Abstract
Failure of one parent's chromosomes to organize nucleoli in an interspecific hybrid is an epigenetic phenomenon known as nucleolar dominance. Selective gene silencing on a scale of millions of bp is known to be involved, but the full extent to which nucleolus organizer region (NOR)-bearing chromosomes are inactivated beyond the NORs is unknown. Aided by genome sequence data for Arabidopsis thaliana, we have mapped the extent of nucleolar dominance-induced silencing in Arabidopsis suecica, the allotetraploid hybrid of A. thaliana and Arabidopsis arenosa. Using a sensitive reverse transcription PCR assay, we show that the four A. thaliana NORs, each ≈4 Mbp in size, are ≈99.5% silenced in A. suecica vegetative leaves, whereas the NORs inherited from A. arenosa remain fully active. The two A. thaliana NORs, NOR2 and NOR4, abut the telomeres on chromosomes 2 and 4, thus there are no genes distal to the NORs. The three protein-coding genes nearest NOR4 on its centromere-proximal side, the closest of which is only 3.1 kb from rRNA gene sequences, are shown to be transcribed in the hybrid despite the silencing of the adjacent ≈4-Mbp NOR. These data argue against hypotheses in which NOR inactivation is attributed to the spread of silencing from adjacent chromosomal regions, but favor models in which NORs or rRNA genes are the targets of regulation.
Keywords: epigenetic phenomena‖ribosomal RNA gene‖nucleolus organizer‖polyploidy‖genetic hybrid
Nucleolus organizer regions (NORs) are genetic loci (1) in eukaryotes at which the genes encoding the precursor of the three largest ribosomal RNAs (rRNAs) are clustered in hundreds to thousands of copies, collectively spanning millions of bp along the chromosome (2–4). Transcription of the rRNA genes by RNA polymerase I initiates the formation of a nucleolus (5), the subnuclear region where ribosomes are assembled (6–8). In interspecies (interspecific) hybrids, the NORs of one progenitor species are often inactivated, regardless of whether that species served as the maternal or paternal parent, whereas NORs inherited from the other progenitor species remain fully active (9–12). Silenced rRNA genes are efficiently derepressed by chemical inhibitors of DNA methylation or histone deacetylation, implicating chromatin modifications in the enforcement (maintenance) of nucleolar dominance (13–15). The same chromatin-mediated repression mechanisms may be responsible for controlling the effective dosage of rRNA genes in “pure” (nonhybrid) species in which not all rRNA genes are expressed at any one time (see ref. 11 for an expanded discussion). However, the mechanisms that discriminate among parental rRNA genes and that lead to the establishment of nucleolar dominance in hybrids remain obscure.
Studies of nucleolar dominance in hybrid frogs and cultured mammalian cells have suggested that parental rRNA gene types might compete for one or more RNA polymerase I transcription factors, with the dominant genes being those that can best recruit the factor(s) (12, 16). However, in plants, transient expression and in vitro transcription studies have failed to detect any preferential expression of dominant rRNA genes as would be predicted if dominant genes have a higher binding affinity for transcription factors (17). The latter studies have also shown that plasmid-borne rRNA genes can be efficiently transcribed in hybrid cells in which their endogenous chromosomal counterparts are repressed, indicating that nucleolar dominance is strictly a chromosomal phenomenon (17). Chromosome rearrangements that move NORs to new chromosomal environments, or that delete sequences adjacent to NORs without altering rRNA gene sequences, can also induce or negate nucleolar dominance (18–21). Collectively, these data suggest that the chromosomal context of an NOR is an important determinant of nucleolar dominance, for reasons that are currently unknown.
One hypothesis to explain the effects of chromosomal context on NOR function could be that NORs are passively inactivated as a byproduct of silencing much larger chromosome segments, with silencing then spreading to the NOR. A prediction is that genes adjacent to the NORs should also be silenced when nucleolar dominance occurs. A model system ideally suited to address this possibility is Arabidopsis suecica, an allotetraploid species that arose by combining diploid genome equivalents from Arabidopsis arenosa (also known as Cardaminopsis arenosa) and Arabidopsis thaliana. Ribosomal RNA genes of both progenitors are maintained in A. suecica, but S1 nuclease protection assays detect only A. arenosa rRNA gene transcripts; A. thaliana rRNA transcripts are typically not detected (14). There are two NORs in A. thaliana, NOR2 and NOR4, located at the distal northern ends of chromosomes 2 and 4, respectively (22, 23). The NORs are capped directly by consensus telomere repeats, thus there are no genes distal to the NORs (23). Each NOR is composed of ≈350–400 rRNA genes (each ≈10–10.5 kb) organized in uninterrupted head-to-tail arrays; thus, no sequences other than rRNA genes are thought to be embedded within the NORs (24). On the centromere-proximal side of the A. thaliana NORs, the flanking chromosome sequences have been determined by the whole-genome sequencing effort of the Arabidopsis Genome Initiative (25–27). A complex jumble of incomplete retrotransposable element sequences spans the ≈50-kb region between NOR2 and the first predicted protein-coding gene on chromosome 2 (25). However, the analogous region flanking NOR4 is comparatively simple and amenable to study, with predicted protein coding genes located within 3.1 kb of rRNA gene sequences (Fig. 1). In this paper, we show that transcripts can be detected from the three predicted protein-coding genes adjacent to NOR4 and that these genes remain transcriptionally active in A. suecica despite the inactivation of the adjacent ≈4-Mbp NOR. These data argue against the possibility that silencing spreads to the NOR from adjacent regions. Instead, the data favor models in which the rRNA genes or some other feature of an NOR are the specific targets of silencing in nucleolar dominance. Furthermore, the fact that silencing is restricted to the NOR suggests that a barrier may exist to prevent repressive chromatin structures from being propagated from silenced rRNA genes to flanking genes.
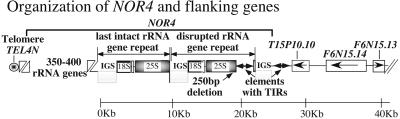
Figure 1.
Organization of the northern end of A. thaliana chromosome 4. Consensus telomere repeats at locus TEL4N are added directly to the first of ≈350–400 rRNA genes, each of which consists of coding sequences for 18S, 5.8S, and 25S rRNA and is separated from the next rRNA gene by an intergenic spacer (23). The most centromere-proximal 25S rRNA gene coding sequence is disrupted by a 2.2-kb sequence bearing 141-bp terminal inverted repeats (TIRs); we predict this to be a nonautonomous retrotransposon. The disrupted rRNA gene is followed by several kilobases of repetitive intergenic spacer (IGS) sequences. The nearest predicted protein-coding gene, T15P10.10 is located ≈3.1 kb downstream of the final IGS repeats. A predicted 1.6-kb nonautonomous retrotransposon, characterized by discontinuous 221-bp TIRs, is present in the ≈3.1-kb interval between the IGS repeats and T15P10.10. Arrows denote the direction of transcription for loci T15P10.10, F6N15.14, and F6N15.13.
Materials and Methods
Plant Material and Nucleic Acid Isolation.
A. suecica accession LC1, A. arenosa accessions 3651 and 9810 (28, 29), and A. thaliana accession Col-0 (Arabidopsis Biological Resource Center, Ohio State University, Columbus) were grown in growth chambers or a glasshouse. Nucleic acids were typically isolated from leaf tissue pooled from 5–10 plants. Tissues were frozen in liquid nitrogen, ground to a powder, and mixed vigorously with 3 vol (wt/vol) of extraction buffer (250 mM Tris⋅HCl, pH 8.5/375 mM NaCl/25 mM EDTA, pH 8.0/1% SDS/1% β-mercaptoethanol/0.5 mg/ml heparin). The homogenate was extracted twice with phenol/chloroform, and total nucleic acids were precipitated from the aqeous phase with 2 vol of ethanol. After centrifugation, pellets were resuspended in sterile water and total RNA was precipitated with 3 M LiCl. Genomic DNA in the supernatant was recovered by ethanol precipitation.
PCR Assays.
PCR of genomic DNA used ≈100 ng of DNA in a 50 μl reaction with 20 pmols of each primer. Reverse transcription (RT)-PCR was performed by using RNA that had been treated with RQ1 DNase I (Promega) to eliminate any contaminating genomic DNA. Total RNA was used in all cases except in the case of F16N15.13, for which poly(A)+ RNA was isolated on oligo(dT) paramagnetic beads according to the manufacturer's instructions (Dynal, Great Neck, NY). RT reactions (20 μl) typically contained 4 μg RNA and 200 units of reverse transcriptase (GIBCO Superscript II) by using conditions recommended by the supplier. Subsequent PCR used RT product resulting from 100–400 ng of input RNA. For PCR amplification of the rRNA gene ITS1 region, 29 cycles (94°, 30 s; 59°, 30 s; 72°, 60 s) were performed with 5′-GCGCTACACTGATGTATTCAACGAG-3′ as the forward primer and 5′-CGCACCTTGCGTTCAAAGACTCGA-3′ as the reverse primer. For F6N15.13 PCR amplification, 35 cycles were performed (94°, 15 s; 57°, 30 s; 72°, 90 s) with 5′-TCGAACAAGCTGCTCGAT CTTCGCG-3′ as the forward primer and 5′-GGCGGTCGTCCCAAACAAGCCTGCC3-′ as the reverse primer. For F6N15.14 PCR amplification, 35 cycles were performed (94°, 30 s; 62°, 30 s; 72°, 60 s) with 5′-CTCAGGAAGCACATTGTTCTGATAG-3′ as the forward primer and 5′-TTGTACTTGAAATCACTGCCTCTAG-3′ as the reverse primer. A single nucleotide polymorphism at the 3′ terminus of the F6N15.14 forward primer prevented amplification of a paralogous gene on A. thaliana chromosome V. Primer specificity was verified by using the primer for genomic cycle sequencing. Two forward primers were used to amplify the T15P10.10 gene, one specifically designed to amplify only the A. thaliana gene copy of interest on chromosome IV: 5′-GATTCAGAAATACTTTACTTACATGCG-3′ (A.t. primer in Fig. 5). A second forward primer amplified the A. arenosa gene and a paralogous gene on A. thaliana chromosome V: 5′-GATTCAGAAACACTTTACTTACATGCC-3′ (A.a. primer in Fig. 5). Single-nucleotide polymorphisms at the very 3′ end of the primers were exploited to achieve these specificities, which were verified by using the primers for genomic cycle sequencing. The reverse primer, used with both forward primers, was 5′-GGATATGGGCACTGCTGGTTTTG3-′). A total of 31 cycles of PCR were performed in each case (94°, 30 s; 60°, 30 s; 72°, 60 s).
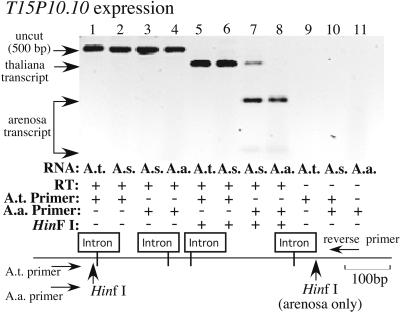
Figure 5.
A. thaliana locus T15P10.10, located only 3.1 kb from rRNA gene sequences, is expressed in A. suecica. A specific forward primer (A.t. primer) was designed to amplify A. thaliana T15P10.10 transcripts but not homologous A. arenosa transcripts. A second primer (A.a primer) was designed to amplify the A. arenosa homolog. The A.a primer also amplifies an A. thaliana paralog located on chromosome V. A common reverse primer was used for all PCR amplifications. Lanes 1–4 show uncut RT-PCR products using the two primer combinations. Lanes 5–8 show HinfI-digested RT-PCR products. A. thaliana products are cut once by HinfI (lanes 5 and 6) whereas A. arenosa RT-PCR products are cut twice (lanes 7 and 8), allowing them to be discriminated in A. suecica (lanes 6 and 7). Note that T15P10.10 is expressed in A. suecica (lane 6) as is its paralog on A. thaliana chromosome V (lane 7, upper-most band) and its A. arenosa homolog (lane 7, bottom 2 bands). Negative control reactions from which reverse transcriptase was omitted was run in lanes 9–11. Black and white were reversed in the digital image to improve the visibility of the weak bands at the bottom of lanes 7 and 8.
PCR amplification products were resolved on 2% agarose gels in TBE (50 mM Tris⋅HCl/50 mM boric acid/1 mM EDTA, pH 8.0) buffer and gels were stained with ethidium bromide by using standard procedures (28). Images of stained gels were obtained by digital photography.
Accession Numbers.
Overlapping annotated bacterial artificial chromosome clones that represent the junction region where NOR4 meets the centromere-proximal arm of chromosome IV are F6N15 (GenBank accession no. AF069299) and T15P10 (GenBank accession no. AF167571). Additional sequence data obtained by the Arabidopsis Genome Initiative but not released in GenBank (at the time of this writing) are available on request from C.S.P.
Results and Discussion
Quantitation of rRNA Silencing in Nucleolar Dominance.
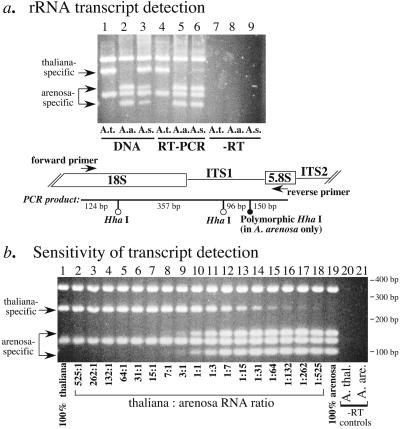
In a previous study (14), we showed that A. arenosa rRNA genes are highly expressed in A. suecica but that A. thaliana rRNA transcripts are not detectable by using S1 nuclease protection assays with end-labeled, species-specific probes. To quantify the degree to which A. thaliana rRNA genes are repressed in A. suecica, a sensitive RT-PCR assay was devised (Fig. 2). This assay exploits a polymorphic HhaI site in the internal transcribed spacer 1 (ITS1) region that allows A. thaliana and A. arenosa rRNA genes and their transcripts to be discriminated in their allotetraploid hybrid, A. suecica (Fig. 2a, lanes 1 and 2). Both A. thaliana and A. arenosa rRNA genes are detected in A. suecica after PCR of genomic DNA and digestion with HhaI (Fig. 2a, lane 3). Using the same primers for RT-PCR followed by HhaI digestion, rRNA transcripts from the A. arenosa genes are readily detected in A. suecica (Fig. 2a, compare lanes 5 and 6). By contrast, A. thaliana rRNA genes are repressed in A. suecica (Fig. 2a, compare lanes 4 and 6), as expected based on previous results (14). However, A. thaliana transcripts can be detected in trace amounts by using RT-PCR (Fig. 2a lane 6). In control reactions (Fig. 2a lanes 7– 9), purified RNA not reverse transcribed into cDNA, because of the omission of reverse transcriptase from the reaction, yielded no PCR products. These control reactions show that contaminating genomic DNA is absent from the purified RNA samples and is not contributing to the signals in Fig. 2a, lanes 4–6.
Figure 2.
A. thaliana rRNA genes are >99% repressed relative to A. arenosa rRNA genes in the allotetraploid hybrid, A. suecica. (a) Ethidium-bromide stained agarose gel showing genomic DNA (lanes 1–3) or reverse-transcribed (RT) total RNA (lanes 4–6) amplified by PCR using primers flanking internal transcribed spacer 1 (ITS1; see diagram) and then subjected to digestion with HhaI. An extra HhaI site in ITS1 of A. arenosa rRNA genes allows A. arenosa (A.a.) and A. thaliana (A.t.) genes and their transcripts to be discriminated in A. suecica (A.s). Both progenitors' rRNA genes are present in A. suecica (lane 3; note that A. arenosa-specific bands are under-represented when in competition with A. thaliana), but only the transcripts from the A. arenosa rRNA genes are abundant in the hybrid (lane 6). RNA samples from which reverse transcriptase was omitted before PCR show that RNA samples are free of contaminating DNA (lanes 7–9). (b) Titration experiment to determine the sensitivity of the RT-PCR assay for detection of under-dominant A. thaliana rRNA gene transcripts. RT product resulting from 100 ng of input total RNA was subjected to PCR in each reaction, but the ratio of A. thaliana to A. arenosa RT product was changed by symmetrical serial dilution. Conditions were identical to those used in a. Note that A. thaliana transcripts are detectable at levels as low as 1:525 relative to A. arenosa rRNA transcripts.
To determine the sensitivity of detection using the PCR assay for nucleolar dominance and to obtain a quantitative estimate for the trace amounts of A. thaliana rRNA gene transcripts detected in A. suecica (Fig. 2a, lane 6), a titration experiment was performed (Fig. 2b). In this experiment, nucleic acid concentrations and all other reaction conditions were identical to those used in Fig. 2a, but the ratio of A. thaliana to A. arenosa RT product was varied systematically. Based on this titration experiment, the limit of detection using these PCR conditions is approximately 1 A. thaliana transcript per 525 A. arenosa rRNA transcripts (Fig. 2b, lane 18). Note that PCR favors the amplification of A. thaliana PCR products (also true in Fig. 2a, lane 3) when A. thaliana and A. arenosa rRNA gene sequences are both present in the reaction. The reasons for this bias are not clear given that the primer-annealing sites are of identical sequence in the two progenitors and the PCR products are the same length. Regardless, the increased sensitivity of detection for A. thaliana rRNA genes is advantageous in this case given that these genes are the under-dominant (repressed) class of rRNA gene in A. suecica. The trace amount of the A. thaliana-specific band in lane 18 of Fig. 2b is equivalent to the trace amount detected in A. suecica (Fig. 2a, lane 6), suggesting an ≈500-fold repression of A. thaliana rRNA gene transcription relative to A. arenosa rRNA gene transcription in the hybrid. This level of repression is equivalent to expressing only ≈3 of the ≈1,500 A. thaliana rRNA genes (24) that are present in A. suecica. Note that there are four A. thaliana-derived NORs in A. suecica: two each of NOR2 and NOR4, suggesting that, on average, there is less than one rRNA gene active per NOR. At present, we have no method to determine whether the small number of active A. thaliana rRNA genes are located at a single NOR or are dispersed among the four NORs. Considering both possibilities, we estimate that each of the ≈4-Mbp A. thaliana NORs in A. suecica are 99.2%–99.8% repressed in the vegetative leaves from which the RNA tested in Fig. 2a was purified.
Expression Analysis of Genes Flanking NOR4.
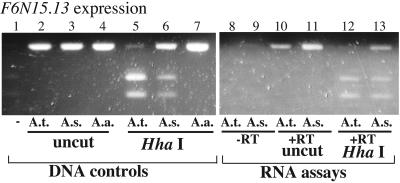
F6N15.13 is a predicted gene locus for which a corresponding cDNA clone (expressed sequence tag W43412) has been partially sequenced. The gene is located ≈12.3 kb from the 3′ end of NOR4 (see Fig. 1). To determine whether F6N15.13 is silenced in A. suecica, coincident with the silencing of the NOR, a primer pair that amplifies a single exon was used (Fig. 3). The primer pair amplifies A. thaliana, A. arenosa, and A. suecica genomic DNA sequences with similar efficiency, yielding a PCR product of identical size (Fig. 3, lanes 2–4). HhaI cleaves the A. thaliana PCR product (lane 5) but not the A. arenosa product (Fig. 3, lane 7), allowing transcripts of both progenitor species to be discriminated and detected in the hybrid. Using genomic DNA for conventional PCR, both progenitors' genes are present in A. suecica, as expected (Fig. 3, lane 6). To determine whether RNA transcripts from the A. arenosa and A. thaliana orthologs are made in A. suecica, RT-PCR followed by HhaI digestion was used. Purified RNA not reverse-transcribed into cDNA yielded no PCR products (Fig. 3, lanes 8 and 9) whereas reverse-transcribed RNA yielded the expected products (Fig. 3, lanes 10 and 11; note that RT-PCR and genomic DNA PCR products are identical in size because a single exon has been amplified). Digestion of RT-PCR products with HhaI showed that A. thaliana F6N15.13 transcripts are readily detected in A. suecica (Fig. 3, compare lanes 12 and 13; also compare these lanes to the genomic DNA controls in lanes 5 and 6) as are transcripts from the A. arenosa orthologs (top band in Fig. 3, lane 13; see genomic DNA controls in lanes 6 and 7). These data indicate that silencing in nucleolar dominance does not include silencing of locus F6N15.13.
Figure 3.
A. thaliana locus F6N15.13 is expressed in A. suecica. PCR using genomic DNA (lanes 1–7) or reverse-transcribed poly(A)+ RNA (lanes 8–13) was performed by using a primer pair that amplifies a single exon. HhaI cuts the A. thaliana (A.t.) PCR products but not the A. arenosa (A.a) products, allowing the progenitors' genes and transcripts to be discriminated in A. suecica (A.s) (lanes 5–7). Note that A. thaliana and A. arenosa transcripts are both detected in A. suecica (lane 12). RNAs not subjected to RT served as negative control in lanes 8 and 9.
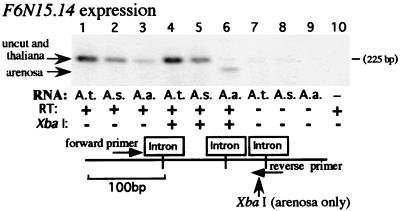
F6N15.14 is a predicted gene with similarity to the human breast cancer susceptibility gene BRCA2; it is located ≈10 kb from the NOR (see Fig. 1). A forward primer was designed that could amplify both F6N15.14 and its A. arenosa homolog(s) but not a paralogous gene located on A. thaliana chromosome 5. The primer was made paralog-specific by having the 3′ terminal deoxynucleotide of the primer correspond to a position at which a single nucleotide polymorphism is located, such that perfect complementarity is achieved only with F6N15.14 on chromosome 4. Specificity was verified by using the oligonucleotide to prime PCR-based sequencing reactions (cycle sequencing), yielding only the sequence of the chromosome 4 paralog (data not shown). The reverse primer used for PCR spanned an exon–exon junction such that only spliced mRNA could be amplified by RT-PCR. A derived cleaved amplified polymorphic sequence (29) marker was engineered by taking advantage of a single nucleotide polymorphism between A. thaliana and A. arenosa to create an XbaI site only in the A. arenosa PCR product (Fig. 4). In the absence of XbaI digestion, RT-PCR using the forward and reverse primers yielded products that were the same size in the progenitor species and in the hybrid (Fig. 4, lanes 1–3). After XbaI digestion, the uncut A. thaliana product was readily detected in A. suecica (compare Fig. 4, lanes 4 and 5). Interestingly, the expected A. arenosa product that is clearly detected in the A. arenosa control (Fig. 4, lane 6) was not detected in the hybrid (Fig. 4, lane 5), suggesting that the arenosa gene(s) is not expressed in A. suecica. This observation might be explained by the results of a recent study showing that in A. suecica many orthologous genes are expressed from one or the other progenitor's genomes, but not both (30).
Figure 4.
Expression of A. thaliana locus F6N15.14 in A. suecica. Using the primer pair shown in the diagram, an XbaI site is created in the A. arenosa RT-PCR product, allowing the A. thaliana (lanes 1 and 4) and A. arenosa (lanes 3 and 6) transcripts to be discriminated in A. suecica. Note that A. thaliana transcripts are detected in A. suecica (lane 5) but the A. arenosa-derived gene's transcripts are not detected (compare lanes 5 and 6). Negative control reactions are shown in lanes 7–10. Black and white were reversed in the digital image to improve the visibility of the weak bands in lanes 3 and 6.
The predicted gene nearest to NOR4, T15P10.10, is located only 3.1 kb from the most centromere-proximal rRNA gene sequences. A specific forward primer (A.t. primer) was designed to specifically amplify T15P10.10 but not an A. thaliana chromosome 5 paralog, nor the A. arenosa homolog(s) (Fig. 5). Primer specificity was again based on a single nucleotide polymorphism that could be exploited at a site complementary to the 3′ terminal position of the oligonucleotides and was verified by DNA cycle sequencing (data not shown). A second forward primer (A.a. primer) was designed to amplify the A. arenosa gene copy. This second primer also amplifies the A. thaliana paralog on chromosome 5 (verified by sequencing; data not shown). Fortunately, after PCR with a common reverse primer, PCR products from the A. arenosa and A. thaliana chromosome-5 homologs can be distinguished because the A. arenosa PCR product contains a distinctive HinfI site missing in A. thaliana homologs. In the absence of HinfI digestion, PCR products of the same size were generated by using the two specific forward primers and reverse transcribed RNA of A. thaliana (Fig. 5, lane 1), A. suecica (Fig. 5, lanes 2 and 3), or A. arenosa (Fig. 5, lane 4) as template. When the primer specific for A. thaliana T15P10.10 was used on chromosome 4, a HinfI-trimmed band was detected in both A. thaliana and A. suecica, indicating that T15P10.10 is expressed in the hybrid (Fig. 5, lanes 5 and 6). When the A. arenosa primer was used, A. arenosa transcripts were also detected in A. suecica by virtue of their distinctive HinfI digestion pattern (compare Fig. 5, lanes 7 and 8). The A. thaliana paralog located on chromosome V was also expressed in A. suecica, and its transcripts were detected by using the A. arenosa primer (upper-most band in Fig. 5, lane 7).
Mechanistic Implications of NOR-Restricted Chromosome Silencing.
Collectively, our data show that genes located as near as 3.1 kb from a repressed NOR remain active, despite the complete, or almost complete, silencing of the adjacent ≈4 Mbp of chromosomal DNA. We conclude that silencing in nucleolar dominance is restricted to the NORs and is not part of a larger chromosome-silencing phenomenon, an example of which is the inactivation of one X chromosome in somatic cells of female mammals (31). Thus, NORs are unlikely to be repressed because of the propagation of repressive chromatin structures or silencing signals that are initiated in chromosomal regions adjacent to the NORs. Likewise, the data suggest that the nucleolar dominance-induced assembly of rRNA genes into repressive chromatin structures, as implicated by their decreased DNase sensitivity (32, 33) and by their derepression if cytosine methylation or histone deacetylation are inhibited (13), does not spread from the NOR to adjacent genes. The latter finding invites the speculation that a chromatin boundary element or insulator (34) might be located at or near the junction between the NOR and adjacent genes, thus allowing their independent regulation as separate chromatin domains. Examples of such boundary elements include sequences flanking heat-shock loci, clustered homeotic genes, and gypsy transposable elements in Drosophila, silent mating type loci in yeast, and globin and lysozyme genes in chicken (for reviews see refs. 35–38). An intriguing possibility is that the retrotransposon sequences found in numerous copies flanking NOR2 and in two copies at the 3′ end of NOR4 (see Fig. 1) may have insulator functions in plants analogous to the properties of gypsy elements in flies. Intergenic spacer sequences of the rRNA genes themselves may also have the properties of boundary elements or insulators based on studies of Xenopus rRNA genes (39). These speculations will need to be tested experimentally in transgenic plants.
Several possibilities concerning the establishment of nucleolar dominance are suggested by the finding that silencing is restricted to the NORs. One hypothesis is that rRNA genes themselves are the targets of regulation and that sequence differences, presumably in the rapidly evolving intergenic spacer regions, allow dominant and under-dominant genes to be discriminated from one another. Such sequence differences in the rRNA gene spacers, which include the promoter and other regulatory sequences, could influence RNA polymerase I transcription factor binding affinities. Consistent with this hypothesis, nucleolar dominance in Xenopus can be mimicked in oocytes by coinjecting dominant and under-dominant rRNA genes, and spacer sequences have been shown to be responsible for this competitive effect (16). However, competitive transient expression or in vitro transcription experiments have thus far failed to reveal any differences in competitive strengths of rRNA gene promoters in Arabidopsis or its relatives in the genus Brassica (14, 17), making it unclear whether transcription-factor competition can explain nucleolar dominance in plants. It is possible that other properties of NORs are influenced by rRNA gene sequences in ways that affect nucleolar dominance even if dominant and under-dominant genes have identical transcription factor binding affinities. For instance, in yeast, every rRNA gene has a potential origin of replication in its intergenic spacer, although only a fraction of these origins are used in any given S phase of the cell cycle (40). Hence, a possibility is that rRNA gene sequence differences might influence replication timing such that dominant genes have first access to the transcription machinery.
A caveat to all speculations that invoke rRNA gene sequence differences as the basis for discriminating parental rRNA gene types in nucleolar dominance is that one would predict that rRNA genes would remain dominant or under-dominant independent of their chromosomal context. However, in Drosophila (18) and cereals (20, 21, 41), chromosome rearrangements that are not thought to alter rRNA gene sequences can have dramatic consequences, suggesting that rRNA gene sequences alone are not sufficient to explain nucleolar dominance. These considerations lead us to think that a combination of factors is likely to influence the establishment of nucleolar dominance. These might include intrinsic structural features of the NORs defined by rRNA gene sequences, sequences immediately flanking the rRNA genes (e.g., telomeres, retrotansposons, sequences between NORs and flanking genes), and broader chromosomal effects that might include signals for positioning within the three-dimensional space of the nucleus (42).
Acknowledgments
We are grateful to Larry Parnell, Richard McCombie, and Rob Martienssen for providing information regarding bacterial artificial chromosome clones T15P10 and F6N15 before their complete annotation. This work was supported by National Institutes of Health Grant R01 GM60380 (to C.S.P.). Publication costs were defrayed by a research prize from the Jean Lowenhaupt Botany Fund (to M.S.L.).
Abbreviations
- NOR
nucleolus organizer region
- RT
reverse transcription
- ITS1
internal transcribed spacer 1
- rRNA
ribosomal RNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McClintock B. Zeit Zellforsch Mik Anat. 1934;21:294–328. [Google Scholar]
- 2.Ritossa F M, Spiegelman S. Proc Natl Acad Sci USA. 1965;53:737–745. doi: 10.1073/pnas.53.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace H, Birnstiel M L. Biochem Biophys Acta. 1966;114:296–310. doi: 10.1016/0005-2787(66)90311-x. [DOI] [PubMed] [Google Scholar]
- 4.Phillips R L, Kleese R A, Wang S S. Chromosoma. 1971;36:79–88. [Google Scholar]
- 5.Karpen G H, Schaefer J E, Laird C D. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- 6.Birnstiel M L. Annu Rev Plant Physiol. 1967;18:25–58. [Google Scholar]
- 7.Scheer U, Weisenberger D. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 8.Shaw P J, Jordan E G. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 9.Navashin M. Cytologia. 1934;5:169–203. [Google Scholar]
- 10.Pikaard C S. Plant Mol Biol. 2000;43:163–177. doi: 10.1023/a:1006471009225. [DOI] [PubMed] [Google Scholar]
- 11.Pikaard C S. Trends Genet. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- 12.Reeder R H. J Cell Biol. 1985;101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z J, Pikaard C S. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z J, Comai L, Pikaard C S. Proc Natl Acad Sci USA. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viera A, Morais L, Barao A, Mello-Sampayo T, Viegas W S. Genome. 1990;33:707–712. [Google Scholar]
- 16.Reeder R H, Roan J G. Cell. 1984;38:39–44. doi: 10.1016/0092-8674(84)90524-5. [DOI] [PubMed] [Google Scholar]
- 17.Frieman M, Chen Z J, Saez-Vasquez J, Shen L A, Pikaard C S. Genetics. 1999;152:451–460. doi: 10.1093/genetics/152.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durica D S, Krider H M. Genetics. 1978;89:37–64. doi: 10.1093/genetics/89.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neves N, Silva M, Heslop-Harrison J S, Viegas W. Chromosome Res. 1997;5:125–131. doi: 10.1023/a:1018470208730. [DOI] [PubMed] [Google Scholar]
- 20.Schubert I, Kunzel G. Chromosoma. 1990;99:352–359. [Google Scholar]
- 21.Nicoloff H. Theor Appl Genet. 1979;55:247–251. doi: 10.1007/BF00265358. [DOI] [PubMed] [Google Scholar]
- 22.Copenhaver G P, Doelling J H, Gens J S, Pikaard C S. Plant J. 1995;7:273–286. doi: 10.1046/j.1365-313x.1995.7020273.x. [DOI] [PubMed] [Google Scholar]
- 23.Copenhaver G P, Pikaard C S. Plant J. 1996;9:259–272. doi: 10.1046/j.1365-313x.1996.09020259.x. [DOI] [PubMed] [Google Scholar]
- 24.Copenhaver G P, Pikaard C S. Plant J. 1996;9:273–282. doi: 10.1046/j.1365-313x.1996.09020273.x. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 26.Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian K D, Terryn N, et al. Nature (London) 1999;402:769–777. [Google Scholar]
- 27.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Neff M M, Neff J D, Chory J, Pepper A E. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee H S, Chen Z J. Proc Natl Acad Sci USA. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. . (First Published May 22, 2001; 10.1073/pnas.121064698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avner P, Heard E. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 32.Macleod D, Bird A. Cell. 1982;29:211–218. doi: 10.1016/0092-8674(82)90105-2. [DOI] [PubMed] [Google Scholar]
- 33.Thompson W F, Flavell R B. J Mol Biol. 1988;204:535–548. doi: 10.1016/0022-2836(88)90353-1. [DOI] [PubMed] [Google Scholar]
- 34.Kellum R, Schedl P. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 35.Eissenberg J C, Elgin S C R. Trends Genet. 1991;7:335–340. doi: 10.1016/0168-9525(91)90424-o. [DOI] [PubMed] [Google Scholar]
- 36.Udvardy A. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geyer P K. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 38.Gerasimova T I, Corces V G. Curr Opin Genet Dev. 1996;6:185–192. doi: 10.1016/s0959-437x(96)80049-9. [DOI] [PubMed] [Google Scholar]
- 39.Robinett C C, O'Connor A, Dunaway M. Mol Cell Biol. 1997;17:2866–2875. doi: 10.1128/mcb.17.5.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linskens M H K, Huberman J A. Mol Cell Biol. 1988;8:1210–1213. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viera R, Mello-Sampayo T, Viegas W. Genome. 1990;33:713–718. [Google Scholar]
- 42.Csink A K, Henikoff S. Nature (London) 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]