SUMMARY
During wound injury, efferocytosis fills the macrophage with a metabolite load nearly equal to the phagocyte itself. A timely question pertains to how metabolic phagocytic signaling regulates the signature anti-inflammatory macrophage response. Here we report the metabolome of activated macrophages during efferocytosis to reveal an interleukin-10 (IL-10) cytokine escalation that was independent of glycolysis yet bolstered by apoptotic cell fatty acids and mitochondrial β-oxidation, the electron transport chain, and heightened coenzyme NAD+. Loss of IL-10 due to mitochondrial complex III defects was remarkably rescued by adding NAD+ precursors. This activated a SIRTUIN1 signaling cascade, largely independent of ATP, that culminated in activation of IL-10 transcription factor PBX1. II-10 activation by the respiratory chain was also important in vivo, as efferocyte mitochondrial dysfunction led to cardiac rupture after myocardial injury. These findings highlight a new paradigm whereby macrophages leverage efferocytic metabolites and electron transport for anti-inflammatory reprogramming that culminates in organ repair.
In Brief
Zhang et al. show that the phagocytic engulfment of dying cells by macrophages (efferocytosis) results in elevated cellular fatty acids, which fuel mitochondrial respiration and activate an NAD+-dependent signal transduction cascade. This metabolic signaling pathway promotes an anti-inflammatory response for wound healing and organ repair following myocardial infarction.
Graphical Abstract
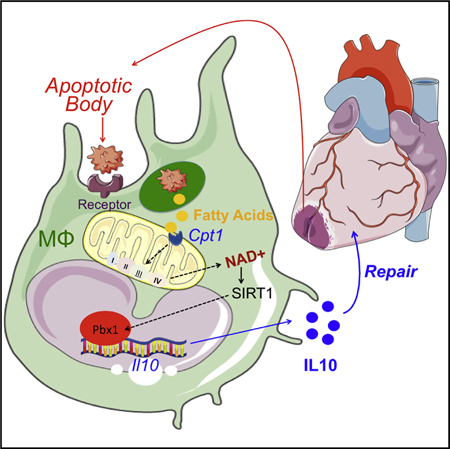
INTRODUCTION
Macrophages preserve systemic self-tolerance and promote inflammation resolution and tissue repair through efferocytosis (Poon et al., 2014). In addition to the recognition and engulfment of dying cells, efferocytosis triggers the production of anti-inflammatory and tissue-reparative cytokines (Fadok et al., 1998) as well as inflammation-resolving bioactive lipids (Serhan and Savill, 2005). While these host-protective responses act in part through lipid-activated nuclear receptor transcription factors (A-Gonzalez et al., 2009), the signal transduction role of metabolites during efferocytic reprogramming is largely untested. This is likely important during syndromes of metabolite imbalance, where the anti-inflammatory potential of macrophages is often depressed and contributes to disease progression (Tabas and Glass, 2013).
It is now well appreciated that intracellular metabolism is integrated with the balance of cell activation. In macrophages, glycolysis is required for both pro-inflammatory polarization and the mobilizing of biosynthetic precursors to combat bacterial infection (Palsson-McDermott et al., 2015). Elevated glucose utilization is also necessary for alternative macrophage activation, the latter initiated by the cytokine interleukin-4 (IL-4) and further accompanied by increased oxidative phosphorylation (Huang et al., 2016). In the case of efferocytosis, biosynthetic precursors are in abundant supply within the phagocytic body, raising the prospect that apoptotic cell catabolism could provide substrates for macrophage reprogramming. Early studies of efferocytosis implicate cellular metabolism for the energetic currency that is necessary for actin-mediated engulf-ment of external bodies (Oren et al., 1963). Separately, mitochondrial uncoupling proteins (Park et al., 2011) and components of mitochondrial fission machinery (Wang et al., 2017) are necessary for multiple rounds of efferocytosis; this is of particular importance during tissue injury, which is characterized by heightened cell turnover. Taken together, these examples emphasize a conserved interplay between efferocytosis and cellular and mitochondrial metabolism; however, what is left unsolved is whether the catabolism of dying cells is integrated to the signature macrophage anti-inflammatory response.
To comprehend relationships between cellular metabolism, inflammation, and tissue repair, we report the metabolome of macrophages during efferocytosis. We implemented unbiased global metabolic pathway analyses to reveal a unique association between fatty acid oxidation (FAO), mitochondrial respiration, and inflammation during the catabolism of apoptotic cells. From our investigation, we discovered that efferocytosis significantly elevated long-chain fatty acid content in macrophages, activated the respiratory chain, and was required to stimulate a macrophage anti-inflammatory response through the generation of metabolic signaling intermediates, particularly NAD+. This non-canonical mitochondrial response was found to also be important during tissue injury, supporting its significance to the broad pathophysiology of wound healing.
RESULTS
Evidence that Anti-inflammatory Efferocytosis Exerts a Mitochondrial Bias
Clearance of dying cells (efferocytosis) during tissue injury associates the metabolism of apoptotic cells by macrophages with gene expression of anti-inflammatory cytokines (Serhan and Sa-vill, 2005). Surprisingly, the underlying metabolic relationships of efferocytosis to basic mechanisms of gene expression remain mysterious. To address potential connections, we first searched for in vivo evidence of metabolic polarization in tissue injury macrophages that were both anti-inflammatory and efferocytic. We employed myocardial infarction (MI) as a clinically relevant model of injury in which defects in efferocytosis lead to heart failure (Wan et al., 2013). To isolate efferocytes, we induced MI in transgenic MHC-Cre mCherry reporter mice. Using this previously validated approach (DeBergeet al., 2017), we could distinguish CD11b+Ly6c-Ly6g-F4/80+CD64+ macrophages that accumulated either high (HI) or low (LO) mCherry signal, consistent with efferocytosis of high versus low levels of injured cardiac cells, respectively. CD64+ mCherry-HI cardiac macrophages expressed heightened levels of the canonical anti-inflammatory cytokine II-10 (Figure 1A), relative to their mCherry-LO counterparts. We further queried for unique signs of metabolic activation. Relative to tissue-sorted monocytes and neutrophils, Mᶲs consumed more oxygen, and in particular, mCherry-HI (efferocytic) Mᶲs (which also expressed II-10) exhibited the highest oxygen consumption rate (OCR) (Figure 1B). This heightened OCR was not incompatible with the hypoxic stress that occurs during MI. For instance, we injected PIMOnidazole (Varia et al., 1998), which labels free sulfhydryls of hypoxic cells, and could identify both hypoxic (PIMO+) and non-hypoxic (PIMO-) cardiac Mᶲs (Figure 1C), the majority of which were, interestingly, PIMO negative at 12 hr post MI. To test associations between hypoxia and efferocytic IL-10, we sorted PIMO+mCherry-,PIMO+mCherry+, PIMO-mCherry-, and PIMO-mCherry+ cardiac macrophages (Figure S1) and measured IL-10 gene expression by qPCR. PIMO-mCherry+ (non-hypoxic efferocytic) macrophages registered the highest IL-10 mRNA levels (Figure 1C), consistent with the notion that cardiac macrophages in areas of oxygen availability have the highest capacity to secrete IL-10. Thus, in a model where efferocytosis is necessary for both inflammation resolution and organ function (Howangyin et al., 2016; Wan et al., 2013), the anti-inflammatory polarization of efferocytic Mᶲs directly coincided with heightened oxidative respiration.
Figure 1. Tissue Injury Phagocytes in the Process of Efferocytosis Produce IL-10 and Exhibit a Mitochondrial Bias.
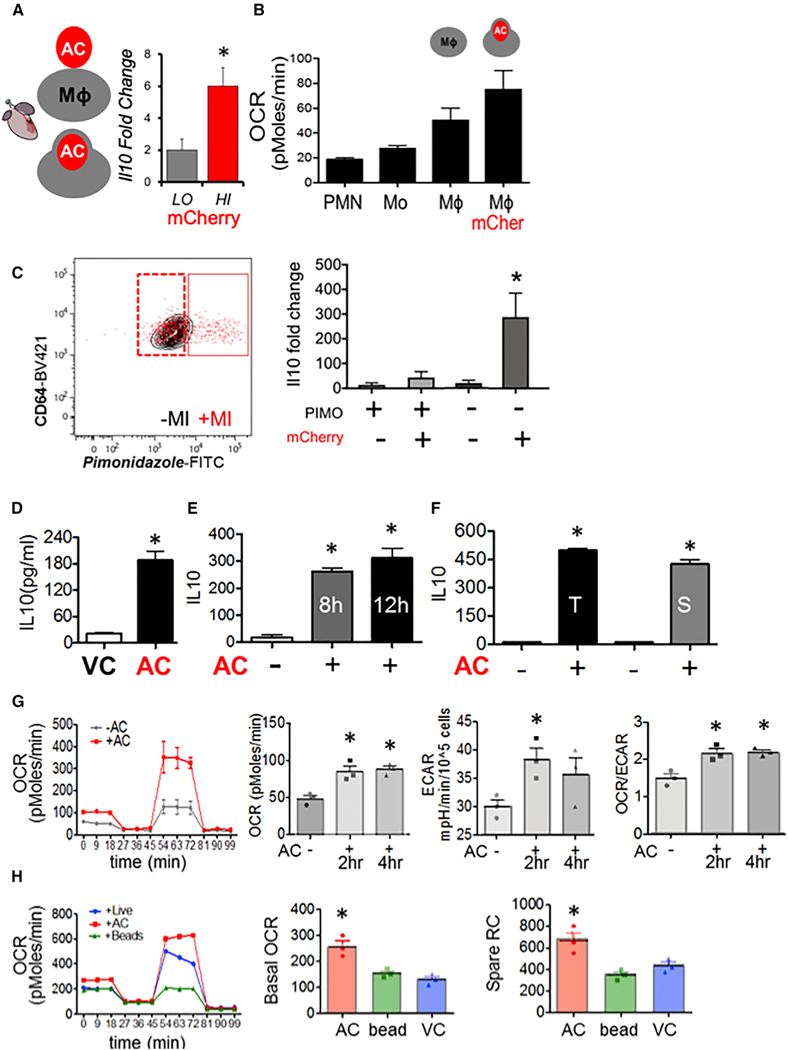
(A) In a model of tissue injury, B6 mice transgenic for cardiac-specific (Myh6-Cre) expression of mCherry were subject to myocardial infarction (MI). Cardiac extracts were prepared and flow cytometry employed to isolate cardiac macrophages (Mf s) (CD11b+F4/80+Ly6g—CD64+) that included low (LO) and high (HI) mCherry+ dying and apoptotic cells (ACs). mCherry-LO and -HI phagocyteswere sorted and relative gene expression normalized to β2m as assessed by qPCR and plotted as fold change over IL-10 from Mᶲs from naive hearts. (B) Cardiac Ly6g+ neutrophils (PMN), LycHI, Ly6g— monocytes (Mo), CD64+ Mfs, and Mᶲs staining for mCherry were interrogated for basal OCR by functional respiration analysis. (C) Cardiac Mfs were stained with hypoxia probe PIMOnidazole and mCherry and sorted for qPCR. (D-F) Elicited primary Mᶲs were co-cultivated with early (Annexin V positive, propidium iodide negative) ACs. Non-engulfed cells were removed from adherent phagocytes, and cell culture medium was analyzed for secreted cytokines. (D) IL-10 responsewith viable Jurkat cells (VCs) versusACs. (E) Time course analysis of IL-10 production. (F) IL-10 secretion after adding primary thymocytes (T) or primary splenocytes (S). (G) OCR of primary elicited Mᶲs ± indicated treatments and quantified basal OCR. To ascertain basal and spare respiratory capacity (SRC), efferocytes were administered sequential treatments ofoligomycin, CCCP, and rotenone plus antimycin. Minutes 0–18 is basal OCR and 54–81 min is SRC. Area underthe curve quantifications are displayed. Also measured was the extracellular acidification rate (ECAR) as a reflection of glycolysis. (H) Mᶲs were co-cultivated with inert beads and/or live cells.
To dig deeper into the cell-intrinsic relationships between efferocytic cellular metabolism and cytokine production, we modeled these events in culture. We elicited activated primary peritoneal macrophages. For apoptotic cells we chose human Jurkat T cells, which are standardized targets for macrophage co-cultivation (Fadok et al., 1998); this also permitted us to distinguish the gene expression response in a species-specific manner after RNA sequencing (see also STAR Methods). In culture, we began with a non-biased screen of secreted chemo kines and cytokines. This approach confirmed that apoptotic cells (Annexin V+, PI−) specifically and reproducibly induced macrophages to produce the canonical anti-inflammatory cytokine IL-10 (Fadok et al., 1998) (Figures 1D and 1E). This response was also validated with primary autologous apoptotic thymocytes and primary splenocytes (Figure 1F). Pro-reparative TGF-β was additionally induced after efferocytosis, as previously reported (Xiao et al., 2008) (Figure S2A), while pro-inflammatory TNF-α was suppressed (Figure S2B). Also induced was GM-CSF, which was recently shown to regulate leukocyte accumulation in heart (Anzai et al., 2017). The specificity of the efferocytic cytokine phenotype was underscored by cytokines that were not affected (Figure S2C). Unexpectedly, IL-9 (Figure S2A) and Eotaxin (Figure S2B), the former linked to T-regulatory cell function (Eller et al., 2011) and the latter to eosinophil responses, were also regulated by efferocytosis (Jose et al., 1994).
Consistent with our in vivo data, basal OCR was significantly elevated in Mᶲs that were fed apoptotic cells (Figure 1G). This was in contrast to Mᶲs co-cultivated with either live cells or inert polystyrene beads (Figure 1H). Remarkably, efferocytes exhibited heightened mitochondrial spare respiratory capacity, consistent with an elevated bioenergetics reserve, potentially provided by the introduction of exogenous metabolic substrates. Glycolytic extracellular acidification rate (ECAR) also trended higher during efferocytosis (Figure 1G); however, the ratio of OCR to ECAR was higher during apoptotic cell engulfment relative to non-efferocytes, consistent with a mitochondrial bias during efferocytosis.
The Efferocytosis Metabolome
A mitochondrial bias suggested a role for fatty acid utilization. To directly probe metabolic remodeling during efferocytosis, we performed unbiased liquid chromatography-tandem mass spectrometry (LC-MS/MS). Over 255 total biochemicals were altered with a p < 0.05, with 181 induced and 74 downregulated (Table S1). Unbiased analysis of the LC-MS/MS readings identified reproducible global changes in metabolites (Figure 2A and Table S1). Metabolite set enrichment analysis highlighted noteworthy escalations in long-chain free fatty acids as well as fatty acid metabolism (Figure 2B). Random forest analysis (Lunetta et al., 2004) revealed that lipid metabolites such as 3-hydroxybu-tyrylcarnitine, behenoylcarnitine, and arachidoylcarnitine were top enriched metabolites in efferocytes (Figure S3A). We further examined lipid metabolites that are involved in FAO. Long-chain fatty acids hexadecanedioate, myristoylglycerol, and linoleoyl-carnitine were also increased in efferocytes. Furthermore, CPT substrate palmitoylcarnitine was also elevated (Figure 2C).
Figure 2. Global Metabolomic Profile of Efferocytosis Highlights FAO Utilization.
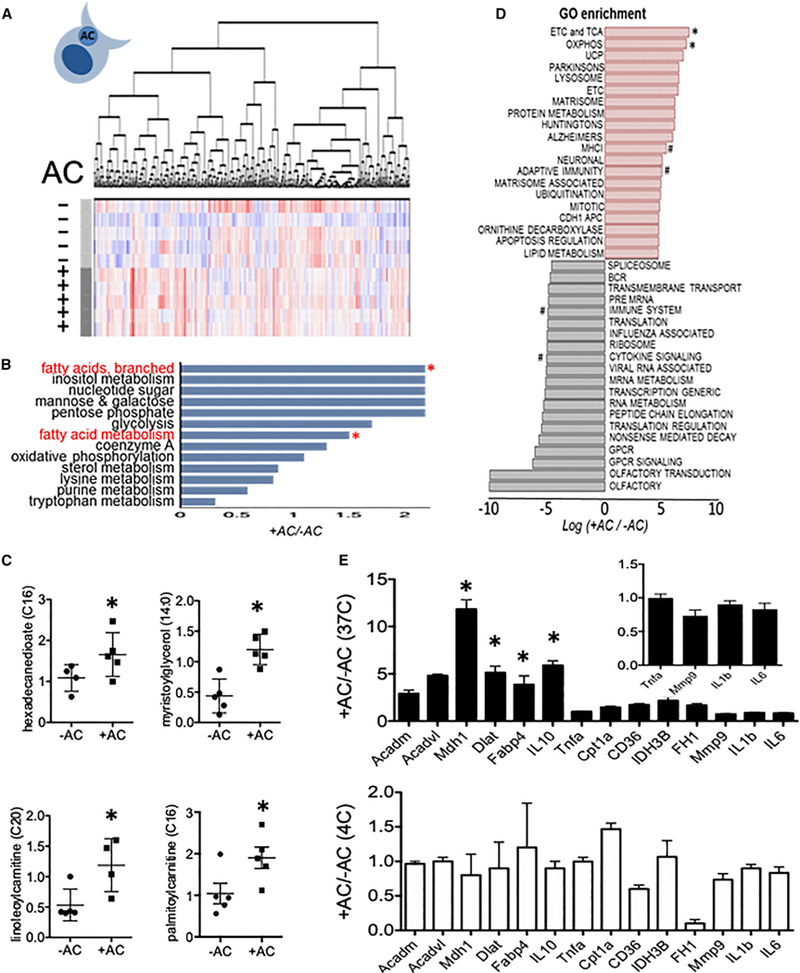
(A) Heatmap after unsupervised hierarchical clustering of metabolitesfrom primary peritoneal Mᶲs, minus (−) versus plus(+) apoptotic cells (n = 5 percondition). Shades of red and blue indicate an increase or decrease of metabolites, respectively. (B) Metabolite set enrichment analysis of significantly altered core pathways. Asterisks indicate pathways of interest. (C) Selected lipid metabolites of interest. (D) Gene ontology analysis after mRNA sequencing screen. # indicates pathways of interest. (E) qPCR validation of selected FAO, lipolysis, TCA, and inflammatory genes versus control (non-filled bars in bottom panel). *p < 0. 05. Inset is magnification of data < 1.
In separate experiments, in which we screened transcriptomic responses to efferocytosis, gene ontology enrichment analysis of canonical pathways from our mRNA-seq analysis revealed the most significantly mobilized pathways; these included mitochondrial electron transport (Figure 2D). Sequencing reads were aligned to murine gene annotation files to distinguish from human sequence (Alvey et al., 2017). Consistent with the metabolomics profile, we also validated that apoptotic cell stimulation of primary macrophages induced expression of genes involved in FAO, the electron transport chain, and oxidative phosphorylation pathways (Figure 2E). In line with these data, qPCR validation did not show contamination from apoptotic cell mRNAs, as control experiments in which apoptotic cells were co-cultivated with macrophages at 4°C did not phenocopy the efferocytosis gene signature. At first approximation, these parallel analyses of principal cytokine, transcriptional, and metabolic modules revealed an efferocytic signature that coupled anti-inflammatory cytokine production to fatty acid breakdown and mitochondrial metabolism.
FAO Is Required for Efferocytosis-Induced IL-10 Production, whereas Glycolysis Is Not
Given the emphasis on fatty acids and the mitochondrion from our functional respiration and metabolite analyses, and given that apoptotic cells are a significant source of external lipid substrate, we hypothesized that apoptotic cells cultured with heightened fatty acids may additively fuel IL-10 production during their catabolism in phagocytes. To this point, we cultivated apoptotic cells with increasing concentrations of fatty acid-supplemented media to elevate fatty acid content (Spector et al., 1979), subsequently induced apoptosis, and overlaid normalized ratios of apoptotic cells to macrophages. Figure 3A demonstrates that levels of IL-10 secretion directly correlated with increasing doses of fatty acid supplementation; this was specific to IL-10, as TNF-α was not affected, as might be suspected under lipotox-icity. Evidence for fatty acid induction of IL-10 in T cells per se was not found (Figure S4A). To address if FAO is required for IL-10 generation in efferocytes, we initially added etomoxir (ETO) to target CPT1 enzymes involved in FAO (Carroll et al., 1978). We found that ETO reduced OCR in macrophages after efferocytosis (Figure 3B), as well as IL-10, but this was independent of TGF-β production (Figure 3C). However, recent findings indicate that ETO disrupts coenzyme A homeostasis (Divakaruni et al., 2018), and so we specifically targeted Cpt1 and Cpt2 gene expression with siRNAs (Figure S6) and then found similar results (Figure 3D). Separately, previous reports elegantly described IL-4-induced macrophage polarization that required glucose utilization (Huang et al., 2016). Efferocytosis was distinctly characterized by the inability of glycolysis inhibitors to blunt IL-10 production (Figures 3E and 3F).
Figure 3. Efferocytic IL-10 Is Fueled by and Requires FAO, Not Glycolysis.
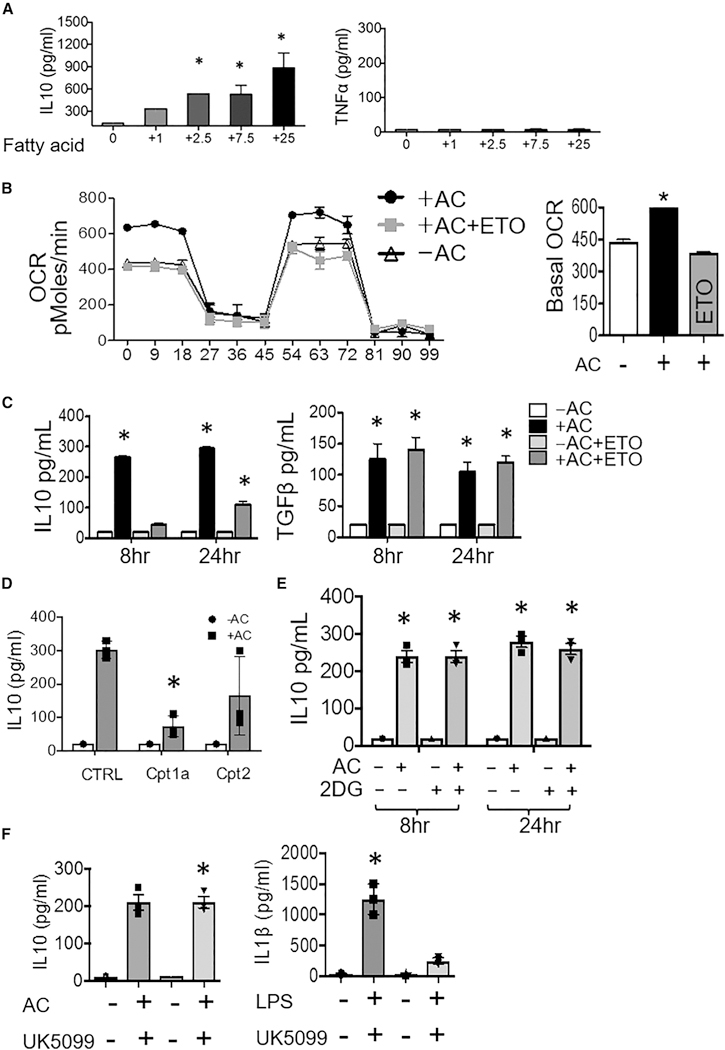
(A) Apoptotic cells (ACs) were previously cultured in increasing concentrations of fatty-acid-supplemented growth media (μg/L fatty acid per liter), prior to co-cultivation with macrophages/Mᶲ at equivalent ACs to Mᶲ ratios and ELISAs performed. (B) FAO-inhibitor etomoxir (ETO) suppresses AC-induced OCR in efferocytes. (C) IL-10 and TGF-β production in AC-treated Mᶲs ± ETO. (D) IL-10 production in AC-treated Cpt1- and Cpt2-deficient Mᶲs. (E) IL-10 production in AC-treated Mᶲs ± 2-deoxyglucose (2DG). (F) IL-10 production in AC-treated Mᶲs ± glucose-inhibitor UK5099 and IL-1 β production in LPS-treated Mᶲs ± UK5099. 5 mM ATP was added 3 hr after LPS addition. *p < 0.05 versus control.
Requirements for the Mitochondrial Electron Transport Chain during Efferocytosis-Induced IL-10 Generation
Consistent with a significant role of the mitochondrion during efferocytic IL-10 regulation, we interestingly documented evidence for spatial localization of mitochondria proximal to phagocytic cups (Figure S4B). Furthermore, elevations in oxygen consumption were consistent with heightened mitochondrial electron transport and mitochondrial oxidative phosphorylation, and the electron transport chain is coupled to mitochondrial FAO (Schönfeld et al., 2010; Wang et al., 2010). Electron transport is catalyzed in part by Rieske iron-sulfur protein, or RISP (Hartl etal., 1986). RISP has been newly implicated in activities beyond electron transport (Ansó et al., 2017); however, such a non-canonical role in macrophages is unclear and of unique significance to inflammation. To test the hypothesis that electron transport contributes to efferocytic anti-inflammatory polarization, we crossed Rispfl/fl mice (Waypa et al., 2013) with a myeloid and macrophage recombinase, LysMcre. Rispfl/fl LysMcre mice developed and matured to both normal body size and dietary intake and did not present with any significant sign of immunocompromise. In primary macrophages, immunoblots confirmed reduction of RISP protein (Figure 4A). RISP is encoded by nuclear gene ubiquinol-cytochrome c reductase (Uqcrfs1), and the failure of RISP to be imported into the mitochondrial inner membrane did not overtly compromise macrophage mitochondrial morphology (Figure 4B). Risp-deficient macrophages also exhibited reduced oxygen consumption at baseline and a compensatory increase in glycolysis, as indicated by moderately elevated extracellular acidification (Figure 4C). Importantly, Risp-deficient macrophages were proficient at phagocytosis (Figure 4D). To test if Risp was required for anti-inflammatory efferocytic polarization, we measured IL-10 production. Super-natants from Risp-deficient macrophages exhibited clear reductions in IL-10 that were specific to efferocytosis (Figure 4E). This was not a generalized defect, as mutant macrophages were competent at producing pro-inflammatory cytokine TNF-α in response to LPS. To address potential off-target effects during cellular differentiation in germline Rispfl/fl LysMcre mice, we reproduced our findings after RNA silencing in mature macrophages (Figure 4F). We next targeted signaling that was downstream of apoptotic cell binding and added Antimycin A after apoptotic cell engulfment, which inhibits complex III activity (Rich, 1983). Figure 4G depicts that Antimycin A also led to IL-10 suppression. We independently corroborated these findings with a structurally dissimilar complex III inhibitor, myxothia-zol (Figure 4H) (Ikeda et al., 1965). Although ATP in Risp-deficient macrophages was acutely reduced during efferocytosis, these levels quickly recovered (Figures S5A and S5B). Moreover, efferocytic IL-10 was not inhibited by interfering with mitochondrial membrane potential (Figure S5C). Separately, complex III and RISP are significant sources of mitochondrial reactive oxygen species (mROS) (Chandel et al., 2000). Efferocytic IL-10 was also not blocked with inhibitors of mROS, interestingly, in contrast to TGF-β (Figure S5D).
Figure 4. Electron Transport Dysfunction Reduces the Capacity of Efferocytes to Produce IL-10.
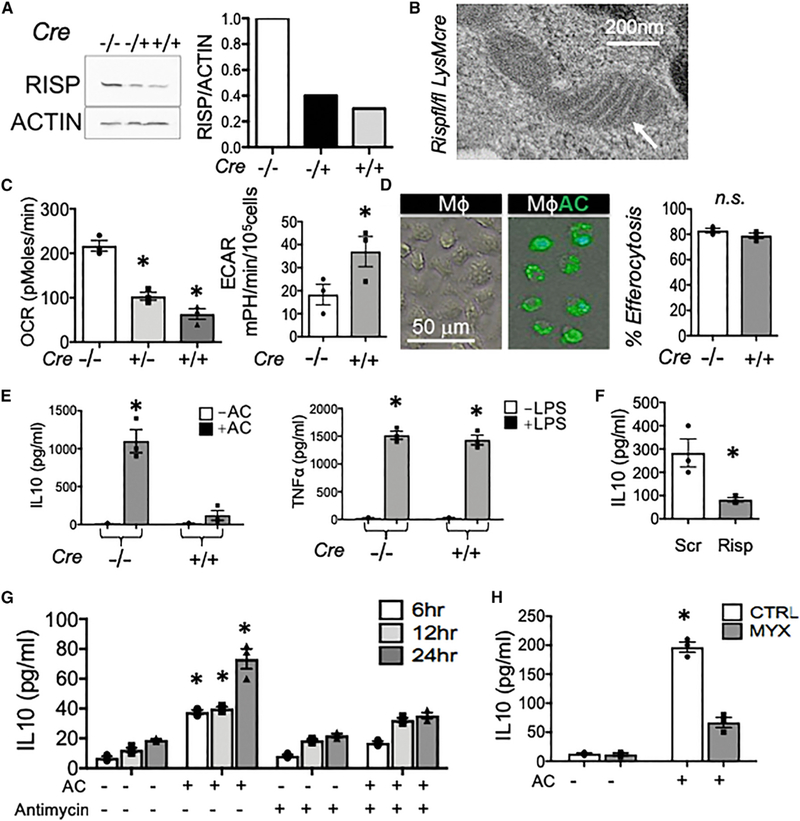
Elicited peritoneal M$s were harvested from Rispfl/fl and Rispfl/fl LysMcre mice. (A) Mᶲs RISP protein was quantified by immunoblot. (B) Transmission electron micrograph of a Risp-deficient Mᶲ. (C) OCR and ECAR profiles of Risp-deficient Mᶲs. (D) Photomicrographs and quantification of efferocytosis. Mᶲs remain unlabeled while apoptotic cells (ACs) are labeled green. n.s., not statistically significant. (E) IL-10 in cell supernatants 12 hr after efferocytosis. TNF-α was measured 6 hr after LPS stimulation. (F) IL-10 production was measured in macrophages after Risp knockdown with siRNA. (G) Acute inhibitor antimycin/AntA added at 100 nM 1 hr post AC feeding. IL-10 was measured with ELISA. (H) Myxothiazol inhibitor of electron transfer to RISP was added to efferocytes and IL-10 measured by ELISA. *p < 0.05.
An Intact Electron Transport Chain Is Necessary for Increased NAD+ Availability and IL-10 Production during Efferocytosis
Metabolic rewiring alters the availability of functional metabolites that regulate macrophage function (Jha et al., 2015). Our nonbiased analysis of Risp deficiency during efferocyotsis revealed specific reductions in nicotinate and nicotinamide metabolism. This included changes in NAD+ metabolites MNA and adenosine 5’-diphosphoribose (ADP-ribose) (Figure 5A), as well as aspartate, which is dependent on a suitable NAD+/NADH ratio within the malate-aspartate shuttle. Nicotinamide and quinolinate were not changed (Figure S3B). Focused pathway analyses corroborated an increased ratio of oxidized coenzyme nicotinamide adenine dinucleotide NAD+ to NADH in efferocytes, which was further reduced with Risp deficiency (Figures 5B and 5C). Thus, aside from its role in metabolic redox reactions, we considered a novel role for NAD+ during efferocytic anti-inflammatory reprogramming. To test this, we added NAD+ precursor NMN to Rispfl/fl LysMcre macrophages during efferocytosis and found that NMN was sufficient to rescue IL-10 production (Figure 5D). Remarkably, NAD-dependent IL-10 was specific to efferocytosis, as NAD+ supplementation alone, in the absence of apoptotic cell feeding, did not lead to elevated IL-10.
Figure 5. NAD+ Supplementation Partially Rescues Efferocytic IL-10 Production from Macrophages Deficient in RISP.
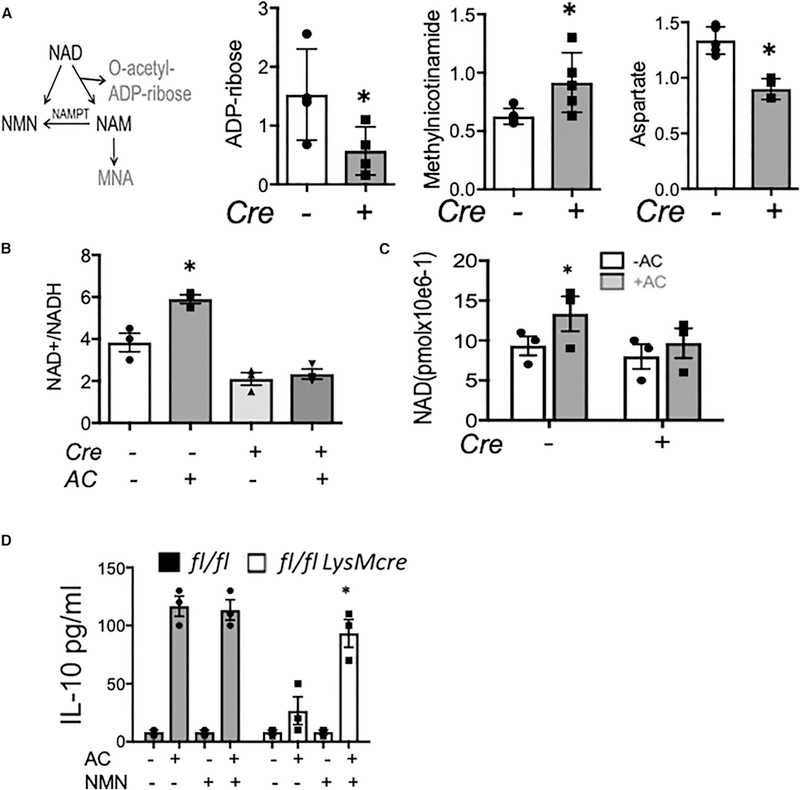
(A) NAD metabolism scheme with a focus on NAD breakdown and buildup of MNA metabolite 1-methylnicotinamide (n-methylnicotinamide). Graphed is the relative change of indicated metabolites in Risp-deficient or control efferocytes, with versus without apoptotic cells. (B) Risp deficiency (LysMcre) reduces the increased NAD+/NADH ratio in Mᶲs during efferocytosis of apoptotic cells (ACs). (C) Mᶲs treated with or without ACs for 3 hr were harvested and used for direct measure of NAD+ levels. (D) Peritoneal efferocytes were administered NAD+ precursor NMN and IL-10 measured by ELISA. *p < 0.05 relative to control.
NAD-Dependent IL-10 Acts through Sirtuins
Elevated NAD+/NADH ratios support the activity of sirtuin deace-tylases (Tanner et al., 2000). We first investigated a role for SIRT2 given previous links to macrophage polarization and bacterial phagocytosis (Ciarlo et al., 2017); however, Sirrt2-deficient phagocytes remained relatively competent at generating IL-10 after efferocytosis (Figure 6A). This was in contrast to Sirt1, which displayed elevated protein levels after efferocytosis (Figure 6B) and was also required relatively more than Sirt2 for IL-10 generation (Figure 6C). SIRT1 protein activity (Chatterjee et al., 2018) was reduced during efferocytosis under Risp deficiency (Figure 6D). Further, and as predicted from a temporal sequence in which NAD+ is upstream of SIRT1 protein activity, Sirt1-deficient macrophages were refractory to IL-10 rescue after supplementation with NAD+ precursor NMN (Figure 6E). To address mechanistic links to IL-10 gene regulation, we explored the role of the transcription factor Pbx-1, or pre-B cell leukemia transcription factor-1 (Pbx-1), which is required for IL-10 expression (Chung et al., 2007). To determine if the SIRT1-IL-10 axis involved Pbx-1, we performed chromatin immunoprecipitation (ChIP) for PBX-1 protein, during efferocytosis and also after knocking down Sirt1. Consistent with our hypothesis, Sirt1 deficiency reduced PBX-1 binding to the apoptotic cell response element of the IL-10 promoter (Figure 6F). Furthermore, in Risp-deficient macrophages, PBX1 binding to the IL-10 promoter was reduced and partially rescued by NMN (Figure 6G). Pbx1-knockdown also abrogated efferocytic IL-10 (Figure 6H), as predicted (Chung et al., 2007). These data support the contention that efferocytic accumulations in NAD+ in turn signal through SIRT1 and PBX1 protein to induce IL-10 gene expression.
Figure 6. Sirtl Is Required for NAD-Dependent IL-10 Production and Activation of the IL-10Transcription Factor PBX.

(A) Sirt2+/+ and Sirt2−/− peritoneal macrophages were challenged with apoptotic cells (ACs), and absolute IL-10 levels were measured by ELISA. (B) Immunoblot of SIRT1 protein in primary Mᶲs during efferocytosis. (C) IL-10 levels were compared between Sirt2 and Sirtl-deficient Mᶲs during efferocytosis. (D) SIRT1 protein activity measured in control versus Risp-deficient Mᶲs that were treated with versus without ACs. (E) NMN supplementation rescues absolute levels of IL-10 production in Risp-deficient efferocytes, but not Sirf1-deficient efferocytes. (F) PBX1 ChIP-IL-10 qPCR was performed in efferocytes treated with Sirf1 siRNA versus scrambled (Scr) control. (G) PBX1 ChIP-IL-10 qPCR before and after efferocytosis in control, Risp-deficient, and Risp-deficient macrophages with NMN supplementation. (H) IL-10 production after Sirt1 siRNA and/or Pbx1 siRNA in primary Mᶲs. *indicates p < 0.05 versus control.
Myeloid Mitochondrial Dysfunction Impairs IL-10-Dependent Wound Healing and Inflammation Resolution
To address the in vivo significance of our findings, we examined Risp-dependent IL-10 in the aforementioned model of tissue injury requiring efferocytosis (Wan et al., 2013) and macrophage IL-10 polarization (Shiraishi et al., 2016) during MI. To isolate myocardial macrophages that were participating in efferocytosis, we induced MI in transgenic mCherry mice, described above, that specifically express red cardiomyocytes (Zhang et al., 2015). Double-positive CD64+ (macrophage) mCherry+ efferocytes exhibited heightened Risp-dependent OCR and expression of genes involved in oxidative phosphorylation (Figures 7A and 7B). In myeloid Risp-deficient mice, MI also led to acute mortality, with more than half of subjects succumbing to death between 4 and 7 days post MI (Figure 7C). Necropsy revealed increased myocardial rupture of the left ventricular free wall and evidence of hemothorax (Figure 7C). Importantly, increases in ventricular rupture were independent of macrophage accumulation, as cardiac flow cytometry revealed similar macrophage levels in the injured myocardium, preceding ventricular rupture (Figure 7D). Within cardiac extracts, IL-10 and TGF-β mRNA were greatly reduced (Figure 7E).
Figure 7. Macrophage Mitochondrial Dysfunction Impairs IL-10-Associated Cardiac Repair.
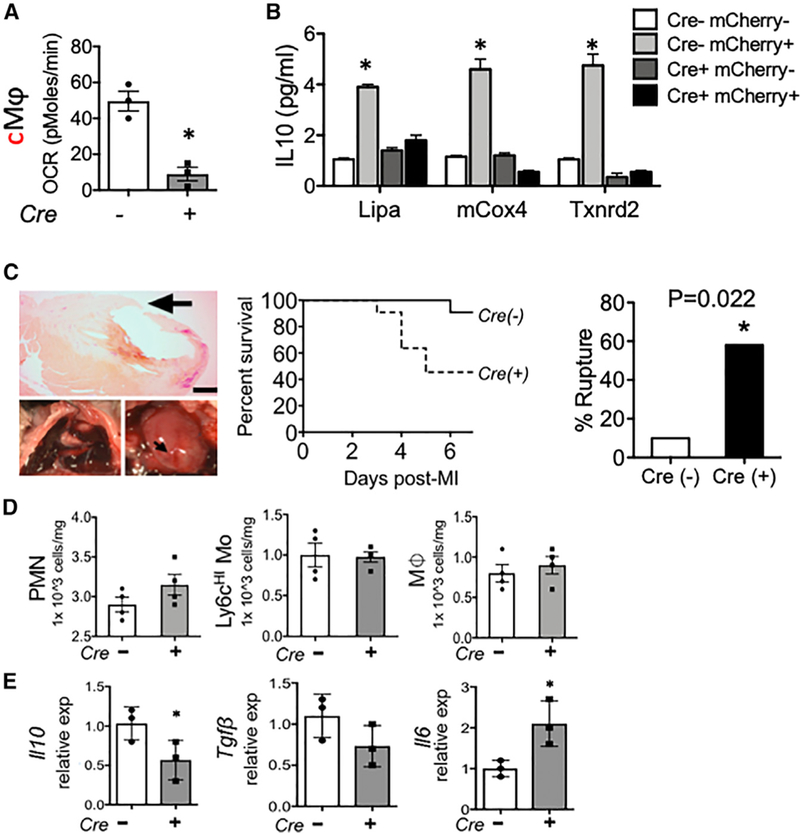
Rispfl/fl and Rispfl/fl LysMcre mice were subjected to experimental myocardial infarction (MI) at the left anterior descending artery, as described in STAR Methods. (A) Cardiac extracts were prepared and flow cytometry employed to isolate left ventricular cardiac Mᶲs (CD45+CD11b+F4/80+Ly6g—CD64+). Mice transgenic for cardiac-specific expression of the fluor mCherry were subjected to MI, and cardiac Mᶲs containing mCherry were interrogated for OCR in Rispfl/fl LysMcre mice. (B) Gene expression by qPCR from indicated cell types, sorted directly from heart. (C) Representative images and Kaplan-Meier survival plot and rupture quantification of indicated mice after MI. n = 10 (Cre—) versus 11 (Cre+) and p = 0.02. (D) Levels of indicated immune cell subsets (PMN, neutrophil; Mo, monocyte; Mᶲ, macrophage) after MI. (E) Gene expression of indicated inflammatory mediators from cardiac extracts. *p < 0.05.
DISCUSSION
Phagocytosis has evolved beyond nutritional uptake to couple with the active synthesis of anti-inflammatory cytokines. An integration of immunity with nutrient sensing would permit efficient energy utilization during anabolic demand. Now, as cellular metabolism is appreciated to contribute to diverse cell functions beyond biosynthetic demand, less appreciated are the fundamental mechanisms to explain how metabolic intermediates participate in non-energetic signal transduction. Mitochondria facilitate immunologic function (Buck et al., 2017) and also act as platforms for superoxide signaling (Chandel et al., 2000; Tait and Green, 2012) and the generation of metabolites that regulate the epigenetic landscape (Jha et al., 2015). A unique aspect of efferocytic catabolism is the potential of metabolic substrates, derived from the apoptotic cell, to directly influence macrophage polarization. In this context, cellular imbalances of lipids and proteins during disease could particularly compromise the integrated metabolic response of macrophages to promote tissue repair.
Early in inflammation, acute-phase monocytes and macrophages are highly glycolytic, particularly under limiting oxygen tensions. Glycolytic remodeling is important for macrophage polarization (Haschemietal., 2012). For example, macrophages that are in contact with Th2-derived cytokines such as IL-4 partially utilize glucose for alternative macrophage polarization (Huang et al., 2016). In our hands, the generation of anti-inflammatory IL-10 was not blocked by inhibitors of glycolysis (Figure 3), although extracellular acidification did trend higher during efferocytosis. One likely explanation is the abundant substrate supply provided by the apoptotic cell, which could preclude needs to mobilize metabolites that are pre-stored in the phagocyte. Relative to modest effects on glycolysis, OCR was significantly elevated during apoptotic cell metabolism, in comparison to that reported for IL-4, where macrophages may use internal lipid stores to fuel mitochondrial metabolism (Huang et al., 2014). IL-4 and apoptotic cells may work together to cooperatively program macrophages for tissue repair (Bosurgi et al., 2017), and it will be informative to examine if IL-4 modulates efferocytic mitochondrial activation.
Changes in macrophage metabolites from our nonbiased me-tabolomics during efferocytosis were both intuitive and less self-evident. For example, we are intrigued by efferocytic-associated escalations in sedoheptulose-7-phosphate (Table S1), which has separately been linked to macrophage polarization in connection with the pentose phosphate pathway/PPP (Ha-schemi et al., 2012). Studies that directly interfere (Köhler et al., 1970) with rate-limiting enzymes of this pathway during efferocytosis would be interesting to validate potential causal relationships of the PPP to cytokine signaling. Furthermore, consistent with increased lipid metabolism, efferocytosis led to accumulations of the monoacylglycerol 1-palmitoleoyl glycerol, which may be formed by lipases during the breakdown of triglycerides or diglycerides (Chandak et al., 2010). Efferocytes also accumulated dephosphocoenzyme A, a direct precursor of coenzyme A. In line with increased gene expression of TCA enzymes, TCA metabolites pyruvate, citrate, and α-ketoglutarate were reduced after apoptotic cell engulfment. Efferocytosis induced itaconate, which can inhibit the accumulation of proin-flammatory succinate during endotoxin challenge (Lampropou-lou et al., 2016). Levels of succinate trended lower in efferocytes (Table S1) but were not statistically different. Efferocytosis may also suppress the macrophage response to lipopolysaccha-ride/LPS (Fadoketal., 1998). Itremainsto be determined if efferocytic metabolites contribute to this suppression. Though the above changes represent just a snapshot of the dynamic flux occurring during efferocytosis, our global analysis provides a foundation from which to test other specific metabolic pathways.
In functional mitochondria, electron transport recycles mitochondrial NADH to NAD+. This occurs together with a secondary oxidation of cytosolic NADH alongside mitochondrial shuttles that transport reducing agents across mitochondrial membranes (Titov et al., 2016). Thus, mitochondrial NAD+ is coupled to NAD+/NADH balance in the cytosol. In macrophages, the synthesis of NAD+ by nicotinamide phosphoribosyl-transferase (NAMPT) promotes cytoskeletal activation during cellular adhesion (Venter et al., 2014). NAD+ accumulation in efferocytes required Risp, and additional reductions in nicotinate and nicotinamide are consistent with efferocytic regulation of the NAD+ pool. Decreases in NAD+/NADH ratio can also lead to increased 2-hydroxyglutarate (2-HG), and this imbalance can impair hematopoietic stem cell differentiation (Ansió et al., 2017). 2-HG, as well as α-ketoglutarate, are further linked to altered epigenetic methylation on key immunoregulatory genes (Liu et al., 2017). Though our preliminary analyses do not reveal significant Risp-dependent changes in 2-HG in the setting of efferocytosis, this does not discount the possibility that other mitochondrial-dependent epigenetic alterations may be functioning, and this is an interesting focus for future studies.
The Mechanism of IL-10 Induction by NAD+ Is through SIRTs
The integration of NAD+ with sirtuin histone deacetylases (Tanner et al., 2000) supports the concept of gene regulation by metabolic signaling. The seven sirtuin isoforms differ in both subcellular localization and function, and SIRT3, SIRT4, and SIRT5 localize to mitochondria (Kim et al., 2010). Given that SIRT1 is cytosolic in its localization, this suggests that alterations in the mitochondrial pool of NAD+ during efferocytosis also affect the cytosolic pool. SIRT1 has also been linked to LXR (Li et al., 2007), and LXR is known to regulate efferocytosis (A-Gonzalez et al., 2009). In skeletal cells, SIRT1 controls the transcription of peroxisome proliferator-activated receptor-gamma co-acti-vator-1α (PGC-1α) (Amat et al., 2009), which separately has been linked to IL-10 induction (Morari et al., 2010) and oxidative metabolism in macrophages (Vats et al., 2006). Elevated oxygen consumption, in conjunction with requirements for SIRT1, also hints that increased mitochondrial biogenesis could in principle be a component of the efferocytic IL-10 response.
Our findings in heart support the physiologic relevance of our cellular working model (Figure S7, Working Model). Within the myocardium, efferocytosis is required to preserve ventricular systolic function after MI (Wan et al., 2013; Dehn and Thorp, 2017). Although MI creates an acutely ischemic microenvironment that dampens oxidative respiration, the resolving phase of inflammation accumulates non-hypoxic macrophages, some of which may generate IL-10 in the myocardium. The significance of Risp-dependent reductions in IL-10 or NAD in vivo may be tested by restoring IL-10 or NMN (Yamamoto et al., 2014) specifically in macrophages with mitochondria dysfunction and during tissue injury. IL-10 is a key protective factor in heart (Krishnamurthy et al., 2009), and its secretion is necessary for wound closure through activation of epithelial proliferation (Quiros et al., 2017). Risp was also required for efferocytic TGF-β, which contributes to early replacement fibrosis. Furthermore, it will be interesting to determine how pattern recognition receptor ligands released from necrotic cells during tissue injury could potentially rewire phagocyte metabolic bias (Krawczyk et al., 2010).
Limitations of Study
A caveat to our findings in heart is that LysMcre may be expressed in certain non-macrophage cells, such as the neutrophil. Neutrophil lipolysis and FAO were recently linked to cellular autophagy, differentiation, and inflammation (Riffelmacher et al., 2017). However, in our hands, LysMcre deletion of Risp did not alter cardiac neutrophil accumulation (Figure 7). Additional aspects of our working model warrant future study. For example, potential signaling axes between apoptotic cell receptors and the mitochondria remain to be fully elucidated. In this context, receptor-mediated signal transduction could cooperatively amplify the mitochondrial response during efferocytosis. In addition, the electron transport chain may function during other core macrophage functions. This includes phagocyte recruitment, phagocyte proliferation and survival, and antigen presentation. In the setting of disease, it will be important to elucidate just how varied metabolite composition contributes to macrophage function and NAD+ generation, such as during cardiometabolic pathophysiology. For example, atherosclerotic progression is characterized by the turnover and pro-inflammatory phagocytosis of cholesterol-laden foam cells and cholesterol crystals (Hansson et al., 2015). Carbon tracing during metabolite flux from apoptotic foam cells to phagocyte has the potential to implicate additional unique immunometabolic cues that may fuel these nonresolving inflammatory disorders. These molecular pathways may also be relevant to pathologies where natural somatic mitochondrial mutations contribute to mitochondrial dysfunction.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pbx-1 | Thermo Fisher | Cat# PA5–17223, RRID:AB_10980993 |
| Risp | Abcam | Cat# Ab14746, RRID:AB_301445 |
| Sirt1 | Abcam | Cat# Ab110304, RRID:AB_10864359 |
| Cpt1 | Abcam | Cat# Ab234111 |
| Cpt2 | Thermo Fisher | Cat# PA5–19222, RRID:AB_10984377 |
| APC anti-mouse Ly-6C | BioLegend | Cat# 128015, RRID:AB_1732087 |
| PerCP anti-mouse Ly-6G | BioLegend | Cat# 127653, RRID:AB_2616998 |
| PE/Cy7 anti-mouse F4/80 | BioLegend | Cat# 123113, RRID:AB_893490 |
| APC/Cy7 anti-mouse/human CD11b | BioLegend | Cat# 101225, RRID:AB_830641 |
| Brilliant Violet 421 anti-mouse CD64 | BioLegend | Cat# 139309, RRID:AB_2562694 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Etomoxir | Sigma-Aldrich | Cat# E1905, CAS: 828934–41-4 |
| Oligomycin | Sigma-Aldrich | Cat# 75351–5MG, CAS: 1404–19-9 |
| Rotenone | Sigma-Aldrich | Cat# R8875, CAS: 83–79-4 |
| Antimycin A | Sigma-Aldrich | Cat# A8674, CAS: 1397–94-0 |
| FCCP | Sigma-Aldrich | Cat# C2920–10MG, CAS: 370–86-5 |
| UK5099 | Sigma-Aldrich | Cat#PZ0160, CAS: 56396–35-1 |
| Fatty acid supplement | Sigma-Aldrich | Cat# F7050–5ML |
| Beta-nicotinamide mononucleotide | Sigma-Aldrich | Cat# N3501–100MG, CAS: 1094–61-7 |
| Brewer thioglycollate medium | Sigma-Aldrich | Cat# B2551–500 g |
| Critical Commercial Assays | ||
| Mouse IL-10 ELISA Set | BD Biosciences | 555252 |
| Mouse TNF ELISA Set | BD Biosciences | 555268 |
| NAD/NADH Assay Kit | Abcam | Ab65348 |
| SIRT activity assay kit | Abcam | Ab156065 |
| Hypoxyprobe Plus Kit | Hypoxyprobe Inc | HP2–100Kit |
| Chromatin Immunoprecipitation Assay | Millipore Sigma | 17–295 |
| ATP Assay Kit | Abcam | Ab83355 |
| TruSeq Stranded mRNA LT sample prep Kit | Illumina | RS-122–2101 |
| Bio-Plex Pro Mouseu Cytokine 23-plex | Bio Rad | M60009RDPD |
| Deposited Data | ||
| RNA-seq | This paper | GEO: GSE121681 |
| Experimental Models: Cell Lines | ||
| Jurkat cells | ATCC | Cat# CRL-8131, RRID:CVCL_U323 |
| Experimental Models: Organisms/Strains | ||
| Rispflox/flox mice | Dr. Schumacker | PMID: 23328522 |
| C57BL/6J mice | The Jackson Lab | RRID: IMSR_JAX:000664 |
| LysmCre mice | The Jackson Lab | Stock No: 004781 |
| Myh6-mCherry mice | The Jackson Lab | Stock No: 021577 |
| Oligonucleotides | ||
| Acadm-F GGATGACGGAGCAGCCAATG Acadm-R ATACTCGTCACCCTTCTTCT |
This paper | N/A |
| Acadvl-F TATCTCTGCCCAGCGACTTT Acadvl-R TGGGTATGGGAACACCTGAT |
This paper | N/A |
| β-actin-F TCCATCATGAAGTGTGACGT TCCATCATGAAGTGTGACGT |
This paper | N/A |
| β-actin-R TACTCCTGCTTGCTGATCCAC | ||
| CD36-F GAGGCATTCTCATGCCAGT GAGGCATTCTCATGCCAGT |
This paper | N/A |
| CD36-R ACGTCATCTGGGTTTTGCAC |
||
| CPT1a-F TGCTTTACAGGCGCAAACTG TGCTTTACAGGCGCAAACTG |
This paper | N/A |
| CPT1a-R GCAGATGTGTCAGGACCGAGT |
||
| Dlat-F TCCTGCAGGTGTCTTCACAG TCCTGCAGGTGTCTTCACAG |
This paper | N/A |
| Dlat-R GACGGAGATTTTCCCTTTCC |
||
| FABP4-F GGATTTGGTCACCATCCGGT GGATTTGGTCACCATCCGGT |
This paper | N/A |
| FABP4-R TTCCATCCCACTTCTGCACC |
||
| FH1-F AGCAATGCATATTGCTGCTG AGCAATGCATATTGCTGCTG |
This paper | N/A |
| FH1-R CGCATACTGGACTTGCTGAA |
||
| IDH3b-F GTTTTTGAGACGGGTGCTCG IDH3b-R TCCCATGTCTCGAGTCCGTA |
This paper | N/A |
| IL1b-F TACGGACCCCAAAAGATGA TACGGACCCCAAAAGATGA |
This paper | N/A |
| IL1b-R TGCTGCTGCGAGATTTGAAG |
||
| IL6-F TAGTCCTTCCTACCCCAATTTCC TAGTCCTTCCTACCCCAATTTCC |
This paper | N/A |
| IL6-R TTGGTCCTTAGCCACTCCTTC |
||
| IL-10-F ATAACTGCACCCACTTCCCA ATAACTGCACCCACTTCCCA |
This paper | N/A |
| IL-10-R GGGCATCACTTCTACCAGGT |
||
| MDH1-F GAAGCCCTGAAAGACGACAG MDH1-R TCGACACGAACTCTCCCTCT |
This paper | N/A |
| Mmp9-F CAAGTGGGACCATCATAACATCA Mmp9-R GATCATGTCTCGCGGCAAGT |
This paper | N/A |
| PDHB-F TCGAAGCCATAGAAGCCAGT |
This paper | N/A |
| PDHB-R AGGCATAGGGACATCAGCAC |
||
| Tnfa-F ACGGCATGGATCTCAAAGAC Tnfa-R AGATAGCAAATCGGCTGACG |
This paper | N/A |
| CHIP pbx binding region-F | This paper | N/A |
| TTTTCTACCAGCAGCAAGCA | ||
| CHIP pbx binding region-R | ||
| ATGCTAGCTGGGTCTTGAGC | ||
| CHIP CTRL-F | This paper | N/A |
| TTCCCTAGGATCAGGGAGGT | ||
| CHIP CTRL-R | ||
| AATGGGCCTCTCTTTCCAGT | ||
| Recombinant DNA | ||
| FlexiTube siRNA risp | QIAGEN | Cat No: GS66694 |
| FlexiTube siRNA SIRT1 | QIAGEN | Cat No: GS93759 |
| FlexiTube siRNA Pbx1 | QIAGEN | Cat No: GS18514 |
| FlexiTube siRNA Cpt1a | QIAGEN | Cat No: GS12894 |
| FlexiTube siRNA Cpt2 | QIAGEN | Cat No: GS12896 |
| Software and Algorithms | ||
| FlowJo X 10.0.7r2 | FlowJo | https://www.flowjo.com/ |
| Prism 7 | GraphPad | https://www.graphpad.com/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the first author Shuang Zhang, shuangzhang2017@u.northwestern.edu.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Models
C57BL/6J mice were bred in our facility and our colonies are refreshed yearly with mice purchased from the Jackson Laboratory. LysmCre mice were purchased from the Jackson Laboratory and bred in house. Uqcrfs1; RISPfl/fl mice were provided by Dr. Paul Schumacker and was generated as described (Waypa et al., 2013). aMHC-mCherry mice were obtained from the Jackson Laboratory. Ten-to sixteen-week-old mice were used for experiments. Investigators were not blinded to the group allocation for some studies. Mice were housed in temperature-and humidity-controlled environments and kept on a 12:12h day/night cycle with access to standard mouse chow and water ad libitum. All studies were approved and reviewed by the Institutional Animal Care and Use Committee at Northwestern University (Chicago, Illinois), protocol #IS00000375.
In Vivo Models of Inflammation and Tissue Repair
For experimental MI: Surgeries were performed on mice 12–16 weeks of age and as described (Wan et al., 2013). Surgeries were performed by an individual blinded to the genotype. Mice were anesthetized with Avertin and secured in a supine position and endotracheal-intubated and ventilated with an Inspira Advanced Safety Single Animal Pressure/Volume Controlled Ventilator (Harvard Apparatus, Holliston, MA) with room air supplemented with oxygen to maintain blood gases within normal physiological limits. The chest wall was shaved and left thoracotomy was performed. With the aid of a dissecting microscope, the left ventricle was visualized and left coronary artery on the anterior wall was permanently ligated with monofilament nylon 8–0 sutures (Ethicon, Somerville, NJ) 2mm distal to the site of its emergence from under the left atrium. Blanching/pale discoloration and hypokinesis of the anterior wall verified LAD ligation. The chest wall was closed with 7–0 nylon sutures and the skin and subcutaneous tissue was closed. Sham operations were performed on animals by passing the suture beneath the LAD without ligating the vessel.
METHOD DETAILS
Efferocytosis Ex Vivo
Peritoneal macrophages were elicited in mice with thioglycollate broth (Sigma-Aldrich) and harvested after lavage. Adherent macrophages were co-cultivated with apoptotic Jurkat T cells. Tcells were induced to apoptosis after ultravioliet irradiation and early apoptotic cells (ACs) were identified as annexin V positive, propidium iodide negative, and overlaid at a ratio of 5 ACs to 1 phagocyte. Non-engulfed Tcells were removed from adherent phagocytes, 1 hr post co-cultivation. Engulfment was confirmed by microscopic and flow cytometric analysis with fluorescently labeled apoptotic cells.
Unbiased Analysis of Efferocytic Secretome
Cell culture supernatant was collected 5 hr post co-cultivation. Chemokines and cytokines were analyzed with a Bio-Plex Pro Mouse cytokine 23-plex from Bio Rad using a Luminex 200 multiplex instrument (Luminex, Austin, TX) and MAGPIX analysis. Additional cytokines, including IL-10, were validated by independent ELISA. For IL-1b measurements, 5mM ATP was added 3 hr after LPS stimulation.
RNA-Sequencing and Bioinformatics
Adherent peritoneal macrophages were stimulated with apoptotic thymocytes for 6 hr, versus control. RNA was isolated from cells with Trizol and RNA quality assessed at the Northwestern Genomics core with RNA integrity values of 10 for a minimum of 100 ng per sample. Library construction was performed in lab and sequencing was performed at the Genomics Core facility at the University of Chicago and Northwestern University. Sequencing libraries were constructed using an Illumina TruSeq Stranded mRNA prep kit LT for mRNA sequencing and constructs sequenced on NextSeq 500 instrument and HiSeq 2500 System from Illumina. For RNaseq, the quality of DNA reads, in FASTQ format, was evaluated using FastQC. Adapters were trimmed and reads were aligned to the Mus musculus genome (mm10) using STAR (Dobin et al., 2013). Read counts for each gene were calculated using HTSeq-count in conjunction with a gene annotation file for mm10 obtained from University of California Santa Cruz at genome.ucsc.edu. Normalization and differential expression were determined using DESeq2. The cutoff for determining significantly differentially expressed genes was an FDR-adjusted p value less than 0.05. Transcriptome analysis was performed at the bioinformatics core at Northwestern University with assistance from Matthew Schipma, PhD.
Metabolomics of Efferocytosis
Metabolomics analyses were carried out in with Metabolon (Durham, NC) as previously described (Evans et al., 2009; Masri et al., 2014) and with the University of Michigan Metabolomics Core. Briefly, control and Risp-deficient peritoneal macrophages were treated as indicated. Cell lysates were harvested, methanol extracted, and analyzed by ultra-high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS; positive mode), UPLC-MS/MS (negative mode) and gas chromatography–mass spectrometry (GC-MS). Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra, and curated by visual inspection for quality control. These data are listed in Table S1.
Respiratory and Glycolytic Analyses
To measure the extracellular acidification rate (ECAR) and OCR, apoptotic cell-treated primary macrophages were plated on an XF24 cell culture microplates coated with CellTak. Experiments were conducted in XF assay medium containing 25 mM glucose, 2 mML-glutamine, and 1 mM Na pyruvate, and analyzed using a Seahorse XF24 extracellular flux analyzer (Agilent Technologies). Where indicated, the following were injected: ATP-synthesis inhibitor oligomycin (1.5 mM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP; 1.5 mM) to uncouple ATP synthesis, rotenone (100 nM) to block complex I, and antimycin A (1 mM) (Sigma) to block complex III. Basal ECAR, OCR, and spare respiratory capacity were generated by Wave Desktop software (Agilent Technologies).
Transmission Electron Microscopy
Macrophages were cultured on sterilized Thermanox plastic coverslips and cultivated with apoptotic cells. Subsequently, coverslip were fixed with 0.1 M sodium cacodylate buffer (pH 7.3) containing 2% paraformaldehyde and 2.5% glutaraldehyde for 30 min at room temperature and were kept at 4_C and then processed for TEM. Coverslips were flat-embedded in resin and cured in a 60_C oven. Samples were sectioned on a Leica Ultracut UC6 ultramicrotome into 70nm sections and collected on 200 mesh copper grids. Samples were processed and imaged on an FEI Tecnai Spirit G2 TEM at the Advantaged Imaging Center at Feinberg School of Medicine with assistance from Lennell Reynolds Jr.
Immunoprecipitation
Whole cell extract was isolated from macrophages treated with or without apoptotic cells. Pbx-1 binding to chromatin was pulled down via chromatin immunoprecipitation. In brief, chromatin was sonicated into 200–1000 bp following crosslinking. After overnight incubation with isotype IgG or PBX-1 antibody, 100ul of protein A beads was added into the solution and incubated for 1hr at 4C. The protein-bound beads were collected, DNA was eluted and reverse cross-linked. DNA was recovered by phenol/chloroform extraction and ethanol precipitation.
Immunoblots
Cells were lysed in RIPA buffer, resolved on 10% polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked using 5% milk and then incubated in the primary antibody overnight. Antibodies were anti-RISP, anti-SIRT1, anti-CPT1A, anti-CPT2, and anti-a-tubulin. Membranes were rinsed 3 times with tween solution and then incubated with secondary antibody. Secondary antibodies utilized were anti-mouse IgG, HRP-linked and anti-rabbit IgG, HRP-linked.
Chromatin Immunoprecipitation and SIRT1 Activity
Chromatin for chromatin immunoprecipitation (ChIP) was prepared and proteins cross-linked to DNA. Chromatin was incubated overnight with antibodies against PBX-1 or isotype control. PCR primers were designed to amplify the IL-10 apoptotic cell response element (Chung et al., 2007) and were performed in primary macrophages after siRNA knockdown of indicated genes. SIRT1 activity was measured with the Sirt1 Activity Assay Kit from Abcam, Cambridge, MA and as described (Chatterjee et al., 2018).
Flow Cytometric Analysis after Experimental MI
Infarcted mice were anesthetized with isoflurane and peripheral blood drawn into citrate anticoagulant solution. Hearts were harvested, perfused with saline to remove peripheral cells, minced with fine scissors, and incubated in a cocktail of collagenase and DNase. Cells were filtered through a 70mm strainer and pelleted at 500×g. Total cell numbers were counted by Trypan blue staining and cell suspensions were rinsed with Hank’s Balanced Salt Solution supplemented with 0.2% (wt/vol) bovine serum albumin and1% wt/vol fetal calf serum.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed with GraphPad Prism (GraphPad Software, La Jolla, CA). Comparisons between two groups were performed using two-tailed, unpaired t test with 95% confidence interval. For comparisons of more than two variables, 1-way ANOVA was utilized with 95% confidence interval. Data are presented as mean ± SEM. For qPCR and murine model of myocardial infarction, power and sample size were estimated based on our previous work. The statistical parameters and criteria for significance can be found in the figure legends.
Supplementary Material
Highlights.
Engulfment of dying cells elevates macrophage fatty acids and oxygen consumption
Mitochondrial respiration during efferocytosis is required for NAD homeostasis
NAD mobilization is required for anti-inflammatory responses in macrophages
Defects in myeloid mitochondrial complex III impair wound healing
ACKNOWLEDGMENTS
Funding support from NHLBI F32HL127958 (M.D.) and HL122309 and HL139812 (E.B.T.), Chicago Biomedical Consortium, AHA predoctoral fellowship to S.Z., NU presidential fellowship to S.Z., and Sidney Bess Memorial Fund. Special thanks to Lennell Reynolds, Jr. at the NU imaging facility. D.R.G. is supported by 2R01CA152601-A1, 1R01CA152799–01A1, 1R01CA168292–01A1, 1R01CA214025–01, the Avon Foundation for Breast Cancer Research, the Lynn Sage Cancer Research Foundation, the Zell Family Foundation, and the Chicago Biomedical Consortium as well the Searle Funds at The Chicago Community Trust.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.cmet.2018.12.004.
DECLARATION OF INTERESTS
Jason Kinchen is an employee of Metabolon.
DATA AND SOFTWARE AVAILABILITY
The RNaseq datasets generated during and/or analyzed during the current study are available at GEO: GSE121681 (WT = Wild-Type, AC = Apoptotic Cell. LPS = lipopolysaccharide).
REFERENCES
- A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Diaz M, Gallardo G, et al. (2009). Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 31, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvey CM, Spinler KR, Irianto J, Pfeifer CR, Hayes B, Xia Y, Cho S, Dingal PCPD, Hsu J, Smith L, et al. (2017). SIRPA-inhibited, marrow-derived macrophages engorge, accumulate, and differentiate in antibody-targeted regression of solid tumors. Curr. Biol. 27, 2065–2077.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, and Villarroya F (2009). SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J. Biol. Chem. 284, 21872–21880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansó E, Weinberg SE, Diebold LP, Thompson BJ, Malinge S, Schumacker PT, Liu X, Zhang Y, Shao Z, Steadman M, et al. (2017). The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 19, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai A, Choi JL, He S, Fenn AM, Nairz M, Rattik S, McAlpine CS, Mindur JE, Chan CT, Iwamoto Y, et al. (2017). The infarcted myocardium solicits GM-CSF for the detrimental oversupply of inflammatory leukocytes. J. Exp. Med. 214, 3293–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosurgi L, Cao YG, Cabeza-Cabrerizo M, Tucci A, Hughes LD, Kong Y, Weinstein JS, Licona-Limon P, Schmid ET, Pelorosso F, et al. (2017). Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 356, 1072–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, and Pearce EL (2017). Metabolic instruction of immunity. Cell 169, 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JE, Brooke MH, DeVivo DC, Kaiser KK, and Hagberg JM (1978). Biochemical and physiologic consequencesofcarnitine palmityltrans-ferase deficiency. Muscle Nerve 1, 103–110. [DOI] [PubMed] [Google Scholar]
- Chandak PG, Radovic B, Aflaki E, Kolb D, Buchebner M, Fröhlich E, Magnes C, Sinner F, Haemmerle G, Zechner R, et al. (2010). Efficient phagocytosis requires triacylglycerol hydrolysis by adipose triglyceride lipase. J. Biol. Chem. 285, 20192–20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, and Schumacker PT (2000). Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism ofO2 sensing. J. Biol. Chem. 275, 25130–25138. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Daenthanasanmak A, Chakraborty P, Wyatt MW, Dhar P, Selvam SP, Fu J, Zhang J, Nguyen H, Kang I, et al. (2018). CD38-NAD+axis regulates immunotherapeutic anti-tumor T cell response. Cell Metab. 27, 85–100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EY, Liu J, Homma Y, Zhang Y, Brendolan A, Saggese M, Han J, Silverstein R, Selleri L, and Ma X (2007). Interleukin-10 expression in macrophages during phagocytosis of apoptotic cells is mediated by homeo-domain proteins Pbx1 and Prep-1. Immunity 27, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlo E, Heinonen T, Théroude C, Herderschee J, Mombelli M, Lugrin J, Pfefferlé M, Tyrrell B, Lensch S, Acha-Orbea H, et al. (2017). Sirtuin 2 deficiency increases bacterial phagocytosis by macrophages and protects from chronic staphylococcal infection. Front. Immunol. 8, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerge M, Yeap XY, Dehn S, Zhang S, Grigoryeva L, Misener S, Procissi D, Zhou X, Lee DC, Muller WA, et al. (2017). MerTK cleavage on resident cardiac macrophages compromises repair after myocardial ischemia reperfusion injury. Circ. Res. 121, 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehn S, and Thorp EB (2017). Myeloid receptor CD36 is required for early phagocytosis of myocardial infarcts and induction of Nr4a1-dependent mechanisms of cardiac repair. FASEB J. 32, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Hsieh WY, Minarrieta L, Duong TN, Kim KKO, Desousa BR, Andreyev AY, Bowman CE, Caradonna K, Dranka BP, et al. (2018). Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 28, 490–503.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller K, Wolf D, Huber JM, Metz M, Mayer G, McKenzie AN, Maurer M, Rosenkranz AR, and Wolf AM (2011). IL-9 production by regulatory T cells recruits mast cells that are essential for regulatory T cell-induced immune suppression. J. Immunol. 186, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, and Milgram E (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification ofthesmall-molecule complement of biological systems. Anal Chem 81, 6656–6667. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, and Henson PM (1998). Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Invest. 101, 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson GK, Libby P, and Tabas I (2015). Inflammation and plaque vulnerability. J. Intern. Med. 278, 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Schmidt B, Wachter E, Weiss H, and Neupert W (1986). Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell 47, 939–951. [DOI] [PubMed] [Google Scholar]
- Haschemi A, Kosma P, Gille L, Evans CR, Burant CF, Starkl P, Knapp B, Haas R, Schmid JA, Jandl C, et al. (2012). The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howangyin KY, Zlatanova I, Pinto C, Ngkelo A, Cochain C, Rouanet M, Vilar J, Lemitre M, Stockmann C, Fleischmann BK, et al. (2016). Myeloid-epithelial-reproductive receptor tyrosine kinase and milkfat globule epidermal growth factor 8 coordinately improve remodeling after myocardial infarction via local delivery of vascular endothelial growth factor. Circulation 133, 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, et al. (2014). Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, and Pearce EJ (2016). Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity 45, 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Tsuji H, Nakamura S, Ichiyama A, Nishizuka Y, and Hayaishi O (1965). Studies on the biosynthesis of nicotinamide adenine dinucleotide. II. A role of picolinic carboxylase in the biosynthesis of nicotinamide adenine dinucleotide from tryptophan in mammals. J. Biol. Chem. 240, 1395–1401. [PubMed] [Google Scholar]
- Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. (2015). Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–30. [DOI] [PubMed] [Google Scholar]
- Jose PJ, Griffiths-Johnson DA, Collins PD, Walsh DT, Moqbel R, Totty NF, Truong O, Hsuan JJ, and Williams TJ (1994). Eotaxin: a potent eosinophil chemoattractant cytokine detected in a guinea pig model of allergic airways inflammation. J. Exp. Med. 179, 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. (2010). SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17,41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler E, Barrach H, and Neubert D (1970). Inhibition of NADP dependent oxidoreductases by the 6-aminonicotinamide analogue of NADP. FEBS Lett. 6, 225–228. [DOI] [PubMed] [Google Scholar]
- Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG, and Pearce EJ (2010). Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P, Rajasingh J, Lambers E, Qin G, Losordo DW, and Kishore R (2009). IL-10 inhibits inflammation and attenuates left ventricular remodeling after myocardial infarction via activation of STAT3 and suppression of HuR. Circ. Res. 104, e9–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC, Griss T, et al. (2016). Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, and Guarente L (2007). SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28,91–106. [DOI] [PubMed] [Google Scholar]
- Liu PS, Wang H, Li X, Chao T,Teav T, Christen S, Di Conza G, Cheng WC, Chou CH, Vavakova M, et al. (2017). α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994. [DOI] [PubMed] [Google Scholar]
- Lunetta KL, Hayward LB, Segal J, and Van Eerdewegh P (2004). Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morari J, Torsoni AS, Anhê GF, Roman EA, Cintra DE, Ward LS, Bordin S, and Velloso LA (2010). The role of proliferator-activated receptor gamma coactivator-1alpha in the fatty-acid-dependent transcriptional control of interleukin-10 in hepatic cells of rodents. Metabolism 59, 215–223. [DOI] [PubMed] [Google Scholar]
- Oren R, Farnham AE, Saito K, Milofsky E, and Karnovsky ML (1963). Metabolic patterns in three types of phagocytizing cells. J. Cell Biol. 17,487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, et al. (2015). Pyruvate kinase M2 regulates Hif-1 α activity and IL-1 β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Han CZ, Elliott MR, Kinchen JM, Trampont PC, Das S, Collins S, Lysiak JJ, Hoehn KL, and Ravichandran KS (2011). Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature 477, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Lucas CD, Rossi AG, and Ravichandran KS (2014). Apoptotic cell clearance: basic biology and therapeutic potential. Nat. Rev. Immunol. 14, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros M, Nishio H, Neumann PA, Siuda D, Brazil JC, Azcutia V, Hilgarth R, O’Leary MN, Garcia-Hernandez V, Leoni G, et al. (2017). Macrophage-derived IL-10 mediates mucosal repair by epithelial WISP-1 signaling. J. Clin. Invest. 127, 3510–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich PR (1983). Electron transfer through the isolated mitochondrial cytochrome b-c1 complex. Biochim. Biophys. Acta 722, 271–280. [DOI] [PubMed] [Google Scholar]
- Riffelmacher T, Clarke A, Richter FC, Stranks A, Pandey S, Danielli S, Hublitz P, Yu Z, Johnson E, Schwerd T, et al. (2017). Autophagy-depen-dent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity 47, 466–480.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfeld P, Wieckowski MR, Lebiedzińska M, and Wojtczak L (2010). Mitochondrial fatty acid oxidation and oxidative stress: lack of reverse electron transfer-associated production of reactive oxygen species. Biochim. Biophys. Acta 1797, 929–938. [DOI] [PubMed] [Google Scholar]
- Serhan CN, and Savill J (2005). Resolution of inflammation: the beginning programs the end. Nat. Immunol. 6, 1191–1197. [DOI] [PubMed] [Google Scholar]
- Shiraishi M, Shintani Y, Shintani Y, Ishida H, Saba R, Yamaguchi A, Adachi H, Yashiro K, and Suzuki K (2016). Alternatively activated macrophages determine repair of the infarcted adult murine heart. J. Clin. Invest. 126, 2151–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AA, Kiser RE, Denning GM, Koh SW, and DeBault LE (1979). Modification of the fatty acid composition of cultured human fibroblasts. J. Lipid Res. 20, 536–547. [PubMed] [Google Scholar]
- Tabas I, and Glass CK (2013). Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SW, and Green DR (2012). Mitochondria and cell signalling. J. Cell Sci. 125, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, and Denu JM (2000). Silent information regulator 2 family of NAD-dependent histone/protein deacetylasesgener-atesa unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97, 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, and Mootha VK (2016). Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varia MA, Calkins-Adams DP, Rinker LH, Kennedy AS, Novotny DB, Fowler WC Jr., and Raleigh JA (1998). Pimonidazole: a novel hypoxia marker for complementary study of tumor hypoxia and cell proliferation in cervical carcinoma. Gynecol. Oncol. 71, 270–277. [DOI] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, and Chawla A (2006). Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter G, Oerlemans FT, Willemse M, Wijers M, Fransen JA, and Wieringa B (2014). NAMPT-mediated salvage synthesis of NAD+ controls morphofunctional changes of macrophages. PLoS One 9, e97378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan E, Yeap XY, Dehn S, Terry R, Novak M, Zhang S, Iwata S, Han X, Homma S, Drosatos K, et al. (2013). Enhanced efferocytosis of apoptotic cardiomyocytes through myeloid-epithelial-reproductive tyrosine kinase links acute inflammation resolution to cardiac repair after infarction. Circ. Res. 113, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mohsen AW, Mihalik SJ, Goetzman ES, and Vockley J (2010). Evidence for physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes. J. Biol. Chem. 285, 29834–29841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Subramanian M, Yurdagul A Jr., Barbosa-Lorenzi VC, Cai B, de Juan-Sanz J, Ryan TA, Nomura M, Maxfield FR, and Tabas I (2017). Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell 171, 331–345.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waypa GB, Marks JD, Guzy RD, Mungai PT, Schriewer JM, Dokic D, Ball MK, and Schumacker PT (2013). Superoxide generated at mitochondrial complex III triggers acute responses to hypoxia in the pulmonary circulation. Am. J. Respir. Crit. Care Med. 187, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YQ, Freire-de-Lima CG, Schiemann WP, Bratton DL, Vandivier RW, and Henson PM (2008). Transcriptional and translational regulation of TGF-beta production in response to apoptotic cells. J. Immunol. 181, 3575–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Byun J, Zhai P, Ikeda Y, Oka S, and Sadoshima J (2014). Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PLoS One 9, e98972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yeap XY, Grigoryeva L, Dehn S, DeBerge M, Tye M, Rostlund E, Schrijvers D, Zhang ZJ, Sumagin R, et al. (2015). Cardiomyocytes induce macrophage receptor shedding to suppress phagocytosis. J. Mol. Cell. Cardiol. 87, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


