan-Chao, which accumulates anthocyanins in petals, that the accumulation increased along with the development of the flower [8,9]. Likewise, in our previous study, we discovered a local buckwheat cultivar, “HHTQ”, in the Guizhou province of China. The cultivar presents red cotyledons, leaves, and flowers during the growth, acting as a potentially favorable material to not only study the molecular biosynthesis of anthocyanin in buckwheat, but also provide the very opportunity to develop anthocyanin-rich buckwheat products. In this study, we have developed the high-throughput RNA sequencing (RNA-seq) in both cotyledon and flowers of the green and red buckwheat cultivars respectively, through which the structural genes involved in anthocyanin biosynthesis were systemically analyzed. Our study may improve understanding of the regulatory mechanism of buckwheat anthocyanin production.
2. Results
2.1. Anthocyanins Quantification
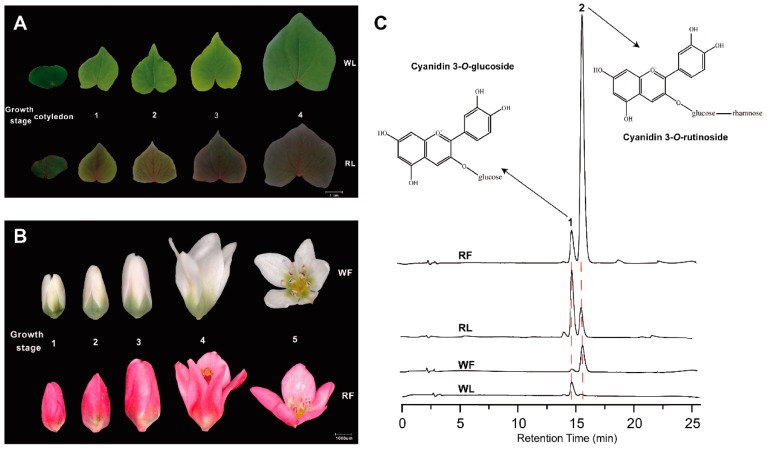
The leaves and flowers from two cultivars at different growth stages are shown in Figure 1. Apparently, cultivar “HHTQ” has red-vein cotyledons, red leaves, and red petals during the different growth stages. Comparatively, cultivar “Beizaosheng” presents green leaves and white flowers (Figure 1A,B. Origin figures can be found in Figure S1). To examine the anthocyanin composition in the buckwheat, the high-performance liquid chromatography (HPLC) was carried out and identified two major kinds of anthocyanins—cyanidin 3-O-gulcoside (peak 1) and cyanidin 3-O-rutinoside (peak 2)—in the cotyledon and flower. The highest levels of cyanidin 3-O-gulcoside and cyanidin 3-O-rutinoside were found in the cotyledon and flower of “HHTQ” (Figure 1C, Table 1). The total anthocyanin contents at flower and cotyledon of the “HHTQ” were 7.51-fold and 5.12-fold higher than that of “Beizaosheng”, respectively, and the red flower of “HHTQ” contained a higher amount of cyanidin 3-O-rutinoside (2.83 mg/g DW) than “Beizaosheng” (0.38 mg/g DW). In the cotyledon of “HHTQ”, cyanidin 3-O-gulcoside showed the highest content (0.97 mg/g DW), obviously higher than cyanidin 3-O-rutinoside, and there were not obvious absorption signals for cyanidin 3-O-rutinoside detected in the cotyledon of “Beizaosheng”. These results suggest that more anthocyanins are accumulated in red cultivar “HHTQ”, and the accumulation of cyanidin 3-O-rutinoside in flowers and the accumulation of cyanidin 3-O-gulcoside in cotyledon are higher.
Figure 1.
The morphology of buckwheat leaves and flowers, and component analysis of anthocyanins in different buckwheat cultivars. (A) The leaves of white cultivar “Beizaosheng” (WL) and red cultivar “HHTQ” (RL) at different growth stages. Scale bar = 1 cm. (B) The flowers of white cultivar “Beizaosheng” (WF) and red cultivar “HHTQ” (RF) at different growth stages. Scale bar = 1000 µm. (C) High-performance liquid chromatography (HPLC) profiles of anthocyanins extracted from the cotyledon and flowers of two buckwheat cultivars (detected at A520).
Table 1.
Anthocyanin levels (mg/g dry weight) in different tissues of the two buckwheat cultivars.
| Peak | Compound | Samples | |||
|---|---|---|---|---|---|
| RF | RL | WF | WL | ||
| 1 | cyanidin 3-O-gulcoside | 0.52 ± 0.01 | 0.97 ± 0.02 | 0.07 ± 0.01 | 0.26 ± 0.01 |
| 2 | cyanidin 3-O-rutinoside | 2.83 ± 0.01 | 0.36 ± 0.01 | 0.38 ± 0.02 | Nd 1 |
| Total | 3.38 ± 0.01 | 1.33 ± 0.02 | 0.45 ± 0.03 | 0.26 ± 0.01 | |
1 Nd means not detected.
2.2. Full-Length Transcriptome Sequencing and RNA-seq
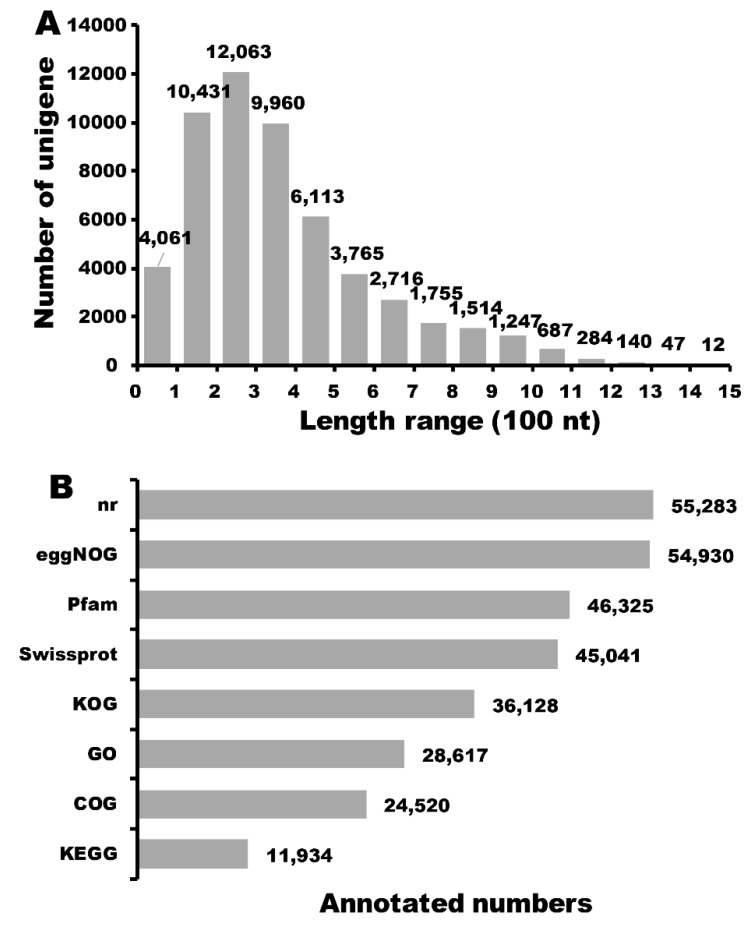
There is a reported genome of buckwheat (Fagopyrum esculentum Moench) [30] which is distantly related to the cultivars used in our study (Fagopyrum tataricum (L.) Gaertn). To fully understand the potential molecular mechanism involved in anthocyanin synthesis, we firstly performed the full-length transcriptome sequencing to three libraries with different size fractions (1–2 K, 2–3 K, and 3–6 K) on five SMRT cells using the PacBio Sequel system (Table S1). A total of 281,566 reads of insert (ROIs), including 126,054 ROIs from two SMRT cells for 1–2 K fractions, 91,798 ROIs from two SMRT cells for 2–3 K fractions, and 63,714 ROIs from a SMRT cell for 3–6 K fractions were generated with an average of 3,536 bp in length, and 123,039 full length reads non-chimeric (FLNC) remained after circular consensus sequence (CCS) generation and filtering for full-length read classification. Secondly, we performed Illumina RNA-Seq to twelve cDNA libraries and there were 22,322,429 (RF), 23,503,180 (WF), 23,413,984 (RL), and 23,218,940 (WL) clean reads left after trimming and filtering (Table S2). The full-length (FL) reads were clustered into consensus clusters based on RS_IsoSeq of SMRT Analysis, then 64,539 high-quality (HQ) and consensus isoforms were merged into 51,673 final consensus sequences in total, and 12,866 low-quality consensus isoforms were rectified using the Illumina RNA-seq data. Subsequently, we utilized CD-HIT for further clustering to obtain the non-redundant transcripts and a total of 56,864 full-length transcripts of Fagopyrum esculentum were generated. There were 37,881 complete ORFs that were obtained by TransDecoder software, and the length of CDS (coding sequence) encoded protein is shown in Figure 2A. Of these CDSs, only a minority (59 CDSs) were more than 1300 nucleotide (nt), and 70.39% of CDSs appeared with a length ranging from 100 to 500 nt. All unigenes were blasted against eight public databases: Nr, Swiss-Prot, KEGG, GO, KOG, Pfam, eggnog, and COG, and the results showed that 55,426 unigenes were annotated in this transcriptome data (Figure 2B). Among the 55,426 unigenes, 24,520 unigenes (44.24%), 36,128 unigenes (65.18%), 46,325 unigenes (83.58%), 55,283 unigenes (99.74%), 54,930 unigenes (99.11%), 45,041 unigenes (81.26%), 28,617 unigenes (51.63%), and 11,934 unigenes (21.53%) were annotated in COG, KOG, Pfam, Nr, eggNOG, Swissprot, GO, and KEGG, respectively.
Figure 2.
Predicted length distribution map of coding sequence (CDS) encoded protein and statistic summary of functional annotation for buckwheat unigenes in public databases. (A) Predicted length distribution map of CDS encoded protein. Nucleotide: (nt). (B) Statistic summary of functional annotation for buckwheat unigenes in eight public databases.
2.3. Identification of Different Expressed Genes
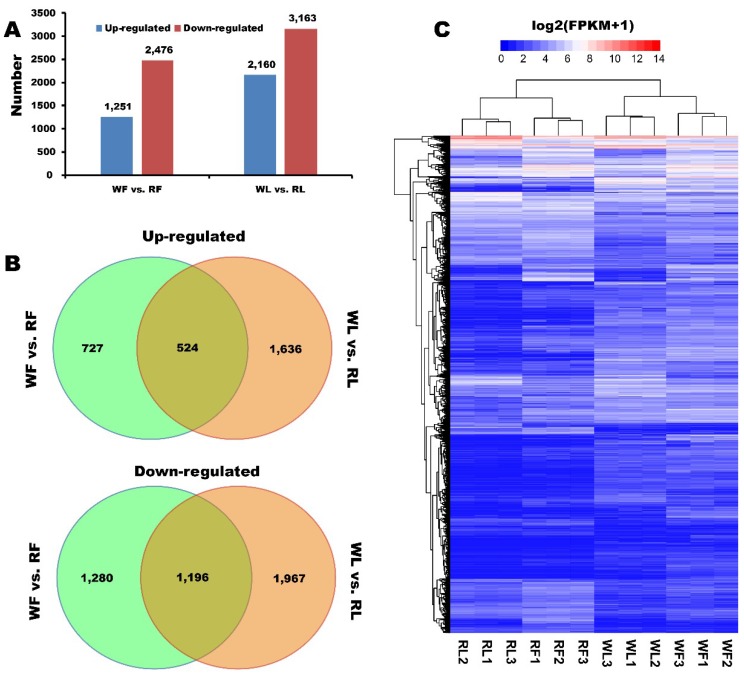
To further reveal the underlying molecular mechanisms of anthocyanin biosynthesis, we performed RNA-seq analysis to the cotyledon and flower materials of white cultivar “Beizaosheng” (WL and WF) and red cultivar “HHTQ” (RL and RF), and each has three replicates (Figure S2). A total of 9050 differently expressed genes (DEGs) in buckwheat were identified though pairwise comparisons, of which 3727 and 5323 DEGs were found between WF and RF, and WL and RL, respectively (Figure 3A). Comparison of the RF with the WF revealed 1251 unigenes that were up-regulated and 2476 unigenes that were down-regulated. In total, 2160 unigenes were up-regulated and 3163 genes were down-regulated under the RL compared with the WL. Five hundred and twenty-four unigenes of 1251 up-regulated DEGs between RF and WF were also among the genes differentially expressed between RL and WL. In addition, 1196 of 2476 down-regulated DEGs between RF and WF were also among the genes differentially expressed between RL and WL (Figure 3B). Furthermore, a general overview of the expression pattern was visualized in a heat map (Figure 3C), which provided an overall understanding of the change in gene expression.
Figure 3.
Changes in gene expression profiles in flowers and leaves of two different cultivars. (A) Numbers of differently expressed genes (DEGs) in pairwise comparisons in flowers and cotyledons of two different cultivars of buckwheat. (B) Venn diagram showing the number of DEGs in flowers and cotyledons of two different cultivars of buckwheat. (C) Heat-map diagrams showing the relative expression levels of total DEGs among the four tissues. WF vs. RF means the flowers sample of white cultivar “Beizaosheng” compared to red cultivar “HHTQ”, and RL vs. RL means the cotyledons samples of white cultivar “Beizaosheng” compared to red cultivar “HHTQ”. Up- and down-regulated means that genes are up- and down-regulated in “Beizaosheng” compared to “HHTQ”.
2.4. GO Annotation, KEGG Pathway, and Enrichment Analyses
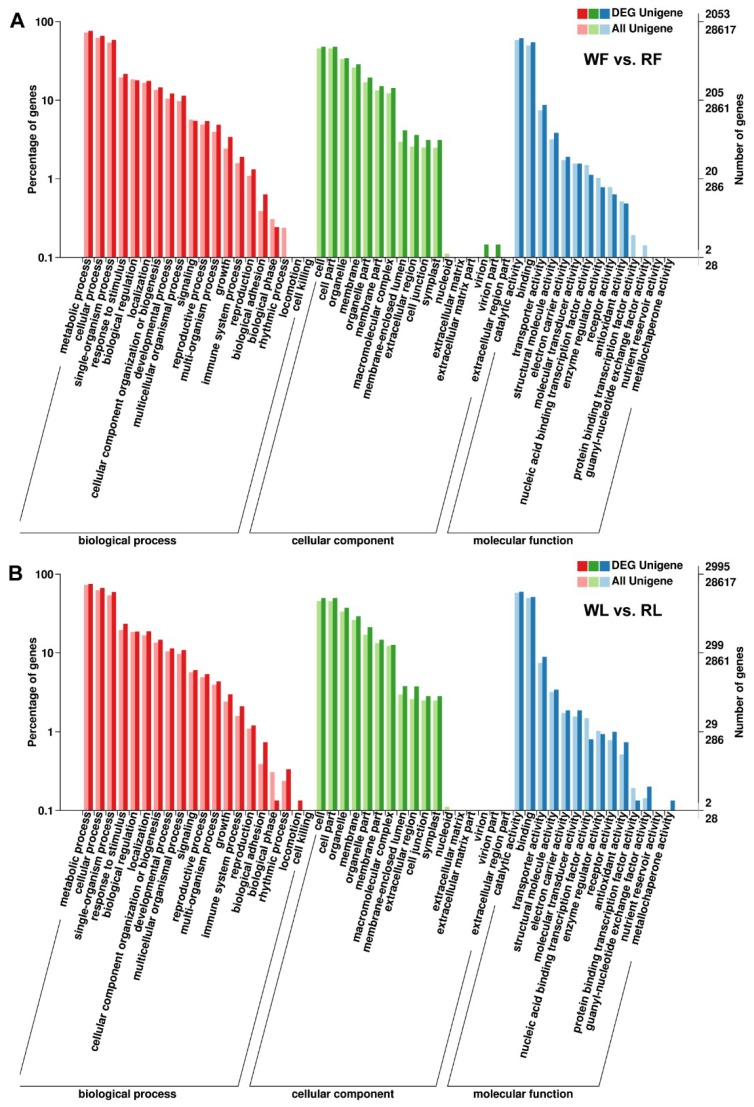
To identify the anthocyanin accumulation genes in the red buckwheat cultivar, the DEGs were evaluated with GO and KEGG pathway analyses. According to GO annotation, the proportions of enriched unigenes were summarized in three main GO categories and distributed into 49 functional terms as follows: 19 terms for biological process, 16 terms for cellular component, and 14 terms for molecular function. These unigenes in the biological process group were mainly involved in the metabolic process, the cellular process, and the single-organism process. The cellular component terms related to cell, cell part, and organelle. Most of the molecular function unigenes were located in the catalytic activity and binding (Figure 4). Meanwhile, an enrichment analysis based on the KEGG database was carried out to further explore the biological functions of the DEGs. The DEGs were significantly enriched in “flavonoid biosynthesis”, “photosynthesis”, and “valine, leucine, and isoleucine biosynthesis” in the WF vs. RF comparison, and “taurine and hypotaurine metabolism”, “photosynthesis”, and “valine, leucine and isoleucine biosynthesis” were significantly enriched in the KEGG pathway analysis in the WL vs. RL comparison (Table S3).
Figure 4.
Gene ontology (GO) categorization of buckwheat unigenes. (A) GO analysis of DEGs between white flowers (WF) and red flowers (RF) in three main categories. (B) GO analysis of DEGs between white cotyledon (WL) and red cotyledon (RL) in three main categories.
2.5. Expression Profile of Genes Responsible for Anthocyanins Production
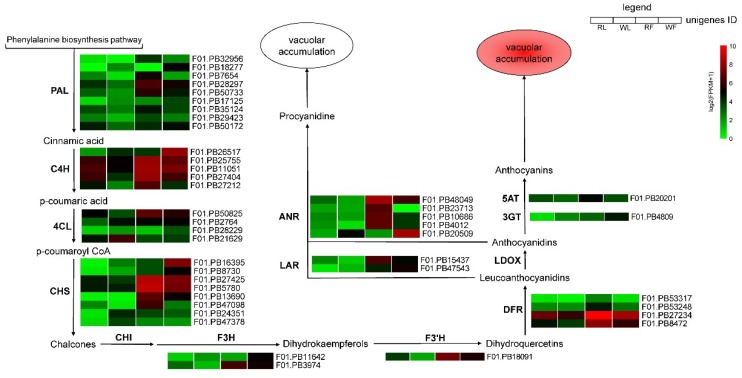
The gene expression levels were normalized to the FPKM (Reads per kilobase of exon model per million mapped reads) to compare the changes in gene expression levels between different colored buckwheat, and the twelve samples were hierarchically clustered based on the log2(FPKM+1), allowing us to observe the overall gene expression pattern (Figure 5). A total of 11 anthocyanin biosynthesis-related structural genes were identified as DEGs, and most of them showed high expression levels in the red buckwheat compared with the white cultivar.
Figure 5.
Heat map diagrams of relative expression levels of anthocyanin biosynthesis-related structural genes in the different colors of buckwheat. PAL: Phenylalanine ammonia-lyase, C4H: Cinnamate 4-hydroxylase, 4CL: 4-coumaroyl:CoA ligase, CHS: Chalcone synthase, CHI: Chalcone isomerase, F3H: Flavanone 3-hydroxylase, F3’H: Flavanone 3’-hydroxylase, DFR: Dihydroflavonol 4-reductase, LDOX: Leucoanthocyanidin dioxygenase, LAR: Leucoanthocyanidin reductase, ANR: Anthocyanidin reductase, 3GT: UDP-glucose: Anthocyanidin 3-O-glucosyltransferase, 5AT: Anthocyanin 5-aromatic acyltransferase.
The predicted proteins encoded by upstream genes included nine PAL (phenylalanine ammonia-lyase) genes that were identified, most of which (except for F01.PB17125 and F01.PB18277) were up-regulated in the RF and RL compared with WF and WL. Five C4H (Cinnamate 4-hydroxylase) genes were identified, most of which (except for F01.PB26517) were up-regulated in the RF and RL compared with WF and WL. Four 4CL (4-coumaroyl:CoA ligase) genes were identified, among which the expression levels of F01.PB50825 was higher in the red cultivar than the white cultivar, and the rest of them were down-regulated. CHS (chalcone synthase) plays an important role in anthocyanin biosynthesis, and eight CHS genes were identified, and half of them were up-regulated in the RF and RL compared with WF and WL. One F3’H (flavanone 3′-hydroxylase) gene and four DFR (dihydroflavonol 4-reductase) genes identified here displayed similar expression profiles, which were up-regulated in the red cultivar compared with the white cultivar. Additionally, there were no LDOX (leucoanthocyanidin dioxygenase) genes identified as DEGs, but there were five ANR (anthocyanidin reductase) genes and two LAR (leucoanthocyanidin reductase) genes, which can promote PA biosynthesis and contribute to the green skinned variant, were identified, and they (except for F01.PB20509) showed expression patterns similar to those of F3’H, except that one ANR gene was up-regulated in the white cultivar compared with the red cultivar.
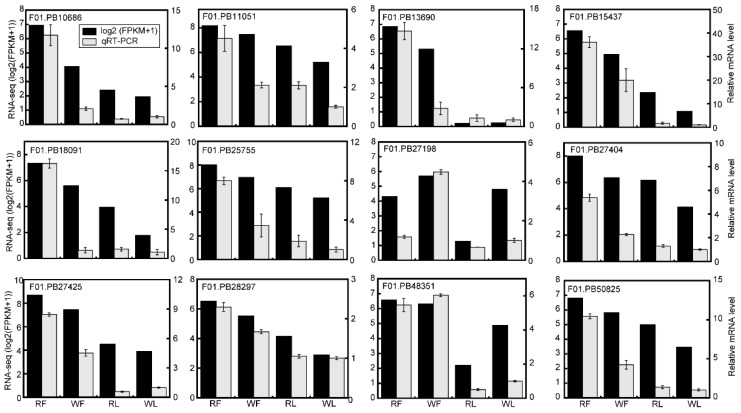
In order to verify the RNA-Seq data, real-time PCR was carried out to validate the candidate genes involved in the anthocyanin biosynthesis. Thirteen candidate DEGs were selected for RT-qPCR assays. The expression pattern of these genes were consistent with those of the transcriptome analysis (Figure 6).
Figure 6.
Expression of anthocyanin biosynthesis-related unigenes quantified by RNA-seq (RNA sequencing) and qRT-PCR (Quantitative real-time PCR) analysis. The left y-axis indicates the log2(FPKM+1) values of unigenes from RNA-seq data (black columns). The right y-axis indicates relative gene expression levels analyzed by qRT-PCR (white columns), and the expression of WL was chosen as the control standardizing all samples, values are presented as mean ± SE (n = 3).
3. Discussion
3.1. Components Variations of Anthocyanin and Color Levels Between “HHTQ” and “Beizaosheng”
The types and quantities of anthocyanins differ among different species. Previous studies have reported that cyanidin 3-O-gulcoside and cyanidin 3-O-rutinoside were identified as the major anthocyanin compound in buckwheat [8,9,31]. In red walnut, the main anthocyanin is cyanidin-3-O-galactoside [32], and malvidin-3,5-O-diglucoside was detected as the major anthocyanin in grapes [33]. In this study, we detected differences in composition and concentrations of anthocaynins between the cotyledons and flowers of buckwheat. “HHTQ” had shown the highest anthocyanins accumulation in cotyledons and flowers, and the accumulation of cyanidin 3-O-rutinoside in flowers and the accumulation of cyanidin 3-O-gulcoside in cotyledon are much higher. Meanwhile, anthocyanins remained the lowest in “Beizaosheng”. This result suggests that the pathways of anthocyanin synthesis differ between the cotyledon and flowers of red common buckwheat and confirmed that the phenotypic differences in coloration between red and green common buckwheat are due to the anthocyanin accumulation. These differences were consistent with the different gene expression pattern in the anthocyanin biosynthesis pathway.
3.2. Genes Involved in the Anthocyanin Biosynthesis Pathway are Differentially Regulated
The anthocyanin biosynthetic pathway is the branch of the phenylpropanoid and flavonoid pathways, and anthocyanins are synthesized in reactions catalyzed by a series of enzymes. Phenylalanine (Phe), the direct precursors of anthocyanin biosynthesis, is catalyzed by PAL, C4H, and 4CL, converting Phe into p-coumaroyl CoA. CHS, the first committed enzyme in anthocyanin biosynthesis [32], catalyzes the p-coumaroyl CoA to chalcone, while chalcone isomerase (CHI) catalyzes the conversion of chalcone to naringenin. F3H, which belongs to the OGD family [10], catalyzes naringenin to form dihydrokaempferol (DHK). The conversion of DHK to (2R,3R)-dihydroquercetin and dihydromyricetin is catalyzed by flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H), respectively. F3′H and F3′5′H are the key enzymes that determine the structures of anthocyanins and affect color formation [20]. Dihydroflavonol-4-reductase (DFR) catalyzes dihydroflavonols to leucoanthocyanidins, which then are converted to corresponding colored anthocyanidins by LDOX/ANS. Finally, UFGT catalyzes anthocyanidin to anthocyanin, and then transports it to the vacuole. In many plants, the expression levels of certain anthocyanin biosynthetic genes were strongly associated with the anthocyanin accumulation [34,35,36,37]. In this study, we identified 35 DEGs that were annotated as certain anthocyanin biosynthetic genes that were up-regulated in RF and RL. F3′H, which leads to the production of the red cyanidin-based pigments [38,39,40], and DFR, which is a key enzyme committed to anthocyanin biosynthesis in the flavonoid biosynthetic pathway [7,41], displayed significantly increased expression levels in RF (Figure 5). Notably, these results suggested that these unigenes are responsible for color formation in the cotyledons and flowers of “HHTQ”.
However, none of the DEGs were annotated as CHI and F3′5′H, except for the DEG, F01.PB27425, which was annotated as CHS, and showed significantly higher expression levels in the RF than WF. Meanwhile, another DEG, F01.PB16395, was annotated as CHS and showed higher transcriptional levels in “Beizaosheng” than in “HHTQ” (Figure 5). Li et al. [32] has also detected three unigenes (annotated as CHS, F3H, and F3’5’H, respectively), demonstrating significantly higher expression levels in green peel than in the red peel of walnut, of which the green tissues are expected to contain more colorless flavonoids.
A critical step in anthocyanin synthesis is mediated by dihydroflavonol-4-reductase (DFR), and overexpression of DRF can increases the accumulation of anthocyanins and produce red flowers [42]. In the present study, four DFR genes were identified and the F01.PB27234 was mostly expressed in the RF (FPKM > 1000), which indicates that it may be crucial for anthocyanin accumulation in the red flowers of buckwheat. Anthocyanidin 3-O-glucosyltransferase (3GT) catalyzes the transfer of the glucosyl moiety form UDP-glucose to the 3-hydroxyl group of the corresponding acceptor molecule. This glycosylation step play an important role in anthocyanin stability and water solubility [43,44]. In our transcriptome analysis, one unigene (F01.PB20201), annotated as 3GT, was found to be lower expressed in the flowers of the “HHTQ” compared with the “Beizaosheng” cultivar. F01.PB20201, annotated as anthocyanin 5-aromatic acyltransferase (5AT), which acrylates the glucose bound at the position of anthocyanidin 3,5-diglucoside [45], was up-regulated in RF. These genes may be involved in the anthocyanin biosynthesis and may cause the formation of red flower buckwheat. These results also point out the direction for further studies on the coloration of vegetables and fruit.
3.3. Components DEGs Related to the Anthocyanin Degradation
Leucoanthocyanidin reductase (LAR) can convert leucoanthocyanidin to 2,3-trans-flavanols, such as (+)-catechin, and its activity correlated with proanthocyanidins accumulation [46,47]. Previous studies indicated that ectopic expression of the apple MdLAR1 gene in tobacco inhibits expression of the late genes in anthocyanin biosynthetic pathways, resulting in a decrease in anthocyanin accumulation in flowers [48]. In our transcriptome data, we also noticed that one LAR gene (F01.PB47543) was up-regulated in the “Beizaosheng” compared with “HHTQ”. The ANR gene encodes an anthocyanin reductase that can reduce anthocyanidins to flavan-3-ols required for the formation of proanthocyanidins on the flavonoids pathway [49,50]. In the present study, four of the ANR genes were up-regulated in the RF, however, one ANR gene (F01.PB20509) was down-regulated in the flowers and cotyledons of “HHTQ”. The previous study showed that experimental down-regulation of ANR by RNAi can induce premature and ectopic anthocyanin formation in Fragaria × ananassa [51]. Whether these genes are involved in the anthocyanin degradation and exert the same effect as the homologous genes still needs further analysis.
3.4. DEGs Involved in the Regulation of Anthocyanin Accumulation
Additionally, anthocyanin accumulation is also regulated by a suite of transcription factors (TFs), including MYB, bHLH, and WD40 families [52,53,54]. The MYB and bHLH proteins combine with the WD40 protein to form WD-repeat/Myb/bHLH transcriptional complexes that activate anthocyanin biosynthetic genes [25,55,56], and the R2R3-MYB can predominantly regulate the accumulation of anthocyanin [18]. In this study a MYB1 homolog (F01.PB20382) and a TT2 homolog (F01.PB13850) were identified and showed a high transcription level in the flower of “HHTQ”. Additionally, three bHLH143 (F01.PB46560, F01.PB12993, and F01.PB52152) were also up-regulation in the RF compared with WF (Table S4). These findings suggest that the MYB and bHLH family may correlate with the regulation of anthocyanin accumulation in the buckwheat. On the other hand, phytohormones, such as abscisic acid (ABA) and jasmonates (JAs), can also stimulate anthocyanin accumulation by regulation anthocyanin biosynthetic genes [57,58,59,60,61]. In sweet cherry (Prunus avium L.), ABA treatment strongly induced anthocyanin accumulation and the colorless phenotype was also observed in the fruits when silencing PacNCED1 (9-cis-epoxycarotenoid dioxygenase 1), the cognate gene encoding a key enzyme in the ABA biosynthesis pathway [59]. In addition, previous studies have shown that JA can strongly induce the LBGs in Arabidopsis [57], and the jasmonate-ZIM-domain (JAZ) proteins, acting as negative regulators of jasmonate-responsive genes, degrade in response to the JA signal, which releases bHLH and MYB TFs to repress JA-regulated anthocyanin accumulation [58,62,63]. In this study, we also detected several NCED1 homologs and JAZ protein homologs (Table S5), and most of the NCED1 homologs showed no significant change between red and white cultivar, however, four JAZ proteins were identified and showed down-regulated in the red buckwheat. These results suggest that the NCED1 genes may not associate with anthocyanin accumulation in the buckwheat, but the JAZ proteins might act as negative regulators and be involved in the anthocyanin biosynthesis pathway. These homologous genes still need further analysis.
4. Materials and Methods
4.1. Plant Materials
Two cultivars, the red common buckwheat ‘HHTQ’ (red leaves and red flowers), and the green common buckwheat ‘Beizaosheng’ (green leaves and white flowers), were used in this study. They were grown in fields at Yangtze University, Jinzhou, China. When the flowers appeared, flower tissues from both cultivars were collected. Cotyledons were collected from 7-day-old aseptic seedlings, which were grown in sugar-free MS medium. All samples were quickly frozen in liquid nitrogen and stored at –80°C until used. To investigate the accumulation pattern of anthocyanin, different growth stages of flowers and leaves were harvested and mixed to measured anthocyanin content. Leaves were classified according to leaf width: “cotyledons”: 4.0–5.0 cm, “Stage 1”: 4.0–5.0 cm, “Stage 2”: 5.0–6.0 cm, “Stage 3”: 6.0–7.0 cm, “Stage 4”: 7.0–9.0 cm, and fresh weights were used to classify the flowers as described by Suzuki et al. [8]: “Stage 1”: 0.0–0.4 mg, “Stage 2”: 0.4–0.8 mg, “Stage 3”: 0.8–1.6 mg, “Stage 4”: 1.6–2.6 mg, “Stage 5” (fully developed petal): 2.6–3.6 mg.
4.2. RNA Preparation
The cotyledons and flowers tissues were collected from red cultivar ‘HHTQ’ and white cultivar ‘Beizaosheng’. The total RNA was extracted from the samples using EASYspin Plant RNA Kit (Aidlab, Wuhan, China) according to the manufacturer’s instructions, and were then treated with DNase I to remove genomic DNA [64]. RNA quality was checked and quantified using a Nanodrop 2000r (Nanodrop Technologies, Wilmington, DE, USA) and Agilent bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), respectively.
4.3. PacBio Iso-Seq Library Preparation and Sequencing
Full-length transcriptome sequencing was carried out at Biomarker Technologies Co. Ltd. (Beijing, China), following manufacturer protocols. Briefly, the high-quality, full-length cDNAs was synthesized from total RNA by using a SMARTer™ PCR cDNA Synthesis Kit (Takara, Dalian, China), and after a round of PCR amplification, the BluePippin™ Size Selection System (Sage Science, Beverly, MA, USA) was used to construct cDNA libraries of different sizes: 1–2 K, 2–3 K, and 3–6 K. Three Iso-Seq libraries were sequenced on five cells with PacBio RS II, and Illumine sequencing data were used to polish the PacBio assembly. The raw sequencing data were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive with a Bioproject ID: PRJNA517031.
4.4. Illumina Sequencing, Data Assembly, and Gene Annotation
Illumina sequencing reads analysis was performed at Biomarker Technologies Co. Ltd. (Beijing, China), following manufacturer protocols. Each sample has three replicates. Briefly, mRNA was separated from total RNA using oligo (dT) magnetic beads in accordance with the Illumina manufacturer’s instructions and fragmented into small pieces using a fragmentation buffer. Then, the first- and second- stand cDNA were synthesized using these fragments as reverse transcription templates. Finally, paired-end adapters were ligated to the ends of the cDNA fragments. The appropriate DNA fragments were used as amplification templates for polymerase chain reaction amplification. The cDNA libraries were sequenced on Illumina HiSeq 2500 platform. The raw reads were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive with a Bioproject ID: PRJNA517026.
Quality parameters, including GC-content, sequence duplication level, and Q30 were used to filter the reads from the Illumina RNA-seq data. Then, the high-quality clean data were obtained from the raw reads after removing the low-quality data, which included adapter sequences and low-quality reads. Cleaned and qualified reads were assembled in Trinity software [65], and the resulting sequences were called Unigenes. BLASTX alignment was performed between these unigene sequences and public protein databases: the NCBI non-redundant (Nr), Swiss-Prot, clusters of orthologous groups of proteins (COG), euKaryotic ortholog groups (KOG), protein family (Pfam), gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG) database, and nucleotide database (NT) [66]. Cut-off value was set as e-value < 1 × 10−5.
4.5. Differentially Expressed Gene Identification
For differential expression tests (DET), we compared two conditions: WF vs. RF, and WL vs. RL. For each comparison, genes with low read counts per million (cpm < 1) that did not represent sufficient statistical significance were excluded from the DET analysis. The libraries were digitally normalized after filtering using the DESeq. Genes with |Fold Change| ≥ 2 and FDR (false discovery rate) < 0.01 were considered as highly differentially expressed genes (DEGs). The gene expression unit was calculated using the FPKM method (fragments per kilobase of transcript per million mapped reads).
4.6. GO Functional Enrichment and Pathway Analysis of DEGs
The DEGs were mapped to GO terms in the GO database (http://www.geneontology.org/) to calculate a gene number for every term. The hypergeometric test was used to find significantly enriched GO terms based on ‘GO::TermFinder’ (http://smd.stanford.edu/help/GOTermFinder/GO_TermFinder_help.shtml/). GO terms conforming top-value through Bonferroni Correction ≤ 0.05 were defined as significantly enriched GO terms. KEGG (http://www.genome.jp/kegg/) was used to perform pathway enrichment analysis of DEGs. MapMan software (version 3.6.0) was used to display expression profiles at the pathway level.
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA from flowers and cotyledons of both cultivars were extracted and subjected to DNase I digestion (Takara, Dalian, China) to remove DNA. The first-strand cDNA for qPCR analysis was synthesized from 500 ng of total RNA using oligo dT (Takara, Dalian, China) according to the manufacturer’s protocols. The primers were designed using Primer Premier 5.0 and beta-Actin was used as a reference gene (Table S6). The qRT-PCR was performed on a CFX 96 real-time PCR system (BioRad, Hercules, CA, USA) using SYBR (TaKaRa), according to manufacturer’s protocols. Each treatment included three replications [67].
4.8. Extraction and HPLC Analysis of Anthocyanins
Anthocyanins were extracted and determined according to published methods with minor modifications [68,69]. Different stages of flower and leaf tissues were collocated and mixed to measure the anthocyanin content. Briefly, 0.1 g of dry samples were extracted with 5 mL of acidic methanol containing 1% HCl (v/v) in the dark at 4 °C overnight, with occasional shaking. Plant materials were sedimented by centrifugation at 4450× g for 15 min, and 500 μL of the supernatant was mixed with 500 μL of MilliQ H2O and 300 μL of chloroform in a 1.5-mL microcentrifuge tube to remove chlorophyll in the extraction. After centrifugation at 12,000× g for 5 min, the supernatants containing anthocyanins were filtered through a Millipore filter (0.22 mm diameter). The HP1260-VWD system (Agilent Technologies) was used to analyze anthocyanins in the extract at 520 nm. A GL Sciences Inertsil ODS-3 C18 column (250 mm × 4.6 mm, 5 μm) was used for separation at 25 °C and eluted using a mobile phase consisting of solvent A (methanol) and solvent B (5% formic acid in water) at a flow rate of 1 mL/min. The elution program was as follows: 0–2 min, 10% A; 2–6 min, 10–20% A; 6–10 min, 20–30% A; 10–15 min, 30–35% A; 15–20 min, 35–50% A; 20–24 min, 50–90% A; 24–29 min, 90–10% A; 29–30 min, 10% A. The anthocyanin standards, cyanidin 3-O-glucoside and cyanidin 3-O-rutinoside were used as controls and to generate standard curves for quantitative analysis of the buckwheat samples.
4.9. Statistical Treatment
All experiments were carried out in triplicate, and data were expressed as mean ± SD (standard deviation). Statistical significance was determined by one-way ANOVA, and statistical significance was accepted at a p-value threshold of <0.05.
5. Conclusions
A high level of anthocyanin accumulation was observed in the cotyledons and flowers of “HHTQ”, and candidate genes involved in the anthocyanin biosynthetic pathway were identified using transcriptome analysis. A total of 3727 and 5323 DEGs were identified in flowers and cotyledons, respectively. The putative structural genes and regulatory genes involved in the regulation of anthocyanin biosynthesis by the different expression patterns, and anthocyanin degradation enzymes, such as ANR and LAR, might play a critical role in degrading anthocyanin. These data will point out the direction for further study on coloration and contribute to a more thorough characterization of anthocyanin biosynthesis at the molecular level in buckwheat. Finally, the red common buckwheat material could be used for hybrid breeding and the genes that are connecting with color formation could be used to improve the anthocyanin content in buckwheat.
Abbreviations
| HPLC | High performance liquid chromatography |
| DET | Differentially expression test |
| DEGs | Differently expressed genes |
| EBGs | Early-stage biosynthetic genes |
| LBGs | Late-stage biosynthetic genes |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| DHK | Dihydrokaempferol |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/6/1493/s1.
Author Contributions
Z.F., Z.H., and J.Y. designed the study and wrote the manuscript. Z.F., S.W., Z.L., S.W., Y.Z., J.S., Z.H., and J.Y participated in experiments. Z.F., S.W., Z.L., S.W., Z.H., and J.Y discussed the results and revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31671755 and No. 31571736) and the Supported Project of Outstanding Doctoral and Master’s Degree Dissertation Cultivation Program of Yangtze University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Shi T., Li R., Chen Q., Li Y., Pan F., Chen Q. De novo sequencing of seed transcriptome and development of genic-SSR markers in common buckwheat (Fagopyrum esculentum) Mol. Breed. 2017;37:147. doi: 10.1007/s11032-017-0743-4. [DOI] [Google Scholar]
- 2.Fang Z., Zhang X., Gao J., Wang P., Xu X., Liu Z., Shen S., Feng B. A buckwheat (Fagopyrum esculentum) DRE-binding transcription factor gene, FeDREB1, enhances freezing and drought tolerance of transgenic Arabidopsis. Plant Mol. Biol. Rep. 2015;33:1510–1525. doi: 10.1007/s11105-015-0851-4. [DOI] [Google Scholar]
- 3.Demidenko N.V., Logacheva M.D., Penin A.A. Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE. 2011;6:e19434. doi: 10.1371/journal.pone.0019434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Q.H., Wang Y.Q., Song J.N., Yang H.B. Transcriptomic identification of salt-related genes and de novo assembly in common buckwheat (F. esculentum) Plant Physiol. Biochem. 2018;127:299–309. doi: 10.1016/j.plaphy.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Gupta N., Naik P.K., Chauhan R.S. Differential transcript profiling through cDNA-AFLP showed complexity of rutin biosynthesis and accumulation in seeds of a nutraceutical food crop (Fagopyrum spp.) BMC Genom. 2012;13:231. doi: 10.1186/1471-2164-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Z.W., Qi R., Li X.F., Liu Z.X. Ectopic expression of FaesAP3, a Fagopyrum esculentum (Polygonaceae) AP3 orthologous gene rescues stamen development in an Arabidopsis ap3 mutant. Gene. 2014;550:200–206. doi: 10.1016/j.gene.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Katsu K., Suzuki R., Tsuchiya W., Inagaki N., Yamazaki T., Hisano T., Yasui Y., Komori T., Koshio M., Kubota S., et al. A new buckwheat dihydroflavonol 4-reductase (DFR), with a unique substrate binding structure, has altered substrate specificity. BMC Plant Biol. 2017;17:239. doi: 10.1186/s12870-017-1200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki T., Kim S.-J., Mohamed Z.I.S., Mukasa Y., Takigawa S., Matsuura-Endo C., Yamauchi H., Hashimoto N., Noda T., Saito T. Structural identification of anthocyanins and analysis of concentrations during growth and flowering in buckwheat (Fagopyrum esculentum Moench) petals. J. Agric. Food Chem. 2007;55:9571–9575. doi: 10.1021/jf071474l. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y.B., Park S.Y., Thwe A.A., Seo J.M., Suzuki T., Kim S.J., Kim J.K., Park S.U. Metabolomic analysis and differential expression of anthocyanin biosynthetic genes in white- and red-flowered buckwheat cultivars (Fagopyrum esculentum) J. Agric. Food Chem. 2013;61:10525–10533. doi: 10.1021/jf402258f. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y., Sasaki N., Ohmiya A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Wang Y., Song Z., Zhang H. Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant. 2016;9:1395–1405. doi: 10.1016/j.molp.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Ren L., Hu Z., Li Y., Zhang B., Zhang Y., Tu Y., Chen G. Heterologous Expression of BoPAP1 in tomato induces stamen specific anthocyanin accumulation and enhances tolerance to a Long-Term low temperature stress. J. Plant Growth Regul. 2014;33:757–768. doi: 10.1007/s00344-014-9423-3. [DOI] [Google Scholar]
- 13.Cao L., Xu X., Chen S., Ma H. Cloning and expression analysis of Ficus carica anthocyanidin synthase 1 gene. Sci. Hortic. 2016;211:369–375. doi: 10.1016/j.scienta.2016.09.015. [DOI] [Google Scholar]
- 14.Shafiq M., Singh Z. Pre-harvest spray application of phenylpropanoids influences accumulation of anthocyanin and flavonoids in ‘Cripps Pink’ apple skin. Sci. Hortic. 2018;233:141–148. doi: 10.1016/j.scienta.2018.01.020. [DOI] [Google Scholar]
- 15.Grotewold E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- 16.He F., Mu L., Yan G.L., Liang N.N., Pan Q.H., Wang J., Reeves M.J., Duan C.Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules. 2010;15:9057–9091. doi: 10.3390/molecules15129057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butelli E., Titta L., Giorgio M., Mock H.P., Matros A., Peterek S., Schijlen E.G., Hall R.D., Bovy A.G., Luo J., et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Su N., Jia L., Tian J., Li H., Huang L., Shen Z., Cui J. Transcriptome analysis of radish sprouts hypocotyls reveals the regulatory role of hydrogen-rich water in anthocyanin biosynthesis under UV-A. BMC Plant Biol. 2018;18:227. doi: 10.1186/s12870-018-1449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quattrocchio F., Wing J.F., Leppen H.T.C., Moi J.N.M., Koe R.E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koes R., Verweij W., Quattrocchio F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 22.Martin C., Prescott A., Mackay S., Bartlett J., Vrijlandt E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991;1:37–49. doi: 10.1111/j.1365-313X.1991.00037.x. [DOI] [PubMed] [Google Scholar]
- 23.Bai S., Saito T., Honda C., Hatsuyama Y., Ito A., Moriguchi T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta. 2014;240:1051–1062. doi: 10.1007/s00425-014-2129-8. [DOI] [PubMed] [Google Scholar]
- 24.Grotewold E., Sainz M.B., Tagliani L., Hernandez J.M., Bowen B., Chandler V.L. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA. 2000;97:13579–13584. doi: 10.1073/pnas.250379897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. J. Plant. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu W., Dubos C., Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Lai B., Hu B., Qin Y.H., Zhao J.T., Wang H.C., Hu G.B. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genom. 2015;16:225. doi: 10.1186/s12864-015-1433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S., Huang Q., Lin C., Lin L., Zhou Q., Lin F., He E. Transcriptome comparison reveals candidate genes responsible for the betalain-/anthocyanidin-production in bougainvilleas. Funct. Plant Biol. 2016;43:278–286. doi: 10.1071/FP15246. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y., Wang J., Qiu Y., Liu T., Song J., Li X. Identification of ‘Xinlimei’ radish candidate genes associated with anthocyanin biosynthesis based on a transcriptome analysis. Gene. 2018;657:81–91. doi: 10.1016/j.gene.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Yasui Y., Hirakawa H., Ueno M., Matsui K., Katsube-Tanaka T., Yang S.J., Aii J., Sato S., Mori M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016;23:215–224. doi: 10.1093/dnares/dsw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S.J., Maeda T., Sarker M.Z.I., Takigawa S., Matsuura-Endo C., Yamauchi H., Mukasa Y., Saito K., Hashimoto N., Noda T., et al. Identification of anthocyanins in the sprouts of buckwheat. J. Agric. Food Chem. 2007;55:6314–6318. doi: 10.1021/jf0704716. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Luo X., Wu C., Cao S., Zhou Y., Jie B., Cao Y., Meng H., Wu G. Comparative transcriptome analysis of genes involved in anthocyanin biosynthesis in red and green Walnut (Juglans regia L.) Molecules. 2017;23:25. doi: 10.3390/molecules23010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L., Fan X., Zhang Y., Jiang J., Sun H., Liu C. Transcriptome analysis of genes involved in anthocyanins biosynthesis and transport in berries of black and white spine grapes (Vitis davidii) Hereditas. 2016;153:17. doi: 10.1186/s41065-016-0021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang H., Fan W., Li H., Yang J., Huang J., Zhang P. Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE. 2013;8:e78484. doi: 10.1371/journal.pone.0078484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soubeyrand E., Basteau C., Hilbert G., van Leeuwen C., Delrot S., Gomes E. Nitrogen supply affects anthocyanin biosynthetic and regulatory genes in grapevine cv. Cabernet-Sauvignon berries. Phytochemistry. 2014;103:38–49. doi: 10.1016/j.phytochem.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 37.Ai T.N., Naing A.H., Arun M., Jeon S.M., Kim C.K. Expression of RsMYB1 in Petunia enhances anthocyanin production in vegetative and floral tissues. Sci. Hortic. 2017;214:58–65. doi: 10.1016/j.scienta.2016.11.016. [DOI] [Google Scholar]
- 38.Ayabe S.I., Akashi T. Cytochrome P450s in flavonoid metabolism. Phytochem. Rev. 2006;5:271–282. doi: 10.1007/s11101-006-9007-3. [DOI] [Google Scholar]
- 39.Huang W., Sun W., Wang Y. Isolation and molecular characterisation of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes from a traditional Chinese medicinal plant, Epimedium sagittatum. Gene. 2012;497:125–130. doi: 10.1016/j.gene.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka Y., Brugliera F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. B. 2013;368:20120432. doi: 10.1098/rstb.2012.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petit P., Granier T., d’Estaintot B.L., Manigand C., Bathany K., Schmitter J.M., Lauvergeat V., Hamdi S., Gallois B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007;368:1345–1357. doi: 10.1016/j.jmb.2007.02.088. [DOI] [PubMed] [Google Scholar]
- 42.Luo P., Ning G., Wang Z., Shen Y., Jin H., Li P., Huang S., Zhao J., Bao M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci. 2015;6:1257. doi: 10.3389/fpls.2015.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshihara N., Imayama T., Fukuchi-Mizutani M., Okuhara H., Tanaka Y., Ino I., Yabuya T. cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase in Iris hollandica. Plant Sci. 2005;169:496–501. doi: 10.1016/j.plantsci.2005.04.007. [DOI] [Google Scholar]
- 44.Sui X., Gao X., Ao M., Wang Q., Yang D., Wang M., Fu Y., Wang L. cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase in Freesia hybrida. Plant Cell Rep. 2011;30:1209–1218. doi: 10.1007/s00299-011-1029-7. [DOI] [PubMed] [Google Scholar]
- 45.Fujiwara H., Tanaka Y., Yonekura-Sakakibara K., Fukuchi-Mizutani M., Nakao M., Fukui Y., Yamaguchi M., Ashikari1 T., Kusumi T. cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J. 1998;16:421–431. doi: 10.1046/j.1365-313x.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- 46.Marles M.A.S., Ray H., Gruber M.Y. New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry. 2003;64:367–383. doi: 10.1016/S0031-9422(03)00377-7. [DOI] [PubMed] [Google Scholar]
- 47.Wang P., Zhang L., Jiang X., Dai X., Xu L., Li T., Xing D., Li Y., Li M., Gao L., et al. Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta. 2018;247:139–154. doi: 10.1007/s00425-017-2771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao L., Vimolmangkang S., Wei G., Zhou H., Korban S.S., Han Y. Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 2015;6:243. doi: 10.3389/fpls.2015.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gargouri M., Manigand C., Mauge C., Granier T., Langlois d’Estaintot B., Cala O., Pianet I., Bathany K., Chaudiere J., Gallois B. Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera. Acta Crystallogr. 2009;65:989–1000. doi: 10.1107/S0907444909025013. [DOI] [PubMed] [Google Scholar]
- 50.Han Y., Vimolmangkang S., Soria-Guerra R.E., Korban S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012;63:2437–2447. doi: 10.1093/jxb/err415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer T.C., Mirbeth B., Rentsch J., Sutter C., Ring L., Flachowsky H., Habegger R., Hoffmann T., Hanke M.V., Schwab W. Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria x ananassa) New Phytol. 2014;201:440–451. doi: 10.1111/nph.12528. [DOI] [PubMed] [Google Scholar]
- 52.de Vetten N., Quattrocchio F., Mol J., Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–1434. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- 53.Albert N.W., Davies K.M., Lewis D.H., Zhang H., Montefiori M., Brendolise C., Boase M.R., Ngo H., Jameson P.E., Schwinn K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014;26:962–980. doi: 10.1105/tpc.113.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie Y., Tan H., Ma Z., Huang J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant. 2016;9:711–721. doi: 10.1016/j.molp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 55.Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y., Liu X., Cai X., Shan X., Gao R., Yang S., Han T., Wang S., Wang L., Gao X. Dihydroflavonol 4-reductase genes from freesia hybrida play important and partially overlapping roles in the biosynthesis of flavonoids. Front. Plant Sci. 2017;8:428. doi: 10.3389/fpls.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shan X., Zhang Y., Peng W., Wang Z., Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009;60:3849–3860. doi: 10.1093/jxb/erp223. [DOI] [PubMed] [Google Scholar]
- 58.Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. The Jasmonate-ZIM-Domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen X., Zhao K., Liu L., Zhang K., Yuan H., Liao X., Wang Q., Guo X., Li F., Li T. A role for PacMYBA in ABA-Regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.) Plant Cell Physiol. 2014;55:862–880. doi: 10.1093/pcp/pcu013. [DOI] [PubMed] [Google Scholar]
- 60.Li Y., Li H., Wang F., Li J., Zhang Y., Wang L., Gao J. Comparative transcriptome analysis reveals effects of exogenous hematin on anthocyanin biosynthesis during strawberry fruit ripening. Int. J. Genom. 2016;2016:1–14. doi: 10.1155/2016/6762731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma Y., Shu S., Bai S., Tao R., Qian M., Teng Y. Genome-wide survey and analysis of the TIFY gene family and its potential role in anthocyanin synthesis in Chinese sand pear (Pyrus pyrifolia) Tree Genet. Genome. 2018;14:25. doi: 10.1007/s11295-018-1240-6. [DOI] [Google Scholar]
- 62.An X., Tian Y., Chen K., Liu X., Liu D., Xie X., Cheng C., Cong P., Hao Y. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015;56:650–662. doi: 10.1093/pcp/pcu205. [DOI] [PubMed] [Google Scholar]
- 63.Liu X., An X., Liu X., Hu D., Wang X., You C., Hao Y. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 2017;68:2977–2990. doi: 10.1093/jxb/erx150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou R., Zhu Y.X., Zhao J., Fang Z.W., Wang S.P., Yin J.L., Chu Z.H., Ma D.F. Transcriptome-wide identification and characterization of potato circular RNAs in response to Pectobacterium carotovorum Subspecies brasiliense infection. Int. J. Mol. Sci. 2018;19:71. doi: 10.3390/ijms19010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., MacManes M.D., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin J.L., Fang Z.W., Sun C., Zhang P., Zhang X., Lu C., Wang S.P., Ma D.F., Zhu Y.X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 2018;8:3086. doi: 10.1038/s41598-018-21489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin J.L., Liu M.Y., Ma D.F., Wu J.W., Li S.L., Zhu Y.X., Han B. Identification of circular RNAs and their targets during tomato fruit ripening. Postharvest Biol. Technol. 2018;136:90–98. doi: 10.1016/j.postharvbio.2017.10.013. [DOI] [Google Scholar]
- 68.Chu H., Jeong J.C., Kim W.J., Chung D.M., Jeon H.K., Ahn Y.O., Kim S.H., Lee H.S., Kwak S.S., Kim C.Y. Expression of the sweetpotato R2R3-type IbMYB1a gene induces anthocyanin accumulation in Arabidopsis. Physiol. Plant. 2013;148:189–199. doi: 10.1111/j.1399-3054.2012.01706.x. [DOI] [PubMed] [Google Scholar]
- 69.Shao Y., Hu Z., Yu Y., Mou R., Zhu Z., Beta T. Phenolic acids, anthocyanins, proanthocyanidins, antioxidant activity, minerals and their correlations in non-pigmented, red, and black rice. Food Chem. 2018;239:733–741. doi: 10.1016/j.foodchem.2017.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.