Abstract
Traumatic temporomandibular joint ankylosis (TMJA) is a common disease and disorder of the temporomandibular joint (TMJ); however, its pathogenesis has yet to be completely elucidated. In the authors' previous studies, the lateral pterygoid muscle (LPM) was confirmed to exert a function in distraction osteogenesis (DO) during the healing of a condylar fracture, which resulted in the formation of excess bone. The aim of the present study was to investigate alterations in the expression of any associated genes via an Affymetrix GeneChip method. The traumatic TMJA model was fabricated by a condylar fracture in the TMJ area of sheep with either a dissected LPM (LPD) or normal (LPN). The untreated sheep served as a control. At 4- and 12 weeks post-surgery, the condylar zone was isolated to perform the gene chip analysis, which was performed according to a standard Affymetrix protocol. The validated genes were further evaluated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The gene chip analysis indicated that the LPN gene expression pattern was similar compared with the DO process, while LPD was similar to that of normal bone fracture healing. The validated genes were collagen type II α1 chain, C-type lectin domain family 3 member A, interleukin 1A, cartilage oligomeric matrix protein, chondromodulin (LECT1), calcitonin receptor (CALCR), transforming growth factor (TGF)-β1, Fos proto-oncogene (FOS), bone γ-carboxyglutamate protein and bone morphogenic protein (BMP)7, among which, BMP7, LECT1, CALCR and FOS were confirmed by RT-qPCR. In conclusion, the present study demonstrated that LPM exerts a DO effect during the pathogenesis of traumatic TMJA, which may provide a novel target for preventing TMJA.
Keywords: condylar fracture, traumatic temporomandibular joint ankylosis, lateral pterygoid muscle, gene chip, distraction osteogenesis
Introduction
The temporomandibular joint (TMJ) is crucial for normal mandibular movement, which is associated with numerous vital physiological processes, including chewing, talking and functions that require the opening of the mouth (1). TMJ ankylosis (TMJA) restricts the movement of the mandible, which results in severe health problems in patients (2). TMJA can result from various factors including trauma, infection, congenital issues and arthritis, among these, trauma in the TMJ region is the most common cause, such as a condylar fracture (3). There are several hypotheses concerning the pathogenesis of traumatic TMJA. For example, it has been suggested that the abnormal healing of the fractured condyle is the principal reason for TMJA in the bone (4). Specifically, injury in the TMJ region may result in haematoma, fibrosis and the abnormal formation of excessive bone (5). A previous study hypothesized that the intra-articular bone fragment may help to increase the extent of ankylosis (6). Based on the anatomy of the lateral pterygoid muscle (LPM), it can be inferred that the contraction of the LPM will generate sufficient distraction force to the condyle neck during the opening of the mouth and confirm the ‘distraction osteogenesis (DO) effect of LPM’ hypothesis, which is that the LPM may exert a DO effect during the healing of the fractured condyle (7). In addition, the LPM exerts an important function in the reconstruction of the sagittal fractured condyle (8) and a previous study has reported that TMJA will be significantly alleviated if the LPM is dissected (9). Furthermore, the study of Dai et al (10) also suggested that local injection of botulinum toxin A would temporarily block impulses from the LPM's and may effectively prevent the occurrence of traumatic TMJA. Consequently, blocking the LPM may serve as a promising therapeutic strategy for the prevention of traumatic TMJA. Unfortunately, the associated molecular mechanisms of traumatic TMJA and the effects of LPM have yet to be completely elucidated, which has limited the development of suitable therapeutic strategies.
In the present study, a gene chip technique was used to systematically investigate any alterations in the expression of whole mRNA during the development of traumatic TMJA caused by a condylar fracture. In addition, the influence of LPM in the gene expression profile was also screened, in an attempt to identify the key molecules associated with this process.
Materials and methods
Animal model establishment and examination
Small-tailed Han sheep were provided by the Laboratory Animal Center of the Fourth Military Medical University (Xi'an, China) and all animal experiments were approved by the animal welfare ethics committee of the School of Stomatology, the Fourth Military Medical University. A total of 6 male sheep (aged 6 months, 25–35 kg) were kept under 20°C, 40–60% humidity with a circadian rhythm (12:12 h light:dark cycle). The animals were fed with a standard diet and had free access to water. The traumatic TMJA model was constructed according to the protocol outlined in previous publications (9,11,12). Briefly, the bilateral TMJ capsule was exposed from an incision to the anterior tragus. The joint capsule was incised and the condyle was exposed, and underwent a sagittal fracture by piezosurgery. Meanwhile, the outside quarter of the joint disc was dissected and a ‘#’ shaped groove was created by piezosurgery on the fossa surface. In addition, the right side LPM was dissected (LPD) while the left side was normal (LPN) i.e. without dissection. The tissue was tightly sutured apart from the capsule and penicillin (1 mg pure penicillin G sodium per sheep for 3 days) was used to prevent infection. Sheep who did not receive surgery i.e. untreated normal sheep (UTR) were used as a blank control. The surgical procedures are illustrated in Fig. S1. A total of 6 sheep were used and randomly divided into 2 groups of 3, which comprised the 4- and 12 weeks evaluation groups. In each group, one sheep was randomly selected as the UTR and the remaining sheep were subjected to surgery. For each animal in the group of surgical sheep, the right side of the TMJ was the LPD group while the left side was the LPN group.
Gene chip RNA preparation
At either 4 or 12 weeks (8), the sheep were sacrificed using an overdose injection of pentobarbital sodium, and the bone tissues within the condylar fractured zones were separated and immediately frozen in liquid nitrogen (−195.79°C). Then the samples were subjected to total RNA extraction, purification and quality testing according to a standard Affymetrix protocol by Shanghai Biotechnology Corporation (Shanghai, China) as reported elsewhere (13). The sample quality test is provided in Table SI.
Gene chip analysis
The prepared RNA samples were subjected to gene chip hybridization, scanning and normalization according to the standard Affymetrix protocol. The differential genes were screened by a threshold method and the genes with a fold-change >2 were considered as differential genes (e.g. the ratio >2 or ratio <0.5). In addition, the data was further analyzed by cluster heat map (Cluster 3.0; http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm), scatter plot (R 3.5.0; http://www.r-project.org/), Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8 Platform (https://david.ncifcrf.gov/) and Gene Ontology (GO) term enrichment (Bioconductor 3.8 in R 3.5.0; http://bioconductor.org/), according to the respective manufacturer's protocol. The abbreviations for the biological processes included in the GO term enrichment analysis were listed in Table SII. Genes were identified as significantly differentially expressed by GO term enrichment analysis (P<0.05).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) RNA preparation
The samples were frozen in liquid nitrogen for 48 h prior to being ground in a cold mortar. The total RNA was extracted by miRCURY RNA Isolation kit-tissue (Exiqon; Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. The RNA concentration was measured by NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1 µg total RNA was used for RT by the PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's protocol. Briefly, RT was conducted at 37°C for 15 min, followed by 5 sec at 85°C to terminate RT. Samples were then stored at 4°C.
RT-qPCR
The cDNA was amplified by SYBR Premix Ex Taq II (Takara Bio, Inc., Otsu, Japan) in order to perform the RT-qPCR procedure according to the manufacturer's protocol. qPCR was performed using an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) under the following conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, and finally the melt curve stage (95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec). The associated primers were synthesized by Takara Bio, Inc., and RPL19 was used as endogenous reference, which are listed in Table SIII. The relative gene expression was calculated by the 2−ΔΔCq method (14). Three independent experiments were repeated.
Statistical analysis
The quantitative data are presented as the mean ± standard deviation of at least three experimental repeats. The one-way analysis of variance with Student-Newman-Keuls post hoc test was used to compare differences between groups within a certain gene (GraphPad Prism 5; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results and Discussion
Cluster heat map
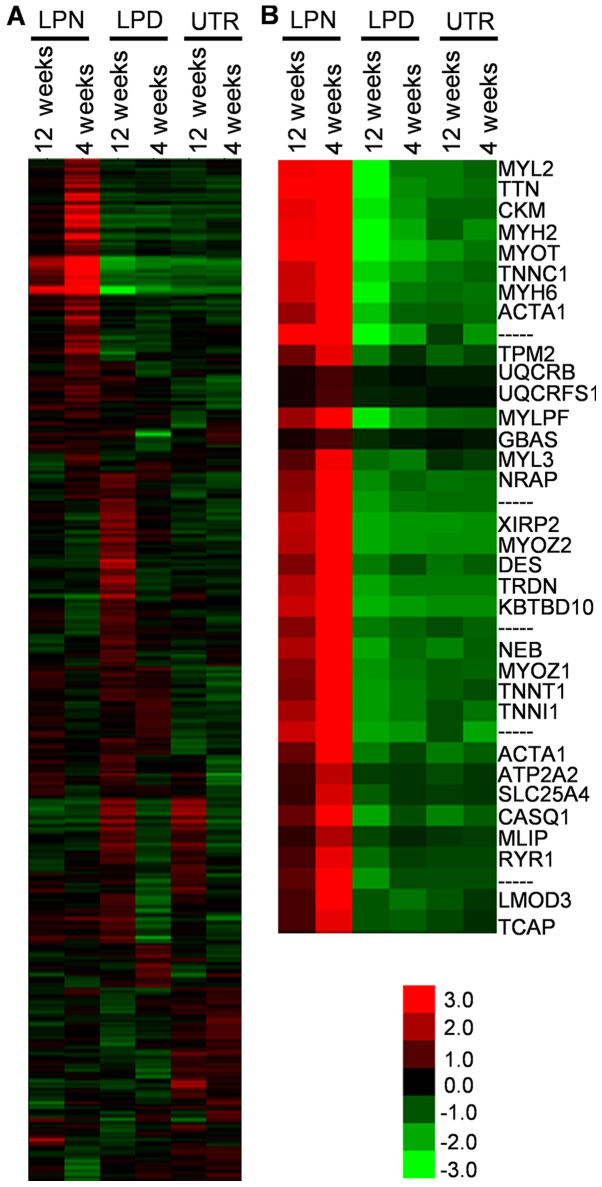
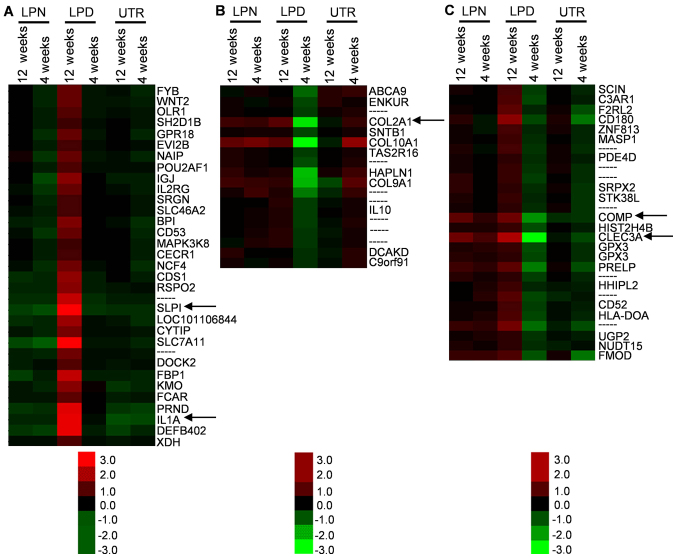
The overall gene expression was classified by heat map analysis and several regions of interest (ROI; the most different areas) were selected from the complete map (Fig. 1A). The cluster similarity was similar for the results obtained at 12 and 4 weeks in the same treatment groups in the majority of clusters (Fig. 1), suggesting that the gene expression pattern was similar in animals in the same treatment groups. Of note, the gene expression in LPD was up- or downregulated in the opposite direction to LPN, at 4 and 12 weeks (Fig. 1B). In addition, the genes associated with osteogenesis, including interleukin (IL)1A, collagen type II α1 chain (COL2A1), cartilage oligomeric matrix protein (COMP) and C-type lectin domain family 3 member A (CLEC3A) were included in these differential zones (Fig. 2A-C).
Figure 1.
Cluster heat maps of genes expression comparisons. (A) The complete map of the scanned genes. (B) The region of interests were selected as the most significant alteration regions from the complete map. LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment.
Figure 2.
Representative osteogenic associated genes regions. The regions of interest were selected based on alterations in LPD, which was (A) upregulated, (B) downregulated or (C) exhibited an average level. The black arrows indicate the osteogenic associated genes of SLPI, IL1A, COL2A1, COMP and CLEC3A. LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment. IL, interleukin; C; COL2A1, collagen type II α1 chain, COMP, cartilage oligomeric matrix protein; CLEC3A, C-type lectin domain family 3 member A.
The cluster heat map can directly display the general information about the expression levels of genes and the ROI can be easily selected from the differential genes zone. The data revealed that the cluster similarity was nearly identical between 4 and 12 weeks in each of the LPN, LPD and UTR groups, indicating that the different treatments can distinctively alter the gene expression patterns. There are certain specific muscle contraction associated genes including myosin light chain 2, myosin heavy chain (MYH)2, troponin C1 (TNNC1), MYH6 and actin α1 that were downregulated in LPD, suggesting that the LPM may serve an important role in the pathogenesis of traumatic TMJA. As bony ankylosis was the principal focus of the present study, the genes associated with osteogenesis were of particular interest. Based on the heat map, the IL1A, COL2A1, COMP and CLEC3A were preliminarily selected as potential critical genes.
Scatter plot
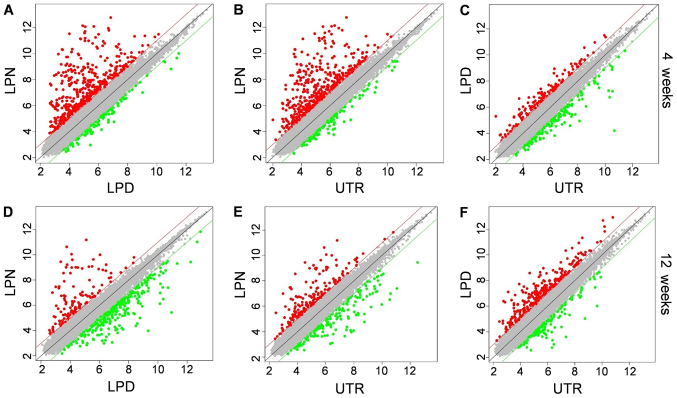
The scatter plot was used to illustrate the genes expression distribution between two different groups. At 4 weeks, it was evident that the majority of genes in LPN were significantly upregulated, irrespective of whether it was being compared with LPD or UTR (Fig. 3A and B). However, the LPD and UTR exhibited near-identical expression patterns (Fig. 3C). Additionally, at 12 weeks, the LPN group exhibited an increased number of downregulated genes when compared with the LPD group (Fig. 3D) and there were more upregulated genes in the LPN group compared with the UTR group (Fig. 3E). Whereas the LPD group had more upregulated genes compared with UTR at the 12 weeks stage (Fig. 3F).
Figure 3.
Scatter plot of relative genes expression. The comparisons between (A) LPN vs. LPD, (B) LPN vs. UTR and (C) LPD vs. UTR groups in 4 weeks. The comparisons between (D) LPN vs. LPD, (E) LPN vs. UTR and (F) LPD vs. UTR groups at 12 weeks. Red dots indicate the upregulated genes while the green dots indicate the downregulated genes. Grey dots indicate genes for which alterations in expression are within threshold. LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment.
The scatter plot can give direct insight into the number of up- and downregulated genes between two groups. Generally, in the present study, the LPN group exhibited a higher expression of active genes in the early stage, whereas more genes were activated in the LPD group at the later stage. Although the exact number of altered genes cannot be ascertained from the scatter plot, it corroborated the authors' hypothesis that genes associated with osteogenesis are dominant. During the process of DO, rapid osteogenesis is occurring in the first month, which may limit the degree to which the mouth can open and decrease the force of LPM (15). As a result, the DO effect of LPM is weakened and returns to a stable state. Therefore, most genes are upregulated at an early stage and are then subsequently downregulated. Conversely, the dissection of LPM makes it similar to the normal bone fracture healing process and the majority of the osteogenesis associated genes are activated in a relatively late stage. However, the exact differential genes involved in this process require further analysis.
DAVID and GO enrichment analysis
In order to determine the specific differential genes in differing treatments, DAVID gene classification and GO enrichment analysis were performed. The DAVID Bioinformatics Resources can be used to categorize the differential genes through different filters, which is extensively used for systematical analysis of large gene lists (16–18). The GO term enrichment analysis can group the differential genes by different biological processes and determine which genes are significant (19,20).
The genes were categorized by DAVID Bioinformatics Resources and more than 2-fold altered genes were considered different. Then the representative specific osteogenesis associated genes were listed. In LPN vs LPD at 4 weeks, the COL2A1, COMP, CLEC3A, calpain 3 and fibromodulin genes were upregulated up to >3-fold (Table I). From the GO term enrichment data, the bone morphogenesis and endochondral bone growth were the most notable, in which COMP and COL2A1 were important genes (Table II). It has previously been reported that the bone formed by DO is formed via intramembranous ossification (21), in which COL2A1 is involved (22). Taken together, these data indicate that the LPN group may be similar to the DO process and the genes associated with intramembranous ossification are activated at early stages. In addition, muscle contraction was also significantly upregulated in the LPN compared with the LPD group (P<0.05; Table II), which may be attributed to the function of LPM.
Table I.
Representative differential genes in 4 weeks by DAVID.
| LPN vs. LPD | LPN vs. UTR | LPD vs. UTR | |||
|---|---|---|---|---|---|
| Genes | Fold-change | Genes | Fold-change | Genes | Fold-change |
| COL2A1 | 10.08 | CD180 | 2.26 | FOS | 5.12 |
| COMP | 4.93 | COMP | 2.19 | MMP12 | 7.77 |
| CLEC3A | 13.95 | CLEC3A | 3.11 | IL1RN | 2.44 |
| CAPN3 | 9.19 | TLR7 | 2.03 | CLEC3A | 0.22 |
| FMOD | 3.00 | HRC | 6.90 | COMP | 0.44 |
| S100A12 | 0.20 | RYR1 | 11.48 | COL2A1 | 0.10 |
| OCM | 0.43 | S100A8 | 0.41 | BMP3 | 0.49 |
| SULF1 | 0.49 | ADIPOQ | 0.32 | LECT1 | 0.31 |
| MMP12 | 0.26 | BMP3 | 0.37 | CALCR | 0.36 |
LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment.
Table II.
The representative genes of GO-term enrichment in 4 weeks.
| BP | Genes |
|---|---|
| LPN vs. LPD | |
| BM | COMP, COL2A1, BGLAP, TGF-β1 |
| CEBM | COMP |
| EBG | COMP, COL2A |
| MC | ADRB2, TNNT3, RYR1, PGAM2, CHRNB1, MYL2, TNNC1, MYH2, MYOM2, CSRP3, TNNT1, TCAP, MYOM1, PLN, ACTA1, TNNI1, CASQ1, TNNC2, DES, MYLK2, CAV3, MYH6, NEB, CKMT2, FXYD1 |
| LPN vs. UTR | |
| NF-kB TF | PRDX3, S100A8, S100A12, AR, CAPN3 |
| OBM | RYR1, THBS3 |
| WNT pathway | PPP2R3A, SFRP2 |
| CRCS | AQP1, NR4A2, GATA6, CD180, S100A9, SOD2, RYR1, FOSB, PRDX3, SLC2A4, YBX3, GOT1, AQP4, ACKR4, ADIPOQ, THBS4, TUBA4A, ALDH1A2, EGLN3, FABP4, PON3, FBLN5, SFRP2, FZD7, LHCGR, FOS, ACSS1, ADCY2, LEPR, FMOD, ACADVL, RXRG, NR4A1, ESRRG, S100A8, S100A12, ANKRD1, TNMD, COMP, AR, RPS6, CAPN3, CAV3, MYH6, CD36, PGR |
| LPD vs. UTR | |
| EBG | COL2A1 |
| NRO | IGFBP5, CALCR, LRP4, SOST, MEPE |
| Ossification | IGFBP5, CYR61, LRP4, HGF, SFRP2, SOST, THBS3,TP53INP2, COL2A1, MEPE |
| CD | COMP, LOLX2, CYR61, BMP3, CNMD, SFRP2, EFEMP1,COL2A1 |
| RES | NR4A2, PLAU, CCR5, VEGFC, VDR, ADIPOQ, CHL1,LYZ, APOD, HGF, SERPINE1, FABP4, CCNB1, SLC12A2,KNG1, SEMA3C, SEMA3A, SFRP2, VCAM1, AQP3, SOST, FOS, LPL, NR4A3, PLAUR, FABP7, NR4A1, DCLK1, S100A8, IFIT2, COL4A1, EDNRB, COL2A1, ABL2, TLR7 |
Biological process abbreviations are defined in Table SII. BP, biological process; LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment.
In LPN vs. UTR at 4 weeks, not only COMP and CLEC3A were upregulated but cluster of differentiation (CD)180, Toll-like receptor 7, histidine rich calcium binding protein and ryanodine receptor 1 (RYR1) genes were also significantly increased, while the expression of S100 calcium binding protein A8 (S100A8), adiponectin (ADIPOQ) and BMP3 were downregulated (Table I). In the GO-term enrichment data, the nuclear factor-κB transcription factor activity and ossification in bone mature and cellular response to chemical stimulus were significantly different, which also contained the S100A8, CD180, RYR1 and ADIPOQ genes (P<0.05; Table II).
In LPD vs. UTR at 4 weeks, the upregulated genes were FOS, matrix metalloproteinase 12 and IL1 receptor antagonist while the CLEC3A, COMP, COL2A1, BMP3, LECT1 and CALCR were markedly downregulated (Table I). In the GO-term enrichment data, the ossification, cartilage development and response to external stimulus were more evident (Table II). The COMP and COL2A genes were also revealed to be important genes in endochondral bone growth, ossification and cartilage development (Table II). Since the cartilage development and endochondral ossification are the main processes during normal bone fracture healing (23,24), it may be inferred that LPD is similar to the normal healing of condylar fractures, which repair slowly over 4 weeks and therefore this explains why genes associated with osteogenesis including COMP, COL2A1, and BMP3 were downregulated.
In LPN vs. LPD at 12 weeks, the ADIPOQ, myosin light chain, phosphorylatable, fast skeletal muscle, TNNC1 and calsequestrin 1 were upregulated while the CD180, FOS, LECT1 and IL1A were downregulated (Table III). In this stage, the response to stress, the immune response, cellular response to chemical stimulus and response to external stimulus were significant, among which the FOS and IL1A exhibited multiple functions in the regulation of this process (Table IV). Lewinson et al (25) reported that the expression of FOS is in line with the DO process, which is upregulated in the early stages and attenuated when the bone trabecula is formed. Consequently, the downregulation of FOS in LPN may suggest that the DO process is entering a stable stage.
Table III.
The representative differential genes in 12 weeks by Database for Annotation, Visualization and Integrated Discovery.
| LPN vs. LPD | LPN vs. UTR | LPD vs. UTR | |||
|---|---|---|---|---|---|
| Genes | Fold-change | Genes | Fold-change | Genes | Fold-change |
| ADIPOQ | 2.22 | COMP | 2.99 | COL2A1 | 2.48 |
| MYLPF | 23.22 | SERPINE1 | 2.67 | IL1A | 12.39 |
| TNNC1 | 29.48 | CLEC3A | 2.87 | CLEC3A | 4.76 |
| CASQ1 | 8.44 | LECT1 | 2.89 | FOS | 4.25 |
| CD180 | 0.43 | LEPR | 0.41 | CD180 | 2.51 |
| TLR7 | 0.48 | BMP7 | 0.40 | COMP | 3.38 |
| IL6 | 0.42 | ADIPOQ | 0.18 | LECT1 | 7.73 |
| FOS | 0.20 | S100A9 | 0.23 | CALCR | 0.49 |
| LECT1 | 0.37 | EDNRB | 0.41 | ADIPOQ | 0.08 |
| IL1A | 0.11 | ||||
LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment.
Table IV.
The representative genes of Gene Ontology-term enrichment in 12 weeks.
| BP | Genes |
|---|---|
| LPN vs. LPD | |
| RS | CHI3L1, TNFAIP3, MB21D1, IL1B, NR4A2, LYZ2, S100A9, SOD2, NOS2, CCL5, TLR2, MX2, EIF2AK2, DTX3L, IL1A, FAS, IER3, BPI, CD27, LAP, GBP5, RSAD2, CXCL16, IL1RN, ADIPOQ, CCL20, PARP9, DERL3, XDH, KMO, MT2A, LYZ, PTAFR, PLAC8, GADD45B, AQP9, MYF6, LY86, MYH2,TUBA4A, DOCK2, HERC5, FGF10, STC2, PML, SLC7A11,VCAM1, PRKCB, FOS, TCAP, TRIB1, NR4A3, ATF3, CCRL2,NR4A1, PTGS2, CASQ1, S100A8, S100A12, CD2, IFIT2, CYSLTR1, ACOD1, CCL7, CYBB, ISG15, DDX58, RFC3, ITGB2, CXCR4, PARPBP, CXCL8, MX1, TLR7, TLR8 |
| IR | TNFAIP3, MB21D1, IL1B, S100A9, NOS2, CCL5, TLR2, MX2, IL1A, FAS, GBP5, RSAD2, CXCL16, IL7R, CCL20, ITK, TNFSF15, AQP9, LY86, MYH2, DOCK2, HERC5, FGF10, PML, SAMSN1, VCAM1, PRKCB, FOS, CCR8, NR4A1, SEMA4A, S100A8, S100A12, IFIT2, ACOD1, CCL7, CYBB, ISG15, DDX58, ITGB2,CXCL10, IL6, CXCL8, MX1, TLR7, TLR8 |
| TLR pathway | TNFAIP3, TLR2, RSAD2, FOS, ACOD1, ITGB2, TLR7, TLR8 |
| CRCS | CHI3L1, TNFAIP3, CTSL, MB21D1, IL1B, MT1A, CX3CR1,NR4A2, CD180, S100A9, SOD2, NOS2, RYR1, CCL5, TLR2, FOSB, EIF2AK2, FBP1, IFIT3, IL1A, FAS, GBP5, CXCL16, IL1RN, ADIPOQ, IL7R, DERL3, XDH, MT2A, CNMD, PTAFR, AQP9, LY86, TUBA4A, HERC5, FGF10, STC2, WNT2, PML, VCAM1, LHCGR, MZB1, JUNB, PRKCB, FOS, TR1B1, CCR8,NR4A3, ATF3, CCRL2, NR4A1, PTGS2, S100A8, S100A12, IFIT2, ACOD1, CCL7, ISG15, ITGB2, MYH6, CXCR4, EGR2, CXCL8, EGR1 |
| RES | CHI3L1, TNFAIP3, MB21D1, IL1B, CX3CR1, NR4A2, LYZ2, CD180, S100A9, NOS2, CCL5, TLR2, MX2, EIF2AK2, FAS, IER3, BPI, LAP, RSAD2, CXCL16, ADIPOQ, CHL1, LYZ, PTAFR, PLAC8, LY86, DOCK2, SDS, HERC5, FGF10, STC2, CSRP3, PML, VCAM1, CDS1, JUNB, FOS, TCAP, TGF-β1, TRIB1, CCR8, NR4A3, CCRL2, NR4A1, ACTA1, PTGS2, SEMA4A, S100A8, S100A12, IFIT2, CYSLTR1, ACOD1, CCL7, ISG15, DDX58, ITGB2, CXCR4, EGR2, CXCL8, TLR7, TLR8, BST-2A, BST-2B |
| LPN vs. UTR | |
| RES | DUSP10, S100A9, STAT1, MX2, EIF2AK2, RSAD2, ADIPOQ, CHL1, CCL21, PLAC8, PGLYRP1, HGF, SERPINE1, LTF, SCN1B, HERC5, SEMA3C, TGF-β1, CSRP3, PML, SFRP2, STAR, VCAM1, TCAP, PLAUR, HP, ACTA1, PTGS2, S100A8, S100A12, IFIT5, IFIT2,LCN2, EDNRB, ISG15, DDX58, BMP7, CAPN3, RELN |
| EBM | COMP, PML |
| CD | COMP, LOXL2, CNMD, PML, SFRP2, BMP7 |
| BD | COMP, HAS2, FREM1, PML, SFRP2 |
| EBG | COMP, PML |
| LPD vs. UTR | |
| EBG | COMP |
| Ossification | CALCR, LRP4, SFRP1, GF, SFRP2, RSPO2, JUNB, PTGS2, BMP5, COL2A1, EGR2 |
| CEBM | COMP, COL2A1 |
| BM | COMP, HAS2, FREM1, SFRP2, COL2A1 |
| RS | CHI3L1, TNFAIP3, IL1B, DUSP10, CAMP, NR4A2, GATA6, LYZ2, S100A9, SOD2, NOS2, CCL5, ITIH4, TLR2, EDN1, CXCL13, IL1A, FAS, IER3, LYZ1, BPI, CD27, LAP, CXCR6, CXCL16, IL1RN, ADIPOQ, HAS2, DERL3, XDH, CCL21, KMO, MT2A, CATHL2, LYZ,C7, PGLYRP1, SFRP1, HGF, SERPINE1, TNFSF8, LY86,LTF, MYH2, EGLN3, FABP4, DOCK2, TBX3, FOXF1,SELP, STC2, SFRP2, STAR, SLC7A11, FOS, CLEC7A, OLR1, TRIB1, NR4A3, DEFB128, LGMN, PLAUR, ATF3, C5AR1, HP, NR4A1, PTGS2, SGK1, S100A8, S100A12,CTSS, EDNRB, ACOD1, CCL7, CYBB, IFNA21, CAPN3, ITGB2, CXCR4, MYOCD, CD163, EYA4, LIPA, TLR10, CXCL8, CD14 |
Biological process abbreviations are defined in Table SII. BP, biological process; LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment.
In LPN vs. UTR at 12 weeks, the osteogenesis associated genes of COMP, serpin family E member 1, CLEC3A, LECT1 were upregulated, while LEPR, BMP7, ADIPOQ, S100A9 and endothelin receptor type B were downregulated (Table III). The genes of response to external stimulus, endochondral bone growth, cartilage development and bone development were significant, among which COMP was involved in multiple bone morphogenesis processes (Table IV). This indicates that although the DO effect at 12 weeks is at a stable stage, the bone morphogenesis activity in LPN remains increased compared with in normal sheep.
In LPD vs. UTR at 12 weeks, the osteogenesis associated genes COL2A1, IL1A, CLEC3A, FOS, CD180, COMP and LECT1 were upregulated and CALCR and ADIPOQ were downregulated (Table III). The GO-term enrichment also suggested that endochondral bone growth, ossification and bone morphogenesis were significant processes (Table IV). COMP and COL2A1 were revealed to be crucial genes in these biological processes (Table IV). The upregulation of genes associated with osteogenesis indicates that during this stage, the LPD bone metabolism is more active, which exerts similarities to a normal bone fracture.
In conclusion, through the gene chip analysis, the COL2A1, CLEC3A, IL1A, COMP, LECT1, CALCR, FOS and BMP7 genes were preliminarily selected as potentially critical genes during the pathogenesis of traumatic TMJA. However, due to the precision limitations of the gene chip technique, it should be taken into consideration that these validated genes may only be partially activated, or not the key vital genes during the establishment of traumatic TMJA. Therefore, the screened genes still require further analysis.
Confirmation of validated genes by RT-qPCR
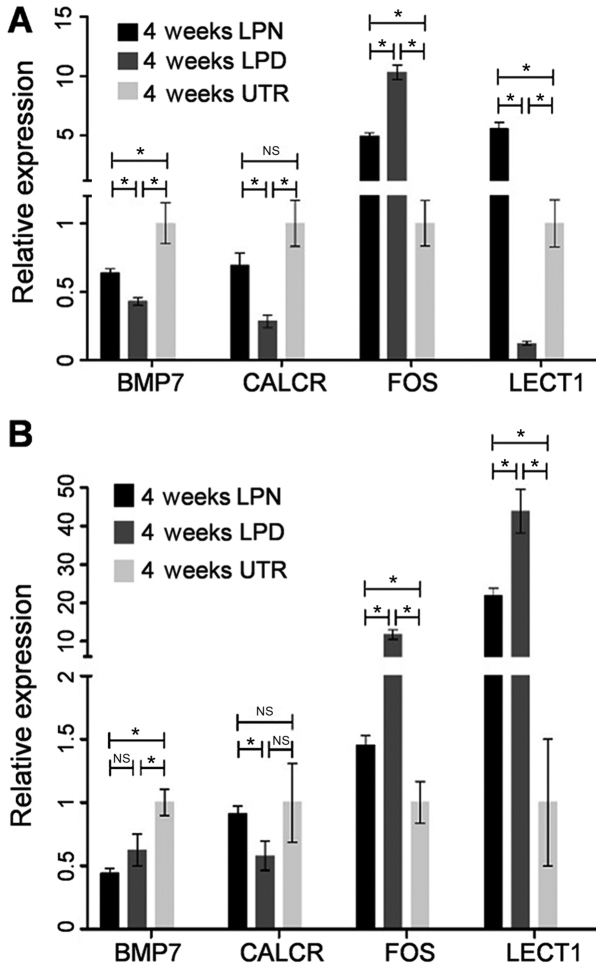
The genes of interest i.e. BMP7, CALCR, FOS and LECT1 were confirmed by RT-qPCR. In LPN vs. LPD at 4 weeks, BMP7 (~1.5-fold), CALCR (~2.5-fold) and LECT1 (>5-fold) were significantly upregulated while FOS (~0.5-fold) was downregulated (P<0.05; Fig. 4A). In LPN vs. UTR at 4 weeks, BMP7 (~0.75-fold) was significantly reduced while the FOS (~5-fold) and LECT1 (~5-fold) were upregulated (P<0.05; Fig. 4A). The expression of CALCR exhibited no statistically significant difference between the LPN and UTR group (Fig. 4A). However, for LPD vs. UTR at 4 weeks, BMP (<0.5-fold), CALCR (~0.2-fold) and LECT1 (~0.1-fold) were decreased while the FOS (~10-fold) was significantly improved (P<0.05; Fig. 4A).
Figure 4.
Reverse transcription-quantitative polymerase chain reaction analysis of BMP7, CALCR, FOS and LECT1. (A) The comparisons between different groups at 4 weeks; (B) The comparisons between different groups at 12 weeks. *P<0.05. LPN, the lateral pterygoid muscle was normal; LPD, the lateral pterygoid muscle was dissected; UTR, Sham surgery without treatment. BMP, bone morphogenic protein; FOS, Fos proto-oncogene, AP-1 transcription factor subunit; CALCR, calcitonin receptor; LECT1, chondromodulin; NS, not significant.
In LPN vs. LPD at 12 weeks, the CALCR (<2-fold) was upregulated whereas the FOS (~0.1-fold) and LECT1 (~0.5-fold) were significantly inhibited (P<0.05; Fig. 4B). The BMP7 results exhibited no statistically significant difference in the LPN group compared with the UTR (Fig. 4B). In LPN vs. UTR at 12 weeks, BMP7 (~0.5-fold) was significantly downregulated whereas FOS (~1.5-fold) and LECT1 (>20-fold) were upregulated (P<0.05; Fig. 4B). There was no difference in the expression of CALCR in LPN vs. UTR at 12 weeks (Fig. 4B). However, in LPD vs. UTR at 12 weeks, BMP7 (<0.5-fold) was downregulated while the expression of FOS (>10-fold) and LECT1 (>40-fold) were markedly upregulated.
In conclusion, the expression of BMP7 and LECT1 confirm the result from the chip data while the CALCR and FOS are slightly different. It is not uncommon that differences between Affymetrix GeneChip data and RT-qPCR measurements are detected. This may be due to the non-specific hybridization of genes and the fact that these processes rely on different algorithms (26). Nevertheless, the gene chip data is considered accurate (27) and a previously study has also suggested that the gene chip data should be coupled with RT-qPCR analysis (28).
In conclusion, the present study demonstrated that normal LPM can significantly increase the expression of osteogenesis associated genes, which is similar to DO gene expression profiles. However, the dissection of LPM can significantly delay the expression of these genes, which is similar to the results observed in normal bone fracture healing. Numerous genes including COL2A1, CLEC3A, IL1A, COMP, LECT1, CALCR, FOS and BMP7 may serve vital roles during the pathogenesis of traumatic TMJA, and warrant further investigation in future studies.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Yang Xue (Department of Oral Surgery, School of Stomatology, The Fourth Military Medical University, Xi'an, China), for her help during the experiment design and data analysis.
Funding
The present study is supported by the China National Key Research and Development Plan Project (grant no. 2016YFC1102903).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KH designed the study and provided financial support. JZ and XS participated in all the animal experiments as well as the gene chip analysis. SJ was involved in interpreting the data and drafted the manuscript. XJ, TD and PL contributed to the animal surgery and data analysis. All authors reviewed and approved the manuscript for submission.
Ethics approval and consent to participate
All the animal experiments were approved by the animal welfare ethics committee of the School of Stomatology, the Fourth Military Medical University (Xi'an, China) in accordance with the relevant guidelines and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
References
- 1.Chantaracherd P, John MT, Hodges JS, Schiffman EL. Temporomandibular joint disorders' impact on pain, function, and disability. J Dent Res. 2015;94(Suppl 3):S79–S86. doi: 10.1177/0022034514565793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baykul T, Aydin MA, Nasir SN, Toptas O. Surgical treatment of posttraumatic ankylosis of the TMJ with different pathogenic mechanisms. Eur J Dent. 2012;6:318–23. [PMC free article] [PubMed] [Google Scholar]
- 3.Arakeri G, Kusanale A, Zaki GA, Brennan PA. Pathogenesis of post-traumatic ankylosis of the temporomandibular joint: A critical review. Br J Oral Maxillofac Surg. 2012;50:8–12. doi: 10.1016/j.bjoms.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 4.He D, Cai Y, Yang C. Analysis of temporomandibular joint ankylosis caused by condylar fracture in adults. J Oral Maxillofac Surg. 2014;72:763.e1–e9. doi: 10.1016/j.joms.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Kaban LB, Perrott DH, Fisher K. A protocol for management of temporomandibular joint ankylosis. J Oral Maxillofac Surg. 1990;48:1145–1151. doi: 10.1016/0278-2391(90)90529-B. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto H, Kurita K, Ogi N, Ishimaru J, Goss AN. The effect of an intra-articular bone fragment in the genesis of temporomandibular joint ankylosis. Int J Oral Maxillofac Surg. 2000;29:290–295. doi: 10.1016/S0901-5027(00)80031-4. [DOI] [PubMed] [Google Scholar]
- 7.Meng FW, Zhao JL, Hu KJ, Liu YP. A new hypothesis of mechanisms of traumatic ankylosis of temporomandibular joint. Med Hypotheses. 2009;73:92–93. doi: 10.1016/j.mehy.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Liu CK, Liu P, Meng FW, Deng BL, Xue Y, Mao TQ, Hu KJ. The role of the lateral pterygoid muscle in the sagittal fracture of mandibular condyle (SFMC) healing process. Br J Oral Maxillofac Surg. 2012;50:356–360. doi: 10.1016/j.bjoms.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Deng TG, Liu CK, Liu P, Zhang LL, Wu LG, Zhou HZ, Ding YX, Hu KJ. Influence of the lateral pterygoid muscle on traumatic temporomandibular joint bony ankylosis. BMC Oral Health. 2016;16:62. doi: 10.1186/s12903-016-0220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai J, Yu H, Zhu M, Shen SG. Injection of botulinum toxin A in lateral pterygoid muscle as a novel method for prevention of traumatic temporomandibular joint ankylosis. J Med Hypoth Ideas. 2015;9:5–8. doi: 10.1016/j.jmhi.2014.06.002. [DOI] [Google Scholar]
- 11.Cheung LK, Shi XJ, Zheng LW. Surgical induction of temporomandibular joint ankylosis: An animal model. J Oral Maxillofac Surg. 2007;65:993–1004. doi: 10.1016/j.joms.2006.06.279. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto H, Kurita K, Ishimaru JI, Goss AN. A sheep model for temporomandibular joint ankylosis. J Oral Maxillofac Surg. 1999;57:812–817. doi: 10.1016/S0278-2391(99)90822-4. [DOI] [PubMed] [Google Scholar]
- 13.Skvortsov D, Abdueva D, Curtis C, Schaub B, Tavaré S. Explaining differences in saturation levels for Affymetrix GeneChip arrays. Nucleic Acids Res. 2007;35:4154–4163. doi: 10.1093/nar/gkm348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Lopes SL, Costa ALF, Gamba TdO, Flores IL, Cruz AD, Min LL. Lateral pterygoid muscle volume and migraine in patients with temporomandibular disorders. Imaging Sci Dent. 2015;45:1–5. doi: 10.5624/isd.2015.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang DaW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classifcation Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0-a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 21.Runyan CM, Gabrick KS. Biology of bone formation, fracture healing, and distraction osteogenesis. J Craniofac Surg. 2017;28:1380–1389. doi: 10.1097/SCS.0000000000003625. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Simpson AH, Triffitt JT. The role of chondrocytes in intramembranous and endochondral ossification during distraction osteogenesis in the rabbit. Calcif Tissue Int. 1999;64:310–317. doi: 10.1007/s002239900625. [DOI] [PubMed] [Google Scholar]
- 23.Shen G, Rabie AB, Zhao ZH, Kaluarachchi K. Forward deviation of the mandibular condyle enhances endochondral ossification of condylar cartilage indicated by increased expression of type X collagen. Arch Oral Biol. 2006;51:315–324. doi: 10.1016/j.archoralbio.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Bradaschia-Correa V, Barrence FA, Ferreira LB, Massa LF, Arana-Chavez VE. Effect of alendronate on endochondral ossification in mandibular condyles of growing rats. Eur J Histochem. 2012;56:e24. doi: 10.4081/ejh.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewinson D, Rachmiel A, Rihanibisharat S, Kraiem Z, Schenzer P, Korem S, Rabinovich Y. Stimulation of Fos- and Jun-related genes during distraction osteogenesis. J Histochem Cytochem. 2003;51:1161–1168. doi: 10.1177/002215540305100906. [DOI] [PubMed] [Google Scholar]
- 26.Mieczkowski J, Tyburczy ME, Dabrowski M, Pokarowski P. Probe set filtering increases correlation between Affymetrix GeneChip and qRT-PCR expression measurements. BMC Bioinformatics. 2010;11:104. doi: 10.1186/1471-2105-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii M, Hashimoto S, Tsutsumi S, Wada Y, Matsushima K, Kodama T, Aburatani H. Direct comparison of GeneChip and SAGE on the quantitative accuracy in transcript profiling analysis. Genomics. 2000;68:136–143. doi: 10.1006/geno.2000.6284. [DOI] [PubMed] [Google Scholar]
- 28.Juang JL, Chen TC, Jiang SS, Hsiung CA, Chen WC, Chen GW, Lin SM, Lin JH, Chiu SC, Lai YK. Coupling multiplex RT-PCR to a gene chip assay for sensitive and semiquantitative detection of severe acute respiratory syndrome-coronavirus. Lab Invest. 2004;84:1085–1091. doi: 10.1038/labinvest.3700136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.